Abstract
In commercial poultry production, there is a lack of natural flora providers since chickens are hatched in the clean environment of a hatchery. Events occurring soon after hatching are therefore of particular importance, and that is why we were interested in the development of the gut microbial community, the immune response to natural microbial colonization, and the response to Salmonella enterica serovar Enteritidis infection as a function of chicken age. The complexity of chicken gut microbiota gradually increased from day 1 to day 19 of life and consisted of Proteobacteria and Firmicutes. For the first 3 days of life, chicken cecum was protected by increased expression of chicken β-defensins (i.e., gallinacins 1, 2, 4, and 6), expression of which dropped from day 4 of life. On the other hand, a transient increase in interleukin-8 (IL-8) and IL-17 expression could be observed in chicken cecum on day 4 of life, indicating physiological inflammation and maturation of the gut immune system. In agreement, the response of chickens infected with S. Enteritidis on days 1, 4, and 16 of life shifted from Th1 (characterized mainly by induction of gamma interferon [IFN-γ] and inducible nitric oxide synthase [iNOS]), observed in younger chickens, to Th17, observed in 16-day-old chickens (characterized mainly by IL-17 induction). Active modification of chicken gut microbiota in the future may accelerate or potentiate the maturation of the gut immune system and increase its resistance to infection with different pathogens.
INTRODUCTION
Salmonella enterica is one of the major causes of human food-borne gastroenteritis worldwide, and since poultry are considered to be the most important source of S. enterica for humans, measures of how to limit S. enterica prevalence in poultry are continuously being sought. The already-in-use measures aiming at the reduction of S. enterica prevalence in poultry include strict hygienic standards and vaccination with attenuated or inactivated vaccines. However, although hygienic standards are important for S. enterica control, at the same time, this represents an issue. Chickens for commercial production are hatched in a clean environment, and unlike all other farm animals, chickens will never get into contact with adult birds to become colonized by the healthy microflora of adults. Colonization of mucosal surfaces in newly hatched chickens is therefore a matter of coincidence, and if a bacterial pathogen appears in the environment, the sterile intestinal tract of a newly hatched chicken represents an empty ecological niche enabling such a pathogen essentially unrestricted multiplication followed by prolonged colonization. This is the reason why the use of competitive exclusion (CE) products enabling early rapid colonization of chickens with healthy adult gut microbiota has been successfully tested in poultry (18, 20). The positive effect of CE products has been explained by the ability of bacteria present in these products to compete directly with pathogens and also to stimulate maturation of the gut immune system of newly hatched chickens.
The interaction between the immune system of the gut and commensal microbiota in chickens starts immediately after hatching and leads to a low level of inflammation characterized by increased interleukin-8 (IL-8) expression (2). This results in the infiltration of heterophils and lymphocytes into the lamina propria or the gut epithelium and normalization of the gut immune system (3, 16, 30). Infiltrating lymphocytes develop further, depending on the gut flora composition, either in terms of a decreasing ratio of αβ to γδ T lymphocytes in the lamina propria or the gut epithelium (15) or in terms of changes in αβ T-cell receptor repertoires (21). In mice, but not in chickens so far, gut microflora has been reported to induce the Th1 and Th17 arms of the immune response, with IL-17 playing an important role in the maturation of the murine gut immune system (9, 12). Interestingly, IL-17 has been shown to be important for defense against bacterial and fungal pathogens (5), and in mice, IL-17 is induced also after S. enterica serovar Typhimurium infection (25). This means that the patterns of immune response to commensals and pathogens may overlap, the former being a subset of the latter. A detailed understanding of these responses may then allow active modification of gut microbiota composition, especially in the early days of life, and potentiation of the immune response against particular bacterial or fungal pathogens.
In this study, we were therefore interested in the development of chicken cecum microflora and the corresponding maturation of the gut immune system. We found out that chickens responded to the natural colonization of cecum by an increased expression of IL-8 and IL-17 in the first week of life. In the second part of this study, we infected chickens with Salmonella enterica serovar Enteritidis (S. Enteritidis) before, during, and after the IL-8 and IL-17 induction and found out that chickens infected before and during microbiota-induced IL-8 and IL-17 responded to S. Enteritidis infection more through Th1, while birds infected after this time point, on day 16 of life, responded more through the Th17 branch of the immune response. We therefore concluded that the transient induction of proinflammatory cytokines by gut microbiota resulted in an activation and normalization of the innate immune system in the chicken gut and also a modified immune response and increased resistance to S. Enteritidis infection.
MATERIALS AND METHODS
Experimental animals and infection.
In all experiments, male ISA Brown newly hatched chickens were used. The chickens were reared in wire cages and allowed free access to water and pathogen-free feed. In the first experiment, three chickens each were sacrificed on days 1, 4, 7, 10, 13, 16, and 19 of life, and the development of natural gut microflora cytokine and gene expression in cecum was monitored. In the second experiment, three chickens each were sacrificed on days 1, 2, 3, 4, 5, and 7 of life, and cytokine gene expression in cecum was monitored. In the last experiment, 1-, 4-, and 16-day-old birds (22 birds in each group) were infected orally with 1 × 106 CFU of the wild-type S. Enteritidis strain 147 spontaneously resistant to nalidixic acid (19) and sacrificed 3 days (6 birds), 10 days (6 birds), and 42 days (10 birds) postinfection, respectively. In addition, in this experiment, three noninfected chickens used as negative controls were sacrificed on days 4, 7, 10, 14, 15, 16, and 19, and six noninfected chickens used as negative controls were sacrificed on days 26, 47, and 58.
Cecal contents were stored at −20°C prior to DNA purification, and cecal wall samples were placed in RNALater solution (Qiagen) and stored at −70°C prior to RNA purification. S. Enteritidis counts in the cecum, liver, and spleen were determined after tissue homogenization in peptone water and plating of 10-fold serial dilutions on xylose-lysine-deoxycholate (XLD) agar supplemented with nalidixic acid. Samples negative after direct plating were subjected to enrichment in modified semisolid Rappaport Vassiliadis medium for qualitative S. Enteritidis determination. Counts of S. Enteritidis samples positive after direct plating were logarithmically transformed. Samples positive only after preenrichment were assigned a value of 1, and negative samples were assigned a value of 0.
16S rRNA gene T-RFLP.
For 16S rRNA gene terminal restriction fragment length polymorphism (T-RFLP) analysis, cecal contents were homogenized by using zirconia silica beads (BioSpec Products) in a MagNALyzer (Roche), and DNA was extracted by a QIAamp DNA stool minikit according to the manufacturer′s instructions (Qiagen). The purified DNA was used as a template in PCR amplification with fluorescently labeled universal primers for 16S rRNA genes 27F (6-carboxyfluorescein [FAM]-5′ AGA GTT TGA TCM TGG CTC AG 3′) and 1492R (5′ GGY TAC CTT GTT ACG ACT T 3′). Following PCR, the amplification products were digested with HaeIII restriction endonuclease, and the resulting fragments were separated and identified by capillary electrophoresis using an ABI310 genetic analyzer (Applied Biosystems).
Quantitative real-time PCR.
Cecal wall samples stored in RNALater were homogenized in exactly the same way as the cecal content samples, and total RNA was purified using TRI reagent (Molecular Research Center). The purity and concentration of RNA were determined spectrophotometrically, and 1 μg of RNA was immediately reverse transcribed by using Moloney murine leukemia virus (MMLV) reverse transcriptase (Invitrogen) and oligo(dT) primers. The resulting cDNA was 10× diluted in sterile distilled water and used as a template in real-time PCR or stored at −20°C until used. Real-time PCR was performed in microplates with a LightCycler II (Roche) and QuantiTect SYBR green PCR kit (Qiagen). The mRNA levels of 18 cytokines, chemokines, or immune-associated proteins (IL-2, -4, -5, -6, -10, -12p35, and -12p40; granulocyte-macrophage colony-stimulating factor [GM-CSF]; NRAMP [natural resistance macrophage-associated protein]; IL-15, -1β, -8, -17, -18, and -22; tumor necrosis factor alpha [TNF-α; also called LITAF]; gamma interferon [IFN-γ]; inducible nitric oxide synthase [iNOS]; and gallinacins 1, 2, 4, and 6) were determined. In addition, expression of three housekeeping genes, coding for GAPDH (glyceraldehyde-3-phosphate dehydrogenase), TATA-binding protein (TBP), and ubiquitin (UB), was determined and used for data normalization (6, 13). Primers for cytokine genes were either described previously (4, 26) or newly designed with Primer3 software (http://frodo.wi.mit.edu/primer3/), and all primer sequences are listed in Table 1. Each sample was subjected to reverse transcription-PCR (RT-PCR) in duplicate, and the mean values were used for subsequent analysis. The threshold cycle (CT) values of genes of interest were normalized to an average CT value of the housekeeping genes, and the relative expression of each representative was calculated as 2−ΔCT. Cytokine expression in the cecum was determined in three independent animal infections, and in the figures, we present results combined from all of these experiments.
Table 1.
List of primers used for the chicken cytokine mRNA quantification
| Primer | Sequence (5′→3′) | Source or reference |
|---|---|---|
| IL-βFor | GAAGTGCTTCGTGCTGGAGT | This study |
| IL-βRev | ACTGGCATCTGCCCAGTTC | |
| IL-8For | ATGAACGGCAAGCTTGGAGCT | 26 |
| IL-8Rev | GCAGCTCATTCCCCATCTT | |
| IL-12βFor | TGGTCCACGCTTTGCAGAT | 4 |
| IL-12βRev | AAGGTTAAGGCGTGGCTTCTTA | |
| IL-17For | TATCAGCAAACGCTCACTGG | This study |
| IL-17Rev | AGTTCACGCACCTGGAATG | |
| IL-18For | ACGTGGCAGCTTTTGAAGAT | 26 |
| IL-18Rev | GCGGTGGTTTTGTAACAGTG | |
| IL-22For | CAGACTCATCGGTCAGCAAA | This study |
| IL-22Rev | GGTACCTCTCCTTGGCCTCT | |
| IFNγFor | GCCGCACATCAAACACATATCT | 4 |
| IFNγRev | TGAGACTGGCTCCTTTTCCTT | |
| TNF-αFor | AATTTGCAGGCTGTTTCTGC | 26 |
| TNF-αRev | TATGAAGGTGGTGCAGATGG | |
| iNOSFor | GAACAGCCAGCTCATCCGATA | 4 |
| iNOSRev | CCCAAGCTCAATGCACAACTT | |
| Gal1For | GAAATGCTCAAGATTTTACCTCTG | This study |
| Gal1Rev | GAAGTTTCCAGAAGCGAGAAG | |
| Gal2For | AGGATTCTTTACCTGCTTTTCTCT | This study |
| Gal2Rev | AGCTTCCGACTTTGATTAGATG | |
| Gal4For | GTGTCTGTAGGTGGACAACATCT | This study |
| Gal4Rev | ATTGCATGTGATATCTTGGAGAAC | |
| Gal6For | TACTGGAGAAGGGAGACAGAAG | This study |
| Gal6Rev | TGATATCTACAAGCATGAATAGGG | |
| GAPDHFor | CCTGCATCTGCCCATTT | 6 |
| GAPDHRev | GGCACGCCATCACTATC | |
| TBPFor | TAGCCCGATGATGCCGTAT | 13 |
| TBPRev | GTTCCCTGTGTCGCTTGC | |
| UBFor | GGGATGCAGATCTTCGTGAAA | 6 |
| UBRev | CTTGCCAGCAAAGATCAACCTT |
ELISA detection of anti-Salmonella LPS antibodies.
To detect Salmonella anti-lipopolysaccharide (anti-LPS) antibodies, the commercial Flockscreen Salmonella enterica serovar Enteritidis antibody enzyme-linked immunosorbent assay (ELISA) kit (x-OvO, United Kingdom) was used to detect serum antibodies in birds infected with S. Enteritidis on day 1, 4, or 16 of life. Forty-two days postinfection, serum was obtained from the birds, and the ELISA was performed exactly as recommended by the manufacturer, except we diluted the serum only 1:5 in the dilution solution provided by the kit.
Statistics.
One-way analysis of variance (ANOVA) was used to analyze cytokine expression levels at different chicken ages with Tukey's post hoc test. Student's t test was used for the analysis of cytokine expression levels in Salmonella-infected and noninfected chickens on any particular day. All statistical analyses were performed using SPSS v. 14.0 software. For clarity, in Fig. 2, 4, and 5, we do not show standard deviation; instead, we indicate statistically significant differences.
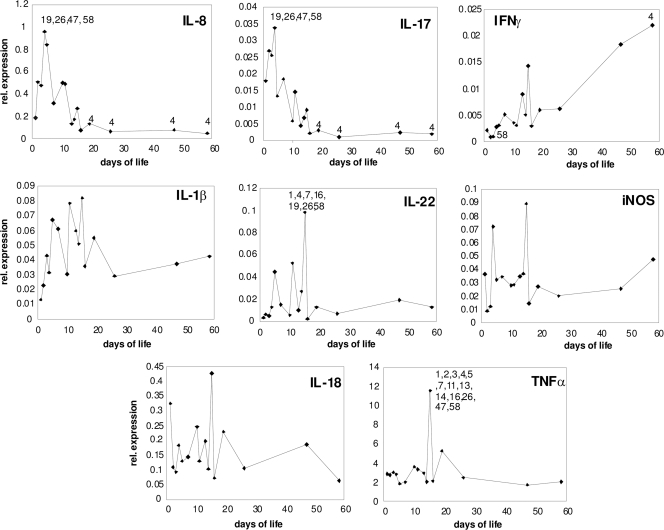
Fig. 2.
Cytokine gene expression in the cecum of noninfected chickens. For days 1, 19, 26, 47, and 58, each dot is a mean value of relative (rel.) expression from six chicks. For days 2, 3, 5, 10, 11, 13, 14, 15, and 16, each dot is a mean value from three chicks. For days 4 and 7, each dot represents a mean value from nine chicks. Numbers in the individual panels indicate significant differences in cytokine expression between particular days of life by one-way ANOVA at P < 0.05.
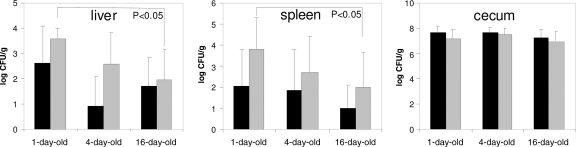
Fig. 4.
S. Enteritidis counts in liver, spleen, and cecum. Black columns represent average S. Enteritidis counts in organs from chickens 3 days postinfection. Gray columns represent average S. Enteritidis counts in organs from chickens 10 days postinfection. S. Enteritidis counts from spleen, liver, and cecum of infected chickens 42 days postinfection were all negative and therefore are not shown.
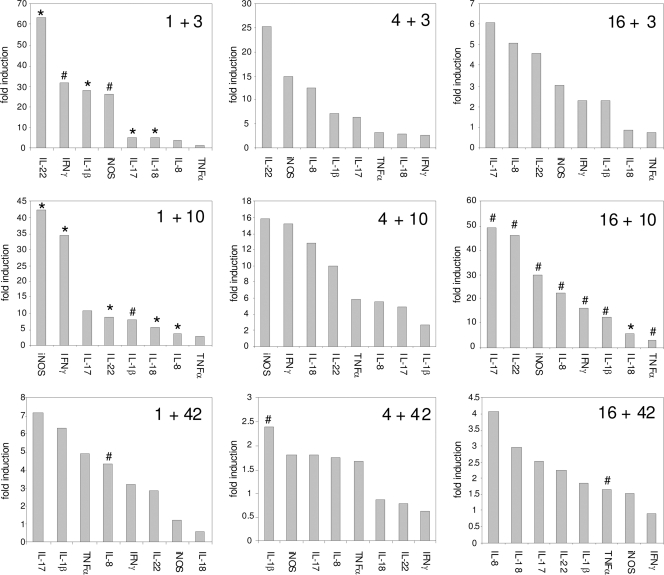
Fig. 5.
Relative increase in fold inductions of selected cytokines after infection with S. Enteritidis. The following format was used: “1 + 3” represents infection of 1-day-old chickens and determination of cytokine gene expression 3 days postinfection. *, value by t test significantly different from that for noninfected controls at P < 0.01; #, value by t test significantly different at P < 0.05.
RESULTS
Chicken gut flora development.
Using a 16S rRNA gene T-RFLP, we determined the development of gut microbiota in chickens, from the day of hatching until day 19 of life (Fig. 1A). The complexity of gut microbiota gradually increased, with the most dramatic development occuring during the first week of life. The lowest complexity of cecal microbiota was observed on day 1, when the flora usually consisted of five different species. Three days later, the number of cecal microbiota increased to 14, which gradually increased to approximately 42 peaks characterizing microbiota in 19-day-old chickens (Fig. 1B). When the numbers of all of the different peaks observed in the three birds tested at each sampling time were counted and the ratios of all different peaks to an average number of peaks were calculated, the highest ratio, and therefore largest number of differences in gut flora composition of individual birds, was observed during the first 10 days of life. On the other hand, the ratio decreased in the second week of life, indicating that the gut microbiota composition in 19-day-old chickens was closely related (Fig. 1B and C).
Fig. 1.
Cecal microflora development in chickens. (A) T-RFLP analysis; (B) number of T-RFLP peaks. Solid symbols, total number of unique peaks identified in three different birds at each time point; open symbols, average number of peaks ± standard deviation (SD) per individual bird. (C) Ratio of the total number of peaks to an average number of peaks.
Cytokine response to cecum colonization by normal gut flora.
Simultaneously with microflora development, we determined the cytokine immune response in chicken cecum. Initially we determined the gene expression of 20 different cytokines and immune-relevant genes. Out of these, expression of IL-2, IL-4, IL-5, IL-6, IL-10, IL-12p35, IL-12p40, and GM-CSF was quite low in the cecum, with CT values of >30, and the genes coding for these cytokines were therefore excluded from further detailed analysis. Expression of IL-15 and NRAMP could be detected easily in cecal samples; however, their expression levels did not change significantly, neither in response to natural gut flora colonization nor in response to S. Enteritidis infection. Expression of IL-1β, IL-8, IL-17, IL-18, IL-22, TNF-α, IFN-γ, and iNOS changed either in response to normal gut colonization or in response to S. Enteritidis infection.
Specific expression profiles in response to gut flora development were observed for IL-8 and IL-17 and partially also for IFN-γ. Expression of IL-8 and IL-17 was characterized by peak expression on day 4 of life followed by a decrease in gene expression, and from day 10, the expression of these two cytokines did not change any more. The expression profile of IFN-γ in the chicken cecum was different from that of the previous two cytokines since it had been increasing gradually, from the day of hatching until the last measurement on day 58 (Fig. 2).
Expression of chicken β-defensins in cecum in a response to colonization by normal gut flora.
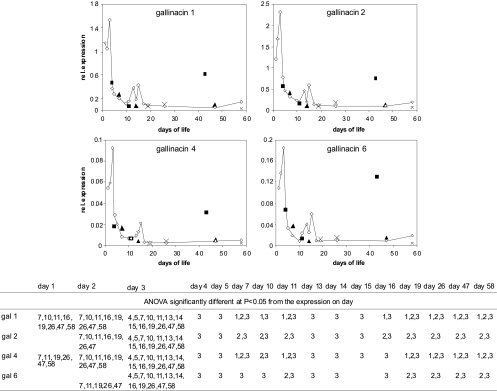
Since IL-17 is the cytokine characteristic for the Th17 arm of the immune response, which controls immune response on mucosal surfaces, in the next experiment, we were interested in whether chicken β-defensins (i.e., gallinacins 1, 2, 4, and 6) will follow the same expression profiles as IL-17. All of the gallinacins exhibited very similar expression profiles, which, however, did not overlap with that of IL-17. The gallinacins were highly expressed on the first 3 days of the chicken's life, but then on day 4, their expression dropped considerably (Fig. 3).
Fig. 3.
Chicken β-defensin gene expression in noninfected chickens (open diamonds) and chickens infected with S. Enteritidis at 1 day old (solid squares), 4 days old (solid triangles), and 16 days old (crosses) and sacrificed 3, 10, and 42 days postinfection. rel., relative. The table under the figure indicates significantly different levels of expression of individual gallinacins (gal 1, 2, 4, or 6) versus those of noninfected chickens at different days of life.
Salmonella infection and counts in cecum.
Next we were interested in the influence of gut microflora on chicken resistance to S. Enteritidis infection. Following previous results, we infected chickens 1, 4, and 16 days old (i.e., before, during, and after the physiological inflammatory response to normal gut flora) and monitored S. Enteritidis counts and cytokine responses 3, 10, and 42 days postinfection. When counts of S. Enteritidis were determined in the cecum, liver, and spleen of infected chickens, the counts in the cecum did not differ among birds infected at different ages, thus showing that gut microbiota did not influence S. Enteritidis cecum colonization. On the other hand, S. Enteritidis counts in the liver and spleen showed a negative correlation with the age of infected chickens as the highest S. Enteritidis counts were observed in chickens infected at 1 day old, and the lowest S. Enteritidis counts in the liver and spleen were observed in chickens infected on day 16 of life (Fig. 4).
Cytokine response of 1-day-old chickens.
The infection of 1-day-old chickens led to significantly increased expression of IL-1β, IL-8, IL-17, IL-18, IL-22, IFN-γ, and iNOS in the cecum compared with that of the noninfected controls at either 3 or 10 days postinfection. Except for IFN-γ and TNF-α, all of the remaining cytokines were maximally expressed 3 days postinfection, and their expression gradually decreased between 10 and 42 days postinfection. Three days postinfection, the highest induction was observed for IL-22 (approximately 60-fold induction), followed by IL-1β, IFN-γ, and iNOS (each induced around 5-fold) compared with that of the noninfected chickens. Ten days postinfection, IFN-γ and iNOS were expressed at levels 35 to 40 times higher in Salmonella-infected chickens than those in noninfected chickens, and expression of IL-22 and IL-1β was upregulated ∼10-fold. Forty-two days postinfection, IL-17 and IL-1β still remained expressed approximately 7 times more than in noninfected control chickens, although due to increased variation, this upregulation was no longer statistically significant (Fig. 5).
Cytokine response of 4-day-old-chickens.
The infection of 4-day-old chickens led to increased expression of IL-1β, IL-8, IL-17, IL-18, IL-22, TNF-α, IFN-γ, and iNOS in the cecum compared with the level in the noninfected controls both 3 and 10 days postinfection; however, due to a high variation in gene expression in both infected and noninfected chickens, none of these upregulations has come out as significant. Despite this, IL-1β, IL-8, IL-17, and IL-22 were maximally expressed 3 days postinfection, and their expression gradually decreased between 10 and 42 days postinfection. IL-18, IFN-γ, iNOS, and TNF-α reached their maximal expression 10 days postinfection. Three days postinfection, the highest induction, compared with noninfected chickens, was observed for IL-22 and iNOS (around 25- and 15-fold, respectively). Ten days postinfection, iNOS and IFN-γ were expressed 15 times higher in S. Enteritidis-infected chickens than in noninfected chickens. Forty-two days postinfection, none of the cytokines in S. Enteritidis-infected chickens remained expressed more than 2.5 times higher than the levels in noninfected controls (Fig. 5).
Cytokine response of 16-day-old chickens.
The infection of 16-day-old chickens led to significantly increased expression of IL-1β, IL-8, IL-17, IL-22, IFN-γ, iNOS, IL-18, and TNF-α in the cecum compared with that of the noninfected controls, which was restricted only to a sampling time of 10 days postinfection. The highest induction was observed for IL-17 and IL-22 (nearly 50-fold), followed by iNOS (30-fold), IL-8 (25-fold), IFN-γ (15-fold), and IL-1β (10-fold) (Fig. 5).
Salmonella infection and expression of β-defensins.
Independent of the age of the infected chicken or type of cytokine response to the infection, chickens never responded to S. Enteritidis by modified expression of gallinacins 1, 2, 4, and 6. The high expression of all of the gallinacins in 1-day-old chickens 42 days postinfection is unlikely to be associated with S. Enteritidis infection since the chickens were virtually free of S. Enteritidis at that time, and they did not respond to S. Enteritidis infection by increased gallinacin expression 4 and 10 days postinfection (Fig. 3).
Salmonella LPS ELISA.
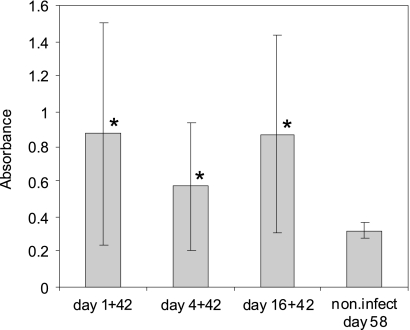
Independent of the age of infected chickens, their infection with S. Enteritidis resulted in a significant increase in the titer of anti-LPS antibodies determined in blood serum 42 days postinfection. High differences in responses of individual birds could be observed as characterized by quite a high standard deviation. The high variation observed in individual bird responses was also a reason why the numerically lower titer of antibodies observed in birds infected on day 4 of life came out as insignificant compared with the titer in those infected at day 1 or day 16 of life (Fig. 6).
Fig. 6.
Salmonella LPS ELISA 42 days postinfection of 1-, 4-, and 16-day-old chickens, with 58-day-old noninfected (non.infect) chickens used as a control. The following format was used: “1 + 42“ represents infection of 1-day-old chickens and detection of serum antibodies 42 days later. *, value by t test significantly different from noninfected controls at P < 0.05.
DISCUSSION
In this study, we have shown that complexity of chicken microbiota gradually developed from day 1 to day 19, and that, among younger birds, there was greater individual variation in microbiota composition than among older birds from the same flock. These observations are similar to the other reports on gut flora development in broiler chickens or pigs (17, 29). These observations also indicate that the birds were initially randomly colonized by bacterial species present in the hatchery and sharing the same environment, which then resulted in a convergence of gut microbiota of individual members belonging to the same flock. Our preliminary characterization of individual components of gut microbiota based on the cloning and sequencing of 16S rRNA amplification products (unpublished data) showed that the peak of 34 nucleotides in size was associated with the presence of Enterobacteriaceae (phylum Proteobacteria), while all the other peaks were associated with the presence of different representatives of the phylum Firmicutes, in agreement with previous reports (1, 17, 24, 33).
Chickens that are hatched as immunologically immature only weakly respond to vaccination with an inactivated antigen (11), and their splenic T cells do not proliferate if treated with various mitogens (16). In both of these studies, the maturation of the immune system was observed to start between days 2 and 4 of life. In agreement with these studies, we observed that for the first 3 days of life, chicken cecum was protected from incoming antigens by increased expression of gallinacins 1, 2, 4, and 6. On day 4, peak expression of IL-8 and IL-17 has been observed that indicated minor, transient, inflammation followed by normalization of the gut immune system and suppression of gallinacin expression. Upregulation of IL-8 in response to the colonization by gut microbiota has been reported also by Bar-Shira and Friedman (2). Although we did not investigate this in our study, it is likely that the same physiological inflammation happened also in studies which determined cellular infiltrates in chicken intestinal tract soon after hatching and in which minor granulocyte and T-lymphocyte infiltrates to cecal lamina propria in chickens on day 4 of life were observed (3, 15, 30). The observed transient inflammation, despite the continuous increase in complexity of gut microbiota, could be caused either by the appearance of gut microbiota with anti-inflammatory activity (9, 28) or, more likely in our opinion, by an adaptation of the gut immune system to contact with microbiota. We also considered whether the transient inflammation could be influenced by nonbacterial antigens present in feed. However, we concluded that this hypothesis is also less likely because in germ-free mice, which uptake feed normally, the inflammation signs and influx of granulocytes and lymphocytes appear only after experimental bacterial colonization (9).
Compared with the response to natural gut colonization, infection by S. Enteritidis induced IL-8 and IL-17, however, to a significantly higher level than that induced by normal gut microbiota. Chickens infected by S. Enteritidis responded also by increased expression of IL-1β, IL-22, iNOS, IFN-γ, and IL-18. Except for IL-17 and IL-22, increased expression of the remaining cytokines in chickens in a response to S. enterica infection has been reported earlier (4, 31). Although not in chickens, IL-17 and IL-22 have been described as highly inducible by Salmonella infection in mice or a human cell line (10, 25, 27). Rather unexpectedly, none of the four gallinacins tested increased in expression in a response to S. Enteritidis infection, further indicating that these gallinacins are not part of IL-17 and the Th17 arm of immune response in chickens.
IL-17 and IL-22 are frequently expressed simultaneously (7, 14), although under specific conditions, IL-17 and IL-22 might be expressed independently by different effector T cells (8, 23). Such different conditions were observed also in our study when IL-22 was induced only by S. Enteritidis infection, while IL-17 was induced both by S. Enteritidis and common gut microbiota. Furthermore, IL-22 was highly induced when the chicks were infected on days 1, 4, and 16 of life, while maximal IL-17 induction in a response to S. Enteritidis infection was observed in 16-day-old chickens. IL-17 may regulate immune response since IL-17 receptors have been found on dendritic cells, macrophages, and T lymphocytes (23). On the other hand, the IL-22 receptor has been observed only on nonimmune cells, stimulating them for production of antimicrobial peptides and also stimulating their growth and regeneration during infection (7, 23). Normal microbiota therefore stimulate the proinflammatory response, which due to the absence of IL-22 upregulation, does not result in tissue damage. On the other hand, infection with S. Enteritidis resulted in significant inflammation, which also required a strengthening of epithelial cell resistance and healing controlled by IL-22 and independent of primarily the Th1 response observed in 1- and 4-day-old chicks or rather the Th17 response observed in 16-day-old chickens.
We did not observe any direct effect of gut microbiota on S. Enteritidis cecum colonization because the same S. Enteritidis counts were observed in chickens infected on days 1, 4, and 16 of life. However, gut microbiota clearly had an effect on immune system maturation, which resulted in increased resistance of older birds to S. Enteritidis infection. IL-17 induced during the first week of the chicken's life likely resulted in an influx and differentiation of Th17 cells in lamina propria as described in the response to microbiota colonization in mice (12, 22). In response to S. Enteritidis infection, mature Th17 cells then produced IL-17 and IL-22 and increased the resistance of epithelial cells by increasing the production of antimicrobial peptides (14) and by recovery of damaged tissue stimulated by IL-22 (32). The increased Th17-dependent resistance in 16-day-old chickens then protected them from extensive S. Enteritidis dissemination to the liver and spleen. In addition, the matured Th17 arm of the gut immune system also caused (i) a delayed maximal cytokine response to S. Enteritidis from day 3 postinfection in 1-day-old chicks to day 10 postinfection in 16-day-old chickens and (ii) reduced absolute levels of cytokine expression in birds infected on day 16 of life compared with the chickens infected immediately after hatching, as well as (iii) shifted the IFN-γ and iNOS responses of 1- and 4-day-old chickens into the IL-22 and IL-17 responses of 16-day-old chickens.
In this study, we have shown that although gut flora gradually developed in young chickens, the most important events occurred during the first week of life when transient gut inflammation could be detected. For the first 3 days of life, chicken cecum was protected by a high level of expression of chicken β-defensins, and this type of cecum protection was replaced from day 4 of life by normalization of the gut immune system induced by transient expression of proinflammatory cytokines IL-8 and IL-17. This physiological inflammation delineates newly hatched chickens responding primarily by the Th1 type of immune response from birds older than 10 days, which respond to S. Enteritidis by the Th17 arm of the immune response. Although microbiota development must have been a rather specific and uncontrolled process in our experiments, it is likely that the same maturation happens in general, as many studies reported either increased cytokine signaling or cellular infiltrates in early days posthatching (2, 3, 15, 30). Such a maturation of the chicken gut immune system leads to increased resistance to S. Enteritidis but likely also to other bacterial or protozoan pathogens. It would be interesting to perform similar experiments with controlled colonization of newly hatched chickens, either with complex gut flora preparations originating from healthy birds modeling the natural conditions or with monocultures with known probiotic potential such Lactobacillus or Bifidobacterium.
ACKNOWLEDGMENTS
This work has been supported by projects MZE0002716202 and QH91238 of the Czech Ministry of Agriculture and AdmireVet project CZ.1.05/2.1.00/01.0006—ED0006/01/01 from the Czech Ministry of Education.
We thank Peter Eggenhuizen for English language corrections.
Footnotes
Published ahead of print on 9 May 2011.
REFERENCES
- 1. Amit-Romach E., Sklan D., Uni Z. 2004. Microflora ecology of the chicken intestine using 16S ribosomal DNA primers. Poult. Sci. 83:1093–1098 [DOI] [PubMed] [Google Scholar]
- 2. Bar-Shira E., Friedman A. 2006. Development and adaptations of innate immunity in the gastrointestinal tract of the newly hatched chick. Dev. Comp. Immunol. 30:930–941 [DOI] [PubMed] [Google Scholar]
- 3. Bar-Shira E., Sklan D., Friedman A. 2003. Establishment of immune competence in the avian GALT during the immediate post-hatch period. Dev. Comp. Immunol. 27:147–157 [DOI] [PubMed] [Google Scholar]
- 4. Berndt A., et al. 2007. Chicken cecum immune response to Salmonella enterica serovars of different levels of invasiveness. Infect. Immun. 75:5993–6007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Curtis M. M., Way S. S. 2009. Interleukin-17 in host defence against bacterial, mycobacterial and fungal pathogens. Immunology 126:177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Boever S., Vangestel C., De Backer P., Croubels S., Sys S. U. 2008. Identification and validation of housekeeping genes as internal control for gene expression in an intravenous LPS inflammation model in chickens. Vet. Immunol. Immunopathol. 122:312–317 [DOI] [PubMed] [Google Scholar]
- 7. Eyerich S., Eyerich K., Cavani A., Schmidt-Weber C. 2010. IL-17 and IL-22: siblings, not twins. Trends Immunol. 31:354–361 [DOI] [PubMed] [Google Scholar]
- 8. Eyerich S., et al. 2009. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J. Clin. Invest. 119:3573–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gaboriau-Routhiau V., et al. 2009. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31:677–689 [DOI] [PubMed] [Google Scholar]
- 10. Godinez I., et al. 2008. T cells help to amplify inflammatory responses induced by Salmonella enterica serotype Typhimurium in the intestinal mucosa. Infect. Immun. 76:2008–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Holt P. S., Gast R. K., Porter R. E., Jr., Stone H. D. 1999. Hyporesponsiveness of the systemic and mucosal humoral immune systems in chickens infected with Salmonella enterica serovar enteritidis at one day of age. Poult. Sci. 78:1510–1517 [DOI] [PubMed] [Google Scholar]
- 12. Ivanov I. I., et al. 2008. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4:337–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Y. P., Bang D. D., Handberg K. J., Jorgensen P. H., Zhang M. F. 2005. Evaluation of the suitability of six host genes as internal control in real-time RT-PCR assays in chicken embryo cell cultures infected with infectious bursal disease virus. Vet. Microbiol. 110:155–165 [DOI] [PubMed] [Google Scholar]
- 14. Liang S. C., et al. 2006. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 203:2271–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lillehoj H. S., Chung K. S. 1992. Postnatal development of T-lymphocyte subpopulations in the intestinal intraepithelium and lamina propria in chickens. Vet. Immunol. Immunopathol. 31:347–360 [DOI] [PubMed] [Google Scholar]
- 16. Lowenthal J. W., Connick T. E., McWaters P. G., York J. J. 1994. Development of T cell immune responsiveness in the chicken. Immunol. Cell Biol. 72:115–122 [DOI] [PubMed] [Google Scholar]
- 17. Lu J., et al. 2003. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 69:6816–6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Methner U., Barrow P. A., Berndt A., Steinbach G. 1999. Combination of vaccination and competitive exclusion to prevent Salmonella colonization in chickens: experimental studies. Int. J. Food Microbiol. 49:35–42 [DOI] [PubMed] [Google Scholar]
- 19. Methner U., Barrow P. A., Gregorova D., Rychlik I. 2004. Intestinal colonisation-inhibition and virulence of Salmonella phoP, rpoS and ompC deletion mutants in chickens. Vet. Microbiol. 98:37–43 [DOI] [PubMed] [Google Scholar]
- 20. Methner U., Barrow P. A., Martin G., Meyer H. 1997. Comparative study of the protective effect against Salmonella colonisation in newly hatched SPF chickens using live, attenuated Salmonella vaccine strains, wild-type Salmonella strains or a competitive exclusion product. Int. J. Food Microbiol. 35:223–230 [DOI] [PubMed] [Google Scholar]
- 21. Mwangi W. N., et al. 2010. Regional and global changes in TCRalphabeta T cell repertoires in the gut are dependent upon the complexity of the enteric microflora. Dev. Comp. Immunol. 34:406–417 [DOI] [PubMed] [Google Scholar]
- 22. Neish A. S., Denning T. L. 2010. Advances in understanding the interaction between the gut microbiota and adaptive mucosal immune responses. F1000 Biol. Rep. 2:27 doi: 10.3410/B2-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nograles K. E., et al. 2008. Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br. J. Dermatol. 159:1092–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qu A., et al. 2008. Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome. PLoS One 3:e2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raffatellu M., et al. 2009. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5:476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rychlik I., et al. 2009. Virulence potential of five major pathogenicity islands (SPI-1 to SPI-5) of Salmonella enterica serovar Enteritidis for chickens. BMC Microbiol. 9:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schulz S. M., et al. 2008. Protective immunity to systemic infection with attenuated Salmonella enterica serovar enteritidis in the absence of IL-12 is associated with IL-23-dependent IL-22, but not IL-17. J. Immunol. 181:7891–7901 [DOI] [PubMed] [Google Scholar]
- 28. Sokol H., et al. 2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U. S. A. 105:16731–16736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thompson C. L., Wang B., Holmes A. J. 2008. The immediate environment during postnatal development has long-term impact on gut community structure in pigs. ISME J. 2:739–748 [DOI] [PubMed] [Google Scholar]
- 30. Van Immerseel F., et al. 2002. Dynamics of immune cell infiltration in the caecal lamina propria of chickens after neonatal infection with a Salmonella enteritidis strain. Dev. Comp. Immunol. 26:355–364 [DOI] [PubMed] [Google Scholar]
- 31. Withanage G. S., et al. 2004. Rapid expression of chemokines and proinflammatory cytokines in newly hatched chickens infected with Salmonella enterica serovar Typhimurium. Infect. Immun. 72:2152–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wolk K., et al. 2004. IL-22 increases the innate immunity of tissues. Immunity 21:241–254 [DOI] [PubMed] [Google Scholar]
- 33. Zhu X. Y., Zhong T., Pandya Y., Joerger R. D. 2002. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol. 68:124–137 [DOI] [PMC free article] [PubMed] [Google Scholar]