Abstract
Porphyromonas gingivalis is a pathogen in severe periodontal disease. Able to exploit an intracellular lifestyle within primary gingival epithelial cells (GECs), a reservoir of P. gingivalis can persist within the gingival epithelia. This process is facilitated by manipulation of the host cell signal transduction cascades which can impact cell cycle, cell death, and cytokine responses. Using microarrays, we investigated the ability of P. gingivalis 33277 to regulate microRNA (miRNA) expression in GECs. One of several miRNAs differentially regulated by GECs in the presence of P. gingivalis was miRNA-203 (miR-203), which was upregulated 4-fold compared to uninfected controls. Differential regulation of miR-203 was confirmed by quantitative reverse transcription-PCR (qRT-PCR). Putative targets of miR-203, suppressor of cytokine signaling 3 (SOCS3) and SOCS6, were evaluated by qRT-PCR. SOCS3 and SOCS6 mRNA levels were reduced >5-fold and >2-fold, respectively, in P. gingivalis-infected GECs compared to controls. Silencing of miR-203 using a small interfering RNA construct reversed the inhibition of SOCS3 expression. A dual luciferase assay confirmed binding of miR-203 to the putative target binding site of the SOCS3 3′ untranslated region. Western blot analysis demonstrated that activation of signal transducer and activator of transcription 3 (Stat3), a downstream target of SOCS, was diminished following miR-203 silencing. This study shows that induction of miRNAs by P. gingivalis can modulate important host signaling responses.
INTRODUCTION
Periodontitis is one of the most prevalent human diseases, and it has been reported that greater than 50% of adults over 30 years of age in the United States have a manifestation of the disease (2). Persistent bacterial infection of the periodontal tissues leads to increased inflammation and in severe cases culminates in the destruction of the alveolar bone and tooth loss. The disease is initiated by consortia of pathogenic bacteria, of which one of the most important members is the Gram-negative anaerobe Porphyromonas gingivalis (12, 22, 30). P. gingivalis expresses several virulence factors that enable colonization of oral surfaces and facilitate tissue destruction (18, 30, 61). Moreover, P. gingivalis can internalize and survive within host gingival epithelial cells (GECs) (29, 58) and move intercellularly without exposure to the extracellular environment (62). Invasion is effectuated through manipulation of host cell phosphorylation-dependent signaling pathways and remodeling of the microfilament and microtubule cytoskeleton (21, 56, 57). P. gingivalis also modulates host cell gene expression and affects pathways that control cytokine production and apoptotic cell death (20, 38). Indeed, infection of epithelial cells by P. gingivalis can stimulate the production of some cytokines, such as interleukin-1β (IL-1β) (53), and suppress the production of others, such as IL-8 (9). The differential regulation of cytokine responses may allow P. gingivalis to orchestrate innate and adaptive immunity. P. gingivalis also inhibits apoptotic cell death in gingival epithelial cells through activation of the Janus kinase (JAK)/signal transducer and activator of transcription (Stat) and Akt pathways (38, 63).
MicroRNAs (miRNAs) are small (≈22-nucleotide [nt]) single-stranded noncoding RNAs that can suppress gene expression by binding to the 3′ untranslated region (UTR) of target mRNA, leading to mRNA degradation and translational repression. In mammals, they are involved in many regulatory processes, including the regulation of differentiation/proliferation (1) and host cell immune responses (43, 51). The expression of miRNAs is often temporally and spatially specific (40, 60), and the differential expression of miRNAs has been directly linked to disease (36, 37). Host cell responses to pathogens have also been shown to be regulated in part by miRNAs. Human monocytes exposed to lipopolysaccharide (LPS) upregulate miRNA-146a (miR-146a) and miR-9, resulting in the downregulation of cytokines involved in the innate immune response (5, 54). Furthermore, miR-146a is essential for endotoxin tolerance in monocytes in vitro (41). P. gingivalis can also regulate miRNA expression, and epithelial cells exposed to nonviable P. gingivalis showed increase miR-105 expression, which in turn suppressed levels of Toll-like receptor 2 (TLR-2) (7).
We hypothesized that live P. gingivalis can affect expression of miRNAs involved in networks that control innate immunity and apoptosis and that extend beyond TLR regulation. Array and in silico analyses were undertaken to identify miRNAs differentially regulated in P. gingivalis-infected primary GECs. P. gingivalis induced the upregulation of miR-203, which directly inhibited suppressor of cytokine signaling 3 (SOCS3) and resulted in increased Stat3 activation. These results suggest a role of miRNA in the pathogenesis of P. gingivalis, contributing to the modulation of host cell immune responses and facilitating bacterial persistence.
MATERIALS AND METHODS
Bacterial and eukaryotic cell culture.
P. gingivalis 33277 was cultured in Trypticase soy broth, supplemented with yeast extract (1 mg ml−1), hemin (5 μg ml−1), and menadione (1 μg ml−1), anaerobically at 37°C. Primary cultures of GECs were generated as described previously (44). Briefly, healthy gingival tissue was collected from patients undergoing surgery for removal of impacted third molars and following Institutional Review Board guidelines. Basal epithelial cells were separated and cultured in keratinocyte growth medium (DermaLife basal medium; Lifeline, Walkersville, MD) in the absence of antibiotics. GECs were used at passage 4. Cells of the HTR8/SV neo trophoblast cell line (HTR8 cells) were provided by Charles Graham (Kingston, Ontario, Canada). HTR8 cells were routinely cultured in RPMI 1640 medium (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum. Eukaryotic cells were cultured at 37°C in 5% CO2.
miRNA array profiling and analysis.
GECs were infected with P. gingivalis at a multiplicity of infection (MOI) of 100 for 6 h, and total RNA was isolated using TRIzol extraction, followed by purification with a Qiagen miRNeasy kit, according to the manufacturer's instructions. RNA quality and concentration were confirmed using an Agilent Bioanalyzer 2100 apparatus. Experiments were performed in triplicate using primary epithelial cells isolated from three different patient sources. Microarray experiments were performed using a service provider (LC Sciences, Houston, TX). Total RNA was size fractionated using a YM-100 Microcon centrifugal filter (Millipore, Billerica, MA), and the small RNAs (<300 nt) were 3′ extended with a poly(A) tail using poly(A) polymerase. An oligonucleotide tag was ligated to the poly(A) tail for later fluorescent dye staining; two different tags were used for the two RNA samples. Hybridization was performed overnight on a μParaflo microfluidic chip using a microcirculation pump (Atactic Technologies, Houston, TX) (13, 66). On the microfluidic chip, each detection probe consisted of a chemically modified nucleotide coding segment complementary to target microRNA (from miRBase, http://microrna.sanger.ac.uk/sequences/, version 13.0) and a spacer segment of polyethylene glycol to extend the coding segment away from the substrate. The detection probes were made by in situ synthesis using photogenerated reagent chemistry. The hybridization melting temperatures were balanced by chemical modifications of the detection probes. After RNA hybridization, tag-conjugating Cy3 and Cy5 dyes were circulated through the microfluidic chip for dye staining. Fluorescence images were collected using a laser scanner (GenePix 4000B; Molecular Devices, Sunnyvale, CA) and digitized using Array-Pro image analysis software (Media Cybernetics, Bethesda, MD). Data were analyzed by first subtracting the background and then normalizing the signals using a Lowess filter (8). The ratio of the two sets of detected signals (log2 transformed, balanced) and P values of the t test were calculated, and those with P values of <0.01 were designated differentially expressed.
Quantitative RT-PCR (qRT-PCR) of miR-203.
Total RNA was isolated and purified from control and infected GECs and size fractionated as described above. Equal amounts (200 ng/reaction mixture) were used to synthesize cDNA employing a TaqMan microRNA reverse transcription (RT) kit and primers specific for miR-203 and RNU-48 (control) (Applied Biosystems, Carlsbad, CA). Negative RT reactions were included in all experiments. Real-time PCR was performed using TaqMan Fast universal PCR master mix (Applied Biosystems). A Bio-Rad iCycler with iCycler IQ (version 3.1) software (Bio-Rad, Hercules, CA) was employed, with the autocalculated threshold cycle (CT) selected. The cycle threshold values were determined, and miRNA expression values were determined relative to the value for RNU-48 following the 2−ΔΔCT method.
Quantitative RT-PCR for mRNA.
To determine expression levels of SOCS3 and SOCS6, RNA was isolated essentially as described above without size fractionation. cDNA was synthesized (2 μg/reaction mixture RNA) using a high-capacity RNA-cDNA kit (Applied Biosystems). Real-time PCRs used TaqMan Fast universal PCR master mix and gene expression assays for SOCS3 or SOCS6 (Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a housekeeping control, and negative RT reactions were included in each assay. Expression values of mRNA were quantified as described above after normalization to the value for GAPDH.
Transfection of GECs with miRNA.
Cells were transfected, using siPORT NeoFX transfection agent, with 100 nM miR-203 mimic, silence miR-203 (si-miR-203; antagomir), si-miR-146b (unrelated control), or a nonsilence control (a nontargeting RNA-negative control with no significant homology to any known human gene sequence) (Applied Biosystems). Following 24 h in transfection medium, the medium was replaced with regular culture medium for a further 24 h. Transfected cells were infected with P. gingivalis at an MOI of 100 for 6 h, and total RNA was isolated for real-time PCR analysis of SOCS3 expression, as described above.
Dual luciferase reporter assay.
Confirmation of miR-203 binding to the putative 3′ UTR binding site of SOCS3 utilized a dual luciferase reporter assay. HTR8 cells at 80% confluence were transfected with 100 ng reporter vector pSGG-SOCS3 3′UTR, empty vector pSGG-EMPTY-3UTR (positive transfection control), or pSGG-GAPDH-3UTR, a vector containing the 3′ UTR region of GAPDH (housekeeping gene control). All vectors were from Switchgear Genomics. Reporter vectors were cotransfected with miR-203 mimic (100 nM) or miR-146 mimic (unrelated negative control) (Applied Biosystems). DharmaFECT Duo (Dharmacon, Lafayette, CO) was used as the transfection reagent in serum-free medium. Following 24 h of incubation in transfection medium, Steady-Glo luciferase assay reagent (Promega, Madison, WI) was added to each well and the plate was incubated at room temperature in the dark for 30 min. The luciferase signal was measured using a Wallac Victor3 1420 multilabel counter luminometer (Waltham, MA). Knockdown was assessed by calculating luciferase signal ratios for specific miRNA/nontargeting control, using data for housekeeping and empty reporter vector groups to control for nonspecific effects.
Western immunoblotting.
Whole-cell lysates of uninfected or P. gingivalis-infected GECs were collected, separated by SDS-PAGE, and transferred onto nitrocellulose membranes by electroblotting. Blots were blocked in 10% skim dry milk in Tris-buffered saline (TBS) overnight at 4°C. Primary antibody was rabbit anti-Stat3 or rabbit anti-phospho-Stat3 (Sigma) at 1:1,000 for 2 h at room temperature. Antigen-antibody binding was detected using horseradish peroxidase-conjugated species-specific secondary antibodies followed by enhanced chemiluminescence Western blotting detection reagents (Perkin-Elmer, Waltham, MA). Densitometric analysis was performed and phospho-Stat3/Stat3 ratios were calculated following normalization to the value for GAPDH.
Microarray data accession number.
The array results have been deposited in the GEO repository (http://www.ncbi.nlm.nih.gov/geo), accession number GSE28635.
RESULTS
miRNAs differentially regulated following P. gingivalis infection of epithelial cells.
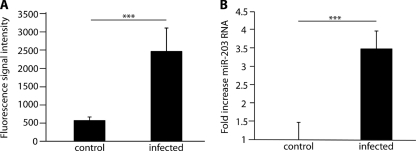
We investigated the potential involvement of miRNA expression in the primary gingival epithelial cell response to P. gingivalis infection using chip-based miRNA profiling. Expression levels of GEC miRNAs are listed in the data set in the supplemental material. Following background subtraction, data were normalized to the statistical mean of detectable transcripts. Clustering analysis by analysis of variance (ANOVA) identified differentially regulated miRNAs (see Fig. S1 and the data set in the supplemental material). Fourteen miRNAs differentially expressed between infected and uninfected groups gave both strong signals and high significance (Table 1). We focused on miR-203, which was upregulated 4-fold in P. gingivalis-challenged cells compared with controls (Fig. 1A) and which is involved in the regulation of a number of epithelial cell properties (51, 60). Increased miR-203 expression was confirmed using qRT-PCR. Expression was upregulated >3.5-fold in infected groups compared with uninfected controls (P < 0.001) (Fig. 1B).
Table 1.
MicroRNAs differentially expresseda in P. gingivalis-infected gingival epithelial cells
| hsa-miR | miRNA expression |
P value | Log2 (infected/control) | |||
|---|---|---|---|---|---|---|
| Control cells |
Infected cells |
|||||
| Mean | SD | Mean | SD | |||
| 149* | 398 | 74 | 2,117 | 363 | 0.0014 | 2.41 |
| 203 | 564 | 96 | 2,437 | 635 | 0.0037 | 2.11 |
| 205 | 27,191 | 1,159 | 22,661 | 621 | 0.0086 | −0.26 |
| 181a | 2,765 | 265 | 1,789 | 171 | 0.012 | −0.63 |
| 1308 | 2,777 | 263 | 4,582 | 559 | 0.012 | 0.72 |
| 107 | 4,427 | 270 | 3,316 | 228 | 0.012 | −0.42 |
| 26a | 6,092 | 243 | 7,229 | 278 | 0.012 | 0.25 |
| 221 | 13,308 | 1,327 | 9,476 | 765 | 0.019 | −0.49 |
| 200a | 1,310 | 214 | 782 | 132 | 0.029 | −0.74 |
| 22 | 2,558 | 227 | 1,176 | 254 | 0.03 | −1.12 |
| 31 | 18,946 | 2,004 | 13,466 | 1,461 | 0.03 | −0.49 |
| 141 | 1,395 | 285 | 434 | 156 | 0.046 | −1.68 |
| 1826 | 7,883 | 622 | 13,721 | 2,715 | 0.047 | 0.80 |
| 575 | 46 | 16 | 458 | 383 | 0.048 | 3.32 |
miRNAs with expression levels above 500 (mean fluorescence signal intensity in arbitrary units) and P values of <0.05 are shown, and data are sorted by P values.
Fig. 1.
Upregulation of miR-203 in GECs following P. gingivalis infection. GECs were infected with P. gingivalis for 6 h at an MOI of 100, and RNA was extracted from infected and control cells. (A) miR-203 expression by fluorescent signal intensity (arbitrary units) in a microarray; (B) qRT-PCR of miR-203. Data were normalized to RNU-48 expression. Error bars represent SDs (n = 12). ***, P < 0.001 by unpaired t test.
Identification of downstream targets of miR-203.
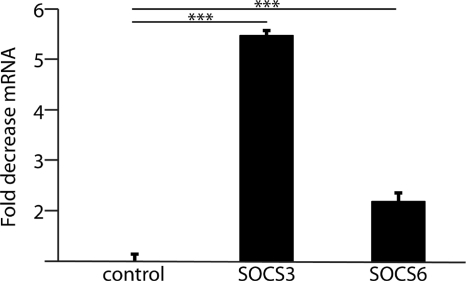
To begin to address the role of miR-203 in GEC responses to P. gingivalis, potential targets of miR-203 were investigated using a bioinformatic approach. The microRNA database (www.mirbase.org) identified over 4,000 predicted targets for miR-203. Several proteins involved in host immune responses had potential miR-203 target sites and were also predicted by 3 different search databases (microRNA.org; Targetscan, version 5.1; and Pictar-vert). In particular, members of the SOCS family proteins, including SOCS2, -3, -5, and -6, were targets for miR-203 in all the databases. Furthermore, SOCS3 has been shown to be a target of miR-203 in skin keratinocytes (51). We thus investigated whether SOCS expression differed in P. gingivalis-infected gingival cells using qRT-PCR for SOCS3 and SOCS6. Levels of SOCS3 and SOCS6 mRNA were reduced in P. gingivalis-infected GECs; SOCS3 was downregulated >5-fold in infected groups (P < 0.001), with SOCS6 expression decreased 2-fold compared with uninfected groups (P < 0.001) (Fig. 2).
Fig. 2.
SOCS mRNA levels are decreased in GECs following P. gingivalis infection. SOCS3 or SOCS6 expression was measured by qRT-PCR on mRNA from GECs infected with P. gingivalis for 6 h at an MOI of 100. Results are representative of 3 independent assays. Data were normalized to GAPDH expression (control). Error bars represent SDs (n = 12). ***, P < 0.001 by unpaired t test.
SOCS3 expression is correlated with miR-203 levels in epithelial cells.
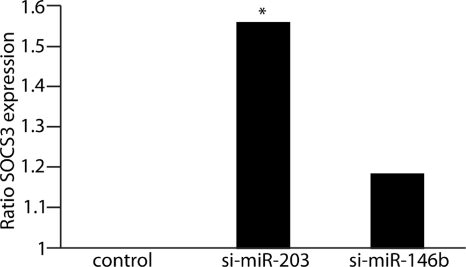
To determine if SOCS3 expression was regulated by miR-203, we knocked down miR-203 levels with si-miR-203, an antagomir. Cells were subsequently infected with P. gingivalis, and SOCS3 expression levels were measured by qRT-PCR (Fig. 3). Knockdown of miR-203 significantly affected SOCS3 expression compared with controls, with a 50% increase in SOC3 expression in the miR-203-silenced group (P < 0.05). The results support the concept that suppression of SOCS3 mRNA levels by P. gingivalis is mediated by miR-203.
Fig. 3.
SOCS3 expression is regulated by miR-203. qRT-PCR for SOCS3 expression in GECs challenged with P. gingivalis at an MOI of 100 for 6 h. Cells were transfected with si-miR-203 (antagomir), si-miRNA-146b (control antagomir), or nonsilence control. Results are representative of 3 independent assays (n = 12) and are shown as SOCS3 expression ratios relative to the nonsilence control. Data were normalized to GAPDH expression. *, P < 0.05 by one-way ANOVA.
miR-203 binds the 3′ UTR region of SOCS3.
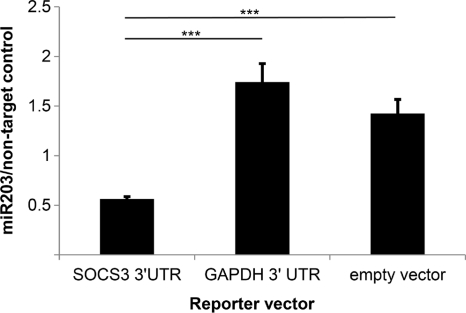
Next we sought to establish direct binding of miR-203 to the UTR of SOCS3. The luciferase reporter vector pSGG-SOCS3 3′UTR, containing the putative miR-203-binding site, was transfected into HTR8 cells. Trophoblasts were selected for this assay, as nonepithelial cells express relatively low levels of miR-203 and qRT-PCR confirmed that they express lower levels of miR-203 than GECs (data not shown). An empty vector served as a positive transfection control (pSGG-EMPTY-3UTR), and pSGG-GAPDH-3UTR, a vector containing the 3′ UTR of GAPDH, was an unrelated gene control. Cells were then dually transfected with either miR-203 mimic or miR-146 mimic (unrelated control), and luminosity was measured. Results were calculated as the ratio of miR-203-transfected to miR control-transfected luciferase activity. Cells transfected with pSGG-SOCS3 3′UTR and miR-203 mimic showed significantly reduced luciferase activity than empty vector and housekeeping vector controls (P < 0.001) (Fig. 4). These results establish direct binding of miR-203 to the 3′ UTR of SOCS3.
Fig. 4.
Binding of miR-203 to SOCS3 3′ UTR binding site. HTR8 cells were transfected with luciferase reporter vector pSGG-SOCS3 3′UTR, an empty vector (pSGG-EMPTY-3UTR), or a vector containing the 3′ UTR region of glyceraldehyde-3-phosphate dehydrogenase (pSGG-GAPDH-3UTR). Reporter vectors were cotransfected with a miR-203 mimic or unrelated negative-control miR mimic (miR-146b). Following 24 h of incubation, luciferase activity was measured. Results are representative of two independent assays (n = 4). Error bars indicate SDs. ***, P < 0.001 by Tukey-Kramer multiple comparison test.
Regulation of SOCS by miR-203 leads to increased Stat3 activation.
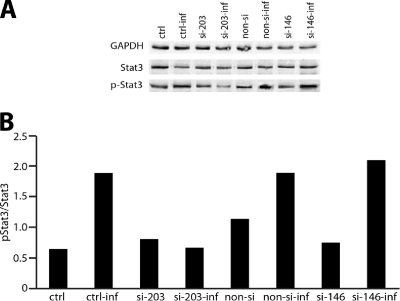
Stat3 is known to be a target of SOCS proteins, including SOCS3. Increased SOCS expression is correlated with reduced Stat3 activation, resulting in an abrogated host cell cytokine response (11). We therefore investigated whether the observed miR-203-dependent decrease in SOCS3 levels in GECs following P. gingivalis infection would regulate Stat3 activation. GECs were transfected with si-miR-203 or nonsilencing/nonrelated miR controls. Stat3 activation was assessed by Western blot analysis with Stat or phospho-specific Stat antibodies. As shown in Fig. 5, P. gingivalis infection increased the level of activated Stat3. Silencing of miR-203 resulted in abrogated Stat3 activation in infected cells compared with transfection control groups. These results confirmed that miR-203 regulates the pathway that leads to activation of Stat3 in GECs infected with P. gingivalis.
Fig. 5.
Stat3 activation by P. gingivalis is inhibited by miR-203 silencing. GECs were transfected with si-miR203 (si-203), si-miR-146 (si-146), or nonsilence control (non-si). (A) Lysates of P. gingivalis-infected (inf) or uninfected cells were examined by Western blotting with antibodies to Stat3 or phosphorylated Stat3. GAPDH antibodies were used as a loading control. (B) Blots were analyzed by scanning densitometry, and ratios of phospho-Stat3/Stat3 were determined. Data are representative of three independent experiments. Controls (ctrl) were cells treated with transfection agent only; control infected (ctrl-inf) cells were transfection-agent-only-treated cells subsequently exposed to P. gingivalis.
DISCUSSION
miRNAs are abundant in the human genome and have emerged as a key component of pathway and network control in a wide range of important cellular processes, including inflammation and apoptosis (3, 25, 64). miRNAs also contribute to host responses to bacterial and viral infections (6, 32, 35). In this work we show that P. gingivalis significantly affected the expression of 14 miRNAs in primary cultures of gingival epithelial cells. While previous studies have shown that heat-killed P. gingivalis can modulate the expression of miRNAs in epithelial cells (7), the current study is the first to show the impact of live, invasive P. gingivalis on the miRNA profile of GECs.
Among the miRNAs significantly regulated by GECs were miR-149*, miR-575, and miR-203. miR-149* has been shown to regulate apoptosis in neuroblastoma cells (33), and miR-575 can be downregulated by lipid peroxidation products in leukemic cells (46). In this study we focused on miR-203, which has defined targets and is predicted to regulate the cytokine signaling pathway, a property with direct relevance to the disease status of the periodontal tissues. One potential downstream target of miR-203 is SOCS, a family of molecules that are intracellular, cytokine-inducible proteins and that inhibit cytokine production as part of a negative-feedback loop (42). SOCS family molecules are also involved in regulating the activity of both T cells and antigen-presenting cells (APCs), such as dendritic cells (DCs), and SOCS3 is thought to regulate the cytokine signaling that controls receptor activator of nuclear factor κB ligand (RANKL)-mediated osteoclastogenesis (55). Binding of miR-203 to SOCS3 in skin keratinocytes has been shown (51); however, other studies failed to show a direct interaction between miR-203 and SOCS3 (31). To contribute to the clarification of this issue, we utilized a dual luciferase assay which confirmed direct binding of miR-203 to the putative target binding site of SOCS3 3′ UTR. SOCS proteins function through inhibition of JAKs, which in turn prevents phosphorylation and activation of Stats, transcription factors that regulate the expression of a number of cytokines. Consistent with this, P. gingivalis infection decreased levels of SOCS3 and -6 mRNA, coincident with induction of miR-203. RNA interference established that regulation of SOCS3 by P. gingivalis was dependent on miR-203. Reduction of SOCS levels also increased the activation status of Stat3, a transcription factor that binds to IL-6-responsive promoter elements (27). P. gingivalis can stimulate the production of IL-6 from gingival epithelial cells (47), and IL-6 levels are elevated in gingival crevicular fluid in periodontal disease (14, 26). Hence, induction of miR-203 by P. gingivalis has the potential to contribute to the inflammation associated with periodontitis. Furthermore, SOCS3 has been shown to play a critical role in modulating cytokine signaling involved in RANKL-mediated dendritic cell-derived osteoclastogenesis, leading to alveolar bone loss in mice (65). Increased miR-203 expression has also been reported in psoriasis and rheumatoid arthritis, indicating that miR-203 may play a role in a number of other chronic inflammatory diseases (51, 52).
In addition to a role in cytokine signaling, targets of Stat3 include antiapoptotic effectors such as Bcl-2, Bcl-XL, and survivin, along with proteins involved in cell cycle progression, such as Fos, cyclin D, and c-Myc (10, 23). Our previous studies have shown that Stat3 is part of the antiapoptotic responses induced in GECs by P. gingivalis, and the current study provides a mechanistic basis for this through the involvement of miR-203. Moreover, SOCS3 overexpression has been shown to reduce cell proliferation and induce apoptosis and partial G0/G1 arrest via pathways that include JAK/Stat3 (24). As P. gingivalis can accelerate cell cycle progression in GECs (28), miR-203 may also contribute to this process through inhibition of SOCS3. The regulation of cell cycle by miR-203 may partially explain the pathological expression in some carcinomas (16, 17, 48).
The mechanism by which P. gingivalis modulates miR-203 levels is currently under investigation but is as yet unknown. However, as stimulation of epithelial cells with heat-killed P. gingivalis does not regulate miR-203 (7), it would appear that viable bacteria are required and that induction may be related to an aspect of the process of internalization or intracellular adaptation. Transcription factors AP-1, SP1, and NF-κB can control expression of miR-203 (39, 50), and although viable P. gingivalis cells do not activate NF-κB in GECs (57), the organism can activate AP-1 in HeLa cells (59) and SP1 in monocytes (15). Activation of AP-1 can occur through the protein kinase C (PKC) pathway, and inhibitors of PKC abrogate expression of miR-203 (50). Both fimbriae (45) and LPS (49) of P. gingivalis can stimulate PKC, suggesting the potential for P. gingivalis to upregulate miR-203 through PKC activation of AP-1.
P. gingivalis selectively modulates multiple pathways in epithelial cells to effectuate invasion, differential cytokine responses, and inhibition of cell death. The results of the current study show that miRNAs such as miR-203 make important contributions to host cell responses to P. gingivalis. As miRNAs are considered attractive targets for therapeutic intervention (4, 19, 34), such an approach may be beneficial for P. gingivalis-induced diseases such as periodontitis.
Supplementary Material
ACKNOWLEDGMENT
We gratefully acknowledge support by NIH/NIDCR DE11111.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 2 May 2011.
REFERENCES
- 1. Aberdam D., Candi E., Knight R. A., Melino G. 2008. miRNAs, ‘stemness’ and skin. Trends Biochem. Sci. 33:583–591 [DOI] [PubMed] [Google Scholar]
- 2. Albandar J. M. 2011. Underestimation of periodontitis in NHANES surveys. J. Periodontol. 83:337–341 [DOI] [PubMed] [Google Scholar]
- 3. Ambros V. 2004. The functions of animal microRNAs. Nature 431:350–355 [DOI] [PubMed] [Google Scholar]
- 4. Bader A. G., Brown D., Winkler M. 2010. The promise of microRNA replacement therapy. Cancer Res. 70:7027–7030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bazzoni F., et al. 2009. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc. Natl. Acad. Sci. U. S. A. 106:5282–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Belair C., Darfeuille F., Staedel C. 2009. Helicobacter pylori and gastric cancer: possible role of microRNAs in this intimate relationship. Clin. Microbiol. Infect. 15:806–812 [DOI] [PubMed] [Google Scholar]
- 7. Benakanakere M. R., et al. 2009. Modulation of TLR2 protein expression by miR-105 in human oral keratinocytes. J. Biol. Chem. 284:23107–23115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bolstad B. M., Irizarry R. A., Astrandand M., Speed T. P. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193 [DOI] [PubMed] [Google Scholar]
- 9. Darveau R. P., Belton C. M., Reife R. A., Lamont R. J. 1998. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect. Immun. 66:1660–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dauer D. J., et al. 2005. Stat3 regulates genes common to both wound healing and cancer. Oncogene 24:3397–3408 [DOI] [PubMed] [Google Scholar]
- 11. Dominguez E., Mauborgne A., Mallet J., Desclaux M., Pohl M. 2010. SOCS3-mediated blockade of JAK/STAT3 signaling pathway reveals its major contribution to spinal cord neuroinflammation and mechanical allodynia after peripheral nerve injury. J. Neurosci. 30:5754–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feng Z., Weinberg A. 2006. Role of bacteria in health and disease of periodontal tissues. Periodontol. 2000 40:50–76 [DOI] [PubMed] [Google Scholar]
- 13. Gao X., Gulari E., Zhou X. 2004. In situ synthesis of oligonucleotide microarrays. Biopolymers 73:579–596 [DOI] [PubMed] [Google Scholar]
- 14. Geivelis M., Turner D. W., Pederson E. D., Lamberts B. L. 1993. Measurements of interleukin-6 in gingival crevicular fluid from adults with destructive periodontal disease. J. Periodontol. 64:980–983 [DOI] [PubMed] [Google Scholar]
- 15. González O. A., Li M., Ebersole J. L., Huang C. B. 2010. HIV-1 reactivation induced by the periodontal pathogens Fusobacterium nucleatum and Porphyromonas gingivalis involves Toll-like receptor 2 [corrected] and 9 activation in monocytes/macrophages. Clin. Vaccine Immunol. 17:1417–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gottardo F., et al. 2007. Micro-RNA profiling in kidney and bladder cancers. Urol. Oncol. 25:387–392 [DOI] [PubMed] [Google Scholar]
- 17. Greither T., et al. 2010. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int. J. Cancer 126:73–80 [DOI] [PubMed] [Google Scholar]
- 18. Hajishengallis G. 2009. Porphyromonas gingivalis-host interactions: open war or intelligent guerilla tactics? Microbes Infect. 11:637–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hammond S. M. 2006. MicroRNA therapeutics: a new niche for antisense nucleic acids. Trends Mol. Med. 12:99–101 [DOI] [PubMed] [Google Scholar]
- 20. Handfield M., et al. 2005. Distinct transcriptional profiles characterize oral epithelium-microbiota interactions. Cell. Microbiol. 7:811–823 [DOI] [PubMed] [Google Scholar]
- 21. Hasegawa Y., et al. 2008. Role of Porphyromonas gingivalis SerB in gingival epithelial cell cytoskeletal remodeling and cytokine production. Infect. Immun. 76:2420–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Holt S. C., Kesavalu L., Walker S., Genco C. A. 1999. Virulence factors of Porphyromonas gingivalis. Periodontol. 2000 20:168–238 [DOI] [PubMed] [Google Scholar]
- 23. Hsieh F. C., Cheng G., Lin J. 2005. Evaluation of potential Stat3-regulated genes in human breast cancer. Biochem. Biophys. Res. Commun. 335:292–299 [DOI] [PubMed] [Google Scholar]
- 24. Iwahori K., et al. 14 October 2010. Overexpression of SOCS3 exhibits preclinical antitumor activity against malignant pleural mesothelioma. Int. J. Cancer [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 25. Jamaluddin M. S., et al. 2011. miRNAs: roles and clinical applications in vascular disease. Expert Rev. Mol. Diagn. 11:79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kamma J. J., Giannopoulou C., Vasdekis V. G., Mombelli A. 2004. Cytokine profile in gingival crevicular fluid of aggressive periodontitis: influence of smoking and stress. J. Clin. Periodontol. 31:894–902 [DOI] [PubMed] [Google Scholar]
- 27. Kojima H., Nakajima K., Hirano T. 1996. IL-6-inducible complexes on an IL-6 response element of the junB promoter contain Stat3 and 36 kDa CRE-like site binding protein(s). Oncogene 12:547–554 [PubMed] [Google Scholar]
- 28. Kuboniwa M., et al. 2008. P. gingivalis accelerates gingival epithelial cell progression through the cell cycle. Microbes Infect. 10:122–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lamont R. J., et al. 1995. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 63:3878–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lamont R. J., Jenkinson H. F. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lena A. M., et al. 2008. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 15:1187–1195 [DOI] [PubMed] [Google Scholar]
- 32. Li L. M., et al. 2010. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 70:9798–9807 [DOI] [PubMed] [Google Scholar]
- 33. Lin R. J., Lin Y. C., Yu A. L. 2010. miR-149* induces apoptosis by inhibiting Akt1 and E2F1 in human cancer cells. Mol. Carcinog. 49:719–727 [DOI] [PubMed] [Google Scholar]
- 34. Liu Z., Sall A., Yang D. 2008. MicroRNA: an emerging therapeutic target and intervention tool. Int. J. Mol. Sci. 9:978–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Z., et al. 2010. Up-regulated microRNA-146a negatively modulates Helicobacter pylori-induced inflammatory response in human gastric epithelial cells. Microbes Infect. 12:854–863 [DOI] [PubMed] [Google Scholar]
- 36. Lu M., et al. 2008. An analysis of human microRNA and disease associations. PLoS One 3:e3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lui W. O., Pourmand N., Patterson B. K., Fire A. 2007. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 67:6031–6043 [DOI] [PubMed] [Google Scholar]
- 38. Mao S., et al. 2007. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell. Microbiol. 9:1997–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Melar-New M., Laimins L. A. 2010. Human papillomaviruses modulate expression of microRNA-203 upon epithelial cell differentiation to control levels of p63 proteins. J. Virol. 84:5212–5221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miska E., Alvarez-Saavedra E., Townsend M., Yoshii A., Šestan N., Rakic P., Constantine-Paton M., Horvitz H. 2004. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 5:R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nahid M. A., Pauley K. M., Satoh M., Chan E. K. 2009. miR-146a is critical for endotoxin-induced tolerance: implication in innate immunity. J. Biol. Chem. 284:34590–34599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nicola N. A., Greenhalgh C. J. 2000. The suppressors of cytokine signaling (SOCS) proteins: important feedback inhibitors of cytokine action. Exp. Hematol. 28:1105–1112 [DOI] [PubMed] [Google Scholar]
- 43. O'Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D. 2007. Micro-RNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U. S. A. 104:1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oda D., Watson E. 1990. Human oral epithelial cell culture I. Improved conditions for reproducible culture in serum-free medium. In Vitro Cell. Dev. Biol. 26:589–595 [DOI] [PubMed] [Google Scholar]
- 45. Ogawa T., Uchida H. 1995. A peptide, ALTTE, within the fimbrial subunit protein from Porphyromonas gingivalis induces production of interleukin 6, gene expression and protein phosphorylation in human peripheral blood mononuclear cells. FEMS Immunol. Med. Microbiol. 11:197–206 [DOI] [PubMed] [Google Scholar]
- 46. Pizzimenti S., et al. 2009. MicroRNA expression changes during human leukemic HL-60 cell differentiation induced by 4-hydroxynonenal, a product of lipid peroxidation. Free Radic. Biol. Med. 46:282–288 [DOI] [PubMed] [Google Scholar]
- 47. Sandros J., et al. 2000. Cytokine responses of oral epithelial cells to Porphyromonas gingivalis infection. J. Dent. Res. 79:1808–1814 [DOI] [PubMed] [Google Scholar]
- 48. Schetter A. J., et al. 2008. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 299:425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shapira L., et al. 1997. Bacterial lipopolysaccharide induces early and late activation of protein kinase C in inflammatory macrophages by selective activation of PKC-epsilon. Biochem. Biophys. Res. Commun. 240:629–634 [DOI] [PubMed] [Google Scholar]
- 50. Sonkoly E., et al. 2010. Protein kinase C-dependent upregulation of miR-203 induces the differentiation of human keratinocytes. J. Investig. Dermatol. 130:124–134 [DOI] [PubMed] [Google Scholar]
- 51. Sonkoly E., et al. 2007. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One 7:e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stanczyk J., et al. 27 October 2010. Altered expression of miR-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stathopoulou P. G., Benakanakere M. R., Galicia J. C., Kinane D. F. 2010. Epithelial cell pro-inflammatory cytokine response differs across dental plaque bacterial species. J. Clin. Periodontol. 37:24–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. 2006. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U. S. A. 103:12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takayanagi H. 2007. Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat. Rev. Immunol. 7:292–304 [DOI] [PubMed] [Google Scholar]
- 56. Tribble G. D., Mao S., James C. E., Lamont R. J. 2006. A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proc. Natl. Acad. Sci. U. S. A. 103:11027–11032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Watanabe K., Yilmaz O., Nakhjiri S. F., Belton C. M., Lamont R. J. 2001. Association of mitogen-activated protein kinase pathways with gingival epithelial cell responses to Porphyromonas gingivalis infection. Infect. Immun. 69:6731–6737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Weinberg A., Belton C. M., Park Y., Lamont R. J. 1997. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 65:313–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yanti L., Kim D., Hwang J. K. 2009. Inhibitory effect of panduratin A on c-Jun N-terminal kinase and activator protein-1 signaling involved in Porphyromonas gingivalis supernatant-stimulated matrix metalloproteinase-9 expression in human oral epidermoid cells. Biol. Pharm. Bull. 32:1770–1775 [DOI] [PubMed] [Google Scholar]
- 60. Yi R., et al. 2006. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet. 38:356–362 [DOI] [PubMed] [Google Scholar]
- 61. Yilmaz O. 2008. The chronicles of Porphyromonas gingivalis: the microbium, the human oral epithelium and their interplay. Microbiology 154:2897–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yilmaz O., Verbeke P., Lamont R. J., Ojcius D. M. 2006. Intercellular spreading of Porphyromonas gingivalis infection in primary gingival epithelial cells. Infect. Immun. 74:703–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yilmaz O., Jungas T., Verbeke P., Ojcius D. M. 2004. Activation of the phosphatidylinositol 3-kinase/Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infect. Immun. 72:3743–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang R., Su B. 2009. Small but influential: the role of microRNAs on gene regulatory network and 3′UTR evolution. J. Genet. Genomics 36:1–6 [DOI] [PubMed] [Google Scholar]
- 65. Zhang X., Alnaeeli M., Singh B., Teng Y. A. 2009. Involvement of SOCS3 in regulation of CD11c+ dendritic cell-derived osteoclastogenesis and severe alveolar bone loss. Infect. Immun. 77:2000–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhu Q., et al. 2007. Microfluidic biochip for nucleic acid and protein analysis. Methods Mol. Biol. 382:287–312 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.