Abstract
The roles of different ribonucleotide reductases (RNRs) in bacterial pathogenesis have not been studied systematically. In this work we analyzed the importance of the different Pseudomonas aeruginosa RNRs in pathogenesis using the Drosophila melanogaster host-pathogen interaction model. P. aeruginosa codes for three different RNRs with different environmental requirements. Class II and III RNR chromosomal mutants exhibited reduced virulence in this model. Translational reporter fusions of RNR gene nrdA, nrdJ, or nrdD to the green fluorescent protein were constructed to measure the expression of each class during the infection process. Analysis of the P. aeruginosa infection by flow cytometry revealed increased expression of nrdJ and nrdD and decreased nrdA expression during the infection process. Expression of each RNR class fits with the pathogenicities of the chromosomal deletion mutants. An extended understanding of the pathogenicity and physiology of P. aeruginosa will be important for the development of novel drugs against infections in cystic fibrosis patients.
INTRODUCTION
A critical step in the life cycles of all types of organisms is cell division with high-fidelity duplication of the DNA. Ribonucleotide reductases (RNRs) are essential enzymes for this step, since they control the sole de novo pathway to the deoxyribonucleotides (dNTPs) required for DNA synthesis and repair. Three major classes of RNRs (I, II, and III) are known, differing in their cofactor requirements and quaternary structures (23, 31). Class I RNRs are generally homodimeric proteins (α2β2) with a larger subunit (α) containing the catalytic and the allosteric domains and a small subunit (β) carrying a stable tyrosyl radical linked to a diiron-oxo center required for radical generation. Since this process requires oxygen, class I enzymes function only aerobically. This class is found in almost all eukaryotes and some microorganisms (20). Class II RNRs are monomeric or dimeric (α/α2), require 5′-deoxyadenosylcobalamin (AdoCbl) (vitamin B12 coenzyme) for radical generation, and are oxygen independent. This class is mainly found in eubacteria, archaea, and some eukaryotic unicellular microorganism (20). Class III RNRs are homodimeric (α2) and contain a stable oxygen-sensitive glycyl radical formed by S-adenosylmethionine (SAM) and a second protein (β2) that it is not required for catalysis once the radical has been generated. This class is functional only under strict anaerobic conditions, and it is found in eubacteria, archaea, and a few unicellular eukaryotes (20). Interestingly Pseudomonas aeruginosa contains functional genes for all three RNR classes (I, II, and III) transcribed in separate operons; the three classes are encoded by the nrdAB, nrdJab, and nrdDG genes, respectively (15). In previous work we have characterized the aerobic expression of the nrdA and nrdJ genes as well as their biochemical and biophysical properties (29, 32). However, the roles of the different RNR classes in P. aeruginosa pathogenesis remain unknown.
P. aeruginosa is a Gram-negative bacterium and a major opportunistic pathogen in diverse hosts. It has the ability to change metabolism and properties to survive various environmental conditions both outside and inside infected organisms. In nature, P. aeruginosa frequently inhabits environments where anaerobic niches develop and often grows in biofilms (33). In humans it is the major cause of nosocomial infections and is frequently associated with infections in immunocompromised patients and with chronic lung infections in cystic fibrosis (CF) patients, where it is the principal cause of morbidity and mortality (21). Recent evidence suggests that the microbial environment in the CF lung is largely anaerobic and that cells of P. aeruginosa persist in the CF lung in very robust anaerobic or hypoxic biofilms (4). Under these conditions, the activity of at least one RNR is crucial for the constant supply of dNTPs. As there is increasing evidence that genes involved in anaerobic respiration are linked to virulence of bacteria (2, 9, 11), the role of each RNR during infection is of prime interest and RNR could be considered a good target to inhibit the growth of P. aeruginosa in chronic infections.
In this work we have studied the expression of each RNR class during aerobic and anaerobic growth of P. aeruginosa as well as their roles during infection. There is a shift in nrd expression during infection, with induction of nrdJ and nrdD expression and concomitant repression of nrdA transcription.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli and P. aeruginosa strains were cultivated in Luria-Bertani medium (LB) at 37°C. Antibiotics and chromogenic substrates were added at the following concentrations: ampicillin, 50 μg/ml; chloramphenicol, 30 μg/ml; gentamicin, 10 μg/ml (E. coli) and 100 μg/ml (P. aeruginosa), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, 30 μg/ml; carbenicillin, 500 μg/ml; and tetracycline, 50 μg/ml.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | recA1 endA1 hsdR supE44 thi-1 gyrA96 relA1 ΔlacU169 deoR φ80dlacZM15 | Stratagene |
| S17-I | recA1 thi-1 pro hsdR <RP4: 2-Tc: Mu-: Km:Tn7, Tpr Smr, integrated into the chromosome | 3 |
| P. aeruginosa | ||
| PAO1 | Wild type (ATCC 6872) | Lab strain |
| ETS102 | PAO1 nrdJ::ΩGm; Gmr | This study |
| ETS103 | PAO1 nrdD::ΩGm; Gmr | This study |
| Plasmids | ||
| pGEM-T-easy | TA cloning vector; Ampr | Promega |
| pEX18Gm | Knockout vector; Gmr | 13 |
| pUTminiTn5Tc | Source of ΩTc cassette; Ampr Tcr | 8 |
| pJN105 | Source for pBBR1MCS-5 (broad host range) | 22 |
| pMIG(Gm) | Source for the promoterless GFP; Gmr | 14 |
| pBBR1MCS-5 | Broad-host-range cloning vector; Gmr | 17 |
| pETS130-GFP | Broad host range, promoterless GFP; Gmr | This study |
| pETS134 | pETS130 derivative carrying nrdA promoter | This study |
| pETS135 | pETS130 derivative carrying nrdJ promoter | This study |
| pETS136 | pETS130 derivative carrying nrdD promoter | This study |
| pETS147 | pEX18Gm derivative carrying ΔnrdA::ΩTc | This study |
| pETS148 | pEX18Gm derivative carrying ΔnrdJ::ΩTc | This study |
| pETS149 | pEX18Gm derivative carrying ΔnrdD::ΩTc | This study |
| pETS159 | pBBR1 derivative carrying nrdJab operon | This study |
| pETS160 | pBBR1 derivative carrying nrdDG operon | This study |
Recombinant DNA and PCR methods.
Plasmid and chromosomal DNAs were isolated using the QIAprep Miniprep or DNeasy tissue kit (Qiagen), respectively. Isolation of DNA fragments from agarose gels was performed using the QIAquick gel extraction kit (Qiagen). PCRs were carried out using High-Expand Taq polymerase (Roche) according to the manufacturer's instructions. Other molecular biology techniques were carried out by standard procedures (24).
Growth curves.
Strains were grown aerobically or anaerobically in minimal M9 medium (24) and monitored spectrophotometrically at an optical density of 540 nm. Anaerobic growth was performed as described previously for E. coli (10) with the addition of 10 g/liter KNO3.
Construction of P. aeruginosa nrd mutants.
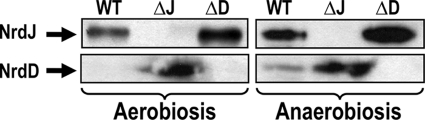
For all three genes a pair of DNA fragments was obtained using primer pair 1mut-2mut and 3mut-4mut, corresponding to DNA fragments at the 5′ and the 3′ ends, respectively (Table 2). After gel purification, each band was digested with BamHI, ligated with T4 DNA ligase (Fermentas), and PCR amplified using only primers 1mut and 4mut to generate a deleted version of each nrd gene. The DNA fragments for the nrdA (1,034 bp), nrdJ (900 bp), and nrdD (830 bp) deletion genes were cloned into the pGEM-T vector. The tetracycline resistance (Tcr) gene was retrieved from the plasmid pUTminiTn5-Tc (8) by digestion with SmaI and inserted into each deleted nrd gene previously digested with BamHI and end filled with Klenow (Fermentas). The nrd deletion alleles with tetracycline resistance were cloned into the pEX18Gm vector previously digested with HindIII and SacI, generating plasmids pETS147, pETS148, and pETS149, and introduced into P. aeruginosa PAO1 by conjugation (29). Corresponding chromosomal deletions were identified following counterselection on tetracycline- and sucrose-containing medium (13). True P. aeruginosa mutants ETS102 (ΔnrdJ) and ETS103 (ΔnrdD) were verify by Southern blot and Western blot analyses (Fig. 1). Complementation plasmids (pETS159 and pETS160) were constructed by cloning the nrdJab and nrdDG operons under the control of their respective native promoters into plasmid pBBR1MCS-5 (Table 1).
Table 2.
Primers used in this study
| Purpose and primer name | Sequencea | Restriction endonuclease |
|---|---|---|
| Mutant construction | ||
| 1mutAHIIIup | 5′-AAAGCTTCGTCGCAGGATCTGG-3′ | HindIII |
| 2mutABIlw | 5′-AAAAGGATCCTTCGCAGGCTTCGCTGATG-3′ | BamHI |
| 3mutABIup | 5′-AAAAGGATCCAACATCACCGGCGTCTCG-3′ | BamHI |
| 4mutASIlw | 5′-GAGCTCGATGCTGCACGCCTGCGG-3′ | SacI |
| 1mutJHIIIup | 5′-AAAGCTTCCCGTCAGGTACGGATAAC-3′ | HindIII |
| 2mutJBIlw | 5′-AAAAGGATCCATGGAGTCCTGGATGGTCC-3′ | BamHI |
| 3mutJBIup | 5′-AAAAGGATCCTATTACGGCAAGTACTGAGG-3′ | BamHI |
| 4mutJSIlw | 5′-AGAGCTCGACAAGGAAGGTGCAGTC-3′ | SacI |
| 1mutDHIIIup | 5′-AAAGCTTCGATCCCGGCAAGATC-3′ | HindIII |
| 2mutDBIlw | 5′-AAAAGGATCCAGCCAGTAGTTCGCGG-3′ | BamHI |
| 3mutDBIup | 5′-AAAAGGATCCGCGCATCCTCCAGGCCGG-3′ | BamHI |
| 4mutDSIlw | 5′-AGAGCTCCTTGCTGGTGGATGAAAGG-3′ | SacI |
| Promoter-probe cloning | ||
| PnrdAGFP-up | 5′-AGGATCCGAATTCTTGCTCCACACAGCCTC-3′ | BamHI |
| PnrdAGFP-lw | 5′-ACCCGGGTTCTCGCGTGTGGTGTCG-3′ | XmaI |
| PnrdJGFP-up | 5′-GGATCCGAATTCTGAACAACCTGGCAGGC-3′ | BamHI |
| PnrdJGFP-lw | 5′-ACCCGGGCCACCGTACGCAACATC-3′ | XmaI |
| PnrdDGFP-up | 5′-AGGATCCGAATTCGCCCGCCTCGCCCAGG-3′ | BamHI |
| PnrdDGFP-lw | 5′-ATCGATTTCAACTTCTCCACAAC-3′ | ClaI |
| nrdJab and nrdDG cloning | ||
| PfullJ-up | 5′-AAGGATCCTGAACAACCTGGCAGGCGAA-3′ | BamHI |
| PfullJ-low | 5′-AAGGATCCGTTCGCCCTTCAAAACGCATCC-3′ | BamHI |
| PfulDG-up | 5′-AAGGATCCTGGACAACTACGTCGTCTTCGC-3′ | BamHI |
| PfulDG-low | 5′-AAGGATCCTGAGTCTTGTGAAGGACAGGCC-3′ | BamHI |
Underlining indicates restriction sites.
Fig. 1.
Relative NrdJ and NrdD expression in wild-type P. aeruginosa and nrd mutants. Western blot analysis of total protein extracts from wild-type and mutant strains (30) is shown. Polyclonal antisera against NrdJ and NrdD were used at a 1:500 dilution.
Southern blot and Western blot analyses.
Genomic DNA was digested with PstI for the nrdJ mutants and with NcoI for the nrdD mutants. The nrdJ and nrdD probes were obtained by PCR using primers 3mutJBHIup-4mutJSllw and 3mutDBIup-4mutDSIlw, respectively (Table 2), and labeled with [γ-32P]dATP. Membranes were hybridized with the nrdJ and nrdD probes as described previously (28).
Western blot analysis was carried out as described previously (30) using NrdJ and NrdD polyclonal antibodies (Agrisera, Sweden) at a 1:500 dilution.
Fly infections.
Drosophila melanogaster flies were maintained on standard corn meal agar medium in vials at 25°C. OregonR flies were used as a wild-type host strain. P. aeruginosa overnight cultures were washed once in phosphate-buffered saline (PBS) (pH 7.2) and diluted to an A540 of 0.1. The bacterial suspension was injected into the dorsal thorax of flies (age 3 to 4 days) using a glass capillary with a microinjector (TriTech Research, CA). P. aeruginosa injection inocula (≈30 cells) were quantified by transferring cells from the tip of the needle to a LB agar plate. At 21 h after infection and at the end of the experiment, the number of bacteria from 10 flies from each condition (wild type, ΔnrdJ, and ΔnrdD) was analyzed.
Construction of the promoter-probe vector (pETS130-GFP) and nrd promoter fusions.
The promoter-probe vector pETS130-GFP was made by cloning the BamHI-KpnI fragment containing the green fluorescent protein (GFP) promoterless part of pMIG(Gm) (14) and the fragment from pJN105 (22), which contains a broad-host-range origin of replication, a conjugative region, and gentamicin resistance. Each nrd promoter region was amplified by PCR using a combination of forward/reverse primers (Table 2). Each fragment (772 bp, 184 bp, and 189 bp for nrdA, nrdJ, and nrdD, respectively) was gel purified and cloned in the pGEM-T vector. After digestion with BamHI-ClaI for nrdA and nrdJ and with BamHI-XmaI for nrdD, fragments were ligated into pETS130-GFP, generating the pETS134, pETS135, and pETS136 plasmids.
GFP measurements.
Five to 10 flies infected with P. aeruginosa cells containing plasmids (pETS130-GFP, pETS134, pETS135, or pETS136) were homogenized at the designated time points in 500 μl PBS, and serial dilutions were plated on LB agar plates and incubated at 37°C for 24 h to determine the number of bacterial cells per fly. GFP values from the homogenates were measured using a FACSCalibur flow cytometer (Becton Dickinson), while data collection and analysis were performed using the CellQuest software (Becton Dickinson).
Statistical analysis.
Survival curves were plotted using Kaplan-Meier analysis, and differences in survival rates were analyzed by the log rank test (GraphPad Prism 5.0). Differences with P values of <0.001 were considered statistically significant.
RESULTS
Construction and characterization of the P. aeruginosa nrd mutants.
We constructed ΔnrdJ and ΔnrdD mutants by inserting a tetracycline cartridge into the wild-type genes. Successful chromosomal integration was verified by Southern blot hybridization demonstrating the integration of the tetracycline resistance gene in the nrd genes (data not shown). Additional verifications of the ΔnrdJ and ΔnrdD mutations were done by Western blotting of aerobic and anaerobic cultures (Fig. 1). NrdJ is expressed in the wild type under both aerobic and anaerobic conditions, whereas NrdD is expressed only weakly compared to NrdJ during anaerobiosis. Interestingly, in the ΔnrdJ mutant NrdD is expressed even under aerobic conditions. None of the mutants had any growth defects based on aerobic growth curves in LB medium (data not shown). Using the same mutation strategy described in Materials and Methods, we did not obtain any viable mutation in the nrdA gene. Some colonies were obtained when the conjugation was performed anaerobically, but they died in traces of oxygen (data not shown). The nrdA and nrdB genes are essential when P. aeruginosa is growing aerobically in LB medium (29). This result is also supported by the fact that no nrdAB mutants were found in a nonredundant transposon insertion in P. aeruginosa PA14 (19), demonstrating the essentiality of the nrdAB genes under aerobic conditions.
Importance of the different nrd genes under aerobic and anaerobic growth conditions.
We grew the wild-type strain and nrd mutants in synthetic minimal medium (M9) aerobically and anaerobically (see Fig. S1 in the supplemental material) with and without hydroxyurea (HU) and 5′-deoxyadenosylcobalamin (AdoCbl). Hydroxyurea is a potent class I RNR inhibitor with a tyrosyl radical-scavenging activity (31), and it mimics class I RNR deficiency in P. aeruginosa (15, 29). AdoCbl is a cofactor required for class II RNR activity (29). Aerobically, the wild type and the ΔnrdJ and ΔnrdD mutants alone and/or with AdoCbl showed the same growth rate, reaching an A540 of ≈2.5 after 21 h (Table 3; see Fig. S1 in the supplemental material). In this case all aerobic RNRs (NrdA and NrdJ) would be functional. The NrdD enzyme is completely inactive under these conditions. A significant difference was observed when HU was included in the medium (HU inhibits class I RNRs and thus mimics NrdA deficiency), with some growth decrease (A540 ≈ 1.5) in the wild-type and ΔnrdD mutant strains; in both cases the class I RNR is inactivated and the only way to produce dNTPs is by the NrdJ enzyme. Western blot analysis of some growth curve samples from the wild-type strain showed an increased expression of NrdJ (Table 3; see Fig. S1 in the supplemental material). On the other hand, a more drastic effect was found in the ΔnrdJ mutant in the presence of HU, where hardly any growth was observed aerobically (A540 ≈ 0.2), as expected since none of the three RNRs are functional under this condition.
Table 3.
Growth and expression characteristics of wild-type P. aeruginosa and the nrd mutants under aerobic and anaerobic conditions
| Addition(s) | Wild type |
ΔnrdJ mutant |
ΔnrdD mutant |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aerobic, 22 h |
Anaerobic, 21 h |
Aerobic, 22 h |
Anaerobic, 21 h |
Aerobic, 22 h |
Anaerobic, 21 h |
|||||||||
| A540 | Predicted RNR activity | NrdJ expressiona | A540 | Predicted RNR activity | NrdJ expression | A540 | Predicted RNR activity | A540 | Predicted RNR activity | A540 | Predicted RNR activity | A540 | Predicted RNR activity | |
| 2.08 | NrdA, NrdJ | 5,908 (1.00) | 0.87 | NrdJ, NrdD | 2,838 (1.00) | 2.09 | NrdA | 0.54 | NrdD | 2.17 | NrdA, NrdJ | 0.8 | NrdJ | |
| HU | 1.20 | NrdJ | 7,840 (1.32) | 1.25 | NrdJ, NrdD | 2,403 (0.85) | 0.22 | 0.48 | NrdD | 1.43 | NrdJ | 1.22 | NrdJ | |
| AdoCbl | 2.51 | NrdA, NrdJ | 1.58 | NrdJ, NrdD | 3,885 (1.37) | 2.13 | NrdA | 0.39 | NrdD | 2.21 | NrdA, NrdJ | 1.16 | NrdJ | |
| HU, AdoCbl | 2.13 | NrdJ | 1.32 | NrdJ, NrdD | 7,722 (2.72) | 0.24 | 0.34 | NrdD | 2.08 | NrdJ | 1.28 | NrdJ | ||
Western blot arbitrary units, with relative expression in parentheses.
P. aeruginosa carries two RNR operons that have the potential to work anaerobically (NrdJ and NrdD). Anaerobically, both the wild type and the ΔnrdD mutant reached the same density (≈1) (Table 3). Under this condition NrdJ and NrdD can function in the wild-type strain, but only NrdJ is active in the ΔnrdD mutant strain. However, the ΔnrdJ mutant had an imbalanced growth (A540 ≈ 0.5), and only NrdD can function anaerobically in the absence of NrdJ and NrdA. The addition of HU had no effect anaerobically, and AdoCbl enhanced the growth of the wild type and the ΔnrdD mutant but did not affect the growth of the ΔnrdJ mutant (Table 3). Western blot analysis of some samples shows NrdJ overexpression (Table 3; see Fig. S1 in the supplemental material), corroborating our previous results (29) and demonstrating that NrdJ is the RNR working under anaerobic conditions. The results of the Western blotting shown in Fig. 1 corroborate these results and show that NrdJ is overexpressed in the ΔnrdD mutant and that NrdD is overexpressed in the ΔnrdJ mutant.
nrdJ and nrdD are important for virulence in P. aeruginosa.
Although the roles of the different nrd genes were tested in minimal medium and with different oxygen tensions, their importance in virulence in P. aeruginosa has not yet been established. We decided to analyze the pathogenicity of P. aeruginosa nrd mutant strains using the D. melanogaster systemic model of infection. Flies are well-established hosts for studying bacterial virulence mechanisms (1, 6).
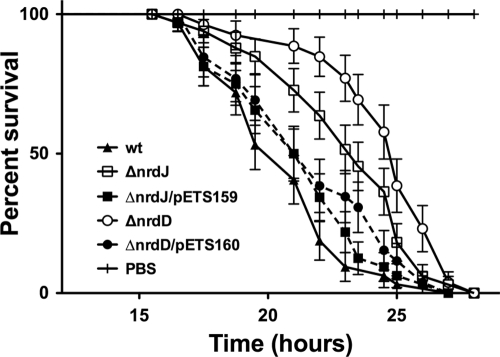
Injection of the same number of wild-type or mutant bacteria (ΔnrdJ or ΔnrdD) showed different killing behavior in flies. Control flies injected with PBS quickly recovered from injury. Wild-type P. aeruginosa caused 50% of the flies to perish at 21 h postinfection, and all flies were dead by 27 h, as has been shown by others (1). In contrast, 50% mortality of the ΔnrdJ and ΔnrdD mutant-infected flies did not occur until 23.5 and 25 h postinfection, respectively, increasing their survival by 2 h 30 min and 4 h, respectively, compared to the wild-type infection (Fig. 2). These results are statistically significant (P < 0.001) by the log rank test (1). To correlate loss of function of nrdJ and nrdD to pathogenicity, these mutations were complemented with corresponding wild-type alleles (pETS159 and pETS160), restoring the wild-type phenotype (Fig. 2). The wild-type strain transformed with pBBR1MCS-5 did not show any change in the survival curve (data not shown). At 21 h after infection, flies infected with wild-type P. aeruginosa contained approximately 4 times more bacteria than flies infected with ΔnrdJ or ΔnrdD bacteria. However, at the end of the experiments, flies from each condition contained comparable levels of bacteria, clearly showing that the mutations in the nrdJ and nrdD genes display attenuated pathogenicity in P. aeruginosa during the infection due to reduced multiplication efficiency.
Fig. 2.
Virulence attenuation of the nrd mutants in D. melanogaster. D. melanogaster was infected with wild-type (wt) P. aeruginosa and with the nrdJ ETS102 and nrdD ETS103 mutant strains. Mutations were complemented with pETS159 and pETS160. Fly survival was monitored for 48 h. As a control, flies were infected with sterile PBS. Flies that died within 3 h of injection (<2%) were excluded from the survival analysis. Each experiment was done in triplicate on different days. Survival rates were expressed as means and standard deviations for 4 groups of 50 flies in three independent experiments. For all comparisons, the difference between wt/ΔnrdJ, wt/ΔnrdD, and ΔnrdJ/ΔnrdD was statistically significant (P < 0.001) using the log rank test (GraphPad Prism 5.0). At 21 h the numbers of bacteria in the flies were 2.65 × 107 ± 2.3 × 107 CFU/ml (wild type), 6.35 × 106 ± 4.9 × 105 CFU/ml (ΔnrdJ), and 6.8 × 106 ± 4.2 × 105 CFU/ml (ΔnrdD), and at the end of the experiment they were 4.7 × 108 ± 0.9 × 108 CFU/ml (wild type), 5.8 × 108 ± 4.0 × 108 CFU/ml (ΔnrdJ), and 5.2 × 108 ± 3.3 × 108 CFU/ml (ΔnrdD).
Shift in nrd gene expression during P. aeruginosa infection.
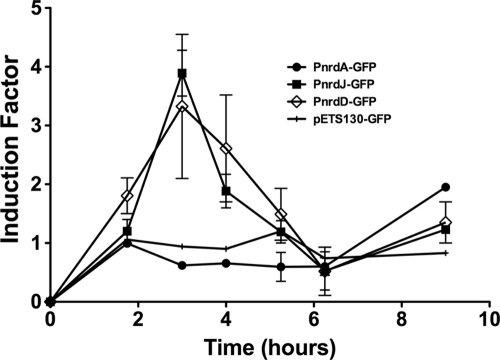
To measure the expression of each nrd operon during the course of infection, we injected flies with the wild-type P. aeruginosa strain carrying the GFP control plasmid or the nrdA, nrdJ and nrdD promoter fusions. At different time points of infection we homogenized a pool of flies (5 to 10, depending on the time point) and measured the GFP fluorescence. We used a high-intensity GFP variant (GFPmut3) that, together with flow cytometry, allows optimal separation of green-fluorescing bacteria from background noise and insect cells (12). Figure 3 shows induction of expression of different nrd genes during the infection. The control plasmid without a promoter fusion showed no changes in GFP intensity during the experiment. There was no induction in nrdA expression in the first hours, with the same level of expression throughout the infection, until the last time point, when it was doubled. However, expression of both nrdJ and nrdD was highly induced, approximately four times compared to nrdA, during the first hours of infection. Significantly, there is a clear shift in nrd expression during P. aeruginosa infection in flies.
Fig. 3.
Expression of nrd genes in D. melanogaster. GFP intensities from each nrd promoter fusion were corrected by the number of bacteria (CFU/ml) present in each sample, and induction factors were calculated by comparison to the GFP intensity for the control plasmid. For determination of viable bacteria, infected flies were collected at various time intervals, ground in PBS, and plated on LB agar plates. The results reflect data from two independent experiments.
DISCUSSION
Recently, large genome-sequencing projects have highlighted a tremendous variation of RNR combinations in bacteria, but an understanding of this redundancy is still lacking (20). In Gammaproteobacteria, for example, different pathogens carry different sets of RNR genes. All Enterobacteriaceae species encode two aerobic RNRs (Ia and Ib) and one anaerobic RNR (III), Actinobacillus pleuropneumoniae encodes two Ia RNRs and one III RNR, Vibrio cholerae encodes Ia and III RNRs, and Legionella pneumophila encodes a Ia RNR. P. aeruginosa encodes three different RNR classes (15), one aerobic (Ia), one B12 dependent (II) (29, 32), and one anaerobic (III). No information is known about their genetic regulation and the functionality of the different RNR classes in P. aeruginosa.
In this study we provide novel insights on the functions of different RNRs in the human pathogen P. aeruginosa and especially in its infection process in a model system. The presence of three different RNR classes possibly reflects its needs to adapt its growth under specific conditions or during infection.
Which RNR classes are normally expressed in P. aeruginosa when it is growing aerobically or anaerobically? Under aerobic conditions the situation is easy to explain, as class I and II RNRs can function aerobically and drastically reduced growth was observed when both classes were inactivated (Table 3). Class II can complement a class I deficiency (generated by the addition of HU, which inactivates class I RNRs), especially when the class II-specific cofactor AdoCbl is supplemented. This result is in agreement with previous observations that addition of AdoCbl allows the class II RNR to be fully functional (29). The situation under anaerobic conditions is more complex and probably reflects the synergistic effects of the anaerobic RNRs (NrdJ and NrdD). The nrdJ mutant showed some growth, but to a lesser extent than the nrdD mutant, which indicates a major role for NrdJ in anaerobic metabolism. Since both enzyme systems support anaerobic growth, a double mutation in nrdJ and nrdD will be needed to completely knock out anaerobic growth. Unfortunately, several attempts to obtain such a mutant failed. As a major conclusion, the class II RNR is clearly a versatile enzyme able to complement other RNR deficiencies both aerobically and anaerobically.
However, what is the situation when P. aeruginosa infects? Few studies have been done to elucidate the importance of each RNR class during infection when more than one RNR is encoded in a given organism's chromosome (7, 16). During infection, P. aeruginosa is growing and dividing quickly, requiring a high and constant supply of dNTPs necessary for DNA replication, and therefore the different nrd genes need to be regulated to avoid an unbalanced supply of dNTPs, which could be fatal for bacterial viability. Our results indicate that both the nrdJ and the nrdD mutations significantly reduce the pathogenicity of P. aeruginosa during infection in D. melanogaster as an animal model. The mutations did not cause P. aeruginosa avirulence but delayed the death of infected flies due to delayed multiplication of the mutant bacteria. In a previous high-throughput transposon experiment with P. aeruginosa, nrdJa was identified as important for anaerobic growth with a minor importance in lettuce leaf infection (9), but this plant pathogen model may not involve strict anaerobic growth.
The significance of the NrdJ and NrdD RNRs during infection is corroborated by the nrd expression pattern within the fly. nrdJ and nrdD are expressed when Pseudomonas proliferates during the first hours of infection, and the nrdAB operon seems to be repressed within the flies shortly after the infection. Clearly our results indicate a shift in expression of nrd genes initially and during the course of the infection.
A common feature of class II and III RNRs is their ability to function anaerobically. Furthermore, class II seems to respond to DNA damage, which is probably a condition that Pseudomonas encounters during infection (29). Our results point to an additive effect of class II and III RNRs, especially during the first stages of the infection, conceivably because anaerobic conditions are found inside the flies. Pseudomonas establishes a progressive and lethal infection in Drosophila after 24 h, when the levels of bacteria reach 108 CFU/ml. The conditions are probably anaerobic or microaerophilic when the pathogen enters the fly, whereas it may experience more aerobic conditions during progression of the infection when all tissues are disrupted with many cavities (18) where oxygen can diffuse easily. At these points the activation or expression of class II and III RNRs is no longer necessary. The bacteria can invade and proliferate in fly tissues, and the ability to cause maximum lethality correlates with the kinetics of invasion and proliferation (18) and with the shift in expression of the different RNR classes encoded in P. aeruginosa. Surely which transcriptional factors are expressed during the various phases of the infectious process will dictate which RNR genes are expressed.
Bacterial pathogens are exposed to various environments during infection, and all proteins may not function under all physicochemical conditions. One of the environmental conditions that Pseudomonas faces is anaerobiosis. The establishment of chronic Pseudomonas infections correlates with the formation of a biofilm structure with clusters of cells encapsulated in a polymeric matrix (5, 26). Internal parts of the biofilm are deprived of oxygen and nutrients (4). Surely the RNR functionality needs to be adapted to the oxygen availability during an infection. In this sense, the different RNR classes could be defined as ecoparalogs, i.e., paralogous proteins that perform the same enzymatic reaction but are optimized for different ecological niches (25). The genetic complexity helps P. aeruginosa to adapt to different ecological niches (27).
In conclusion, we show that a fine-tuned expression of nrd genes is important for the pathogenicity of Pseudomonas in Drosophila. Identification of which genes are important during infection may help to identify potential targets as novel candidates for antimicrobial therapy. RNR could be considered such a target, as it is a key enzyme for replication of the vast majority of pathogens.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Ylva Engström's group for excellent assistance with the fly work.
This work was supported by the Swedish Research Council and Swedish Cancer Foundation (to B.-M.S.) and by grants from the Fondo de Investigación Sanitaria (PI081062) and Consolider (CSD2008-00013) from the Spanish Ministerio de Ciencia e Innovación, the ERA-NET PathoGenoMics Ramón y Cajal program, and the Catalan Cystic Fibrosis Foundation (to E.T.).
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 18 April 2011.
REFERENCES
- 1. Apidianakis Y., Rahme L. G. 2009. Drosophila melanogaster as a model host for studying Pseudomonas aeruginosa infection. Nat. Protoc. 4:1285–1294 [DOI] [PubMed] [Google Scholar]
- 2. Baek S. H., Rajashekara G., Splitter G. A., Shapleigh J. P. 2004. Denitrification genes regulate Brucella virulence in mice. J. Bacteriol. 186:6025–6031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bagdasarian M., et al. 1981. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237–247 [DOI] [PubMed] [Google Scholar]
- 4. Costerton J. W. 2002. Anaerobic biofilm infections in cystic fibrosis. Mol. Cell 10:699–700 [DOI] [PubMed] [Google Scholar]
- 5. Costerton J. W., Stewart P. S., Greenberg E. P. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 6. D'Argenio D. A., Gallagher L. A., Berg C. A., Manoil C. 2001. Drosophila as a model host for Pseudomonas aeruginosa infection. J. Bacteriol. 183:1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dawes S. S., et al. 2003. Ribonucleotide reduction in Mycobacterium tuberculosis: function and expression of genes encoding class Ib and class II ribonucleotide reductases. Infect. Immun. 71:6124–6131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Lorenzo V., Herrero M., Jakubzik U., Timmis K. N. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Filiatrault M. J., Picardo K. F., Ngai H., Passador L., Iglewski B. H. 2006. Identification of Pseudomonas aeruginosa genes involved in virulence and anaerobic growth. Infect. Immun. 74:4237–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garriga X., et al. 1996. nrdD and nrdG genes are essential for strict anaerobic growth of Escherichia coli. Biochem. Biophys. Res. Commun. 229:189–192 [DOI] [PubMed] [Google Scholar]
- 11. Ge Z., et al. 2000. Fumarate reductase is essential for Helicobacter pylori colonization of the mouse stomach. Microb. Pathog. 29:279–287 [DOI] [PubMed] [Google Scholar]
- 12. Hansen M. C., Palmer R. J., Jr., Udsen C., White D. C., Molin S. 2001. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383–1391 [DOI] [PubMed] [Google Scholar]
- 13. Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 14. Izallalen M., Levesque R. C., Perret X., Broughton W. J., Antoun H. 2002. Broad-host-range mobilizable suicide vectors for promoter trapping in gram-negative bacteria. Biotechniques 33:1038–1040, 1042–1043 [DOI] [PubMed] [Google Scholar]
- 15. Jordan A., et al. 1999. Ribonucleotide reduction in Pseudomonas species: simultaneous presence of active enzymes from different classes. J. Bacteriol. 181:3974–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kirdis E., et al. 2007. Ribonucleotide reductase class III, an essential enzyme for the anaerobic growth of Staphylococcus aureus, is a virulence determinant in septic arthritis. Microb. Pathog. 43:179–188 [DOI] [PubMed] [Google Scholar]
- 17. Kovach M. E., et al. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 18. Lau G. W., et al. 2003. The Drosophila melanogaster Toll pathway participates in resistance to infection by the gram-negative human pathogen Pseudomonas aeruginosa. Infect. Immun. 71:4059–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liberati N. T., et al. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103:2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lundin D., Torrents E., Poole A. M., Sjöberg B.-M. 2009. RNRdb, a curated database of the universal enzyme family ribonucleotide reductase, reveals a high level of misannotation in sequences deposited to Genbank. BMC Genomics 10:589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lyczak J. B., Cannon C. L., Pier G. B. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051–1060 [DOI] [PubMed] [Google Scholar]
- 22. Newman J. R., Fuqua C. 1999. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203 [DOI] [PubMed] [Google Scholar]
- 23. Nordlund P., Reichard P. 2006. Ribonucleotide reductases. Annu. Rev. Biochem. 75:681–706 [DOI] [PubMed] [Google Scholar]
- 24. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25. Sanchez-Perez G., Mira A., Nyiro G., Pasic L., Rodriguez-Valera F. 2008. Adapting to environmental changes using specialized paralogs. Trends Genet. 24:154–158 [DOI] [PubMed] [Google Scholar]
- 26. Singh P. K., et al. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762–764 [DOI] [PubMed] [Google Scholar]
- 27. Stover C. K., et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 28. Torrents E., Jordan A., Karlsson M., Gibert I. 2000. Occurrence of multiple ribonucleotide reductase classes in gamma-proteobacteria species. Curr. Microbiol. 41:346–351 [DOI] [PubMed] [Google Scholar]
- 29. Torrents E., Poplawski A., Sjöberg B.-M. 2005. Two proteins mediate class II ribonucleotide reductase activity in Pseudomonas aeruginosa: expression and transcriptional analysis of the aerobic enzymes. J. Biol. Chem. 280:16571–16578 [DOI] [PubMed] [Google Scholar]
- 30. Torrents E., Roca I., Gibert I. 2004. The open reading frame present in the nrdEF cluster of Corynebacterium ammoniagenes is a transcriptional regulator belonging to the GntR family. Curr. Microbiol. 49:152–157 [DOI] [PubMed] [Google Scholar]
- 31. Torrents E., Sahlin M., Sjöberg B.-M. 2008. The ribonucleotide reductase family—genetics and genomics, p. 17–77 In Andersson K. K. (ed.), Ribonucleotide reductases. Nova Science Publishers, Hauppauge, NY [Google Scholar]
- 32. Torrents E., Westman M., Sahlin M., Sjöberg B.-M. 2006. Ribonucleotide reductase modularity: atypical duplication of the ATP-cone domain in Pseudomonas aeruginosa. J. Biol. Chem. 281:25287–25296 [DOI] [PubMed] [Google Scholar]
- 33. Xu K. D., Stewart P. S., Xia F., Huang C. T., McFeters G. A. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64:4035–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.