Abstract
Phylogenetic analysis of the crystal structure of the Enterococcus faecalis SlyA (EF_3002) transcriptional factor places it between the SlyA and MarR regulator subfamilies. Proteins of these families are often involved in the regulation of genes important for bacterial virulence and stress response. To gather evidence for the role of this putative regulator in E. faecalis biology, we dissected the genetic organization of the slyA-EF_3001 locus and constructed a slyA deletion mutant as well as complemented strains. Interestingly, compared to the wild-type parent, the ΔslyA mutant is more virulent in an insect infection model (Galleria mellonella), exhibits increased persistence in mouse kidneys and liver, and survives better inside peritoneal macrophages. In order to identify a possible SlyA regulon, global microarray transcriptional analysis was performed. This study revealed that the slyA-EF_3001 locus appears to be autoregulated and that 117 genes were differentially regulated in the ΔslyA mutant. In the mutant strain, 111 were underexpressed and 6 overexpressed, indicating that SlyA functions mainly as an activator of transcription.
INTRODUCTION
Enterococcus faecalis is a natural member of the intestinal tracts of humans and animals. This ubiquitous Gram-positive bacterium is abundant in various ecological niches and is a highly versatile bacterium. E. faecalis can transit from a commensal relationship with the host to being causative agent of invasive infection, and it is now a leading cause of nosocomial infections in the United States and Europe (36). These infections include urinary tract infections, endocarditis, and surgical wound infection and are especially common in immunocompromised patients (14). Because of its innate and acquired resistance to many antibiotics, E. faecalis infections are increasingly difficult to treat (32). Therefore, the virulence of the organism is being examined for new insights into infection prevention and treatment.
A number of E. faecalis putative virulence factors have been identified, including a cytolytic toxin (Cyl), a cell surface adhesion termed aggregation substance (Agg), an antiphagocytic surface capsule (Cps), an enterococcal polysaccharide antigen (Epa), gelatinase (GelE), serine protease (SprE), collagen binding protein (Ace), enterococcal surface protein (Esp), pilus subunits (Ebp), and a general stress protein (Gls24) (12, 15, 20, 21, 29, 31, 33). Enterococcal virulence is subtle and complex, involving both pathogen and host factors, and all of these factors rarely occur in a single strain. In other organisms, virulence genes are controlled by environmental stresses and involve secondary RNA polymerase σ factors and specific transcriptional regulators. E. faecalis lacks a σB-like general stress σ factor, but about 10 regulators have been shown to be involved in virulence and stress response in E. faecalis (6, 13, 25).
In 2003, the crystal structure of an E. faecalis SlyA-like (EF_3002) transcriptional factor (a member of the MarR/SlyA family) was determined by Wu and collaborators (37). SlyA was found to possess the same winged-helix DNA binding motif observed in the Escherichia coli MarR repressor and in the MexR repressor of Pseudomonas aeruginosa. Phylogenetic analysis placed it between the SlyA and MarR regulators based on sequence similarity to members of both subfamilies (37).
No functional studies have been conducted to define the role of this regulator. Proteins of the MarR/SlyA family regulate expression of virulence genes. Examples include the following: SlyA from Salmonella enterica serovar Typhimurium, which is involved in the modification of the bacterial surface that protects cells from the toxic compounds produced by the host; RovA from enteropathogenic Yersinia enterocolitica, which regulates inv, encoding a surface protein important for the invasion; PecS from the phytopathogen Erwinia chrysanthemi, which downregulates genes encoding pectate lyases involved in virulence; and Hor from Erwinia carotovora, AphaA from Vibrio cholerae, and MgrA from Staphylococcus aureus, which directly or indirectly regulate processes related to infection (3, 5, 7, 16, 17, 34).
In the present study, we report a functional analysis of the SlyA regulator (EF_3002) of E. faecalis. Using different infection models, we found that SlyA plays a role in the virulence of E. faecalis as well as in its persistence inside the host. In addition, our global transcriptional analysis revealed that the lack of SlyA up- and downregulated more than 100 loci.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The parental E. faecalis strain used in this work was strain V19, a plasmid-cured derivative of the vancomycin-resistant clinical isolate V583 (38). E. coli XLIBlue was used as the recipient for cloning. Lactococcus lactis IL1403 and E. coli DH5α served as controls for assays of Galleria mellonella infection and survival within macrophages. E. faecalis V19 and its derivates were grown without shaking at 37°C in M17 medium supplemented with 0.5% glucose (GM17), and when required, erythromycin (150 μg/ml) was added. E. coli strains were cultured with shaking at 37°C in LB medium with ampicillin (100 μg/ml), chloramphenicol (20 μg/ml), or erythromycin (150 μg/ml) when required. L. lactis was cultured without shaking at 30°C in GM17. Plasmids used in this study are listed in Table 1.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant properties | Reference or source |
|---|---|---|
| Strains | ||
| E. faecalis | ||
| V19 | Strain V583 (clinical isolate), without plasmid | 38 |
| V19 ΔslyA | V19 isogenic derivative slyA mutant | This study |
| V19 ΔslyA pCU1-slyA | V19 isogenic derivative slyA mutant with pCU1 carrying slyA | This study |
| E. coli | ||
| XL1Blue | recA1 endA1 gyrA96 thi-1 hsdR17supE44 relA1 lac[F′ proABlacIq ZΔM15] Tn10 (Tetr) | Stratagene |
| DH5α | recA endA1 gyrA96 thi-1 hsdR17 [rK− mK+supE44 relA1 lacZΔM15] | 15 |
| Plasmids | ||
| pMad | oripE194ts, Emr, Ampr, bgaB | 1a |
| pMad-ΔslyA | pMad carrying slyA deletion | This study |
| pCU1 | Cmr shuttle vector for E. coli and E. faecalis | 2 |
| pCU1-slyA | pCU1 carrying slyA | This study |
General molecular methods.
PCR was performed with GoTaq polymerase (Promega, Madison, WI). The primers used for this work are listed in Table 2. PCR products and plasmids were purified using the Nucleospin plasmid kit (Macherey-Nagel, Dünen, Germany). Restriction endonucleases, alkaline phosphatase, and T4 DNA ligase were purchased from Amersham Biosciences (Piscataway, NJ), Promega, and Roche Applied Science (Indianapolis, IN), respectively, and used according to the manufacturers' instructions. Genomic DNA extraction and other standard techniques were carried out as described by Sambrook et al. (30).
Table 2.
Primers used in this study
| Primer namea | Sequence (5′→3′)b |
Use | |
|---|---|---|---|
| Forward | Reverse | ||
| EF3002 (slyA) | GTGCGCGAAAATCAACACT | CATTTTTACGCATCCGGACTA | RT-PCR |
| EF3002R1 | CCTTAAATTCAATATTACTG | 5′RACE-PCR | |
| EF3002R2 | CTTTTCTTGGATAATCCCAGG | 5′RACE-PCR | |
| EF3002U | CCGGTCGACATGAGTTTCTAGAGGAGTTCCTA | CGCGGATCCGGCCCGCGCAATCATGCCAATTTCTC | Cloning in pMad, RT-PCR |
| EF3002D | CGCGGATCCGTCAGCGAAGATTGGGAATTTGT | GAAGATCTCGATTATTTGTTCACTTTGAACG | Cloning in pMad, RT-PCR |
| EF3002int | GAGAAATTGGCATGATTGCGCGGGCC | TCTGTGGCATAAATCCGCTTA | RT-PCR |
| EF3001int | TAAATCGTGAGACGGCACAA | AAAGGAAAGAAGCTGGGGAC | RT-PCR |
| EF3002tot | GACGCTATAACTAATTTTATCTTAC | TTAATAATTTCGAGTGTTCCC | Cloning in pCU1 |
| PU/PR | GTAAAACGACGGCCAGT | CAGGAAACAGCTATGAC | Cloning verification |
From the annotated sequence available at http://www.tigr.org. EF3002U and EF3002D are primers used to clone upstream (U) and downstream (D) sequences of slyA in order to construct the deletion mutant by a double-crossover, and EF3002tot is for the in trans-complemented strain.
The underlined sequences are restriction enzyme recognition sites.
Using RNA extracted from strain V19, the transcriptional start point of slyA was determined by the rapid amplification of cDNA ends (RACE) (Roche, Mannheim, Germany) according to the manufacturer's instructions.
For reverse transcriptase PCR (RT-PCR) experiments, two micrograms of RNA was reverse transcribed with random hexamer primers and QuantiTect enzyme (Qiagen, Valencia, CA) according to the manufacturer's recommendations. cDNA and genomic DNA (positive control) were then used as templates for PCRs using primers listed in Table 2.
Construction and complementation of ΔslyA.
For the construction of a slyA deletion mutant (ΔslyA) strain, allelic replacement was used essentially as previously described (19). Seventy-nine percent of the slyA gene was deleted in the mutant strain, starting 42 bp after the GTG start codon and stopping 51 bp before the TAA stop codon. Briefly, a DNA fragment (obtained using chromosomal DNA of E. faecalis V19 as a template) containing ligated upstream (1,011-bp) and downstream (1,020-bp) sequences of the desired deletion were cloned into plasmid pMAD (Table 1), and 1 μg of recombinant plasmid was used to transform competent cells. Single-crossover transformants (dark blue and erythromycin-resistant colonies) were used for temperature shifts in order to release the plasmid. Candidate clones resulting from a double-crossover event were isolated on GM17 agar with or without erythromycin. Antibiotic-susceptible clones were analyzed for the presence of a deleted slyA gene and verified by sequencing.
To complement the slyA gene in trans, a PCR fragment containing slyA and its promoter (which was inferred from the nucleotide sequence and verified by RACE-PCR) were cloned into the plasmid pCU1 (Cmr) (2). The resulting vector (pCUslyA) was then used to transform the ΔslyA mutant. In order to compare the phenotype, of the complemented strain and the wild type, empty pCU1 was also introduced into strain V19 and the ΔslyA mutant strain.
Infection and survival experiments.
Infection of G. mellonella larvae with E. faecalis was accomplished as previously described by Lebreton et al. (20). Briefly, using a syringe pump (KD Scientific, Holliston, MA), larvae (about 0.3 g and 3 cm in length) were infected subcutaneously with washed E. faecalis from an overnight culture in GM17, with 6 × 106 ± 0.6 × 106 CFU per larva administered in 10 μl of sterile saline buffer. In each test, 30 insects were infected, and the experiments were repeated at least three times. L. lactis IL1403 was also tested under the same conditions as an avirulent control. Larval killing was then monitored at 20 h postinfection.
The virulences of V19 and the slyA deletion mutant were also assessed in an intravenous infection model, essentially as described by Gentry-Weeks et al. (11). Briefly, overnight cultures of the strains grown in brain heart infusion broth (BHI) supplemented with 40% heat-inactivated horse serum were centrifuged, and the resulting pellets were resuspended in sterile phosphate-buffered saline (PBS) to achieve final concentrations of 1 × 109 bacteria/ml. Aliquots of 100 μl from each strain were used to inject each of 10 female BALB/c mice (10 weeks old; Harlan Italy S.r.l., San Pietro al Natisone, Udine, Italy) through the tail vein. Infection experiments were repeated three times. Mice were monitored twice daily and were euthanized by CO2 asphyxiation 7 days after infection. Kidneys and livers were then removed aseptically, weighed, and homogenized in 5 ml of PBS using a stomacher (model 80; Pbi International, Milan, Italy) for 120 s at high speed. For CFU determination, serial homogenate dilutions were plated onto Enterococcus selective agar (Fluka Analytical, Switzerland) to avoid environmental contamination, and plating efficiency was comparable to that with a nonselective medium. CFU counts were analyzed by the unpaired t test.
Survival of E. faecalis in mouse peritoneal macrophages was also tested as described previously (10). Briefly, the ΔslyA mutant and V19 were grown as standing cultures at 37°C in BHI for 16 h. The bacteria were then pelleted and resuspended in 5 ml of PBS for injection. Mice were infected with 107 to 108 cells of each strain (estimated by CFU determination) by intraperitoneal injection of 200 μl of the PBS-enterococcus suspension. After 6 h of infection, the peritoneal macrophages were collected by two peritoneal washes, each using 5 ml of PBS. The cell suspension was dispensed into 24-well tissue culture plates and incubated at 37°C under 5% CO2 for 2 h. After exposure to antibiotics (vancomycin at 10 μg/ml and gentamicin at 150 μg/ml) to kill extracellular bacteria (verified by the absence of CFU in supernatant fluids), the infected macrophages were washed, and triplicate wells of macrophages were lysed with detergent to quantify viable intracellular bacteria. No difference in the MICs of these antibiotics was observed for the ΔslyA mutant and wild-type strains. The procedure was repeated three times, and results were analyzed using one-way analysis of variance with a Bonferroni correction posttest with SPSS statistical software (SPSS, Chicago, IL).
All statistical analyses were performed using Prism software (version 5.00) for Windows (GraphPad Software, San Diego, CA). For all comparisons, a P value of less than 0.05 was considered significant.
Sample preparation for microarray analysis.
E. faecalis V19 and ΔslyA strains were cultured in GM17 to mid-exponential phase (optical density [OD] of 0.5). RNA was extracted using the RNeasy Midiprep kit (Qiagen). RNA samples were treated with DNase, and DNA elimination was verified by PCR amplification. Microarray hybridizations were performed three times with three independent RNA preparations. cDNA preparation, fragmentation, labeling, and hybridization were performed as described in the Affymetrix manual (1). After washing, hybridization was assessed at 570 nm with a resolution of 1.56 μm using an Affymetrix GeneChip Scanner 3000 7G. Results were analyzed and compared using GeneChip Operating Software (GCOS) version 1.4. Correlation coefficients (r values) between experiments were from 0.934 to 0.99. RNA species that varied in abundance by greater than 2-fold were included in the analysis.
Microarray data accession number.
Microarray data have been deposited at the EMBL-EBI database, and the accession number is E-MEXP-2903.
RESULTS
Genetic characterization of EF_3002, encoding the SlyA regulator of E. faecalis.
Five potential transcriptional regulators have been annotated in the genome of E. faecalis V583 as being members of the MarR family, and they share moderate sequence identity at the amino acid level (49 to 54% identity). As noted by Wu and collaborators (37), EF_3002 was the most homologous to SlyA-like proteins identified in other species of bacteria and archaea that are involved in virulence and/or stress response. Examination of the currently available sequences of 26 different genomes of E. faecalis shows that the slyA gene is present in each and is part of the core genome of E. faecalis (4, 27, 28).
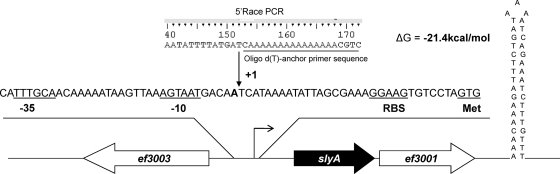
The slyA open reading frame (ORF) appears to begin with a GTG initiator codon and encodes a protein of 150 amino acids with a predicted molecular mass of 17.4 kDa and a pI of 9.5. The start codon is preceded by seven nucleotides by a potential ribosome binding site sequence (RBS), GGAAG. Using 5′ rapid amplification of cDNA ends (5′RACE), a transcriptional start point of the slyA gene was mapped (Fig. 1). We concluded that the A at 31 bp upstream of the GTG codon corresponds to the nucleotide +1 of the transcript. A putative −10 box (AGTAAT) located 5 nucleotides upstream from the transcriptional start point and a sequence near the −35 box consensus TTTGCA separated by 15 bp were identified. Furthermore, as shown in Fig. 1, the slyA locus (including EF_3001) is followed by an inverted repeat (ΔG = −21.4 kcal/mol) that may function as a transcriptional terminator.
Fig. 1.
Genetic organization of the slyA (EF_3002) chromosomal region of E. faecalis. Large arrows represent the genes which compose the ORFs, and their orientation shows the transcriptional direction. The nucleotide sequence of the slyA promoter region is shown. The transcriptional initiation nucleotide (+1) identified by 5′RACE is in bold and marked by an arrow, and a putative ribosome binding site sequence, −10 and −35 motifs, and the transcriptional initiation codon are underlined.
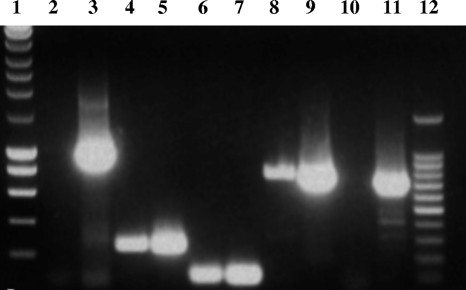
Based on comparison between slyA of E. faecalis and other homologous genes in Gram-positive genomes, as well as our RT-PCR results (see below), we suspected that slyA and downstream gene EF_3001 (encoding a protein homologous to PaiA of Bacillus subtilis, which is an N1-acetyltransferase) were in an operon structure. To test this possibility, we performed RT-PCR using primers able to (i) amplify parts of slyA and EF_3001 individually to demonstrate the presence of transcripts of each in the cell and (ii) amplify from slyA to EF_3001, including the 15-bp intergenic region, to determine whether transcription can proceed from slyA into EF_3001 as in an operon. Additionally, to demonstrate the absence of contaminating DNA in the preparation, sequences upstream of EF_3002 and downstream of EF_3001, which would not be predicted to be cotranscribed, were amplified as well (Fig. 2). As shown in Fig. 2, an amplicon could be generated across slyA-EF_3001, indicating that readthrough does occur.
Fig. 2.
Reverse transcriptase PCR (RT-PCR) analysis of slyA-EF_3001 transcription. The primers used are listed in Table 2. PCRs were performed on chromosomal DNA (lanes 3, 5, 7, 9, and 11) or cDNA (lanes 2, 4, 6, 8, and 10) as a template. To demonstrate the absence of contaminating DNA in the cDNA preparation, sequences upstream of slyA (EF3002U primers, 1.011 bp) (lanes 2 and 3) and downstream of EF_3001 (EF3001int5′ and EF3002D3′ primers, 728 bp) (lanes 10 and 11), which would not be predicted to be cotranscribed, were amplified. The presence of slyA and EF_3001 transcripts was shown by amplifications of internal fragments of slyA (EF3002int primers, 249 bp) (lanes 4 and 5) and of EF_3001 (EF3001int primers, 100 bp) (lanes 6 and 7). To determine whether transcription can proceed from slyA into EF_3001, amplifications of part of slyA-EF_3001 including the 15-bp intergenic region (EF3002int5′ and EF3001int3′ primers, 778 bp) (lanes 8 and 9) were performed. Lanes 1 and 12 correspond to the 10-kb and 100-bp DNA ladders, respectively.
SlyA is involved in virulence and bacterial persistence within mouse organs and peritoneal macrophages.
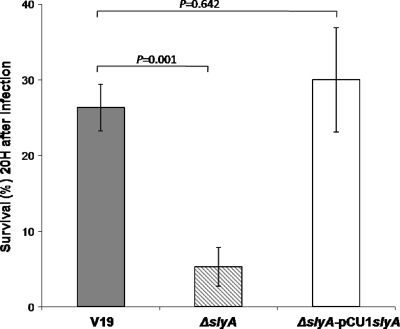
Based on the literature and despite the lack of phenotype for the ΔslyA mutant toward stresses likely encountered during infection, such as H2O2, acidity, lysozyme, urine, or serum (data not shown), SlyA may nevertheless be involved in the virulence of E. faecalis. In order to verify this, we compared the abilities of the wild-type and the ΔslyA mutant strains to kill G. mellonella larvae in infection experiments. As shown in Fig. 3, the rate of killing was significantly higher in larvae infected with the ΔslyA mutant strain than in those infected with wild-type V19. After 20 h of infection, more than 26% of the V19-infected larvae were still alive, whereas fewer than 6% of the animals infected with ΔslyA strain had survived (P = 0.001). The nonpathogenic Gram-positive bacterium L. lactis IL1403 was used as a control; all of the larvae infected with this organism survived. Experiments conducted with the complemented ΔslyA mutant strain, in which virulence was restored to the wild-type level, confirmed that the observed phenotype was due to the deletion of slyA (Fig. 3). In addition, experiments with the strains containing empty pCU1 were performed. Wild-type and mutant phenotypes were not modified by the presence of this vector (data not shown). slyA deletion mutants have been also created in two other backgrounds of E. faecalis (JH2-2 and OG1) and their virulence tested using the G. mellonella model. In both cases, the mutants were 2- to 3-fold more virulent than the corresponding wild-type strains (data not shown).
Fig. 3.
Effect of slyA inactivation on virulence. Percent survival of G. mellonella larvae at 20 h after infection with around 1.2 × 105 CFU of E. faecalis V19, the ΔslyA mutant, and the complemented ΔslyA mutant strain (ΔslyA pCU1-slyA) is shown. Experiments were repeated at least three times, and the results represent the mean ± standard deviation for live larvae.
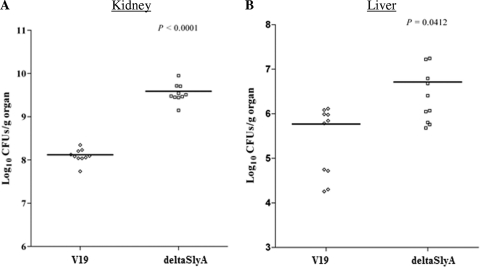
We then examined the fates of the mutant and wild-type strains in the murine systemic infection model, specifically, tissue burdens in kidneys and livers of infected mice. As shown in Fig. 4, the ΔslyA mutant strain showed statistically significantly increases of tissue burden in both organs. The ΔslyA mutant exhibited an increase of 1.6 log units (P < 0.0001) in kidney and of 1 log unit (P = 0.0412) in liver compared to the V19 parental strain, showing that SlyA is involved in the survival of E. faecalis within the host.
Fig. 4.
Bacterial persistence within mouse organs. Enterococcal tissue burdens in kidneys and in livers from BALB/c mice infected intravenously with 1 × 108 cells of E. faecalis wild-type V19 and the ΔslyA mutant strain are shown. Kidney pair and liver homogenates were obtained from groups of 10 mice sacrificed and necropsied at day 7 postinfection. The results, expressed as log10 CFU per gram of tissue, represent values recorded separately for each of the 10 mice. Horizontal bars represent the geometric means. P values of less than 0.05 were considered to be significant.
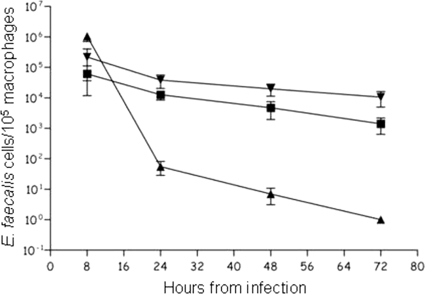
Phagocytic cells constitute an important part of the innate immunity against pathogens. To determine whether the ΔslyA mutant was affected at this stage of infection, cells were tested in a macrophage infection model. The intracellular survival of the ΔslyA mutant and wild-type strain V19 was monitored by determining the number of viable bacteria inside infected mouse peritoneal macrophages over a 72-h time course. No significant difference was observed between the cell counts at 8 h postinfection, suggesting that phagocytosis of the two strains by the macrophages was similar. However, as shown in Fig. 5, at 24 h (P < 0.007), 48 h (P < 0.0019), and 72 h (P < 0.0026) postinfection, the ΔslyA mutant was significantly more abundant intracellularly. At 72 h after infection, the survival rate of the ΔslyA mutant in mouse macrophages was approximately 1 log unit higher than that of the wild-type strain (Fig. 5).
Fig. 5.
Time course of intracellular survival of E. faecalis V19 (▪) and the ΔslyA mutant (▾) within murine peritoneal macrophages. Results for E. coli strain DH5α (▴) are shown as a control of a nonvirulent strain in macrophages. The results represent the mean ± standard deviation of viable intracellular bacteria per 105 macrophages from three independent experiments with three wells.
Identification of the SlyA regulon in E. faecalis.
In order to identify candidate genes that contribute to increased persistence in organs and macrophages, microarray analysis was used to compare gene expression in wild-type and slyA mutant cells (22). Total RNA was isolated from cells harvested in mid-exponential growth phase. Genes whose expression differed by more than 2-fold were analyzed further and are listed as members of the SlyA regulon in Table 3. This study revealed that 117 genes were differentially regulated (with P values of <0.05) in the ΔslyA mutant. Most exhibited reduced expression in the ΔslyA mutant strain (2- to 32.24-fold; 111 genes), with only 6 genes exhibiting increased transcript abundance in the mutant strain (2.04- to 16.62-fold). It is probable that genes with the most increased or decreased transcriptional deregulation have an important impact on the mutant cells. Among these, the transcript encoding EF_3217, which is annotated as encoding a putative helicase, was 32-fold more abundant in the wild-type cells. Other gene products obviously expressed at lower levels in the absence of SlyA included transcriptional regulators of the ArgR family (EF_0102-0103 operon, involved in arginine metabolism) and energy and nucleotide metabolism enzymes (putative EF_3137-3139 and EF_1783-1786 operons, respectively). Expression of EF_3001, the second member of the slyA operon, was more than 16-fold higher in the absence of SlyA. Two other clearly SlyA-repressed genes (overexpressed in the mutant) play roles in protein degradation (EF_2997) and glycosis/glycogenesis reaction (putative EF_0718-0719 operon) (Table 3). Major deregulation results were confirmed by quantitative RT-PCR (qRT-PCR), showing a decrease of 20-fold for EF_3217 in the mutant strain while EF_0718, EF_2997, EF_3203, and EF_3001 expression levels in the ΔslyA mutant were increased by 2.1-, 3.6-, 3.2-, and 124-fold, respectively (results not shown).
Table 3.
Up- and downregulated genes in the ΔslyA mutant
| Category and locus | Gene product function | Fold change in expression |
|---|---|---|
| Protein synthesis | ||
| EF_3203 | Ribosomal protein L33 | +2.04 |
| Protein fate | ||
| EF_2524 | Sortase family protein | −2.55 |
| EF_2997 | Peptidase, M20M25M40 family | +3.81 |
| Phage | ||
| EF_0309 | Excisionase, putative | −2.27 |
| EF_1455 | Terminase, large subunit, putative | −2.04 |
| DNA metabolism | ||
| EF_1435 | Recombination protein U, putative | −2.15 |
| EF_2546 | Recombinase, phage integrase family | −2.48 |
| Cell envelope and cell division | ||
| EF_2533 | FtsK-SpoIIIE family protein | −2.24 |
| Transport and binding protein | ||
| EF_0758 | Cadmium-translocating P-type ATPase | −2.16 |
| EF_0893 | Amino acid ABC transporter | −2.97 |
| EF_3137 | PTS system, IIB component | −2.17 |
| EF_3138 | PTS system, IID component | −2.53 |
| EF_3139 | PTS system, IIC component | −3.06 |
| EF_3208 | ABC transporter, permease protein | −2.53 |
| EF_3209 | ABC transporter, ATP binding protein | −2.18 |
| Regulatory and DNA integration | ||
| EF_0102 | Transcriptional regulator, ArgR family | −2.71 |
| EF_0103 | Transcriptional regulator, ArgR family | −3.49 |
| EF_0307 | Transcriptional regulator, CroCI family | −2.78 |
| EF_0317 | Transcriptional regulator, CroCI family | −2.16 |
| EF_0719 | Transcriptional regulator, DeoR family | +2.19 |
| EF_1423 | Transcriptional regulator, CroCI family | −2.25 |
| EF_1450 | Positive control factor, putative | −2.05 |
| EF_2138 | Transcriptional regulator, CroCI family | −2.53 |
| EF_2141 | Transcriptional regulator, CroCI family | −2.45 |
| EF_2528 | Transcriptional regulator, CroCI family | −2.35 |
| Nucleotide metabolism | ||
| EF_1783 | Phosphoribosylformylglycinamidine synthetase I | −3.55 |
| EF_1784 | Phosphoribosylformylglycinamidine synthase, PurS protein | −3.76 |
| EF_1785 | Phosphoribosylaminoimidazole-succinocarboxamide synthase | −3.80 |
| EF_1786 | Phosphoribosylaminoimidazole carboxylase, ATPase subunit | −2.39 |
| Energy metabolism | ||
| EF_0271 | Glycosyl hydrolase, family 1 | −2.43 |
| EF_0696 | Tagatose 1,6-diphosphate aldolase | −2.03 |
| EF_0718 | 1-Phosphofructokinase | +3.12 |
| EF_1603 | Sucrose-6-phosphate dehydrogenase | −2.07 |
| Other metabolism | ||
| EF_0312 | Aspartate 1-decarboxylase domain protein | −2.54 |
| EF_0759 | SapB protein, putative | −2.31 |
| EF_1434 | DnaD domain protein | −2.06 |
| EF_3001 | Protease synthase and sporulation negative | +16.62 |
| EF_3217 | Helicase, putative | −32.24 |
| Virulence | ||
| EF_0149 | Aggregation substance, putative | −2.10 |
| EF_1426 | VrII protein, putative | −2.70 |
| Unknown | ||
| EF_0131/EF_1085/EF_1451 | Conserved domain protein | −2.01 to 3.07 |
| EF_0135/EF_0142/EF_0308/EF_0315/EF_0326/EF_0328/EF_0329/EF_1081/EF_1456EF_1459/EF_1466/EF_1467/EF_1470/EF_1471/ EF_1485/EF_1533/EF_2140/EF_2220EF_2519/EF_2520/EF_2521/ EF_2529/EF_2534/EF_2536/EF_2951/EF_2996 | Conserved hypothetical protein | ±2.00 to 2.83 |
| EF_0132/EF_0134/EF_0310/EF_0311/EF_0313/EF_0314/EF_0316/ EF_0320/EF_0327/EF_1015EF_1067/EF_1083/EF_1278/EF_1418/ EF_1424/EF_1425/EF_1427/EF_1428/EF_1429/EF_1431EF_1439/ EF_1458/EF_1489/EF_1686/EF_1812/EF_1988/EF_2133/EF_2135/ EF_2136/EF_2139EF_2523/EF_2526/EF_2530/EF_2531/EF_2532/ EF_2537/EF_2538/EF_2539/EF_2540/EF_2541EF_2542/EF_2942/ EF_2944/EF_2945/EF_2949/EF_2950/EF_2952/EF_2953 | Hypothetical protein | −2.00 to 3.36 |
DISCUSSION
Members of the MarR/SlyA transcriptional factor family are widely distributed among prokaryotes. Some of them modulate the transcription of virulence and/or stress genes. That is the case for MarR of E. coli, PecS of E. chrysanthemi, HprR of B. subtilis, RovA of Y. enterocolitica and Y. pseudotuberculosis, SlyA of S. enterica serovar Typhimurium, MarR of Bacillus anthracis, and MgrA of S. aureus (7). From these observations, it appeared that the physiological characterization of this protein would be of interest to increase our understanding about regulators involved in the virulence in this opportunistic pathogen.
We characterized the slyA promoter region, and our results showed that slyA belongs to a likely bicistronic operon with EF_3001, encoding an N1-acetyltransferase enzyme (8). Such an organization is conserved in Bacillus, Listeria, Lactobacillus, and Clostridium genomes, and a gene homologous to EF_3001 is also present near a putative MarR-encoding gene in Streptococcus gordonii.
One important finding of this work is that ΔslyA mutants of three different strains were more virulent than the parental strains in the G. mellonella model. It has been shown that the G. mellonella immune response has a number of structural and functional similarities to the innate immune response of mammals, and this insect was shown to be an excellent infection model to evaluate the involvement of E. faecalis pathogenic factors (i.e., Fsr, GelE, and Ace) in virulence (9, 18, 20). Recently, it has been proved that group A Streptococcus virulence in these wax worms strongly correlates with data obtained in vertebrate models (mice and monkeys) (26). In addition, mutant strain bacterial counts recovered from macrophages and from liver and kidney after peritoneal and systemic infections were higher than those of the wild type. This phenotype does not seem to be due to a better tolerance to environmental stress, since growth was not increased in the presence of several stressing agents likely encountered during infection and was even somewhat more sensitive when bile salts were added to the medium (data not shown). Two E. faecalis mutants affected in the PerR and Err10 transcriptional regulators (a two-component system) also display better survival within macrophages than the parental wild-type strain (23, 35). In contrast to the ΔslyA mutant, a PerR-deficient strain of E. faecalis is more resistant to H2O2, and an err10 mutant survived better with heat and bile salts challenges (23, 35). Nevertheless, we cannot exclude that SlyA may have a role in an as-yet-unidentified stress response encountered during host infection. These observations make SlyA a special case of this regulator family, since marR/slyA mutants of other studied bacteria are less virulent or have no role in infection (3, 5, 7, 16, 24, 34). Despite common folding and similar DNA binding properties of members of the MarR/SlyA family, structural analysis showed that local divergence in sequence allows them to bind to different gene targets (37).
It may be suggested that SlyA could be a repressor for the expression of virulence factors in E. faecalis. It is then tempting to speculate that among genes overexpressed in the ΔslyA mutant, one or more could play a role in the more virulent phenotype observed for the mutant. In this hypothesis, EF_3001 may be an interesting candidate. It is the most induced gene in the ΔslyA mutant and encodes a protein similar to B. subtilis PaiA, which is an N1-spermidine/spermine acetyltransferase (52% identical and 71% similar) (8). This enzyme is involved in polyamine homeostasis, preventing toxicity due to an excess of this molecule. Overexpression of EF_3001 should then be an advantage for the ΔslyA mutant during infection. In order to test this hypothesis, we tried to construct an EF_3001 mutant strain using a double-crossover strategy with a thermosensitive replication plasmid. Unfortunately, all antibiotic-sensitive clones obtained after the second recombination harbored the wild-type gene. These results strongly suggest that EF_3001 may be essential for E. faecalis. In addition, the more modest but significant overexpression of the putative fructose phosphotransferase system (PTS) operon EF_0718-0719, encoding phosphofructokinase and a transcriptional regulator of the DeoR family, should also participate in the better survival of the ΔslyA mutant inside the host compared to the V19 parental strain. Transcription of EF_3217, encoding a putative helicase, is strongly downregulated in the ΔslyA mutant. However, it is unlikely that this reduced expression has an impact in the more virulent phenotype of the mutant, because EF_3217 seems to be part of a mobile element and is present in only 5 of the 26 sequenced genomes (4, 27, 28).
It is of note that 11 loci encoding transcriptional regulators were deregulated in the mutant strain, and it is probable that some members of the SlyA regulon are under several transcriptional controls. Such a regulatory cascade has been shown for S. aureus and Salmonella (17, 24).
In conclusion, in this work we have characterized SlyA of E. faecalis, which is a transcriptional regulator involved in persistence in host cells as well as in virulence. Identification of the SlyA regulon by a global transcriptomic approach is a first step in the identification of new effectors of pathogenicity.
ACKNOWLEDGMENTS
The expert technical assistance of Isabelle Rincé, Marie-Jeanne Pigny, and Evelyne Marchand is greatly appreciated. We thank A. Benachour, S. Gente, J.-M. Laplace, A. Rincé, N. Sauvageot, N. Verneuil, A. Budin-Verneuil, and A. Hanin for helpful discussions.
This study was partly supported by grants from the Agence Nationale de la Recherche in the frame of a transnational ERA-NET PathoGenoMics program (ANR-06-PATHO-008-01) and by grant AI072360.
Footnotes
Published ahead of print on 2 May 2011.
REFERENCES
- 1. Affymetrix, 2006. GeneChip expression analysis technical manual: procaryotic target preparation. P/N 702232 Rev 2 Affymetrix, Santa Clara, CA [Google Scholar]
- 1a. Arnaud M., Chastanet A., Débarbouillé M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Augustin J., et al. 1992. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur. J. Biochem. 204:1149–1154 [DOI] [PubMed] [Google Scholar]
- 3. Bergman N. H., et al. 2007. Transcriptional profiling of Bacillus anthracis during infection of host macrophages. Infect. Immun. 75:3434–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bourgogne A., et al. 2008. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 9:R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buchmeier N., et al. 1997. SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect. Immun. 65:3725–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coburn P. S., Baghdayan A. S., Dolan G. T., Shankar N. 2008. An AraC-type transcriptional regulator encoded on the Enterococcus faecalis pathogenicity island contributes to pathogenesis and intracellular macrophage survival. Infect. Immun. 76:5668–5676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ellison D. W., Miller V. L. 2006. Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 9:153–159 [DOI] [PubMed] [Google Scholar]
- 8. Forouhar F., et al. 2005. Structural and functional evidence for Bacillus subtilis PaiA as a novel N1-spermidine/spermine acetyltransferase. J. Biol. Chem. 280:40328–40336 [DOI] [PubMed] [Google Scholar]
- 9. Gaspar F., et al. 2009. Virulence of Enterococcus faecalis dairy strains in an insect model: the role of fsrB and gelE. Microbiology 155:3564–3571 [DOI] [PubMed] [Google Scholar]
- 10. Gentry-Weeks C. R., Karkhoff-Schweizer R., Pikis A., Estay M., Keith J. M. 1999. Survival of Enterococcus faecalis in mouse peritoneal macrophages. Infect. Immun. 67:2160–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gentry-Weeks C., Estay M., Loui C., Baker D. 2003. Intravenous mouse infection model for studying the pathology of Enterococcus faecalis infections. Infect. Immun. 71:1434–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giard J. C., Rince A., Capiaux H., Auffray Y., Hartke A. 2000. Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotrophic effect on cell morphology, stress sensitivity, and gene expression in Enterococcus faecalis. J. Bacteriol. 182:4512–4520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Giard J. C., et al. 2006. Characterization of Ers, a PrfA-like regulator of Enterococcus faecalis. FEMS Immunol. Med. Microbiol. 46:410–418 [DOI] [PubMed] [Google Scholar]
- 14. Gilmore M., Coburn P., Nallapareddy S., Murray B. 2002. Enterococcal virulence, p. 301–354 In Gilmore M. S. (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance, American Society for Microbiology, Washington, DC [Google Scholar]
- 15. Hancock L. E., Gilmore M. S. 2002. The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proc. Natl. Acad. Sci. U. S. A. 99:1574–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haque M. M., Kabir M. S., Aini L. Q., Hirata H., Tsuyumu S. 2009. SlyA, a MarR family transcriptional regulator, is essential for virulence in Dickeya dadantii 3937. J. Bacteriol. 191:5409–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ingavale S., van Wamel W., Luong T. T., Lee C. Y., Cheung A. L. 2005. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect. Immun. 73:1423–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kavanagh K., Reeves E. P. 2004. Exploiting the potential of insects for in vivo pathogenicity testing of microbial pathogens. FEMS Microbiol. Rev. 28:101–112 [DOI] [PubMed] [Google Scholar]
- 19. La Carbona S., et al. 2007. Comparative study of the physiological roles of three peroxidases (NADH peroxidase, alkyl hydroperoxide reductase and thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis. Mol. Microbiol. 66:1148–1163 [DOI] [PubMed] [Google Scholar]
- 20. Lebreton F., et al. 2009. ace, which encodes an adhesin in Enterococcus faecalis, is regulated by Ers and is involved in virulence. Infect. Immun. 77:2832–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mäkinen P. L., Clewell D. B., An F., Mäkinen K. K. 1989. Purification and substrate specificity of a strongly hydrophobic extracellular metalloendopeptidase (“gelatinase”) from Streptococcus faecalis (strain 0G1-10). J. Biol. Chem. 264:3325–3334 [PubMed] [Google Scholar]
- 22. McBride S. M., Fischetti V. A., Leblanc D. J., Moellering R. C., Gilmore M. S. 2007. Genetic diversity among Enterococcus faecalis. PLoS One 2:e582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muller C., et al. 2008. Characterization of two signal transduction systems involved in intracellular macrophage survival and environmental stress response in Enterococcus faecalis. J. Mol. Microbiol. Biotechnol. 14:59–66 [DOI] [PubMed] [Google Scholar]
- 24. Navarre W. W., et al. 2005. Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol. Microbiol. 56:492–508 [DOI] [PubMed] [Google Scholar]
- 25. Ogier J., Serror P. 2008. Safety assessment of dairy microorganisms: the Enterococcus genus. Int. J. Food Microbiol. 126:291–301 [DOI] [PubMed] [Google Scholar]
- 26. Olsen R. J., Watkins M. E., Cantu C. C., Beres S. B., Musser J. M. 1 March 2011. Virulence of serotype M3 group A Streptococcus strains in wax worms (Galleria mellonella larvae). Virulence [Epub ahead of print.] doi:10.4161/viru.2.2.14338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Palmer K. L., et al. 2010. High-quality draft genome sequences of 28 Enterococcus sp. isolates. J. Bacteriol. 192:2469–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paulsen I. T., et al. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074 [DOI] [PubMed] [Google Scholar]
- 29. Qin X., Singh K. V., Weinstock G. M., Murray B. E. 2001. Characterization of Fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183:3372–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 31. Shankar N., et al. 2001. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect. Immun. 69:4366–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shepard B. D., Gilmore M. S. 2002. Antibiotic-resistant enterococci: the mechanisms and dynamics of drug introduction and resistance. Microbes Infect. 4:215–224 [DOI] [PubMed] [Google Scholar]
- 33. Singh K. V., Nallapareddy S. R., Murray B. E. 2007. Importance of the ebp (endocarditis- and biofilm-associated pilus) locus in the pathogenesis of Enterococcus faecalis ascending urinary tract infection. J. Infect. Dis. 195:1671–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Spory A., Bosserhoff A., von Rhein C., Goebel W., Ludwig A. 2002. Differential regulation of multiple proteins of Escherichia coli and Salmonella enterica serovar Typhimurium by the transcriptional regulator SlyA. J. Bacteriol. 184:3549–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Verneuil N., et al. 2005. Contribution of a PerR-like regulator to the oxidative-stress response and virulence of Enterococcus faecalis. Microbiology 151:3997–4004 [DOI] [PubMed] [Google Scholar]
- 36. Wisplinghoff H., et al. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
- 37. Wu R., et al. 2003. Crystal structure of Enterococcus faecalis SlyA-like transcriptional factor. J. Biol. Chem. 278:20240–20244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao C., et al. 2010. Role of methionine sulfoxide reductases A and B of Enterococcus faecalis in oxidative stress and virulence. Infect. Immun. 78:3889–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]