Abstract
The Porphyromonas gingivalis recombinant VimA can interact with the gingipains and several other proteins, including a sialidase. Sialylation can be involved in protein maturation; however, its role in virulence regulation in P. gingivalis is unknown. The three sialidase-related proteins in P. gingivalis showed the characteristic sialidase Asp signature motif (SXDXGXTW) and other unique domains. To evaluate the roles of the associated genes, randomly chosen P. gingivalis isogenic mutants created by allelic exchange and designated FLL401 (PG0778::ermF), FLL402 (PG1724::ermF), and FLL403 (PG0352::ermF-ermAM) were characterized. Similar to the wild-type strain, FLL402 and FLL403 displayed a black-pigmented phenotype in contrast to FLL401, which was not black pigmented. Sialidase activity in P. gingivalis FLL401 was reduced by approximately 70% in comparison to those in FLL402 and FLL403, which were reduced by approximately 42% and 5%, respectively. Although there were no changes in the expression of the gingipain genes, their activities were reduced by 60 to 90% in all the isogenic mutants compared to that for the wild type. Immunoreactive bands representing the catalytic domains for RgpA, RgpB, and Kgp were present in FLL402 and FLL403 but were missing in FLL401. While adhesion was decreased, the capacity for invasion of epithelial cells by the isogenic mutants was increased by 11 to 16% over that of the wild-type strain. Isogenic mutants defective in PG0778 and PG0352 were more sensitive to hydrogen peroxide than the wild type. Taken together, these results suggest that the P. gingivalis sialidase activity may be involved in regulating gingipain activity and other virulence factors and may be important in the pathogenesis of this organism.
INTRODUCTION
Porphyromonas gingivalis, a Gram-negative anaerobic bacterium, is an important etiological agent of adult chronic periodontitis. In addition, this organism appears to be associated with cardiovascular diseases and an increased risk of preterm birth and low birth weight of infants in pregnant women with infection-induced periodontal disease (4). The ability of P. gingivalis to colonize the periodontal pocket and interact with other organisms such as Tannerella forsythia and Fusobacterium nucleatum is a prerequisite for infection-induced periodontal diseases (34). Several cell surface-associated virulence factors (e.g., gingipains, fimbriae, hemagglutinin, capsule, and lipopolysaccharide) that can directly or indirectly affect the periodontium or facilitate interaction with other periodontopathic pathogens have been characterized in P. gingivalis (16). However, the roles of sialidase and sialopeptidases as potential virulence factors in P. gingivalis are yet to be explored. Because of their importance in the breakdown of carbohydrates and other glycoprotein conjugates, these enzymes could facilitate attachment and enhance commensalism with other oral pathogens, especially biofilm formers (44).
Sialidases (neuraminidases) are glycosylhydrolases that cleave the glycoketosidic linkages of sialic acid (Sia) O-acceptor substrates by an exohydrolytic reaction. Functionally similar to sialidases, the O-sialoglycoprotease hydrolyzes the Sia O-acceptor substrate through an endohydrolytic reaction (36, 55). Sialidase activity has been found in viruses, bacteria, protozoa, fungi, and metazoans (6, 7, 55). Bacterial sialidases have been considered virulence factors in many pathogenic organisms, such as Corynebacterium diphtheriae, Vibrio cholerae, Streptococcus pneumoniae, and group B streptococci, which colonize mucosal surfaces (8). They have been shown to be involved in infection and tissue destruction (57), peroxide scavenging during oxidative stress (22), and the modulation of host innate immunity (55). Furthermore, production of sialidase may be a critical factor in the provision of free sialic acid, a fermentable carbohydrate source for bacterial proliferation (8, 20).
Sialic acid has been predominantly found as the terminal carbohydrate in eukaryotes and prokaryotes. Sialic acid naturally occurs in prokaryotes as nine-carbon keto sugar acids derived from N-acetylneuraminic acid (Neu5Ac) (53). Many commensal and pathogenic bacteria use environmental (host) sialic acids as sources of carbon, nitrogen, energy, and amino sugars for cell wall synthesis (35). The breakdown of sialic acid residues and sialoconjugates by sialidases contributes to a wide range of important biological functions such as cellular interaction and conformational stabilization of glycoproteins in the cell membrane that could expose or mask receptors for ligand binding and other enzymatic interactions (2). While the role of sialidase in sialic acid metabolism has been known in other oral pathogens like Tannerella forsythia (38), it is yet to be explored in P. gingivalis.
Glycosylation is a posttranslational regulatory mechanism that is important in gingipain biogenesis and pathogenicity in P. gingivalis (13, 25). Further, the posttranslational addition of carbohydrates to the gingipains is highly variable, thus implying a role for multiple factors in this process (10). We previously reported that the VimA protein, which can modulate gingipain activity in P. gingivalis, can also interact with a putative sialidase protein (1, 30, 50, 53). Although sialylation can be involved in protein maturation, its role, if any, in gingipain maturation/activation is unknown. Here we report the characterization of a sialidase and two sialoglycoproteases from P. gingivalis.
MATERIALS AND METHODS
Bioinformatics analysis.
The DNA and amino acid sequences were retrieved from the Oralgen database (Los Alamos National Laboratory; http://www.oralgen.lanl.gov) and aligned using Bioedit (http://www.mbio.ncsu.edu/bioedit/bioedit.html). The phylogenetic relationship between the oral pathogens was analyzed using MEGA version 4.0 (49). The distance was calculated using the Kimura 2-parameter model, and for clustering the neighbor-joining method was employed using bootstrap values based on 1,000 replicates (39). The amino acid sequences were analyzed using ClustalW version 2.0 (http://www.ebi.ac.uk/). The secondary structure prediction and modeling of the protein were performed using Modeler 9v8 (41). The models were validated using the WHATIF program (56). The transcription factor binding motifs were identified using PROMO version 8.3 (11). The signal peptide and potential cleavage sites were predicted using both the neural network and hidden Markov model (23).
Bacterial strains and growth condition.
Bacterial strains and plasmids used in this study are listed in Table 1. P. gingivalis strains were grown in brain heart infusion (BHI) broth (Difco, Detroit, MI) supplemented with hemin (5 μg/ml), vitamin K (0.5 μg/ml), and cysteine (0.1%). Experiments with hydrogen peroxide were performed in BHI without cysteine. All cultures were incubated at 37°C unless otherwise stated. P. gingivalis strains were maintained in an anaerobic chamber (Coy Manufacturing, Ann Arbor, MI) in 10% H2, 10% CO2, and 80% N2. Growth rates for P. gingivalis strains were determined spectrophotometrically (optical density at 600 nm [OD600]). Antibiotics were used at the following concentrations: clindamycin, 0.5 μg/ml; erythromycin, 300 μg/ml; tetracycline, 3 μg/ml; carbenicillin, 100 μg/ml.
Table 1.
Strains and plasmids used
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| Strains | ||
| P. gingivalis W83 | Wild type | 31 |
| FLL92 | vimA-defective mutant | 52 |
| FLL401 | PG0778 defective | This study |
| FLL402 | PG1724 defective | This study |
| FLL403 | PG0352 defective | This study |
| FLL401c | FLL401 with pFLL401a | This study |
| FLL402c | FLL402 with pFLL402a | This study |
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ thi-1 gyrA96 relA1 | Invitrogen |
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 recA1 ara139 Δ(ara-leu)7697 galU galK rpsl (Strr) endA1 nupG | Invitrogen |
| Plasmids | ||
| pCR-2.1.TOPO | Apr Kmr | Invitrogen |
| pVA2198 | SprermF-ermAM | 12 |
| pT-COW | AprtetQ | 13a |
| pFLL401a | AprtetQ TC::PG0778 | This study |
| pFLL402a | AprtetQ TC::PG1724 | This study |
| pFLL403c | AprtetQ TC::PG0352 | This study |
| pFLL403a | pCR-2.1-TOPO PG0352-PG0353 | This study |
| pFLL403b | pFLL403a with PG0352 interrupted with ermF-ermAM | This study |
DNA isolation and analysis.
P. gingivalis chromosomal DNA was prepared as previously described (27). For plasmid DNA analysis, DNA extraction was performed by the alkaline lysis procedure previously reported (12). For large-scale preparation, plasmids were purified using the Qiagen (Santa Clarita, CA) Plasmid Maxi kit.
Construction of PG0778- and PG1724-defective mutants.
Construction of isogenic mutants defective in the PG0778 and PG1724 genes was carried out by long-PCR-based fusion of several fragments using overlapping extension PCR as previously described (20). The primers used in the development of deletion mutants are given in Table S1 in the supplemental material. Briefly, 1-kilobase flanking fragments upstream and downstream of the target genes were PCR amplified from the chromosomal DNA of P. gingivalis W83. The ermF cassette was amplified from the pVA2198 plasmid (Table 1) using custom-made oligonucleotide primers containing overlapping nucleotides for the upstream and downstream fragments. These three fragments were fused together using the forward primer of the upstream fragment and the reverse primer of the downstream fragment. The fusion PCR program consisted of 1 cycle of 5 min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 55°C, and 4 min at 68°C, with a final extension of 5 min at 68°C. The PCR-fused fragment was used to transform P. gingivalis W83 by electroporation as previously described (1). The cells were plated on BHI agar containing 10 μg/ml of erythromycin and incubated at 37°C for 7 days. The correct gene replacement in the erythromycin-resistant mutants was confirmed by colony PCR and DNA sequencing.
Generation of PG0352-defective mutant P. gingivalis.
A 2.5-kb fragment carrying the intact PG0352 and downstream gene PG0353 was amplified by PCR using custom-made oligonucleotide primers (see Table S1 in the supplemental material). This fragment was cloned into the pCR-2.1-TOPO plasmid vector (Invitrogen, Carlsbad, CA) and was designated pFLL403a. The ermF-ermAM cassette, which confers erythromycin/clindamycin resistance in Escherichia coli and P. gingivalis (9), was PCR amplified from pVA2198 with Pfu Turbo (Stratagene, La Jolla, CA) and ligated into the HincII restriction site of the PG0352 gene. The resultant recombinant plasmid, pFLL403b, was used as a donor in electroporation of P. gingivalis W83.
Complementation of the PG0778-, PG1724-, and PG0352-defective isogenic mutants.
DNA fragments containing an upstream regulatory region and the open reading frame (ORF) for PG0778, PG1724, or PG0352 were amplified from P. gingivalis W83 chromosomal DNA using the appropriate primer set (see Table S1 in the supplemental material). A BamHI restriction site was designed at the 5′ ends of both primers to facilitate the subcloning of the PCR fragment into the BamHI-digested pT-COW plasmid (17). The purified recombined plasmids, designated pFLL401a, pFLL402a, and pFLL403c, were used to electrotransform P. gingivalis FLL401 (PG0778::ermF), FLL402 (PG1724::ermF), and FLL403 (PG0352::ermF-ermAM), respectively. The transformants were selected on BHI agar plates in the presence of erythromycin and tetracycline.
Electroporation.
Electroporation of cells was performed as previously reported (12). Briefly, 1 ml of an actively growing culture of P. gingivalis was used to inoculate 10 ml of BHI broth and incubated overnight at 37°C. Ninety milliliters of the same prewarmed (37°C) medium was then inoculated with 3 ml of the overnight culture and was incubated for an additional 4 h until an OD600 of 0.7 was obtained. The cells were then harvested by centrifugation at 3,000 × g for 15 min at 4°C and were washed twice in 50 ml of electroporation buffer (10% glycerol, 1 mM MgCl2), filter sterilized, and stored at 4°C. The final cell pellet was resuspended in 0.5 ml of electroporation buffer. A 100-μl sample of resuspended cells and 5 μg of DNA were placed in a sterile electrode cuvette (0.2-cm gap). The cells were then pulsed with a Gene Pulser (Bio-Rad, Hercules, CA) at 2,500 V for 9.5 ms and then were incubated on ice for 5 min. The cell suspension was then added to 0.5 ml of BHI broth and was incubated for approximately 16 h. A 100-μl sample was plated on solid medium containing erythromycin and was incubated anaerobically at 37°C for 5 to 10 days.
Growth analysis during oxidative stress conditions.
Actively growing cultures of P. gingivalis W83, FLL401, FLL402, and FLL403 were incubated at 37°C under anaerobic conditions for 24 h. For adaptation to oxidative stress conditions, the wild type and isogenic mutants were grown in BHI broth (without cysteine) plus 0.25 mM H2O2. Controls were grown in the absence of H2O2. Growth rates were determined spectrophotometrically (OD600).
RT-PCR analysis.
Total RNA was extracted from P. gingivalis strains grown to early stationary phase (OD600 of 1.2 to 1.3) using the RiboPure kit (Ambion, Foster City, CA) according to the manufacturer's instructions. The primers used for reverse transcription-PCR (RT-PCR) analysis are listed in Table S1 in the supplemental material. The RT-PCR mixture (50 μl) contained 1 μg of template RNA in Superscript One-Step RT-PCR mix (Invitrogen, Carlsbad, CA). RT-PCR cycling conditions were 1 cycle of 5 min at 94°C followed by 30 cycles of 30 s at 94°C, 30 s at 54°C, and 1 min at 68°C, with final extension of 5 min at 68°C.
SDS-PAGE and immunoblot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with a 4 to 12% Bis-Tris separating gel in MOPS (morpholinepropanesulfonic acid)-SDS running buffer according to the manufacturer's instructions (NuPAGE Novex gels; Invitrogen, Carlsbad, CA). Samples were prepared (65% sample, 25% 4× NuPAGE lithium dodecyl sulfate [LDS] sample buffer, 10% NuPAGE reducing agent), heated at 72°C for 10 min, and then electrophoresed at 200 V for 65 min in the XCell SureLock minicell system (Invitrogen, Carlsbad, CA). The protein bands were visualized by staining with SimplyBlue SafeStain (Invitrogen, Carlsbad, CA). The separated proteins were then transferred to BioTrace nitrocellulose membranes (Pall Corporation, San Diego, CA) using a semidry Trans-Blot apparatus (Bio-Rad, Hercules, CA) at 15 V for 25 min. The blots were probed with gingipain-specific antibodies (45). Immunoreactive proteins were detected by the procedure described in the Western Lightning Plus chemiluminescence reagent kit (Perkin-Elmer Life Sciences, Fremont, CA). The secondary antibody was goat anti-rabbit or anti-chicken IgG (heavy plus light chains)-horseradish peroxidase (HRP) conjugate (Zymed Laboratories, San Francisco, CA).
Gingipain activity assay.
The presence of Arg-X- and Lys-X-specific cysteine protease activities was determined using a microplate reader (Bio-Rad) by the method of Fletcher et al. (12) for cells grown to exponential phase (OD600 of 0.8). Briefly, activity of arginine gingipains was measured with 1 mM BAPNA (Nα-benzoyl-dl-arginine-p-nitroanilide) in activated protease buffer (0.2 M Tris-HCl, 0.1 M NaCl, 5 mM CaCl2, 10 mM l-cysteine, pH 7.6). Lysine gingipain activity was measured with ALNA (acetyl-Lys-p-nitroanilide-HCl). After the substrate and culture were incubated, the reaction was stopped by the addition of 50 μl of glacial acetic acid. The OD405 was then measured against a blank sample containing no proteinase.
Assay for estimation of total protease activity.
Total protease estimation used a fluorescence resonance energy transfer (FRET)-based method (protease assay kit; Molecular Probes, Carlsbad, CA) in accordance with the manufacturer's instructions. The protease substrate comprises a fluorophore and a quencher moiety separated by an amino acid sequence. Protease cleavage of the amino acid sequence separates the fluorophore from the quencher, resulting in the emission of a detectable fluorescent signal. The magnitude of the resultant signal is proportional to the degree of substrate cleavage and can therefore be used to quantitate the enzyme activity. The assay was performed in a 96-well plate. The fluorescence was measured using the microplate reader FLx800 (BioTek) at 490-nm excitation and 520-nm emission wavelengths.
Sialidase assay.
Sialidase estimation in the P. gingivalis mutants was carried out using the Amplex Red neuraminidase (sialidase) assay kit (Molecular Probes, Carlsbad, CA). Briefly, 50 μl of the P. gingivalis sample was mixed with 50 μl of the Amplex Red reagent, 50 μl of HRP, and 50 μl of the galactose oxidase solution. Finally, 50 μl of the substrate was added to the mixture, and the mixture was incubated at 37°C for 30 min in a light-protected container. The assay utilizes Amplex Red to detect H2O2 generated by galactose oxidase oxidation of desialylated galactose, the end product of neuraminidase activity. The H2O2 in the presence of HRP reacts with Amplex Red reagent to generate resorufin, the red fluorescent oxidation product, which is detected spectrophotometrically (OD492) using the BioTek FLx800 microplate reader. Substrates used in the sialidase assay include 3′sialyllactose, O-sialyl-d-lactose, 3N-acetylneuraminic acid, mucin (bovine submaxillary), feutin, and colomic acid. (Sigma-Aldrich, St. Louis, MO). A neuraminidase and a nonneuraminidase supplied with the kit were used as positive and negative controls, respectively.
Hemagglutination assay.
Hemagglutination activity was determined using sheep, bovine, chicken, and horse red blood cells (RBCs) as previously reported (51). Twenty-four-hour cultures of P. gingivalis W83 and mutants were harvested by centrifugation (10,000 × g for 10 min), washed twice in phosphate-buffered saline (PBS) buffer, and resuspended to a final OD600 of 1.2. Erythrocytes were washed twice with 1× PBS and resuspended in 1× PBS to a final concentration of 1%. An aliquot (100-μl volume) of the bacterial suspension was serially diluted 2-fold with PBS in a round-bottom 96-well microtiter plate. An equal volume (100 μl) of 1% erythrocytes was mixed with each dilution and incubated at 4°C for 3 h. Hemagglutination was visually assessed, and the hemagglutination titer was determined as the reciprocal of the highest dilution showing complete hemagglutination.
Lectin binding hemagglutination assay.
Sialidase production was assayed in microtiter plates using fresh sheep erythrocytes. The erythrocytes were collected and suspended in Alsever's solution (10%) and washed three times with 0.01 M PBS buffer (PBS, Na2HPO4, KH2PO4, NaCl, pH 7.4). Bacterial cells were grown in BHI broth, resuspended to 1.5 × 108 bacteria/ml, and then washed three times by centrifugation (12,000 × g, 10 min) in PBS. An aliquot (1 ml) of bacterial cells was added to 10 ml of the washed erythrocytes to achieve a final concentration of 1%, and the cells were then carefully homogenized and incubated at 37°C for 4 h under anaerobic conditions. Lectins from Agaricus bisporus (ABA; 1.8 mg/ml), Erythrina cristagalli (ECA; 2.3 mg/ml), Limulus polyphemus (LPA; 2.5 mg/ml), Triticum vulgaris (WGA; 2.5 mg/ml), and Arachis hypogaea (PNA; 2.5 mg/ml) and concanavalin A (ConA; 2.1 mg/ml) (Sigma-Aldrich, St. Louis, MO) were serially diluted in PBS. An aliquot (5 μl) of each dilution was then added to the suspension of bacteria and erythrocytes (1:1), which was incubated at room temperature for 1 h followed by 12 h at 4°C. The presence of sialidase was indicated by agglutination (bacteria-erythrocytes-lectin). A mixture of bacteria and erythrocytes without lectin, which results in precipitation, was used as the negative control. A mixture of erythrocytes with PBS was used as the RBC control. The agglutination titer was defined as the reciprocal of the endpoint dilution. The inhibition of the sialidase production was verified in the wild-type W83 strain as previously reported (29). Briefly, bacteria treated with 1 ml of 1 mM galactose and incubated at room temperature for 30 min were washed in PBS and harvested by centrifugation (12,000 × g, 10 min). Five microliters of diluted lectins was mixed with 50 μl of bacterial cells (1.5 × 108 cells/ml), treated with 1 mM galactose, and added to 50 μl of erythrocytes (1% final concentration). Plates were homogenized and incubated at 37°C for 1 h. Enzyme inhibition was indicated by the precipitation of the erythrocytes. Overnight incubation was avoided, as the test system showed lysis of RBCs.
Electron microscopy.
Transmission electron microscopy (TEM) was performed using the FEI Tecnai G2 TEM according to the method of Harris (17). Briefly, Formvar-carbon-coated grids were prepared; the Formvar support was removed by placing the grids in an atmosphere of solvent vapor. The grids were then placed on a wire mesh in a glass petri dish, with carbon tetrachloride below the wire mesh. Five to 10 ml of the sample was placed under the carbon side of a 4- by 5-mm square of mica (approximately twice the size of an EM grid). The grid was washed in 0.5% acetic acid and then with acetone. The carbon film was broken to free the specimen grid, after which the grid was placed in stain solution-neutral 1% aqueous phosphotungstic acid for 30 s. After being blotted dry, the grid was examined using the FEI Tecnai G2 TEM.
Epithelial cell culture.
HeLa cells were grown and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (100 IU/ml), streptomycin (100 IU/ml), and amphotericin B (2.5 mg/ml) (Invitrogen, Carlsbad, CA) at 37°C under a 5% CO2 atmosphere. Confluent stock cultures were trypsinized, adjusted to approximately 5 × 103 cells/ml, seeded into 12-well plates (Nunc, Rochester, NY) at 1 ml per well, and further incubated for 48 h to reach semiconfluence (105 cells per well).
Adherence and standard antibiotic protection assay.
Invasion was quantified using the standard antibiotic protection assay (58). Briefly an isolated colony harvested from solid agar plate was grown to exponential phase in BHI broth. The cells were then centrifuged, washed three times in PBS, and then adjusted to 107 CFU/ml of bacteria (confirmed by colony count) in Dulbecco's modified Eagle's medium. Epithelial cell monolayers were washed three times with PBS, infected with bacteria at a multiplicity of infection (MOI) of 1:100 (105 epithelial cells), and then incubated at 37°C for 90 min under a 5% CO2 atmosphere. Nonadherent bacteria were removed by washing with PBS, while cell surface-bound bacteria were killed with metronidazole (200 μg/ml, 60 min). P. gingivalis W83 and all the isogenic mutants are sensitive to 100 μg/ml of metronidazole. After removal of antibiotic, the internalized bacteria were released by osmotic lysis in sterile distilled water with scraping. Lysates were serially diluted, plated (in duplicate) on BHI agar, and incubated for 6 to 10 days. The number of bacterial cells recovered was expressed as a percentage of the original inoculum. The number of adherent bacteria was obtained by subtracting the number of intracellular bacteria from the total number of bacteria obtained in the absence of metronidazole (6).
RESULTS
In silico analysis.
The genome of P. gingivalis revealed the presence of three sialidase-related genes, of which one (PG0352) is annotated as a “conserved hypothetical protein or putative sialidase” gene. The other two genes (PG0778 and PG1724) are annotated as O-sialoglycoprotease genes (gcp) (www.oralgen.lanl.gov). No upstream or downstream sialidase-related genes are associated with PG0352, PG0778, or PG1724. P. gingivalis was missing homologs similar to a large putative sialic acid utilization and transport locus present in the ge- nome of T. forsythia (oral pathogen sequence databases [www.oralgen.lanl.gov]).
The clad containing PG0352 was more closely related (68% homology) to sialidases from Actinomyces naeslundii (encoded by ANA1493), T. forsythia (encoded by TF0035 and TF2207), Clostridium perfringens, and Bacteroides fragilis (Fig. 1A). PGO352 from P. gingivalis W83 shows 85% homology to PGN1608 from P. gingivalis ATCC 33277. The variations were due to amino acid substitutions at positions 60 (A→V), 87 (L→I), 151(H→N), 152 (N→D), 159 (Q→K), 160 (R→K), 183 (L→H), 185 (E→K), and 189 (H→Y). These variations were predominantly found in the transmembrane domain of the protein.
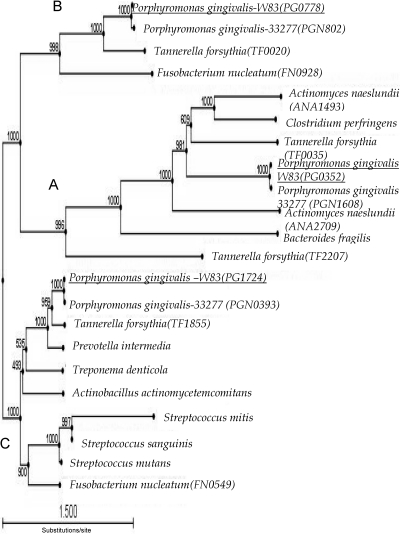
Fig. 1.
Phylogenetic analysis of sialidases and O-sialoglycoprotease from several oral and anaerobic bacteria. The P. gingivalis putative sialidase (PG0352) segregates in clad A with sialidases from other bacteria. One of the P. gingivalis O-sialoglycoproteases (PG0778) segregates in clad B. Clad C segregates into two subclads, one bearing the streptococcal O-sialoglycoprotease and the second bearing the P. gingivalis O-sialoglycoprotease (PG1724) along with those of other periodontal pathogens.
The PG0352 sialidase clad was more closely related to the O-sialoglycoprotease clad, which contains P. gingivalis PG0778, T. forsythia TF0020, and F. nucleatum FN0928 (Fig. 1B). It is noteworthy that, similar to what was found for P. gingivalis, the two O-sialoglycoproteases (encoded by gcp) from T. forsythia (TF0020 and TF1855) segregated into different clads. However, TF0020 was more closely related to the sialidases.
The second cluster containing PG1724 is bifurcated into two subclads. PG1724 is more closely related to the O-sialoglycoproteases from the oral pathogens (Fig. 1C).
Multiple amino acid sequence alignment of the sialidase and two sialidase-related proteins from P. gingivalis revealed unique domains and motifs (see Fig. S1 in the supplemental material). The O-sialoglycoprotease PG0778 revealed an N-terminal metal binding domain followed by a transmembane helix domain consisting of 22 amino acids between positions 17 and 37. The end of the transmembrane domain bears a twin arginine peptide signal followed by a cleavage site (see Fig. S1A in the supplemental material). PG0352 revealed a 13-amino-acid stretch of an inner membrane domain followed by a transmembrane domain consisting of 17 amino acids. A signal peptide consisting of His-Leu-Ser-Gly followed by a potential cleavage site was observed at the end of the transmembrane domain (see Fig. S1B in the supplemental material). This transmembrane domain appears to form the helix encompassing a hydrophobic region of 17 amino acids. The presence of a signal peptide at the N terminus suggests that the sialidase of P. gingivalis PG0352 is membrane bound. The entire protein contains repetitive hydrophobic aspartic acid motifs in six positions and also contains a long catalytic domain between positions 182 and 240. The catalytic domain encompasses the consensus a sequence motif that is characteristic of other sialidases (see Fig. S1B in the supplemental material). It also contains the characteristic SXDXGXTW domain at four positions between amino acid positions 230 and 240, 290 and 300, 360 and 370, 410 and 420, and 468 and 475. However, the signature sequence motif at the fifth position, between 468 and 475, was found to be partial and incomplete (see Fig. S2 in the supplemental material). These signature motifs are well conserved in the bacterial sialidases and also in the other oral pathogens studied (7). A carbohydrate-specific lectin binding domain between amino acids 198 and 218 was predicted.
PG1724 revealed an N-terminal metalloprotease consensus domain with a potential cleavage site. A transmembrane domain was observed between positions 136 and 152. In contrast to PG0778, PG1724 contains only one transmembrane domain (see Fig. S1C in the supplemental material). Two metal binding histidine domains are present, one at the 115th position before the transmembrane domain and the other at the 289th position. Both PG0778 and PG1724 contain metal binding domains at the N-terminal and C-terminal ends, with a cleavage site at the N terminus. However, PG1724 lacks a signal peptide. In PG1724 a lectin binding domain was observed between amino acids 277 and 282.
Multiple sequence alignment of the O-sialoglycoprotease amino acid sequences of oral pathogens with PG0778 and PG1724 revealed a DAXG/DAXD consensus signature motif. Asp boxlike motifs (SXDXTXSV, SXAXGXXW, SXGXSXII, and SXSXGXXTX) similar to the unique Asp box sequence SXDXGXTW of the sialidases were observed (see Fig. S3 in the supplemental material).
In silico modeling of PG0352 confirmed that the protein could be classified under the BNR-neuraminidase class (Pfam structural classification CL434). PG0778 and PG1724 showed the catalytic beta propeller structure in which PG0778 had a unique terminal cleft and a molecular groove structure (see Fig. S4 in the supplemental material).
Construction of PG0352, PG0778, and PG1724 deletion mutants in P. gingivalis W83.
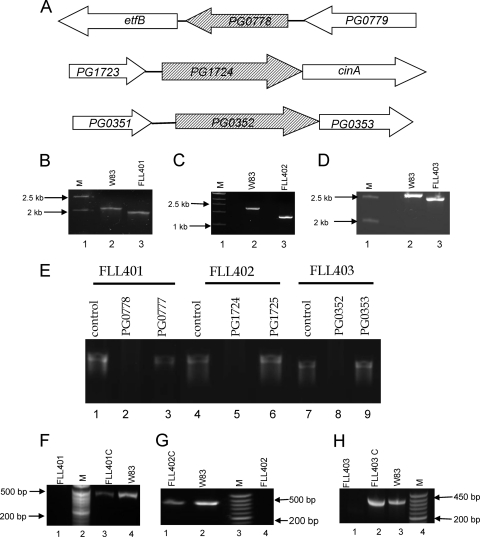
Isogenic mutants of P. gingivalis defective in PG0352, PG0778, and PG1724 were constructed by allelic-exchange mutagenesis. The schematic representation of the genetic structures of PG0352, PG0778, and PG1724 is shown in Fig. 2A. Following electroporation and plating on selective medium, we detected erythromycin-resistant colonies after a 5- to 7-day incubation period. The mutants were confirmed by colony PCR (Fig. 2B, C, and D) and DNA sequencing. To compare their phenotypic properties with those of wild-type strain W83, all mutants were plated on BHI blood agar plates. Similar to the wild-type strain, the PG0352- and PG1724-defective mutants displayed a black-pigmented phenotype. In contrast, the PG0778-defective isogenic mutants were not black pigmented. Mutants designated P. gingivalis FLL401 (PG0778::ermF), P. gingivalis FLL402 (PG1724::ermF), and P. gingivalis FLL403 (PG0352::ermF-ermAM) were randomly chosen for further study. Inactivation of the sialidase gene (PG0352) and sialoglycoprotease genes (PG0778 and PG1724) had no polar effects on the expression of their downstream genes (Fig. 2E). A generation time of approximately 3 h was determined for the wild type and P. gingivalis FLL403 compared to 10 and 13 h for P. gingivalis FLL402 and P. gingivalis FLL401, respectively. Complementation of the PG0778-, PG1724-, and PG0352-defective isogenic mutants was confirmed by RT-PCR analysis (Fig. 2F, G, and H).
Fig. 2.
Construction and confirmation of P. gingivalis isogenic mutants. (A) Chromosomal structures of PG0778, PG1724, and PG0352. Hatched arrows indicate the coding genes, and white arrows indicate genes located downstream. The direction of the arrows indicates the transcription orientation. (B) Colony PCR confirmation of the mutant FLL401.Lane 1, molecular size markers; lane 2, PCR amplification of 2.2-kb fragment from W83; lane 3, 1.8-kb PCR amplicon. The primers used were 1 and 6 (see Table S2 in the supplemental material). (C) Colony PCR confirmation of the mutant FLL402. Lane 1, molecular size markers; lane 2. 1.7-kb PCR amplicon from W83; lane 3, 0.9-kb PCR amplicon from FLL402. The primers used were 7 and 12 (see Table S2 in the supplemental material). (D) Colony PCR confirmation of the mutant FLL403. Lane 1, molecular size markers; lane 2, 2.5-kb PCR amplicon from W83; lane 3, 2.3-kb PCR amplicon from FLL402. The primers used were 13 and 14 (see Table S2 in the supplemental material). (E) RT-PCR confirmation showing the nonpolar effects on the inactivated sialidase (PG0352) and sialoglycoproteases (PG0778 and PG1724) in isogenic defective mutants. Lane 1, PG0778 product using W83 cDNA as the template (472 bp); lane 2, PG0778 product using FLL401 cDNA as the template; lane 3, PG0777 product using FLL401 cDNA as the template (436 bp); lane 4, PG1724 product using W83 cDNA as the template (451 bp); lane 5, PG1724 product using FLL402 cDNA as the template; lane 6, PG1725 product using FLL402 cDNA as the template (447 bp); lane 7, PG0352 product using W83 cDNA as the template (360 bp); lane 8, PG0352 product using FLL403 cDNA as the template; lane 9, PG0353 product using FLL403 cDNA as the template (179 bp). (F) Transcription analysis of P. gingivalis mutant FLL401 by RT-PCR. The primers used were 19 and 20 (see Table S2 in the supplemental material). Lane 1, FLL401 mutant showing no amplification of the transcript; lane 2, molecular size markers; lane 3, complemented FLL401 showing a 472-bp amplified product; lane 4, wild-type W83. (G) Transcription analysis of P. gingivalis mutant FLL402 by RT-PCR. The primers used were 21 and 22 (see Table S2 in the supplemental material). Lane 1, complemented FLL401 showing 451-bp amplified product; lane 2, wild-type W83; lane 3, molecular size markers; lane 4, FLL401 mutant showing no amplification of the transcript. (H) Transcription analysis of P. gingivalis mutant FLL403 by RT-PCR. The primers used were 25 and 26 (see Table S2 in the supplemental material). Lane 1, FLL403 mutant showing no amplification of the transcript; lane 2, complemented FLL403 showing 360-bp amplified product; lane 3, wild-type W83; lane 4, molecular size markers.
PG0778 is the major contributor to sialidase activity in P. gingivalis.
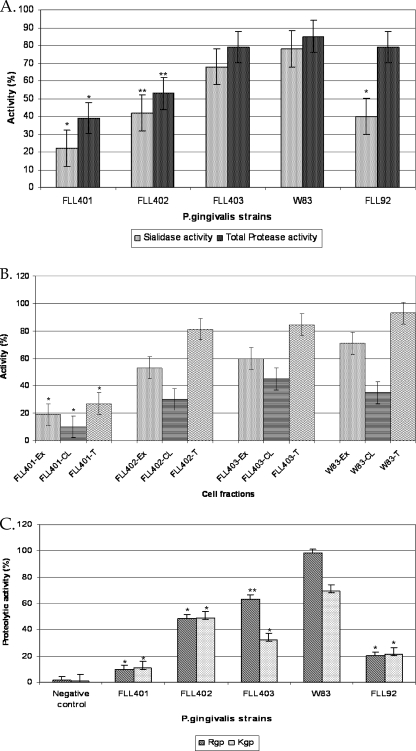
Sialidase activity was determined using whole-cell cultures of the P. gingivalis isogenic mutant strains and their complemented derivatives. As shown in Fig. 3 A, sialidase activity in P. gingivalis FLL401 was reduced by approximately 70% in comparison to those in FLL402 and FLL403, which were reduced by approximately 42 and 5%, respectively. Complementation with the wild-type genes in trans restored sialidase activity to the parent strain levels (see Fig. S5A in the supplemental material). Most of the sialidase activity in P. gingivalis W83 and isogenic mutants was localized to the extracellular fraction (Fig. 3B).
Fig. 3.
Comparison of total protease and sialidase activities in P. gingivalis. P. gingivalis was grown to exponential phase (OD600 of 0.8) in 10 ml of BHI broth supplemented with hemin and vitamin K. (A) Total protease and sialidase activities of P. gingivalis mutants FLL401, FLL402, FLL403, and FLL92. Total protease activity was estimated using a FRET-based assay, and sialidase was estimated using an enzyme-based assay method. *, P < 0.001; **, P < 0.01. (B) Distribution of sialidase activity in P. gingivalis. Sialidase activity using the Amplex Red neuraminidase kit was tested in the total cell fraction (T), the extracellular fraction (Ex), and cell lysate (CL). *, P < 0.001. (C) Gingipain activity of P. gingivalis mutants. Activity against BAPNA (Rgp) and ALNA (Kgp) was tested in whole-cell culture. The results shown are from three independent experiments. *, P < 0.001; **, P < 0.01.
Disruption of the sialidase-related genes affects gingipain activity.
The gingipains, which are both extracellular and cell membrane associated, consist of arginine-specific protease (Arg-gingipain [Rgp]) and lysine-specific protease (Lys-gingipain [Kgp]) (9). Posttranslational modification, including the addition of sialic acid residues, is important in gingipain biogenesis (9, 10, 53). Because the VimA protein can play a role in the regulation of this process and can interact with other proteins, including PG0352 (53), the effect of sialidase/sialoglycoprotease on gingipain activity was determined. Inactivation of PG0778, PG1724, and PG0352 resulted in reduced gingipain activity in the isogenic mutants compared to the wild type (Fig. 3C). It is noteworthy that P. gingivalis FLL401 (PG0778::ermF), which showed background gingipain activity, also had the greatest reduction of sialidase activity (Fig. 3A). P. gingivalis FLL92, a vimA-defective mutant which has been previously shown to have reduced gingipain activity (1, 30, 53), also showed reduced sialidase activity (Fig. 3A). Although there was a dramatic reduction in gingipain activity in P. gingivalis sialidase/sialoglycoprotease isogenic mutants, there were no changes in the expression of the gingipain genes (data not shown).
Among the proteolytic enzymes in P. gingivalis, the gingipains are responsible for at least 85% of the proteolytic activity (24). A FRET-based method was used to determine the effects of sialidase activity on nongingipain proteolytic activity in P. gingivalis. Inactivation of PG0352 had no significant effect on proteolytic activity; however, in PG0778- and PG1774-defective mutants the reductions were 35% and 52%, respectively. It is noteworthy that, although P. gingivalis FLL92 showed reduced gingipain activity, consistent with previous reports (30) (Fig. 3C), nongingipain activity was reduced by only approximately 5% compared to the wild-type W83 strain (Fig. 3A). Taken together, these results suggest that O-sialoglycoprotease and sialidase may be involved in the posttranscriptional regulation of gingipains although we cannot rule out its effects on other nongingipain enzymes. Complementation of the sialidase/sialoglycoprotease isogenic mutant with the wild-type gene restored gingipain activity to wild-type levels (see Fig. S5B in the supplemental material).
Alteration in gingipain biogenesis in sialidase/sialoglycoprotease-defective mutants.
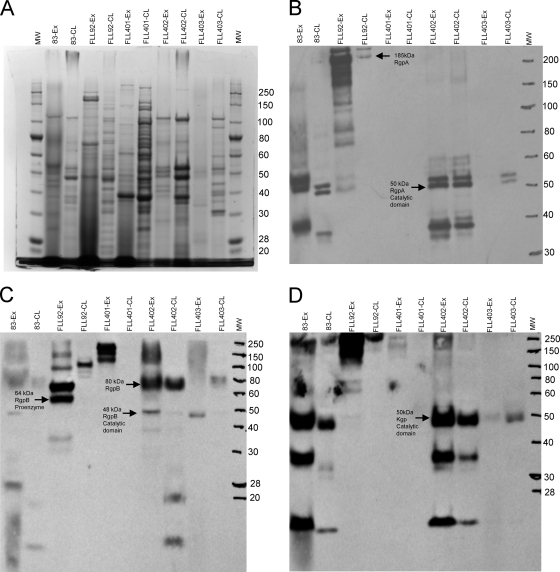
The presence of mRNA transcripts for the gingipain genes in the P. gingivalis sialidase/sialoglycoprotease-defective mutants may suggest that PG0778, PG1724, and PG0352 are involved in posttranscriptional regulation of gingipain maturation and activation. To examine any changes in extracellular or cellular proteins from P. gingivalis FLL401, FLL402, and FLL403 in comparison to the wild-type W83 and vimA-defective mutant FLL92, fractions from the exponential growth phases of all the strains were analyzed by SDS-PAGE. As shown in Fig. 4 A, there were multiple protein bands, in contrast to results for the wild type and P. gingivalis FLL92, that were unique to the sialidase/sialoglycoprotease-defective mutants. The presence of the gingipains was evaluated with antibodies against RgpA, RgpB, and Kgp. As shown in Fig. 4B to D, the sialidase- and sialoglycoprotease-defective mutants had multiple unique immunoreactive bands for RgpB, RgpA, and Kgp in contrast to the wild-type strain and P. gingivalis FLL92, the vimA-defective mutant. There were no immunoreactive bands for RgpA in P. gingivalis FLL401. Immunoreactive bands representing the catalytic domains for RgpA, RgpB, and Kgp were present in P. gingivalis FLL402 and FLL403 (Fig. 4B to D). Similar bands were missing in P. gingivalis FLL401.
Fig. 4.
Gingipain profile of P. gingivalis sialidase mutants compared with vimA mutant (FLL92) and wild-type W83 from exponential-phase cultures. Acetone-precipitated proteins isolated from supernatants from cultures grown to an OD600 of 0.8 were analyzed by SDS-PAGE and stained with SimplyBlue SafeStain (A) or analyzed by immunoblotting using gingipain-specific antibodies (B to D). Approximately 20 to 25 μg of protein was loaded in each lane. Antibodies: chicken anti-RgpA (B), rabbit anti-RgpB (C), and chicken anti-Kgp (D). Ex, extracellular fraction; CL, cell lysate fraction. Arrows indicate the unprocessed, partially processed, or mature gingipains; MW, molecular weight markers (thousands).
Differential recognition of sialic acid subtypes by P. gingivalis.
Sialylated glycoconjugates are common binding sites for both bacteria and other microbes (42, 48). Variation in glycan linkages and the extent of different substitutions in sialic acid are species specific (Table 2). Sialidase-sensitive hemagglutination with RBCs from different species was used to determine differential recognition of sialic acid subtypes by the P. gingivalis sialidase/sialoglycoprotease-defective mutants. As shown in Table 3, there was no hemagglutination of the horse RBCs by the isogenic mutants or wild-type strain. In contrast, a differential pattern of hemagglutination was observed for bovine, chicken, and sheep RBCs. High titers of P. gingivalis FLL401 were required to agglutinate bovine and sheep RBCs (Table 3). It is noteworthy that the wild-type strain and P. gingivalis FLL92 showed the same hemagglutination profile. Complementation of the isogenic mutant with the wild-type gene restored hemagglutination activity to wild-type levels.
Table 2.
Variation in glycan linkages in various species of RBCs
| Source of RBCs | Substitution(s)a |
|---|---|
| Human | 5N-Ac (no Gc), no 9O-Ac |
| Chicken | 5N-Ac, >95%; 5N-Gc, 5% |
| Sheep | 5N-Ac (no Gc), no 9O-Ac |
| Bovine | 5N-Ac |
| Horse | 5N-Gc, 90%; 4O-Ac, 6%; 9O-Ac, 4% |
Gc, glycolyl; Ac, acetyl.
Table 3.
Hemagglutination titers of P. gingivalisa
| Strain | Titer for RBCs from: |
|||
|---|---|---|---|---|
| Bovine | Chicken | Sheep | Horse | |
| FLL401 | 16 | 8 | 16 | 0 |
| FLL402 | 2 | 0 | 16 | 0 |
| FLL403 | 0 | 0 | 8 | 0 |
| W83 | 2 | 0 | 16 | 0 |
| FLL92 | 4 | 0 | 16 | 0 |
Hemagglutination titers of P. gingivalis mutants in bovine, chicken, sheep, and horse RBCs. There was no hemagglutination in horse RBCs by isogenic mutants. FLL401 showed high titers in bovine and sheep RBCs. FLL402 and FLL403 showed highest titers in sheep RBCs. Hemagglutination titer was determined as the reciprocal of the highest dilution showing complete hemagglutination.
Differential substrate specificity of P. gingivalis mutants.
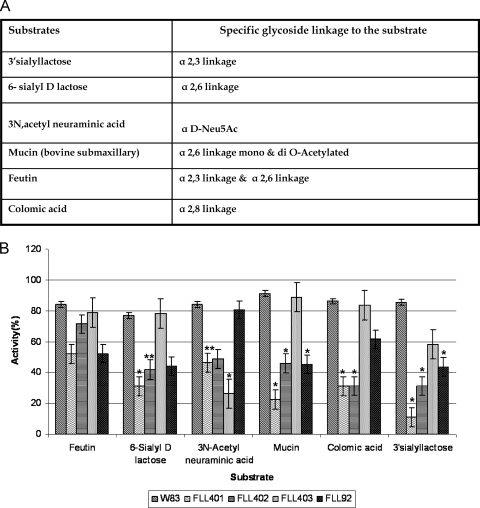
The ability of the P. gingivalis sialidase/sialoglycoprotease-defective mutants to release sialic acid from a variety of natural substrates with specific glycoside linkages was evaluated (Fig. 5 A). As shown in Fig. 5B, P. gingivalis FLL401, FLL402, and FLL403 had reduced activity compared to the wild-type strain against all the substrates tested. P. gingivalis FLL401 showed the lowest activity against mucin and 3′sialyllactose. P. gingivalis FLL403, carrying the defective sialidase, showed the lowest activity against 3N-acetylneuraminic acid. Complementation of the sialidase/sialoglycoprotease isogenic mutant with the wild-type gene restored substrate specificity similar to that of the wild type (see Fig. S5C in the supplemental material).
Fig. 5.
Substrate specificity of P. gingivalis mutants. (A) Table showing substrates used in this study with their respective glycoside linkages to the substrate. (B) Sialidase activity of P. gingivalis W83 and isogenic mutants FLL401, FLL402, and FLL403. The assay was performed using the Amplex Red neuraminidase (sialidase) assay kit (Molecular Probes). The three P. gingivalis mutants showed significant reduction in activity compared with the wild type (*, P < 0.001; **, P < 0.01).
The sialidase/sialoglycoprotease-defective mutants recognized different glycosidic linkages.
The abilities of different lectins (Table 4) to interact with the bacterial cell surface were evaluated using hemagglutination of sheep RBCs. In the presence of ConA, there was no hemagglutination in the wild type compared to the sialidase/sialoglycoprotease-defective mutants, which showed high titers (Table 5). The wild type showed high (256) titers for both LPA and WGA in contrast to the isogenic sialidase/sialoglycoprotease-defective mutants, which had titers of 4 to 16. P. gingivalis FLL403 had the lowest hemagglutination titer for LPA and the highest titer for PNA. Overall the lowest hemagglutination titers were observed for P. gingivalis FLL401, FLL402, and FLL403 for ABA, PNA, and LPA, respectively.
Table 4.
Lectins and their sugar-specific linkages
| Lectin (source) | Sugar specificity |
|---|---|
| ABA (Agaricus bisporus) | Galactose (β 1,3) N-acetylgalactosamine |
| ConA (Canavalia ensiformis) | α-Mannose; α-glucose |
| ECA (Erythrina cristagalli) | Galactose (β 1,4) N-acetylglucosamine |
| LPA (Limulus polyphemus) | Sialic acid (N-acetylneuraminic acid) |
| WGA (Triticum vulgaris) | N-Acetyl-β-d-glucosamine |
| PNA (Arachis hypogaea) | d-Galactose (β 1,3) N-acetylglucosamine |
| d-Galactose (β 1,4) N-acetylglucosamine |
Table 5.
Summary of lectin hemagglutination titersa
| Strain | Titer in the presence of: |
|||||
|---|---|---|---|---|---|---|
| ABA | ECA | LPA | ConA | WGA | PNA | |
| W83 | 32 | 64 | 256 | 0 | 256 | 16 |
| FLL401 | 0 | 2 | 4 | 128 | 4 | 8 |
| FLL402 | 8 | 32 | 16 | 256 | 16 | 0 |
| FLL403 | 8 | 4 | 0 | 256 | 8 | 128 |
Hemagglutination titer is the reciprocal of the highest dilution showing agglutination. P. gingivalis mutants FLL401 and FLL402 showed very low hemagglutination titers in the presence of ECA and LPA.
The cell surface is altered in the sialidase/sialoglycoprotease-defective mutants.
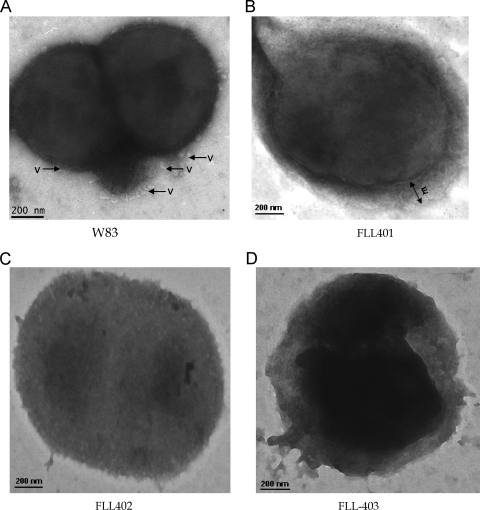
Sialylation can play a role in cell surface modification (28, 43). Electron microscopy was used to evaluate the cell surface ultrastructure of the wild type compared to the sialidase/sialoglycoprotease-defective mutants. While the wild type revealed an intact cell wall showing blebbing vesicles (Fig. 6 A), P. gingivalis FLL401, the O-sialoglycoprotease-defective mutant, revealed a fuzzy and thick external layer of the membrane that was found to be uniform along the whole area of the cell (Fig. 6B). The P. gingivalis FLL402 mutant showed an intact outer covering with minimal change in surface morphology (Fig. 6C). FLL403 showed a diffused outer covering and a densely stained cytoplasm (Fig. 6D). It is noteworthy that the three mutants did not show vesicle formation under the same growth conditions.
Fig. 6.
Electron micrograph showing variation in the surface morphology of P. gingivalis W83 and isogenic mutants FLL401, FLL402, and FLL403. The samples were processed from log-phase cultures of the mutants (OD600 = 0.8) negatively stained on 400 mesh copper grids using 1% phosphotungstic acid and viewed under the transmission electron microscope (FEI Technai G2). (A) W83 showing blebbing of membrane vesicles (V); (B) FLL401 showing fuzzy and thick external layer (E); (C) FLL402 showing intact outer covering; (D) FLL403 showing diffused outer covering with dense cytoplasm. Images represent a consistent view of several fields.
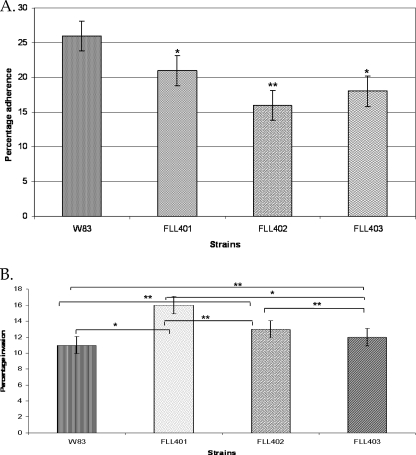
The sialidase/sialoglycoproteases can modulate the invasive capacity of P. gingivalis.
To investigate a putative role of the sialidase (PG0352) and O-sialoglycoproteases (PG0778 and PG1724) in the invasive capacity of P. gingivalis, we compared the wild type and isogenic mutants P. gingivalis FLL401, FLL402, and FLL403. Figure 7 A shows differential adherences to HeLa cells for all 3 mutants compared to the wild type. P. gingivalis FLL401 showed the least adherence. FLL402 showed more adherence than the other two mutants. P. gingivalis FLL401 showed the highest invasion capacity, followed by FLL402 and FLL403 in descending order (Fig. 7B). In comparison with that of the wild-type strain, the invasion capacity was increased by 16% in FLL401, 13% in FLL402, and 11% in FLL401. Complementation of the isogenic mutant with the wild-type gene restored the adhesion and invasion capacity to the wild-type levels.
Fig. 7.
(A) Adherence by P. gingivalis to epithelial cells. Experiments were made in triplicate. Error bars, standard deviations (SD). t tests revealed statistically significant differences between the wild type and mutants and among the three mutants (*, P < 0.001; **, P < 0.01). (B) Invasion by P. gingivalis of epithelial cells. Experiments were made in triplicate. Error bars, SD. t tests revealed statistically significant differences between the wild type and mutants and among the three mutants (*, P < 0.01; **, P < 0.05).
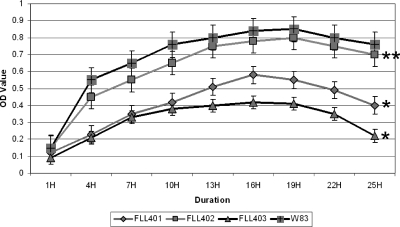
The sialidase/sialoglycoprotease-defective mutants are sensitive to oxidative stress.
Sialic acid can play a protective role against oxidative damage (22). The relative significance of the P. gingivalis sialidase and O-sialoglycoprotease in oxidative stress resistance was evaluated by exposure of the isogenic defective mutants to hydrogen peroxide. As shown in Fig. 8, isogenic mutants defective in PG0778 and PG0352 were more sensitive to hydrogen peroxide than the wild type. P. gingivalis FLL402, carrying the PG1724 inactivated gene, showed a sensitivity pattern similar to that of the wild type.
Fig. 8.
The sialidase/sialoglycoprotease-defective mutants are more sensitive to hydrogen peroxide than the wild type. Actively growing P. gingivalis cells were incubated with 0.25 mM hydrogen peroxide in the absence of cysteine, and growth was measured over 25 h (*, P < 0.001; **, P < 0.05).
DISCUSSION
The microenvironment of the periodontal pocket contains an abundance of sialylated glycoproteins such as fibronectin (33), transferrin, and immunoglobulins (15), found in saliva and gingival crevicular fluid (48). While a number of periodontal bacteria are known to exploit host sialylated glycoproteins as a nutrient source, sialidase activity also plays an important role in their pathogenicity. Sialidase treatment of immunoglobulins can make them more susceptible to proteolytic degradation; sialidases may also reveal cryptic receptors and/or adhesion sites for bacterial binding/interaction and hence tissue and host tropism (5, 19). Our studies have identified three sialidase-related genes in P. gingivalis which have shown a specific pattern of clustering with other related genes from the bacteria. Our in silico analysis and gene knockout studies revealed PG0352 to be a putative sialidase and an ortholog of the products of nanH of T. forsythia (TF0035) and ANA1493 and ANA2709 of A. naeslundii. PG1724 showed similarity to glycoproteases from Bacillus subtilis, Haemophilus influenzae, and E. coli. Further, based on domain similarity, PG0778 and PG1724 were grouped with other metal-dependent proteases and also may have chaperone activity (oral pathogen sequence databases [www.oralgen.lanl.gov]). In our study, the sialoglycoprotease appears to be important for the growth of P. gingivalis. It is likely that these enzymes are important in the breakdown of glycoprotein conjugate to satisfy the asaccharolytic requirements of P. gingivalis. However, we cannot rule out the involvement of these enzymes in other important metabolic activities of the cell, including the generation of sialic acid from host-derived substrates. This sialic acid could be involved in stability of proteins, including the gingipains. Thus, it is likely that important proteins involved in both the metabolic and pathogenic capacity of the organism are modulated by the presence or absence of sialic acid. This is under further investigation.
Amino acid sequence analysis of PG0352, PG0778, and PG1724 revealed the characteristic sialidase Asp signature motif (SXDXGXTW) along with other highly conserved amino acids. It is interesting to note that this Asp box, which is unique to sialidases, was also observed in the O-sialoglycoproteases from other oral pathogens. This may suggest functional relatedness and may highlight their relative significance in microbial colonization of the periodontal pocket with an abundance of sialylated glycoproteins (54). In PG0352, a FRIP region, which is known to bind the carboxylate group of sialic acid (14), was juxtaposed to the exosialidase domain. Our study identified a common motif (DAXG/DAXD [Asp-Ala-X-Gly/Asp-Ala-X-Asp]) that is unique to the P. gingivalis O-sialoglycoproteases. In addition to the conserved catalytic domain, a noncatalytic lectin-like domain, which may be involved in carbohydrate recognition and cell adhesion (14), was conserved in PG0352, PG0778, and PG1724.
Inactivation of PG0778 resulted in a non-black-pigmented phenotype and also a 70 to 90% reduction in sialidase and gingipain activities. Inactivation of the two other sialidase-related genes did not affect the black pigmentation phenotype even though sialidase and gingipain activities were reduced by approximately 50%. Black pigmentation is due to iron accumulation on the bacterial cell surface (26). Gingipain activity has been shown to play a role in iron protoporphyrin accumulation on the bacterial cell surface (45); however, the involvement of a sialoglycoprotease is unclear. Because the level of expression of the gingipain genes was unaltered in the sialidase/sialoglycoprotease isogenic mutants, it is likely that PG0778 and the other sialidase-related gene is involved in the posttranslational regulation of the gingipains. Analysis of the monosaccharide composition of the gingipains indicates the presence of at least nine different sugars, including high levels of sialic acid (37, 40). The level of sialylation and its role in gingipain maturation/activation are unclear. Catalytic domains for the gingipains were observed in P. gingivalis FLL402 and FLL403 but were missing in P. gingivalis FLL401. This may imply that PG0778 plays a more significant role in gingipain biogenesis in P. gingivalis. A specific mechanism for this observation is under further investigation. The sialidase/sialoglycoprotease appeared not to affect nongingipain protease activity.
The P. gingivalis sialidase/sialoglycoproteases recognized different structural subtypes of sialic acid. The PG0778 sialoglycoprotease interacted with the subtypes containing 5N-acetyl (5N-Ac) and 9O-Ac modifications, which are predominantly found in eukaryotic cells (18). The PG0352 sialidase recognized the 5N-Ac substitution. The subtype facilitating attachment of PG1774 contained 5N-Ac with 9O-Ac substitutions. The significantly reduced activity of P. gingivalis FLL403 against 3N-acetylneuraminic acid further confirms PG0352 as a putative sialidase. The cleavage level of the mucin substrate indicated that glycoprotein substrates with O substitutions might be resistant to PG0352 in contrast to the sialoglycoproteases PG0778 and PG1724. Furthermore, PG0778 may be specific to glycoproteins with α2,3 and α2,6 glyco-side linkages. PG1724 may be specific to glycoproteins with α2,3 or α2,8 glycoside linkages. The α2,3 glycoside linkage is one of the most common glycoside linkages among bacteria, including P. gingivalis (3). From our lectin studies, PG0778 showed specificity to galactose (β 1,3) N-acetylgalactosamine and galactose (β1,4) N-acetylglucosamine. PG1724 was specific to d-galactose (β 1,3) N-acetylglucosamine, d-galactose (β 1,4) N-acetylglucosamine, d-Gal-β-1-3dGalNAc, and d-Gal-β-1-4dGlcNAc, while PG0352 was specific to sialic acid.
Taken together, these data could imply that colonization of the periodontal pocket by P. gingivalis may require synergy between the sialidase and sialoglycoproteases. For example, the mucin on the mucin-covered epithelial cells may be first hydrolyzed by PG0778 and/or PG1724. This would expose the terminal sialic acid on epithelial cells, which is then cleaved by PG0352 to mediate/enhance attachment. However, based on the ability of sialoglycoproteases to cleave other substrates, we cannot rule out a role for them in mediating bacterium-host or bacterium-bacterium interactions. The multiple desialylated glycoproteins or glycolipids could provide various receptors for P. gingivalis which would bind to the various exposed glycosidic residues when terminal sialic acid is cleaved. This raises the question of what role the P. gingivalis sialidase and sialoglycoproteases may play in biofilm formation. The sialidases from several bacteria, including the oral pathogen T. forsythia, have a role in mucosal infection by participating in biofilm production by presumably targeting sialylated bacterial exopolysaccharides (32, 38, 46).
Our adhesion/invasion studies using HeLa cells suggested that these functions are affected by the P. gingivalis sialidase and sialoglycoproteases. While sialidase/sialoglycoprotease-deficient isogenic mutant cells were shown to be physically attached to the HeLa cells, the invasion rates of these strains were significantly higher than the wild type. Because highly invasive P. gingivalis subtypes have reduced cell-associated arginine-specific protease activity (47), it is still unclear what direct role the sialidase or sialoglycoproteases play in this process. It is noteworthy that P. gingivalis FLL401, which had the highest reduction in gingipain activity, had the highest invasion rate. The coordinated regulation of sialidase/sialoglycoproteases and gingipain activities is unclear. It is likely that the regulation may involve the VimA protein (52, 53). Preliminary studies suggest that VimA is multifunctional and may be involved in acetylation (31) and in protein sorting (W. Aruni, unpublished data). A posttranslational regulatory mechanism that is VimA dependent is under further investigation.
Release of free monomeric sialic acid when it is cleaved from the sugar chain can detoxify H2O2 (22). This reaction reduces the dangerous oxygen radicals, as H2O2 and NANA are converted into H2O and nontoxic carboxylic acid (21, 22). The ability of the P. gingivalis sialidase/sialoglycoproteases to cleave multiple substrates, which could result in the release of sialic acid, would be consistent with the increased sensitivity of the sialidase/sialoglycoprotease-defective isogenic mutants to H2O2 compared to the wild-type strain. This could be one of multiple oxidative resistance strategies utilized by P. gingivalis to survive in the periodontal pocket.
Collectively, the data from this study suggest a model for P. gingivalis infection and survival in the periodontal pocket. The sialidase and sialoglycoproteases can modify host glycoconjugates and expose potential binding receptors for bacterium-host interactions. Cleavage of desialylated proteins can provide the substrate for the proteases to generate the peptides needed to satisfy the asaccharolytic property of P. gingivalis. The released sialic acid can act as a reactive oxygen species (ROS) scavenger to reduce the oxidative stress in the inflammatory environment of the periodontal pocket. Thus, the sialidase and sialoglycoproteases may be important in the pathogenesis of P. gingivalis. This study represents a starting point in our exploration to further understand the role of sialylation in the pathogenic potential of P. gingivalis. The identification and characterization of genes such as trans-sialidase and sialyltransferase genes will shed light on sialic acid metabolism/utilization in P. gingivalis. This could lead to the development of novel therapeutic approaches for the treatment of P. gingivalis-associated diseases.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Loma Linda University and public health grants DE-13664 and DE-019730 from NIDER (to H.M.F.).
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 18 April 2011.
REFERENCES
- 1. Abaibou H., et al. 2001. vimA gene downstream of recA is involved in virulence modulation in Porphyromonas gingivalis W83. Infect. Immun. 69:325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Angata T., Varki A. 2002. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 102:439–469 [DOI] [PubMed] [Google Scholar]
- 3. Bertozzi C. R., Freeze H. H., Varki A., Esko J. D. 2009. Glycans in biotechnology and the pharmaceutical industry, p. 769–784In Varki A., et al. (ed.), Essentials of glycobiology, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 4. Bobetsis Y. A., Barros S. P., Offenbacher S. 2006. Exploring the relationship between periodontal disease and pregnancy complications. J. Am. Dent. Assoc. 137(Suppl.):7S–13S [DOI] [PubMed] [Google Scholar]
- 5. Cacalano G., Kays M., Saiman L., Prince A. 1992. Production of the Pseudomonas aeruginosa neuraminidase is increased under hyperosmolar conditions and is regulated by genes involved in alginate expression. J. Clin. Invest. 89:1866–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Castaneda-Roldan E. I., et al. 2004. Adherence of brucella to human epithelial cells and macrophages is mediated by sialic acid residues. Cell. Microbiol. 6:435–445 [DOI] [PubMed] [Google Scholar]
- 7. Copley R. R., Russell R. B., Ponting C. P. 2001. Sialidase-like Asp-boxes: sequence-similar structures within different protein folds. Protein Sci. 10:285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corfield T. 1992. Bacterial sialidases—roles in pathogenicity and nutrition. Glycobiology 2:509–521 [DOI] [PubMed] [Google Scholar]
- 9. Curtis M. A., duse-Opoku J., Rangarajan M. 2001. Cysteine proteases of Porphyromonas gingivalis. Crit. Rev. Oral Biol. Med. 12:192–216 [DOI] [PubMed] [Google Scholar]
- 10. Curtis M. A., Thickett A., Slaney J. M., Rangarajan M., duse-Opoku J., Shepherd P., Paramonov N., Hounsell E. F. 1999. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect. Immun. 67:3816–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Farre D., et al. 2003. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 31:3651–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fletcher H. M., et al. 1995. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect. Immun. 63:1521–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gallagher A., duse-Opoku J., Rangarajan M., Slaney J. M., Curtis M. A. 2003. Glycosylation of the Arg-gingipains of Porphyromonas gingivalis and comparison with glycoconjugate structure and synthesis in other bacteria. Curr. Protein Pept. Sci. 4:427–441 [DOI] [PubMed] [Google Scholar]
- 13a. Gardner R. G., Russell J. B., Wilson D. B., Wang G. R., Shoemaker N. B. 1996. Use of a modified Bacteroides-Prevotella shuttle vector to transfer a reconstructed beta-1,4-d-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B(1)4. Appl. Environ. Microbiol. 62:196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gaskell A., Crennell S., Taylor G. 1995. The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll. Structure 3:1197–1205 [DOI] [PubMed] [Google Scholar]
- 15. Grenier D., et al. 2001. Role of gingipains in growth of Porphyromonas gingivalis in the presence of human serum albumin. Infect. Immun. 69:5166–5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hajishengallis G. 2009. Porphyromonas gingivalis-host interactions: open war or intelligent guerilla tactics? Microbes Infect. 11:637–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harris J. R. 2007. Negative staining of thinly spread biological samples. Methods Mol. Biol. 369:107–142 [DOI] [PubMed] [Google Scholar]
- 18. Haverkamp J., Schauer R., Wember M. 1976. Neuraminic acid derivatives newly discovered in humans: N-acetyl-9-O-L-lactoylneuraminic acid, N, 9-O-diacetylneuraminic acid and N-acetyl-2,3-dehydro-2-deoxyneuraminic acid. Hoppe-Seylers Z. Physiol. Chem. 357:1699–1705 [DOI] [PubMed] [Google Scholar]
- 19. Honma K., Mishima E., Sharma A. 2011. Role of Tannerella forsythia NanH sialidase in epithelial cell attachment. Infect. Immun. 79:393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horton R. M., Cai Z. L., Ho S. N., Pease L. R. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528–535 [PubMed] [Google Scholar]
- 21. Iijima R., Takahashi H., Ikegami S., Yamazaki M. 2007. Characterization of the reaction between sialic acid (N-acetylneuraminic acid) and hydrogen peroxide. Biol. Pharm. Bull. 30:580–582 [DOI] [PubMed] [Google Scholar]
- 22. Iijima R., Takahashi H., Namme R., Ikegami S., Yamazaki M. 2004. Novel biological function of sialic acid (N-acetylneuraminic acid) as a hydrogen peroxide scavenger. FEBS Lett. 561:163–166 [DOI] [PubMed] [Google Scholar]
- 23. Johnson L. S., Eddy S. R., Portugaly E. 2010. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kadowaki T., et al. 1998. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. J. Biol. Chem. 273:29072–29076 [DOI] [PubMed] [Google Scholar]
- 25. Lamont R. J., Jenkinson H. F. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lewis J. P., Dawson J. A., Hannis J. C., Muddiman D., Macrina F. L. 1999. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J. Bacteriol. 181:4905–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marmur J. 1961. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3:585–594 [DOI] [PubMed] [Google Scholar]
- 28. Nabi I. R., Raz A. 1987. Cell shape modulation alters glycosylation of a metastatic melanoma cell-surface antigen. Int. J. Cancer 40:396–402 [DOI] [PubMed] [Google Scholar]
- 29. Nakano V., Fontes Piazza R. M., vila-Campos M. J. 2006. A rapid assay of the sialidase activity in species of the Bacteroides fragilis group by using peanut lectin hemagglutination. Anaerobe 12:238–241 [DOI] [PubMed] [Google Scholar]
- 30. Olango G. J., Roy F., Sheets S. M., Young M. K., Fletcher H. M. 2003. Gingipain RgpB is excreted as a proenzyme in the vimA-defective mutant Porphyromonas gingivalis FLL92. Infect. Immun. 71:3740–3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Osbourne D. O., et al. 2010. The role of vimA in cell surface biogenesis in Porphyromonas gingivalis. Microbiology 156:2180–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Parker D., et al. 2009. The NanA neuraminidase of Streptococcus pneumoniae is involved in biofilm formation. Infect. Immun. 77:3722–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pham T. K., et al. 2010. A quantitative proteomic analysis of biofilm adaptation by the periodontal pathogen Tannerella forsythia. Proteomics 10:3130–3141 [DOI] [PubMed] [Google Scholar]
- 34. Pihlstrom B. L., Michalowicz B. S., Johnson N. W. 2005. Periodontal diseases. Lancet 366:1809–1820 [DOI] [PubMed] [Google Scholar]
- 35. Plumbridge J., Vimr E. 1999. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J. Bacteriol. 181:47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Powell L. D., Varki A. P. 2001. Sialidases. Curr. Protoc. Mol. Biol. Chapter 17:Unit17 [DOI] [PubMed] [Google Scholar]
- 37. Rangarajan M., Hashim A., duse-Opoku J., Paramonov N., Hounsell E. F., Curtis M. A. 2005. Expression of Arg-gingipain RgpB is required for correct glycosylation and stability of monomeric Arg-gingipain RgpA from Porphyromonas gingivalis W50. Infect. Immun. 73:4864–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roy S., Douglas C. W., Stafford G. P. 2010. A novel sialic acid utilization and uptake system in the periodontal pathogen Tannerella forsythia. J. Bacteriol. 192:2285–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 40. Sakai E., et al. 2007. Construction of recombinant hemagglutinin derived from the gingipain-encoding gene of Porphyromonas gingivalis, identification of its target protein on erythrocytes, and inhibition of hemagglutination by an interdomain regional peptide. J. Bacteriol. 189:3977–3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sali A., Blundell T. L. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815 [DOI] [PubMed] [Google Scholar]
- 42. Schauer R. 2009. Sialic acids as regulators of molecular and cellular interactions. Curr. Opin. Struct. Biol. 19:507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Severi E., Hood D. W., Thomas G. H. 2007. Sialic acid utilization by bacterial pathogens. Microbiology 153:2817–2822 [DOI] [PubMed] [Google Scholar]
- 44. Sharma A., Inagaki S., Sigurdson W., Kuramitsu H. K. 2005. Synergy between Tannerella forsythia and Fusobacterium nucleatum in biofilm formation. Oral Microbiol. Immunol. 20:39–42 [DOI] [PubMed] [Google Scholar]
- 45. Smalley J. W., Thomas M. F., Birss A. J., Withnall R., Silver J. 2004. A combination of both arginine- and lysine-specific gingipain activity of Porphyromonas gingivalis is necessary for the generation of the micro-oxo bishaem-containing pigment from haemoglobin. Biochem. J. 379:833–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Soong G., et al. 2006. Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. J. Clin. Invest. 116:2297–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Suwannakul S., Stafford G. P., Whawell S. A., Douglas C. W. 2010. Identification of bistable populations of Porphyromonas gingivalis that differ in epithelial cell invasion. Microbiology 156:3052–3064 [DOI] [PubMed] [Google Scholar]
- 48. Takamatsu D., Bensing B. A., Prakobphol A., Fisher S. J., Sullam P. M. 2006. Binding of the streptococcal surface glycoproteins GspB and Hsa to human salivary proteins. Infect. Immun. 74:1933–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 50. Vanterpool E., Aruni A. W., Roy F., Fletcher H. M. 2010. regT can modulate gingipain activity and response to oxidative stress in Porphyromonas gingivalis. Microbiology 156:3065–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vanterpool E., Roy F., Fletcher H. M. 2005. Inactivation of vimF, a putative glycosyltransferase gene downstream of vimE, alters glycosylation and activation of the gingipains in Porphyromonas gingivalis W83. Infect. Immun. 73:3971–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vanterpool E., Roy F., Sandberg L., Fletcher H. M. 2005. Altered gingipain maturation in vimA- and vimE-defective isogenic mutants of Porphyromonas gingivalis. Infect. Immun. 73:1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vanterpool E., et al. 2006. VimA is part of the maturation pathway for the major gingipains of Porphyromonas gingivalis W83. Microbiology 152:3383–3389 [DOI] [PubMed] [Google Scholar]
- 54. Varki A. 2009. Multiple changes in sialic acid biology during human evolution. Glycoconj. J. 26:231–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vimr E. R. 1994. Microbial sialidases: does bigger always mean better? Trends Microbiol. 2:271–277 [DOI] [PubMed] [Google Scholar]
- 56. Vriend G. 1990. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8:52–56, 29 [DOI] [PubMed] [Google Scholar]
- 57. Wang Q., Chang B. J., Riley T. V. 2010. Erysipelothrix rhusiopathiae. Vet. Microbiol. 140:405–417 [DOI] [PubMed] [Google Scholar]
- 58. Yilmaz O., Young P. A., Lamont R. J., Kenny G. E. 2003. Gingival epithelial cell signalling and cytoskeletal responses to Porphyromonas gingivalis invasion. Microbiology 149:2417–2426 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.