Abstract
The social amoeba Dictyostelium expresses a hypoxia inducible factor-α (HIFα)-type prolyl 4-hydroxylase (P4H1) and an α-N-acetylglucosaminyltransferase (Gnt1) that sequentially modify proline-143 of Skp1, a subunit of the SCF (Skp1/Cullin/F-box protein)-class of E3 ubiquitin-ligases. Prior genetic studies have implicated Skp1 and its modification by these enzymes in O2-regulation of development, suggesting the existence of an ancient O2-sensing mechanism related to modification of the transcription factor HIFα by animal prolyl 4-hydroxylases (PHDs). To better understand the role of Skp1 in P4H1-dependent O2-signaling, biochemical and biophysical studies were conducted to characterize the reaction product and the basis of Skp1 substrate selection by P4H1 and Gnt1. 1H-NMR demonstrated formation of 4(trans)-hydroxyproline as previously found for HIFα, and highly purified P4H1 was inhibited by Krebs cycle intermediates and other compounds that affect animal P4Hs. However, in contrast to hydroxylation of HIFα by PHDs, P4H1 depended on features of full-length Skp1, based on truncation, mutagenesis, and competitive inhibition studies. These features are conserved during animal evolution, as even mammalian Skp1, which lacks the target proline, became a good substrate upon its restoration. P4H1 recognition may depend on features conserved for SCF complex formation as heterodimerization with an F-box protein blocked Skp1 hydroxylation. The hydroxyproline-capping enzyme Gnt1 exhibited similar requirements for Skp1 as a substrate. These and other findings support a model in which the protist P4H1 conditionally hydroxylates Skp1 of E3SCFubiquitin-ligases to control half-lives of multiple targets, rather than the mechanism of animal PHDs where individual proteins are hydroxylated leading to ubiquitination by the evolutionarily-related E3VBCubiquitin-ligases.
Keywords: 4-hydroxyproline, cellular slime mold, cytoplasmic glycosylation, peptide NMR
Skp1 is a subunit of E3SCFUbiquitin-ligases and other protein complexes (1). In the social soil amoeba Dictyostelium discoideum, Skp1 contains a novel hydroxyproline (Hyp)1-linked pentasaccharide. The Hyp-glycosylation pathway consists of P4H1, a non-heme Fe(II)-dependent dioxygenase that modifies Skp1 at Pro143, and five sequentially acting cytoplasmic glycosyltransferase activities encoded by three genes (2). P4H1 may serve as an O2 sensor, as the enzyme has a high Km for O2 in vitro (3), elevated O2 levels are required for P4H1-null cells to culminate into fruiting bodies, and reduced O2 is sufficient for P4H1-overexpression cells (4). Reverse genetic analyses suggest that the sequential post-translational glycosylation events modulate the effect of hydroxylation in hierarchical fashion (5). Based on biochemical and genetic studies, Skp1 is the only substrate for P4H1 and the glycosyltransferases in cells, and mediates the role of the pathway enzymes on culmination and other developmental transitions (6). As recently summarized (2), the Hyp-glycosylation pathway appears to be widely expressed in unicellular protists including some significant human and plant pathogens.
Among known prolyl 4-hydroxylases, P4H1 is most related to the sequences of Egl-9 (Eggless-9), an enzyme involved in O2-regulation in C. elegans, and the 3 human homologs that sense O2 in humans. Egl-9 and the human PHDs (prolyl hydroxylase domain proteins; also known as EGLNs or HPHs) modify hypoxia-inducible factor-α (HIFα), a subunit of the HIFα-HIF® transcriptional factor heterodimer whose accumulation directly induces hypoxia response genes that support glycolysis, angiogenesis, and erythropoiesis (7). Hydroxylation of Pro402 or Pro564 of HIFα results in recognition by the von Hippel-Lindau subunit of the E3VBCUb-ligase followed by degradation within the 26S-proteasome. Egl-9 and human PHD1-3 are thought to serve as direct O2-sensors, in part due to their relatively high Km values for O2 as a substrate. Their activity in cells is promoted by the availability of the co-substrate αKG (α-ketoglutarate, a Krebs cycle intermediate), ascorbate, and Fe(II), and inhibited by the product succinate, other Krebs cycle metabolites, and reactive oxygen species (e.g., 8,9). Much remains to be learned about the regulation of PHDs, and the significance of additional PHD targets in animal cells (10).
Whereas HIFα and possibly other proteins mediate PHD/Egl-9-dependent hypoxic responses in animal cells, Dictyostelium apparently lacks HIFα and the VBC-complex that recognizes it, and P4H1-dependent hypoxic responses appear to be mediated by Skp1 (6). Conversely, though Skp1 is highly conserved in eukaryotes, the equivalent of Pro143 is notably absent in chordates. Interestingly, Skp1 is evolutionarily related, not to HIFα, but to elongin C, a subunit of the E3VBCUb-ligase that targets HIFα for degradation. The homologous E3SCFUb ligases, conserved from yeast to humans, contain the SCF subcomplex whose Skp1 adaptor links the scaffold protein cullin-1 to an F-box protein, which presents targets such as cell cycle and other regulatory proteins for polyubiquitination. Analysis of the human and Dictyostelium genomes predicts more than 50 F-box proteins (unpublished data), potentially diversifying the pool of SCF complexes, and thereby suggesting a global role in regulation of the proteome (11). Interestingly, there is no evidence that P4H1 regulates the stability of its Skp1 target (6).
A biochemical analysis of substrate selection by P4H1 was initiated to provide insight into mechanism of this evolutionary shift between protists and animals, and to reveal clues about regulation of its hydroxylase activity. As shown here, adoption of a direct P4H1 assay revealed similar sensitivity to metabolic inhibitors, and 1H-NMR studies demonstrated formation of (2S,4R)-4-hydroxy-L-proline (a.k.a. trans-4-hydroxyproline, Hyp), as for human PHD2 (12–14). However, whereas PHD2 recognizes truncated oxygen-dependent degradation domains (ODDs) of HIFα, and peptides as short as 15-mers have successfully been employed as substrates, P4H1-processing of Skp1 depended on global structural attributes. Remarkably, substrate recognition is conserved in animal Skp1s. The implication that P4H1 recognition depends on conserved features important for SCF-complex formation was supported by loss of substrate activity when Skp1 was complexed with an F-box protein. Such a recognition mechanism could explain P4H1’s apparent high degree of specificity for Skp1, and suggests how Skp1 modification may be regulated in cells. A similar dependence on Skp1 features was observed for the Hyp-capping activity of Gnt1, whose gene is phylogenetically co-distributed with protist P4H1 and in some genomes appears to be encoded as a separate domain of the same protein (2). Whereas animal PHD’s render global effects by regulating the half-life of HIFα with transcriptional consequences on many genes, global effects of Dictyostelium P4H1 and its partner Gnt1 may be rendered by the targeting of Skp1 alone that controls the half-lives of many proteins via the large family of E3SCFUb-ligases.
EXPERIMENTAL PROCEDURES
Prolyl 4-hydroxylase assay
The 14CO2-release assay, modified from ref. 15, contained 5 pmol His6P4H1, 500 pmol Skp1, 50 mM Tris-HCl (pH 7.4), 50 µM [1-14C]αKG (56.8 mCi/mmol; Perkin-Elmer), 5 µM Fe(II)SO4, 1 mM ascorbate, 2.5 mM DTT, 0.2 mg/ml catalase (Sigma bovine liver), in a total volume of 100 µl. His6P4H1 was purified from E. coli as described (3) with modifications according to ref. 15. His6P4H1, Skp1, and other components were deposited as 3 separate aliquots in a 7-ml scintillation vial with a septum lid. For inhibition studies, P4H1 was briefly preincubated with peptides, other Skp1A isoforms, or poly-L-Pro (Mr=10,000–30,000; 20,000 used for concentration calculation; Sigma). The reaction was initiated by introducing a 0.5-ml conical tube containing 200 µl hyamine hydroxide into the vial, incubated on a shaker for 60 min at 29 °C, and terminated by the addition of 200 µl MeOH through the septum to the reaction mix. After 30 min on ice, the 14CO2 reaction product was quantitated by transfer of the hyamine hydroxide solution to 6 ml of Optiphase HiSafe 3 (Wallac), which was analyzed by liquid scintillation counting. 14CO2 values obtained in the absence of P4H1, which were typically <10% but ranged up to 50% for some Skp1 preparations, were subtracted as background.
The one-step coupled 3H-incorporation assay, modified from ref. 3, contained 31 fmol His6P4H1, Dd-His6Gnt1 or DdDp-His6Gnt1, 1–2 µM UDP-[3H]GlcNAc (40 Ci/mmol), 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 2.5 mM DTT, 0.02% Tween-20, 0.25 mg/ml BSA, 0.5 mM αKG, 2 mM ascorbic acid, 5 µM FeSO4, 0.2 mg/ml catalase, in a final volume of 20 µl at 29 °C. The reaction was initiated by the addition of acceptor substrate (typically 6 pmol Skp1A), and terminated and processed as below.
In the two-step 3H-version, the initial P4H1 reaction was conducted in a final volume of 10 µl in the absence of Gnt1 and UDP-GlcNAc. After 15 min, the reaction was supplemented with His6Gnt1, 1 µM UDP-GlcNAc and 1 mM PCA (P4H1 inhibitor), to a final volume 20 µl. After 2 h, 85 µl H2O was added, and 25 µl was mixed with 1 µg soybean trypsin inhibitor in 8 µl 4× Laemmli sample buffer and subjected to SDS-PAGE. Gel slices corresponding to and surrounding the position of trypsin inhibitor were quantitated as before (3). Background levels in the absence of P4H1 were typically <1%. The remaining volume was supplemented with 100 µg bovine serum albumin, diluted to 1 ml with cold 10 mM pyrophosphate in 10% TCA, filtered over a Whatman GF/C filter, and quantitated for 3H in 9 ml BioSafe NA.
Gnt1 assay
Hydroxylated Skp1 was prepared in an in vitro reaction with His6P4H1, and the reaction was verified to be >95% complete based on MALDI-TOF-MS of endo Lys-C peptides as described (6). The standard assay consisted of 0.31 µM Skp1, native purified Dd-Gnt1 (19), Dd-His6Gnt1 or DdDp-His6Gnt1, and 1 µM UDP-[3H]GlcNAc in 50 mM Tris-HCl (pH 7.4), 5 mM DTT, 0.25 mg/ml BSA, 10 mM MgCl2, 0.02% Tween-20, for 15 min at 29°C, as described (19). Incorporation was assayed by TCA precipitation and/or SDS-PAGE gel and scintillation counting as above. Background values (<40 dpm) from zero-time incubations were subtracted.
Chimeric DdDp-His6Gnt1
Previous preparations of Dd-Gnt1 isolated from E. coli contained chaperonins (19) that might interfere with enzyme activity. The related species D. purpureum contains a predicted Gnt1 ortholog that lacks a stretch of 80 Asn residues and other Asn-rich inserts, as commonly found in D. discoideum proteins (20). A hybrid coding sequence was generated from the 16-amino acid exon 1 of Dd-Gnt1 and exon 2 of Dp-Gnt1 (see Supplement). The resulting chimera encodes native Dp-Gnt1 with 5 conservative substitutions in the first 8 amino acids at positions whose amino acids are not well conserved among Dictyostelium Gnt1s. DdDp-Gnt1 was expressed in E. coli Gold cells by induction with 1 mM IPTG for 15 h at 22 °C. A soluble extract was prepared and adsorbed onto a 1.2 ml Ni++-HP Sepharose column (GE Healthcare), which was washed with 5 mM and then 60 mM imidazole in 50 mM Tris-HCl (pH 7.4), 0.5 M NaCl, and eluted with a 60–1000 mM gradient of imidazole in the same buffer. DdDp-Gnt1 was immediately desalted over a PD10 column into 50 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 2 mM DTT, and aliquots were frozen at −80 °C.
Skp1 preparations
The purification of Dd-His10Skp1A from E. coli over a Ni++ column was as described (21). Dd-Skp1A(P143A) was purified from E. coli using DEAE-Sepharose Fast Flow, phenyl-Sepharose fast flow (Hi-Sub), and Source 15Q columns, under non-denaturing conditions, as described (3). Native Skp1A, and the mutants Skp1A1 and Skp1A2 (17) were expressed and purified similarly. Dd-FLAG-Skp1 was purified from the S100 fraction of stationary stage Dictyostelium using a Sepharose M2-antibody column (Sigma) and elution with 3X-FLAG peptide as described (6).
Skp1 Cys mutants were generated by site-directed mutagenesis of pET19b-Skp1A (3), using primers described listed in Table S1, according to the Stratagene QuikChange® protocol. Multiple mutations were generated using multiple primer pairs simultaneously.
Hs-FLAG-OCP-2 (Skp1 isoform b) and FLAG-OCP-2 (E147P) from human, and Skr1 and Skr2 cDNAs from C. elegans (see Supplement), were expressed in E. coli, and purified by virtue of their FLAG tags as above.
Pro- or Hyp-containing Skp1 peptide137–149 and peptide133–155 were previously described (5,18). Peptide123–162 was released from carboxamidomethylated His10Skp1A or in vitro-hydroxylated Skp1A (see NMR section below) by treatment with CNBr as described (16), except that CNBr was dissolved in 70% (v/v) CF3COOH (Pierce), and Skp1 was incubated at 4 °C for 16 h. The sample was diluted in a 5-fold excess of water and then taken to near dryness by vacuum centrifugation. The sample was resolubilized by the addition of 10% (v/v) CF3COOH in water and, after clarification by centrifugation, the supernatant was injected onto a C8 (Discovery Bio Wide Pore, Supelco) column and eluted with a conventional CH3CN gradient in 0.1% CF3COOH. Skp1123–162 eluted as an isolated peak based on A215. Its identity, purity, and absence of chemical modifications other than alkylation of C156, were confirmed by MALDI-TOF-MS (data not shown). Peptide 31–122 was insoluble.
Skp1 pretreatments and alkylation
Skp1 preparations were heated at 60 or 100 °C for 15 or 3 min, respectively, or brought to 6 M urea by addition of crystals and heated to 60 °C for 15 min. Urea was taken to <4 mM by cycles of concentration and dilution with 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 15% (v/v) glycerol, in a Nanosep-10K Omega centrifugal concentration device (PALL). Samples were brought to pH 2.5 by addition of 0.1 M glycine-HCl, incubated at 4 °C for 12 min, restored to neutrality by addition of 1 M Tris-HCl, pH 8.2, and buffer exchanged as above.
For alkylation, DTT was added to 7 mM, iodoacetamide was added to 17.5 mM, and the sample incubated for 1 h at 22 °C in the dark. The reaction was quenched by the addition of DTT to a final concentration of 20 mM added, and buffer-restored as above. Alkylation of urea-treated Skp1 was conducted in the presence of 6 M urea as above.
The Skp1 preparations (500 pmol) were analyzed after HPLC purification on a Supelco C8-column MALDI-TOF-MS, as described (3,6). For further analysis, samples were digested with endo-Lys C (1:100 (w/w); Wako) in 1 mM DTT in 0.1 M Tris-HCl, pH 9.2 at 37 °C for 20 h. The sample was analyzed directly by MALDI-TOF-TOF-MS in positive ion mode using α-cyanocinnamic acid as the matrix, or first fractionated by HPLC as before (6). Except for low abundance ions corresponding to disulfide linked peptides (including to self) attributable to adventitious associations, essentially all observed ions could be assigned as modified or unmodified Skp1 peptides.
Isolation of Skp1A/FbxA complex
pET19b-Skp1-FbxA (see Supplement) was transfected into ER2566 competent cells (NEB). At OD595 of 0.5–0.6 in LB-ampicillin, cultures were shifted from 37 °C to 22 °C and Skp1 and His6⊗N-FbxA coexpression was induced by addition of 1 mM IPTG. After 16 h, cells were recovered in 20 mM Tris-HCl (pH 8.0) and resuspended in cell lysis buffer (100 mM Tris-HCl, pH 8.2, 5 mM benzamidine, 0.5 µg/ml pepstatin A, 5 µg/ml leupeptin and aprotinin, 0.5 mM PMSF and 1 mg/ml lysozyme) using a Dounce tissue grinder. Cells were lysed using a French pressure cell and supplemented with 10 µg/ml DNAse and 50 µg/ml RNAse. The mixture was purified on a Ni++-column as above. In some trials, the Skp1-His6⊗N-FbxA complex was further purified by Q-Sepharose ion exchange chromatography or gel filtration.
Circular dichroism
Skp1A was purified essentially to homogeneity and analyzed at 0.2 mg/ml in 210 µl of 10 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, in a N2-purged 1.0 mm Quartz cuvette on a Jasco J-715 Spectropolarimeter operated at 1.0 nm bandwidth in 1 nm steps. Three spectra were summed for final readout. For secondary structure prediction, the Jasco mdeg data files were converted into molar ellipticity per residue, and analyzed using 3 predictors (SELCON3, CDSSTR, and CONTINLL), in CDPro (http://lamar.colostate.edu/~sreeram/CDPro/main.html), using reference file #4 for soluble, non-membrane proteins. Averages are shown since all predictors were within 7% agreement.
NMR spectroscopy
1.6 mg of Skp1A1 was modified to near completion using His6P4H1 in a reaction containing 0.5 mM αKG, and purified on a Phenomenex C5 column in an ascending gradient of CH3CN in 0.1% CF3COOH. Skp1 was digested with endo Lys-C, and the peptides fractionated on a Phenomenex C12 column on a Pharmacia SmartSystem, yielding 60 nmol (based on calculated extinction coefficient at 260 nm) of the peptide NDFTP*EEEEQIRK, where P* refers to the modified Pro.
The two model dipeptides containing Hyp or hyp were synthesized by condensing Fmoc-Thr(OtBu)-OH with the methyl amide of the appropriate prolyl amine. The Fmoc group was employed to minimize epimerization of the Cα stereogenic center of the Thr residue during this relatively sluggish peptide bond formation. The Fmoc group was subsequently removed by β-elimination and the N-terminus acetylated. The tert-butyl ethers were cleaved under mildly acidic conditions and the dipeptides purified by RP-HPLC. The methods will be published separately2, and structural confirmations based on NMR are shown in Figs. S2 and S3.
1H-NMR spectra were acquired at either 500 MHz on a Varian INOVA instrument with a standard triple-resonance probe or at 900 MHz on a Varian VNMRS Instrument using a triple-resonance cold probe, at the Rocky Mountain regional NMR facility at the University of Colorado Denver, Anschutz Medical Campus. One dimensional spectra were acquired in D2O in a Shigemi 5 mm NMR tube with 32,768 data points over a 14,535 Hz spectral width (900 MHz) or 5636 points over a 2500 Hz spectral width (500 MHz), with a 1.7 s preparatory delay. Gradient COSY spectra at 900 MHz were acquired with a 14,535 Hz spectral width in the direct dimension and 8500 Hz spectral width in the indirect dimension, collecting 32,768 points in the direct dimension and 700 increments. At 500 MHz, gCOSY spectra were acquired with 2500 Hz spectral widths in both dimensions, collecting 5636 points in the direct dimension and 1024 increments. 2D spectra were processed with a sine bell weighting function in both dimensions.
RESULTS
P4H1 assays
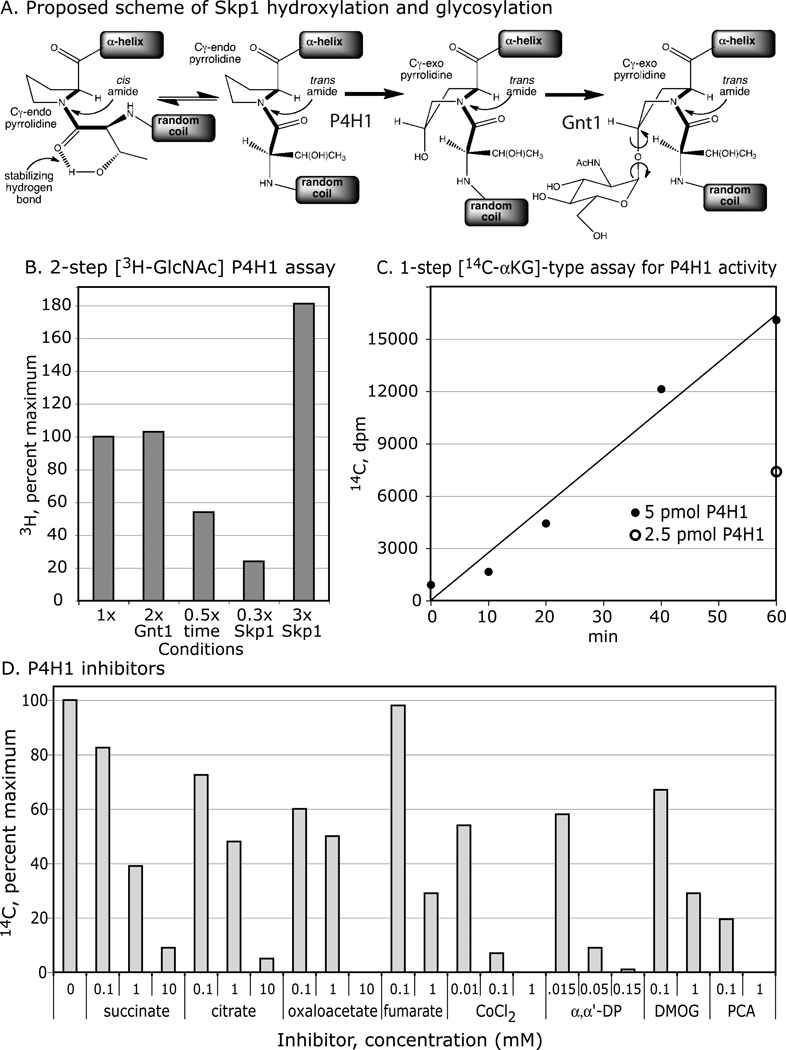
The hydroxylation of Pro143 of Skp1 by P4H1, and its subsequent substitution by GlcNAc, mediated by Gnt1, are illustrated in Fig. 1A. The prediction that the hydroxyl is installed at the 4-position in the trans-configuration (2) is verified below. Potential conformational consequences are inferred from studies on HIFα (14). In a previous study, E. coli expressed His6P4H1 was shown to modify E. coli-expressed Dd-His10Skp1, in a coupled reaction in which the hydroxylated product of the reaction was radioactively labeled by Gnt1 in the presence of UDP-[3H]GlcNAc (3). To avoid potential effects of epitope tags, which can affect hydroxylation in vivo (6), and denaturation when desorbing from antibody affinity columns, Dd-Skp1A was expressed as its native sequence and chromatographically purified to near homogeneity under non-denaturing conditions (Fig. S1). To avoid potential influence of Gnt1 on P4H1, a 2-step version, in which the P4H1 and Gnt1 reactions were conducted sequentially, was implemented and found to depend on incubation time and Skp1 concentration (Fig. 1B). Dd-Skp1A(P143A) was inactive (see below), indicating specificity for P143, the in vivo modification site. Activity was not stimulated or inhibited by addition of desalted cytosolic extract from P4H1-null stationary phase Dictyostelium cells (data not shown).
Figure 1. Prolyl hydroxylase assays and effects of inhibitors.
(A) Proposed reaction scheme of P4H1 and Gnt1. The Thr-Pro143 dipeptide, preceded by a coil and succeeded by an α-helix in Skp1 (see Fig. 3C), is illustrated. At left, the cis-trans equilibrium around the connecting peptide bond is represented. 4(trans)-hydroxylation, catalyzed by P4H1, is expected to reinforce equilibrium in favor of the trans peptide bond and promote the Cγ-exo pyrrolidine conformation, based on studies with HIFα (14). Gnt1 catalyzes addition of GlcNAc, predicted to be in α-linkage (2). (B) Coupled, 2-step 3H-assay. The basic reaction was conducted for 15 min, followed by termination with PCA, supplementation with UDP-[3H]GlcNAc and Gnt1, and incubation for 2 additional h. The reaction product was detected as incorporation of 3H into the Skp1 band after SDS-PAGE. Skp1A concentration, and reaction time were varied as indicated. Doubling the Gnt1 concentration had no effect on activity indicating sufficient enzyme for maximal conversion to product. 100% incorporation under standard conditions represented 19,300 dpm. (C) Time and concentration dependence of the 14C-release assay. The indicated amount of purified His6P4H1 was incubated with purified 10 µM Skp1A in the presence of [1-14C]-αKG, ascorbate and FeCl2, for the time indicated. Activity was assayed as released 14CO2. Background activities of P4H1 alone, and Skp1 substrate alone, were subtracted. (D) Effects of Krebs cycle metabolites, and other known inhibitors of Hs-PHD2, based on the 14C-release assay conducted using 5 µM Skp1A. Results are averaged from 2–4 trials, in which 100% incorporation was typically 24,000 dpm. All compounds were in aqueous solution, except for DMOG, whose EtOH carrier reduced activity by 19% at the highest concentration tested.
Separate assays for Gnt1 (shown later) and P4H1 were developed to analyze effects on each enzyme individually. P4H1 was assayed by conversion of the co-substrate [1-14C]αKG to 14CO2 and succinate. The standard reaction yielded time dependent formation of 14CO2 that was linear at 1 h and dependent on the level of P4H1 (Fig. 1C). Activity was dependent on Pro143 (see below). The specific activity corresponded to one reaction cycle per 10 sec, assuming all protein is active and after correction for suboptimal levels of 14C-αKG, which was comparable to that of the 3H-coupled assay. Consistent with this, addition of stoichiometric levels of Gnt1 and/or UDP-GlcNAc did not affect the 14C-release results (data not shown), indicating the Gnt1 does not activate or inhibit P4H1. In comparison, the 14C-assay was up to 1000-fold less sensitive, and subject to variable background levels presumably attributable to contaminating non-heme dioxygenases in the Skp1 preparations. Although insufficient material was available for systematic kinetic analyses, dependence on Skp1A concentration (data not shown) suggested that the previous Km estimate of 0.2 µM, based on the coupled 1-step 3H assay of Dd-His10Skp1 (3), is about an order of magnitude too low. In the less sensitive 14C-assay, this Skp1 exhibited variable activity, which might be due to Fe+2-chelation by high concentrations of the His10-tag.
P4H1: inhibition by metabolites and small molecules
The effects of small molecules known to inhibit human PHD2 were analyzed using the 14C-assay. In the presence of 0.05 mM αKG, 0.1 mM succinate (enzyme product) and fumarate exhibited minimal effect, whereas modest inhibition was exhibited by citrate and oxaloacetate (Fig. 1D). 50–70% inhibition occurred at 1 mM for each of the four Krebs cycle intermediates, with greater inhibition occurring at 10 mM. These parallel effects on mammalian PHD2 (8), and raise the possibility of metabolic regulation of P4H1 activity as suggested in mammalian cells.
His6P4H1 was also inhibited by the chelators α,α’-dipyridyl and EDTA (not shown), the αKG antagonists DMOG and PCA, and CoCl2 (Fig. 1D). The concentrations are similar to those affecting the mammalian enzymes, and are consistent with inhibitory effects of α,α’-dipyridyl and ethyl-PCA on Skp1 hydroxylation in cells (17).
P4H1-product analysis
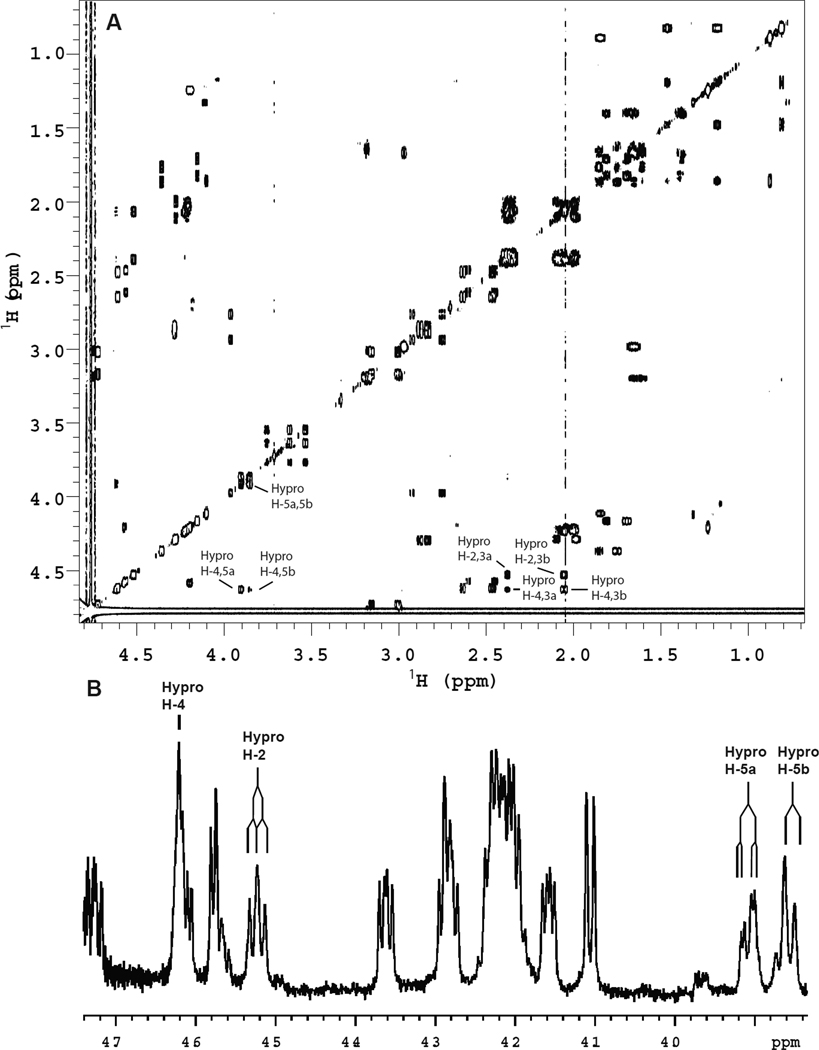
The configuration of hydroxyproline was predicted to be (2S,4R)-4-hydroxy-L-proline (or trans-4-hydroxyproline, Hyp) but, because positional (3- or 4-) and regio (cis- or trans-) isomers can occur, the product of the P4H1 reaction was identified to assess homology with animal PHDs. Skp1 was in vitro hydroxylated as above and, after endo Lys-C digestion, approximately 60 nmol of the NDFTP*EEEEQIRK peptide was purified by conventional HPLC (data not shown). As shown in the gCOSY spectrum (25), the hydroxyproline spin system is unique in that four (H-2, -4, -5a and -5b) of the six 1H signals are downfield in the H-α region of the spectrum (Fig. 2A), indicative of hydroxylation at C4. The H-2 signal of the hydroxyproline did not overlap with other signals (Fig. 2B) and showed two large J couplings to H-3a and H-3b (J2,3a ≈ J2,3b = 8.7 Hz). Similarly, the H-5a and H-5b signals were resolved and showed a large geminal coupling (J5a,5b = −11.8 Hz) as well as two smaller 3-bond couplings to H-4 of 3.6 Hz and ≤ 1.5 Hz. The H-3/4 J-couplings were not possible to extract as the H-4 signal is complex (split by four other 3-bond couplings) and was partially overlapped with another H-α signal; the H-3a and H-3b signals were overlapped with other aliphatic proton signals. The relative stereochemistry of substituents on the pyrrolidine ring was assigned by comparison with diastereomeric synthetic dipeptides that varied in their configuration at C4 of proline: Ac-L-Thr-L-Hyp-NHMe and Ac-L-Thr-L-hyp-NHMe [hyp = (2S,4S)-4-hydroxy-L-proline, a.k.a cis-hydroxyproline]. The N- and C-termini were blocked as amides (Ac and methyl, respectively) to emulate the larger peptide backbone. The spectra of these smaller peptides were accumulated at 500 MHz (1H), both for 1D spectra where relevant regions are shown in Fig. S2, and for 2D gCOSY spectra (Fig. S3). Both stereoisomers showed major and minor forms due to cis/trans isomerization about the peptide bond that is slow on the NMR time scale. For the (2S,4R) dipeptide (Fig. S2A) the 3-bond H-2/H-3 couplings of the hydroxyproline were both large (J2,3a = 7.9 Hz, J2,3b = 9.5 Hz). These coupling constants are typical of the Cγ-exo conformation of the pyrrolidine ring that is favored for Hyp (26,27). The H-5a/H-5b coupling was −11.7 Hz and the two H-4/H-5 couplings were ≤ 1.5 Hz and 3.6 Hz, similar to those of the Skp1 peptide. For the (2S,4S) dipeptide (Fig. S2B) the H-2/H-3 couplings were significantly different (J2,3a = 9.2 Hz, J2,3b = 4.7 Hz). This is consistent with the Cγ-endo conformation of the pyrrolidine ring that is favored for hyp (26,27). The J5a,5b = −11.0 Hz was similar to the (2S,4R) diastereomer, but the H-4/H-5 couplings were different (J4,5a = 5.0 Hz and J4,5b = 3.6 Hz). Thus the hydroxyproline spin system of the Skp1 peptide closely matched that of Ac-L-Thr-L-Hyp-NHMe, not Ac-L-Thr-L-hyp-NHMe, indicative of the (2S,4R) absolute stereochemistry. Interestingly, the Ac-Thr-Hyp-NHMe dipeptide exists as a 6:1 ratio of rotamers about the prolyl amide bond (Fig. S2A) whilst the 13-mer derived from the native Skp1 protein (Fig. 2B) presents as almost a single species. This is attributed to the increased steric requirements of the extended peptide backbone compared to the capping functionality in the model compounds (28).
Figure 2.
Skp1 product analysis by 1H-NMR. A scaled-up version of the reaction shown in Fig. 1C was performed, and the hydroxylated product was recovered and digested with endo Lys-C. 60 nmol of the 13-mer peptide corresponding to NDFTP*EEEEQIRK, where P* refers to the modified Pro, was purified to homogeneity, exchanged to D2O, and analyzed at 900 MHz. (A) 2D gCOSY spectrum; relevant crosspeaks of the hydroxyproline spin system are indicated. (B) 1D 1H spectrum; pertinent signals are marked.
P4H1: low activity of peptides
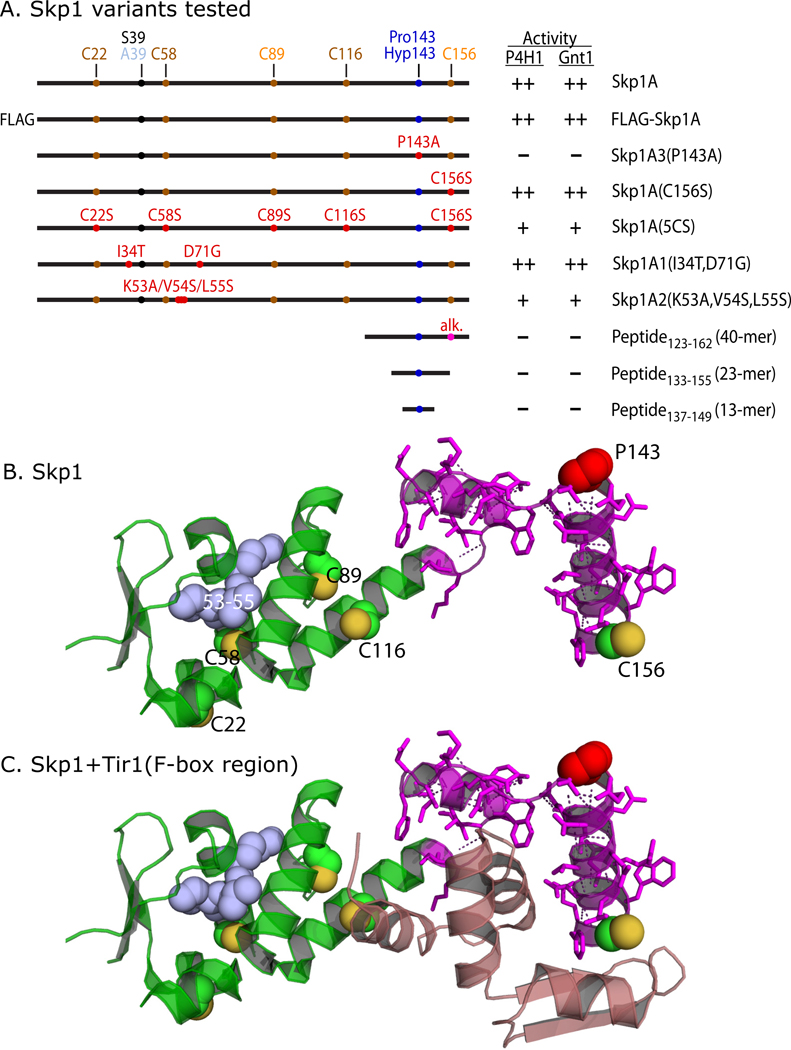
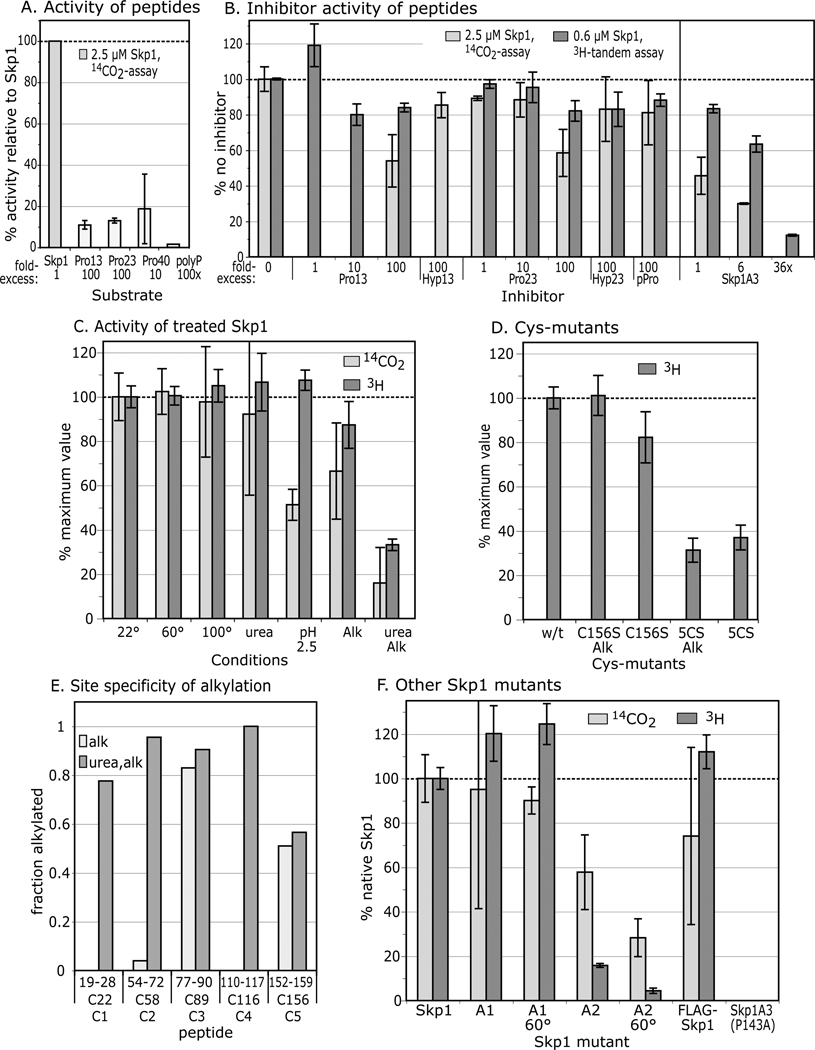
To test if the local region around Pro143 is sufficient for substrate activity, a previously described (18) synthetic peptide corresponding to Skp1133–155 (23-mer), whose position in Skp1 is depicted in Fig. 3A, was examined. Less than 15% activity was detected at 100-fold over the standard Skp1 concentration using the 14C-assay (Fig. 4A). Similar results were obtained using Skp1137–149 (13-mer). Lower concentrations of these peptides had proportionately less activity (data not shown). Finally, a 40-mer centered on P143, corresponding to the magenta-colored region in Fig. 3B, was purified from Skp1 after cleavage with CNBr. At a 10-fold higher concentration, Skp1123–162 also exhibited minimal acceptor activity. These results contrast with human PHD2, which exhibits substantial activity toward peptides of HIF1α (24).
Figure 3. Summary of mutant constructs.
(A) Wild-type, mutant, and truncated Skp1 sequences are illustrated. Positions of the target Pro143, the S39A polymorphism which distinguishes Skp1A from Skp1B, the 5 Cys residues, and mutated amino acids, are indicated (see Fig. S4 for sequence). C89 and C156 are differentially colored to indicate their accessibility in the native protein. Corresponding substrates for Gnt1 were prepared by exhaustive hydroxylation by P4H1. Substrate activities of the Skp1 preparations toward P4H1 and Gnt1, from this study, are summarized: ++ = normal activity; + = low activity; − = no activity detected. (B) Crystal structure of Skp1 excerpted from a complex of Arabidopsis thaliana Ask1 (Skp1) with the TIR1 F-box protein and auxin (32; PDB 2P1N), using PyMol. Pro143 is in red; S-atoms of Cys residues (substituted according to Dictyostelium sequence based on the alignment in Fig. S4) are in yellow; the 40-mer peptide (with polar contacts shown) is in magenta; the consecutive point mutations of Skp1A2 are in blue. (C) Amino acids 8–74 of TIR1, which includes its F-box domain, is included in mauve.
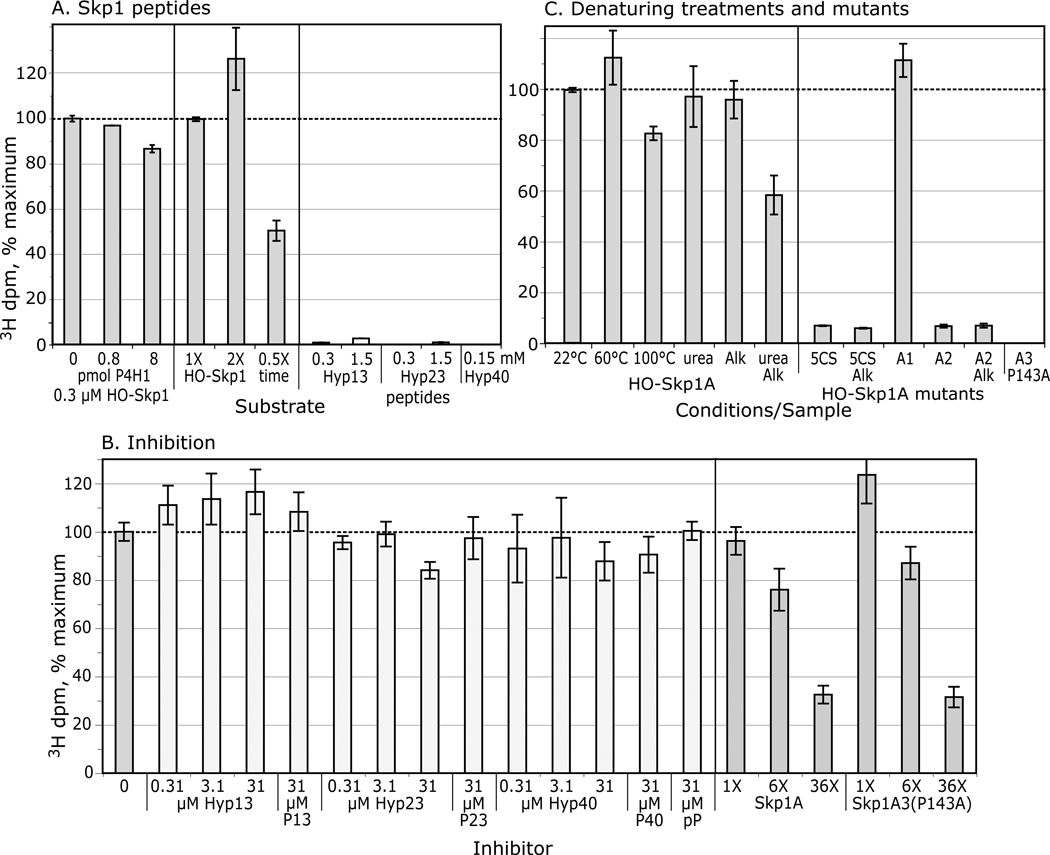
Figure 4. Substrate activity of Skp1 peptides, and denatured, alkylated, and mutant Skp1s.
(A) Comparison of Skp1 peptides and poly-L-Pro with Skp1A, provided at the concentrations indicated, using the 14C-release assay. Average values from 2–3 independent trials, ±SEM, are shown. (B) Effects of peptides and mutant Skp1A3(P143A) on the reaction with Skp1A, using either the 14C-release assay (Skp1A = 5 µM) or the coupled, 2-step 3H-assay (Skp1 = 0.63 µM). (C) Skp1A was subjected to denaturing conditions and/or alkylation with iodoacetamide, and restored to native reaction buffer as required. Recovery was verified by SDS-PAGE (Fig. S1). (D) The all 5Cys-Ser mutant Skp1 (5CS) and Skp1A(C156S) were untreated or alkylated (without urea). Data are presented as average values ± SEM (100% = 12,000–36,000 3H dpm; 8000–16,000 14C dpm). (E) Mapping of alkylation sites. After iodoacetamide treatment in the presence or absence of 6 M urea, Skp1 samples were digested with endo Lys-C and analyzed by MALDI-TOF-MS. Ion currents corresponding to alkylated Cys-peptides, relative to total ions associated with Cys-peptides (non-alkylated and disulfides) are reported. (F) Skp1A1, Skp1A2, FLAG-Skp1 and Skp1A3(P143A) were prepared from E. coli under non-denaturing conditions, and assayed at 2.5 µM using the 14C-release assay (100% activity = 24,600 dpm), or at 0.63 µM using the coupled 3H-GlcNAc two-step assay, at 22 ° or 60 °C as indicated.
A competition assay was employed to address if the peptides can bind P4H1. At a concentration excess of 100-fold, the 13- and 23-mers began to exhibit minimal inhibition of Skp1A hydroxylation using the 14C-assay, not seen with either the corresponding hydroxylated 23-mer or with poly-L-Pro (Fig. 4B). These effects were not as pronounced using the 1-step coupled assay, suggesting a higher sensitivity of the 14C-assay for decreased activity as discussed below. In comparison, a 6-fold excess of Skp1A(P143A), inactive as a substrate, inhibited the reaction by 40–70% (depending on which assay), and activity was reduced to ≤10% by a 36-fold excess of the mutant protein. This suggests that P4H1 recognition involves determinants on Skp1 not presented by the peptides, because they either lie elsewhere in the protein, or require conformational constraints imposed by the full-length protein.
P4H1: denaturing treatments of Skp1
To examine the significance of its folded state, Skp1 was subjected to various denaturing treatments. Remarkably, heating to 60 or 100 °C did not affect activity (Fig. 4C), though it did reduce background attributable to contaminating αKG-dependent dioxygenases (not shown).
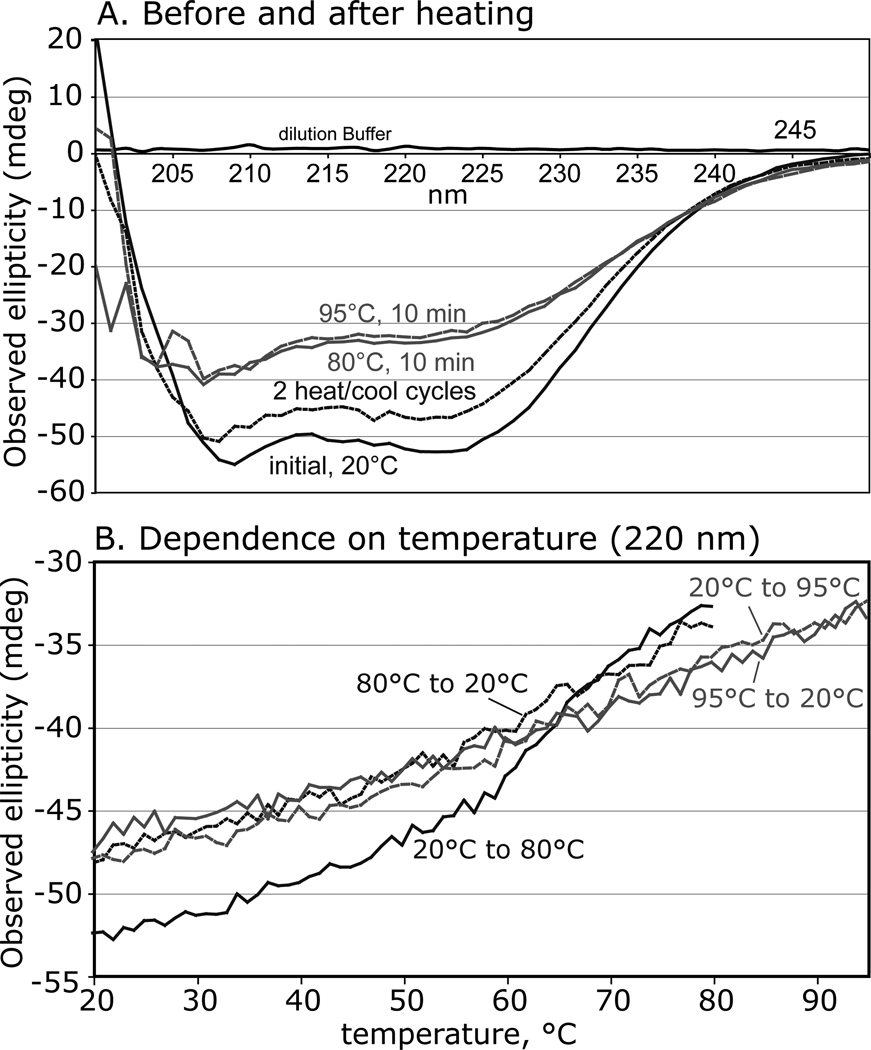
Circular dichroism (CD) studies were employed to address the folded state of Skp1 before and after heating. Skp1 showed substantial secondary structure predicting 51% α-helix, 9% β-sheet, and 17% turn content at 22 °C (Fig. 5A), similar to values of 46% α-helix and 5% β-sheet inferred from a crystal structure with an F-box protein (Fig. 3C). These features were partially lost at 80 °C but mostly restored upon return to 22 °C indicative of substantial refolding. A subsequent heating cycle to 95 °C and back yielded an identical result, and a time course analysis revealed gradual loss and recovery of secondary structure elements during the temperature change regimes (Fig. 5B). After two heating cycles and return to 22 °C, the analysis predicted 15% less α-helix that was partially replaced by increased β-sheet and turn content. This suggested that Skp1 is a structured protein that readily partially refolds and reacquires acceptor activity after heat-induced denaturation.
Figure 5. Circular dichroism analysis of Skp1A.
(A) Far-ultraviolet spectra were collected at 0.2 mg/ml in a 1 mm cuvette at 22 °C, and after heating to 80 °C, recycling to 22 °C, returning to 95 °C, and back again to 22 °C. (B) Time course analysis at 220 nm, with temperature varying at 2 °C/min. Data are representative of trials from 2 independent Skp1 preparations.
Pretreatment of Skp1 with 6 M urea had no effect on acceptor activity using either assay, suggesting refolding after denaturation as observed after heating. Pretreatment with pH 2.5 reduced activity to 50% using the 14C-release assay but not the coupled assay. The reason for the discrepancy is not known but other results also indicate that the 14C-release assay is more sensitive, possibly due to differences in the reaction conditions, which include a suboptimal concentration of αKG, and shaking during the reaction. Thus exposure to pH 2.5, previously used to purify myc-tagged versions of Skp1 by affinity chromatography, had a modest inhibitory effect on Skp1 activity.
P4H1: low activity after Skp1 Cys-modifications
Alkylation of urea-treated Skp1 with iodoacetamide decreased acceptor activity by about 80% (Fig. 4C). Similarly, converting all 5 Cys residues to Ser residues by site-directed mutagenesis (see Fig. 3) reduced activity by about 70% (Fig. 4D), and alkylation rendered no further effect as expected. SDS-PAGE analysis confirmed recovery of Skp1s after treatments (Fig. S1). In contrast, alkylation in the absence of urea only slightly diminished activity (Fig. 4C), and Skp1 bearing a C156S substitution exhibited nearly normal activity even after alkylation. Sites of alkylation were mapped to interpret the significance of these observations.
MALDI-TOF-MS analysis of Skp1A exposed to 7 mM DTT and alkylated in the absence of urea indicated addition of 2–3 acetamide groups per polypeptide, whereas treatment in urea showed quantitative modification of all 5 Cys residues (data not shown). Analysis of endo Lys-C-generated peptides from the non-urea sample by shotgun MALDI-TOF-MS showed that C89 was carboxamidomethylated and C156 was ~50%-alkylated (Fig. 4E). In contrast, all 5 Cys-peptides were alkylated in the urea-treated sample (except that C156 remained half-alkylated suggesting intrinsic resistance to derivatization). Similar results were observed in scans of HPLC-fractionated samples (data not shown), indicating that calculations were not biased by selective suppression of select peptides. Thus C22, C58 and C116 (positions shown in Fig. 3B) are normally protected from alkylation consistent with the CD data that Skp1 is folded in normal buffer. Modification of these same residues by alkylation or mutagenesis selectively inhibited substrate activity, consistent with the peptide studies indicating the importance of regions distant from Pro143.
Since Hs-Skp1 was previously reported to dimerize at high concentrations (29), we considered its potential influence on substrate activity. However, gel filtration and multi-angle light scattering comparisons of native, alkylated, and Cys-mutated Skp1s at concentrations used in the enzyme assays offered no evidence for dimerization (data not shown). Skp1 does have a tendency to form dimers during SDS-PAGE, which was diminished using 50 mM DTT and by the Cys-mutations (data not shown), but Cys-residues important for substrate activity appear to be buried and no evidence for substantial disulfide-bonding was noted in the MALDI-TOF-MS studies.
P4H1: activity of other Skp1 mutants
Previous work showed that mutant Skp1A1-myc, Skp1A2-myc and FLAG-Skp1 are, unlike otherwise normal Skp1A-myc, poorly hydroxylated when expressed in growing cells (30,17,6). As shown in Fig. 3, these differences also map to the N-terminal subdomain of Skp1, which is primarily associated with cullin binding (31). Modestly reduced activity (60%) was observed for Skp1A2 using the 14C-release assay (the lower activity using the 3H-assay was due to a strong effect on Gnt1, as shown later). However, the in vitro reactions showed nearly normal acceptor activity for Skp1A1 and FLAG-Skp1 (Fig. 4F). Thus poor modification of Skp1A2-myc in cells could be explained by weak substrate activity toward P4H1, but the poor in vivo modification of Skp1A1-myc and FLAG-Skp1A requires some other explanation. The amino acid changes map to the opposite end of Skp1 from P143 (see Fig. 3), and imply that hydroxylation in vivo is subject to positive regulation by a mechanism that is distinct from intrinsic P4H1 substrate activity and is inhibited by these N-terminal changes.
Conservation of Skp1 recognition
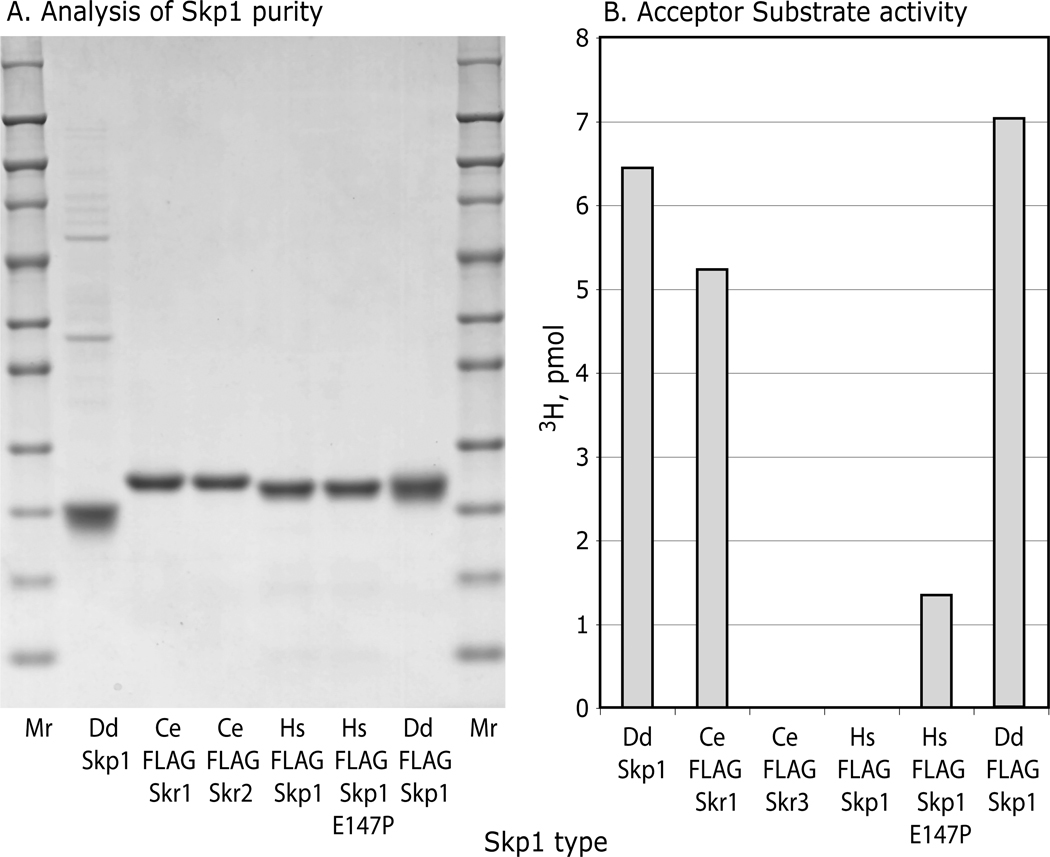
Animal Skp1s were examined to test whether P4H1 recognition determinants are conserved in organisms whose PHDs modify HIFα rather than Skp1, and whose genomes lack downstream glycosyltransferase genes (33). A Skp1 (Skr1) from the roundworm Caenorhabditis elegans which contains the equivalent of Pro143, and Skp1 (OCP2) from human, which lacks Pro143, were expressed with N-terminal FLAG-tags and purified (Fig. 6A). These Skp1s have similar lengths and are 58% and 65% identical, or 76% and 80% similar, respectively, to the Dd-Skp1 sequence (see alignment in Fig. S4). Remarkably, Ce-FLAG-Skr1 was as good as an acceptor using the coupled, 1-step P4H1 assay as Dd-FLAG-Skp1 (Fig. 6B). Modification appeared to be specific for the equivalent of Pro143, because Ce-FLAG-Skr2, an isoform whose Pro is replaced by Ala, is not an acceptor. Although it is not known if FLAG-Skr1 is a substrate for Egl-9 (the C. elegans PHD that hydroxylates HIFα), a crude desalted soluble extract of a mixed culture of C. elegans adults and larvae failed to modify Dd-Skp1A.4 Human FLAG-Skp1, which when overexpressed complements deletion of Saccharomyces cerevisiae Skp1 (34), was not an acceptor. Surprisingly, a mutant form of Hs-FLAG-Skp1 (E147P) in which the Pro was restored was a good acceptor, exhibiting >20% specific activity at the concentration tested. However, Hs-FLAG-Skp1(E147P) was not modified in the presence of a dialyzed rabbit reticulocyte lysate reported to efficiently hydroxylate HIFα (35).4 Since the comparisons were performed using the coupled, 1-step 3H assay, these Skp1s are also substrates for the subsequently acting Gnt1. Thus, determinants important for P4H1 and Gnt1 recognition are conserved in animal Skp1s, despite not being under purifying selection for this activity, which suggested that the enzymes may recognize features important for SCF-complex formation.
Figure 6.
P4H1 and Gnt1 modify animal Skp1s. Skp1 isoforms from C. elegans (Skr1 and Skr2) and H. sapiens (OCP-2, or isoform B), and mutant OCP-2(E147P), were expressed as N-terminally FLAG-tagged proteins in E. coli and purified by means of their FLAG-tags. (A) SDS-PAGE analysis followed by staining with Coomassie blue for total protein. (B) Analysis of P4H1 acceptor activity of 0.31 µM FLAG-Skp1’s using the coupled, 1-step 3H-assay. Data are representative of 2 independent trials at different concentrations.
Skp1-FbxA complex
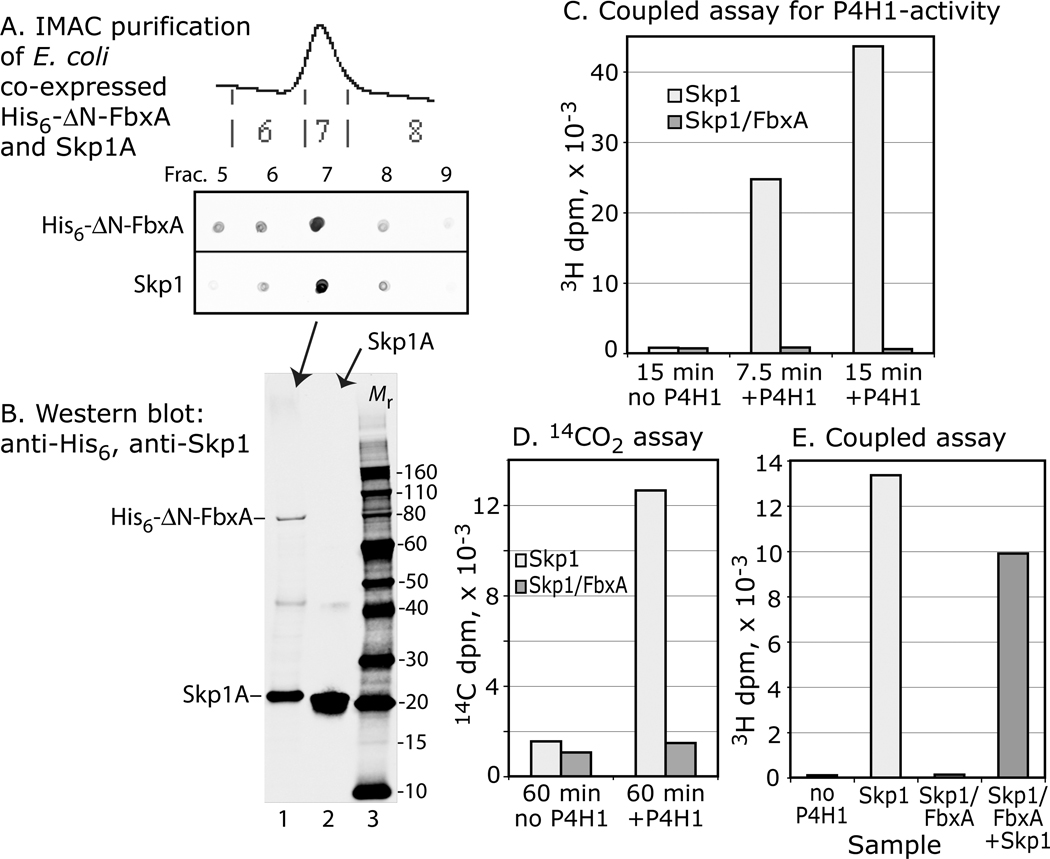
In cells, Skp1 forms a stable complex with F-box proteins (e.g., Fig. 3C), which are generally insoluble when expressed separately from Skp1 (36). To test whether heterodimerization with an F-box protein affected activity, Skp1 was coexpressed in E. coli with His6⊗N-FbxA, a previously described N-terminally truncated form of a Dictyostelium F-box protein important for slug morphogenesis (23,37). As expected, a fraction of His6⊗N-FbxA was soluble when expressed with Skp1, but not in its absence (data not shown), suggesting formation of a possible native complex as occurs in cells. Purification of His6⊗N-FbxA by virtue of its N-terminal His6 tag resulted in copurification of Skp1 (Fig. 7A,B). No acceptor activity was detected using the sensitive coupled 3H assay (Fig. 7C), compared to high activity of an equivalent amount of Skp1 in a parallel assay. Similarly, no activity was detected using the 14C-assay (Fig. 7D), indicating that inhibition occurred at the P4H1 step. Inclusion of additional free Skp1 yielded expected activity (Fig. 7E), excluding the possibility of an adventitious inhibitor. Similar results were obtained after further purification of the complex by anion exchange chromatography or gel filtration (data not shown). The results are consistent with a model in which determinants for P4H1 and F-box recognition overlap, as suggested by the evolutionary comparison above.
Figure 7. Skp1 complexed with FbxA is not a substrate. Skp1A and His6-⊗N-FbxA were co-expressed in E. coli and purified by Ni+2-IMAC.
(A) Dot-blot analyse s show that Skp1A copurified with His6-⊗N-FbxA. (B) Western blot analysis confirmed the presence of His6-⊗N-FbxA and Skp1A in frac. 7 (lane 1), and Skp1A in a Skp1A preparation (lane 2); blots were used to estimate relative levels of Skp1A for the assays. (C) Coupled, 1-step 3H-assay showing absence of acceptor activity for the purified Skp1A/FbxA complex, in comparison with a similar level of free Skp1A in a parallel reaction. (D) Similar analysis using the 14C-release assay. (E) Addition of an equal amount of free Skp1A to the reaction with Skp1A/His6-⊗N-FbxA resulted in near normal acceptor activity. Data are representative of two independent experiments.
Gnt1 studies
The apparent joining of P4H1- and Gnt1-like coding regions as separate domains within the same gene of some protist genomes led to the prediction that the two enzymes may normally act in concert and that Gnt1 has the same or relaxed specificity requirements (33,2). In an earlier study, Gnt1 was assayed using mutant Skp1A1-myc, which when expressed in Dictyostelium accumulates as a mixture of unhydroxylated, hydroxylated but not GlcNAc-modified, and other isoforms (30,18). To generate a wild-type substrate, HO-Skp1A was prepared as above (Fig. 2) and, for comparison, hydroxylated versions of mutant Skp1A1, Skp1A2 and Skp1A(5CS) were prepared. Assay conditions were developed in which the reaction was dependent on time (Fig. 8A), though the Skp1 concentration was close to saturation consistent with the previously estimated Km of 0.56 µM (18). Unlike the coupled assays of P4H1 in which Gnt1 was in excess to promote efficient conversion of P4H1 product, Gnt1 levels were limiting to permit initial velocity measurements before >10% substrate was converted. Native Dd-Gnt1, and Dd-His6Gnt1 and DdDp-His6Gnt1 from E. coli, yielded similar results so the latter chimera, essentially the ortholog from D. purpureum, is shown because it lacks a poly-Asn tract that interferes with expression and purification from E. coli (18). The specific activity toward DdDp-His6Gnt1 was about 0.2/min, lower than the 1/sec value expected for a glycosyltransferase but an improvement over the 1/h reported previously (18). Under these conditions, Gnt1 activity was not affected by the presence of P4H1 (Fig. 8A), or by the addition of desalted cytosolic extracts from Dictyostelium strains that are null for Gnt1 or both Gnt1 and P4H13 (data not shown).
Figure 8. Skp1 substrate activity toward Gnt1.
(A) Skp1A was hydroxylated as in Fig. 2 and Hyp-peptides were prepared as in Fig. 4A. Incorporation in the presence of purified DdDp-His6Gnt1 and UDP-[3H]GlcNAc was assayed as described in Methods. (B) Gnt1 was pretreated with peptides or the non-substrates Skp1A or Skp1A3(P143A) prior to assay as in panel A. (C) Normal and mutant HO-Skp1 samples, left untreated or subjected to denaturing treatments as in Fig. 4, were incubated at 0.31 µM and assayed as above. Data are presented as average values ± SEM.
Although synthetic 13-mer and 23-mer Hyp-peptides (see Fig. 3A) were substrates, their activities were much lower even at 5000-fold higher concentration (Fig. 8A), consistent with a previous study (18). The 40-mer Hyp-peptide was also a poor substrate at the lower concentration tested. These peptides and their Pro-counterparts were also ineffective inhibitors, except for a slight effect of the 23-mer at a 1000-fold concentration excess that was specific for the Hyp-relative to the Pro-form (Fig. 8B). In contrast, two full length Skp1 isoforms lacking Hyp143 each inhibited the reaction by 70% at 36-fold excess (Fig. 8B). Inhibition by non-Hyp containing Skp1 may be mediated by determinants distant from the active site of Gnt1 and Hyp143.
Mutant HO-Skp1A1 (see Fig. 3A), inefficiently modified when expressed as its C-terminally myc-tagged form in cells, was a robust Gnt1 substrate in vitro with activity comparable to that of wild-type HO-Skp1A (Fig. 8C). Since the C-terminal myc-tag did not affect modification of normal Skp1A-myc, an unknown inhibitory factor(s) is implicated in vivo. In contrast, mutant HO-Skp1A2 was a poor in vitro acceptor, which mirrored the weak activity toward His6P4H1 (Fig. 4F). As also noted for the P4H1 reaction, denaturing treatments including heat or urea, or alkylation with iodoacetamide, had minimal effect on substrate activity, whereas alkylation in the presence of urea was modestly inhibitory (Fig. 8C). Consistent with the latter result, replacement of all 5 Cys-residues with Ser was inhibitory albeit to an even greater extent (>90%). In summary, the similar profiles of substrate acceptor sensitivity suggest that P4H1 and Gnt1 have related mechanisms for recognizing and modifying Skp1.
DISCUSSION
P4H1 fulfills many criteria to be considered the Dictyostelium ortholog of HIFα-type PHDs, previously thought to be animal-specific. Earlier studies established the non-heme dioxygenase character of P4H1 including reliance on αKG and ascorbate, high Km for O2, and role in O2-dependent development (3,4). The present study shows that P4H1 catalyzes the stereoselective oxidation of Pro143 to (2S,4R)-4-hydroxy-L-proline (Hyp), by analogy to the product of the mammalian enzyme on HIFα (12–14). Using a direct 14CO2-release assay similar to that employed for HIFα-type PHDs, Skp1-dependent activity is inhibited by the product succinate, and other Krebs cycle intermediates including oxaloacetate, citrate, and fumarate, over concentration ranges that similarly affect Hs-PHD2 (7). As expected, P4H1 is also inhibited by CoCl2 and PCA, consistent with a previous report using the coupled 3H-assay (3), DMOG, and the chelators α,α’-dipyridyl and EDTA. α,α’-dipyridyl and the ethyl ester of PCA were previously observed to inhibit Skp1 hydroxylation in cells (17).
In contrast to these similarities, P4H1 modifies Skp1 rather than HIFα, which is apparently absent from many protist genomes, and the stability of Skp1 is not affected by the presence of P4H1 and the glycosyltransferases in cells (6)3. Whereas human PHD2 may recognize multiple substrates (7,10), current evidence indicates only a single substrate for P4H1 (6). To begin to understand the basis by which P4H1 recognizes Skp1, chemically and genetically modified forms of Dd-Skp1A were analyzed for substrate acceptor activity with the overall conclusion that recognition is highly dependent on structure determinants distant from the Pro target (see summary of activities in Fig. 3A). Based on the direct assay, synthetic and native peptides centered on the target Pro143 and as long as 40 amino acids were poor acceptors (Fig. 4A), and only very weak inhibitors at 100-fold concentration excess (Fig. 4B). In contrast, a 6-fold concentration excess of the Skp1A3(P143A) mutant, not a substrate owing to replacement of the target Pro, inhibited the reaction by 50%. These results suggest that the first 122 amino acids of Skp1 contribute to recognition of Pro143, but do not differentiate whether they present a unique recognition determinant or impose conformational constraints on the C-terminal region allowing recognition by the P4H1 active site.
Analysis of the crystal structure of Hs-PHD2 complexed with a 19-mer peptide from HIF1α shows the importance of a β2β3/loop for covering the substrate-bound active site and conferring substrate selectivity of the PHDs toward the N-ODD or C-ODD (38). This region, whose framework but not specific sequence is conserved in P4H1 (33), may also contribute to P4H1 selection of Pro143. However, studies show that Hs-PHD2 is as active toward a 19-mer peptide centered on Pro564 as the complete HIFα C-ODD domain (24), so the minimal activity of P4H1 toward Skp1 peptides suggests additional complexity.
All Skp1 structures to date have been obtained from complexes with F-box proteins and reveal a consistent picture of stereotyped secondary structure (e.g., Fig. 3C). Our CD studies affirm considerable secondary structure (Fig. 5A) suggesting that free Skp1 is folded in solution too, which is consistent with protection of 3 Cys-residues from alkylation (Fig. 4D). Skp1 reversibly denatures over a broad temperature range (Fig. 5B), implying potential local conformational flexibility that may be relevant to P4H1 recognition as suggested for PHD2 binding to HIF1α C-ODD peptides (38). While loss of local flexibility when complexed with FbxA may contribute to substrate inactivity (Fig. 7), steric hindrance or a conformational change may also contribute.
A role for overall conformation is reinforced by the inhibitory effects of point mutations in a potential actin-binding motif (16) at codons 53–55, and by the similar effects of alkylating all five Cys-residues or their mutation to Ser-residues (Fig. 5C). The positions of the 3 critical residues (C22, C58 and C118), N-terminal to the 40-mer peptide (see Fig. 3), are consistent with a role for distal determinants in Skp1 substrate activity. Though no evidence for involvement of Cys residues in intra- or inter-molecular disulfide bonds emerged from chromatography and MS-analyses (not shown), further studies of this issue are warranted.
A phylogenetic analysis of these 3 Cys residues shows that they are not absolutely conserved in Skp1s known or expected to be P4H1 substrates (2; Fig. S4), suggesting that, if recognition mechanisms of P4H1 orthologs are conserved, these Cys residues indirectly or only partially contribute, with adjacent amino acids, to a critical recognition or folding motif(s). In contrast, the KVL/53–55 motif mutated in Skp1A2 is highly conserved in all Skp1s, and K53 has been implicated in contacting cullin in SCF complexes (31). The significance of cullin binding, which occurs primarily in the N-terminal region of Skp1, remains to be examined.
Features of Skp1 that are important for P4H1 substrate activity are highly conserved in animal Skp1s. C. elegans FLAG-Skr1 was as active as Dd-Skp1A when tested at well below the Km for Dd-Skp1A and apparently depended on the equivalent of Pro143 (Fig. 6). Furthermore, human FLAG-Skp1, which is not a substrate owing to the replacement of the equivalent of Pro143 by Glu in chordates, became a good substrate upon restoration of the Pro. These Skp1s were not hydroxylated when presented to extracts of mixed cultures of C. elegans or to rabbit reticulocyte lysates4, under conditions that yield easily detected substrate activity of P4H1-like activity in extracts of Dictyostelium (6), which is consistent with the apparent absence of sequence or structure similarity between HIFα and Skp1. This remarkable conservation of P4H1 acceptor activity in the absence of purifying selection suggests that P4H1 depends on determinants important for other functions of Skp1 such as binding to F-box proteins or cullins in SCF complexes, consistent with loss of substrate activity when Skp1 was bound to FbxA (Fig. 7).
The KVL/53–55 motif may be important for normal P4H1-processing because ectopically expressed Skp1A2-myc appears to be poorly modified in proliferating cells and at stationary phase (17). However, Skp1A1-myc and FLAG-Skp1A, which are also poorly hydroxylated in vivo, are as active as normal Skp1A in the assay. Other evidence also points to the importance of unknown factors for hydroxylation in vivo. For example, normal Skp1 is poorly hydroxylated when modestly overexpressed in slugs, even when P4H1 is simultaneously overexpressed (6). In addition, the specific activities of P4H1 and Gnt1 are low, consistent with additional factors that enhance activity in cells though attempts to detect such effects by addition of cytosolic extracts to the reactions were unsuccessful. The present results suggest that some Skp1 mutations may affect accessibility rather than processing by P4H1 per se.
The parallel analysis of the substrate activity of HO-Skp1 toward the next enzyme in the pathway, Gnt1, revealed a remarkably similar dependence on Skp1 features (Fig. 8). Therefore P4H1 and Gnt1 appear to depend on overlapping determinants for recognition of Skp1, despite the anticipated preference of Hyp for the Cγ-exo pyrrolidine conformation relative to unmodified Pro (14), as illustrated in Fig. 1A. It was previously noted that several protists are predicted, based on genomic analyses, to express their P4H1- and Gnt1-like enzymes as a fusion protein, a strategy often used to promote processive substrate processing as observed for subsequent enzymes in this pathway (2). Though the two Dictyostelium enzymes may still share the same recognition mechanisms, their separation may diversify regulation of Skp1 processing.
Related findings have been reported for the next 3 enzymes in the pathway. The ®3GalT that modifies the Skp1 product of the Gnt1 reaction was essentially inactive toward GlcNAc-O-peptides, which also exerted only minimal inhibition at very high concentrations (5). The present data validate the predicted 4(trans)-position of the hydroxyl group employed for the glycopeptide synthesis in those studies. The acceptor activity of GlcNAc-O-Skp1 was only mildly inhibited by denaturing treatments, whereas Cys-alkylation inhibited about 80%, comparable to effects on P4H1 and Gnt1. In comparison, the α2FucT activity that modifies the product of the ®3GalT reaction, and is mediated by a separate domain of the same PgtA protein, was not affected by Cys-alkylation, and disaccharide conjugates were much better acceptors than were peptides for the 3 earlier enzymes. In contrast, the subsequently acting α3GalT, which modifies the product of the α2FucT reaction, is dramatically inhibited (>90%) by the denaturing treatments employed here (39). Thus overall, the different enzymes of the pathway appear to employ distinct and complex determinants for the successive modification of Skp1.
The proposed distal determinants may be important for the specificity and regulation of P4H1, Gnt1, PgtA-®3GalT, and AgtA-α3GalT toward Skp1-Pro143 in the cell. Indeed, current biochemical and genetic evidence suggests that each of these enzymes is devoted solely to modification of Skp1 (2,6)8, which contrasts with evidence for multiple functional substrates for animal PHDs (7,10). This raises the question of why complex and, in some cases varied determinants are required by the successive enzymes. Such mechanisms would be expected if the modifications are influenced by Skp1 folding or interactions with other proteins or factors. Mutant strains that are unable to accomplish each of the modifications show distinct dependences on O2 to execute culmination and other developmental steps, indicative of altered signaling associated with each successive processing intermediate in the pathway (2). The present results indicate that Skp1 itself and its interactions with other proteins play a role in this regulation, in addition to potential independent modulation of the pathway enzymes themselves. Structural studies of the enzyme-substrate complexes are needed to further evaluate this model.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Dr. K. Nakayama (Kyushu) for providing the Ce-Skr1 and Ce-Skr2 cDNAs, Dr. Rick Firtel (UCSD) for FbxA strains, Dr. Margaret Nelson (Allegheny) for FbxA plasmids, Dr. Scott Plafker (OUHSC) for Skp1 plasmids, Dr. Christa Feasley (OUHSC-OCMG) for advice, Ruby Rahman and Dr. Karla Rodgers (OUHSC) for advice on the CD experiments, and Bruce Baggenstoss (OUHSC) for performing multi-angle light scattering analyses.
Footnotes
Supported in part by NIH grants R01-GM37359 and R01-GM84383-02S1. KDW was supported by a grant from the OUHSC Graduate College and the Provost’s Office.
Abbreviations: αKG, α-ketoglutarate; CD, circular dichroism; E3, ubiquitin-protein isopeptide ligase; DMOG, dimethyloxalylglycine; GnT1, (Skp1 protein)-hydroxyproline α-N-acetyl-D-glucosaminyltransferase; HIF, hypoxia-inducible factor; Hyp, trans-4-hydroxy-L-proline aka (2S,4R)-hydroxyproline; hyp, cis-4-hydroxy-L-proline aka (2S,4S)-hydroxyproline; ODD, oxygen-dependent degradation domain of HIFα, which may be designated as N- or C-terminal; P4H, prolyl 4-hydroxylase; PCA, protocatechuic acid or 3,4-dihydroxybenzoate; PHD, prolyl hydroxylase domain protein; RP-HPLC, reversed phase high performance liquid chromatography; SCF, Skp1-cullin-Fbox protein complex; Ub, ubiquitin; VBC, protein complex consisting of the von Hippel-Lindau protein, elongin B, elongin C and cullin-2
Karaunaratne, C. V., and Taylor, C. M., unpublished data.
Zhang, D., van der Wel, H., and West, C. M., unpublished data.
Xu, Y., Yu, X., H. van der Wel, H., and West, C. M., unpublished data.
SUPPORTING INFORMATION AVAILABLE
This section includes Supplemental Methods (plasmid constructions), Table S1 (PCR primers), Fig. S1 (protein purity), Figs. S2 and S3 (NMR spectra on synthetic dipeptides), and Fig. S4 (Skp1 sequence alignment). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Willems AR, Schwab M, Tyers M. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 2.West CM, Wang ZA, van der Wel H. A cytoplasmic prolyl hydroxylation and glycosylation pathway modifies Skp1 and regulates O2-dependent development in Dictyostelium. Biochim. Biophys. Acta. 2010;1800:160–171. doi: 10.1016/j.bbagen.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Wel H, Ercan A, West CM. The Skp1 prolyl hydroxylase from Dictyostelium is related to the hypoxia-inducible factor-alpha class of animal prolyl 4-hydroxylases. J. Biol. Chem. 2005;280:14645–14655. doi: 10.1074/jbc.M500600200. [DOI] [PubMed] [Google Scholar]
- 4.West CM, van der Wel H, Wang ZA. Prolyl 4-hydroxylase-1 mediates O2-signaling during development of Dictyostelium. Development. 2007;134:3349–3358. doi: 10.1242/dev.000893. [DOI] [PubMed] [Google Scholar]
- 5.Wang ZA, van der Wel H, Vohra Y, Buskas T, Boons G-J, West CM. Role of a cytoplasmic dual-function glycosyltransferase in O2-regulation of development in Dictyostelium. J. Biol. Chem. 2009;284:28896–28904. doi: 10.1074/jbc.M109.022574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang ZA, Singh D, van der Wel H, West CM. Prolyl hydroxylation- and glycosylation-dependent functions of Skp1 in O2-regulated development of Dictyostelium. Dev. Biol. 2011;349:283–295. doi: 10.1016/j.ydbio.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 8.Koivunen P, Hirsila M, Remes AM, Hassinen IE, Kivirikko KI, Myllyharju J. Inhibition of hypoxia-inducible factor (HIF) hydroxylases by citric acid cycle intermediates: possible links between cell metabolism and stabilization of HIF. J. Biol. Chem. 2007;282:4524–4532. doi: 10.1074/jbc.M610415200. [DOI] [PubMed] [Google Scholar]
- 9.Pagé EL, Chan DA, Giaccia AJ, Levine M, Richard DE. Hypoxia-inducible factor-1alpha stabilization in nonhypoxic conditions: role of oxidation and intracellular ascorbate depletion. Mol. Biol. Cell. 2008;19:86–94. doi: 10.1091/mbc.E07-06-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mikhaylova O, Ignacak ML, Barankiewicz TJ, Harbaugh SV, Yi Y, Maxwell PH, Schneider M, Van Geyte K, Carmeliet P, Revelo MP, Wyder M, Greis KD, Meller J, Czyzyk-Krzeska MF. The von Hippel-Lindau tumor suppressor protein and Egl-9-Type proline hydroxylases regulate the large subunit of RNA polymerase II in response to oxidative stress. Mol. Cell. Biol. 2008;28:2701–2717. doi: 10.1128/MCB.01231-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yen HC, Elledge SJ. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science. 2008;322:923–929. doi: 10.1126/science.1160462. [DOI] [PubMed] [Google Scholar]
- 12.Hon WC, Wilson MI, Harlos K, Claridge TD, Schofield CJ, Pugh CW, Maxwell PH, Ratcliffe PJ, Stuart DI, Jones EY. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVH. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 13.Min JH, Yang H, Ivan M, Gertler F, Kaelin WG, Jr, Pavletich NP. Structure of an HIF-1alpha -pVHL complex: hydroxyproline recognition in signaling. Science. 2002;296:1886–1889. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 14.Loenarz C, Mecinović J, Chowdhury R, McNeill LA, Flashman E, Schofield CJ. Evidence for a stereoelectronic effect in human oxygen sensing. Angew Chem. Int. Ed. Engl. 2009;48:1784–1787. doi: 10.1002/anie.200805427. [DOI] [PubMed] [Google Scholar]
- 15.Hewitson KS, Schofield CJ, Ratcliffe PJ. Hypoxia-inducible factor prolyl-hydroxylase: purification and assays of PHD2. Meth. Enzymol. 2007;435:25–42. doi: 10.1016/S0076-6879(07)35002-7. [DOI] [PubMed] [Google Scholar]
- 16.West CM, Kozarov E, Teng-umnuay P. The cytosolic glycoprotein FP21 of Dictyostelium discoideum is encoded by two genes resulting in a polymorphism at a single amino acid position. Gene. 1997;200:1–10. doi: 10.1016/s0378-1119(97)00194-7. [DOI] [PubMed] [Google Scholar]
- 17.Sassi S, Sweetinburgh M, Erogul J, Zhang P, Teng-umnuay P, West CM. Analysis of Skp1 glycosylation and nuclear enrichment in Dictyostelium. Glycobiology. 2001;11:283–295. doi: 10.1093/glycob/11.4.283. [DOI] [PubMed] [Google Scholar]
- 18.Teng-umnuay P, van der Wel H, West CM. Identification of a UDP-GlcNAc:Skp1-hydroxyproline GlcNAc-transferase in the cytoplasm of Dictyostelium. J. Biol. Chem. 1999;274:36392–36402. doi: 10.1074/jbc.274.51.36392. [DOI] [PubMed] [Google Scholar]
- 19.van der Wel H, Morris HR, Panico M, Paxton T, Dell A, Kaplan L, West CM. Molecular cloning and expression of a UDP-GlcNAc:hydroxyproline polypeptide GlcNAc-transferase that modifies Skp1 in the cytoplasm of Dictyostelium. J. Biol. Chem. 2002;277:46328–46337. doi: 10.1074/jbc.M208024200. [DOI] [PubMed] [Google Scholar]
- 20.Sucgang R, Kuo A, Tian X, Salerno W, Parikh A, Feasley CL, Dalin E, Tu H, Huang E, Barry K, Lindquist E, Shapiro H, Bruce D, Schmutz J, Fey P, Gaudet P, Anjard C, Mohan MB, Basu S, Bushmanova Y, van der Wel H, Katoh M, Coutinho PM, Saito T, Elias M, Schaap P, Kay RR, Henrissat B, Eichinger L, Rivero-Crespo F, Putnam NH, West CM, Loomis WF, Chisholm R, Shaulsky G, Strassmann JE, Queller DC, Kuspa A, Grigoriev I. Comparative genomics of the social amoebae Dictyostelium discoideum and D. purpureum. 2010 doi: 10.1186/gb-2011-12-2-r20. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West CM, Scott-Ward T, Teng-umnuay P, van der Wel H, Kozarov E, Huynh A. Purification and characterization of an α1,2-L-fucosyltransferase, which modifies the cytosolic protein FP21, from the cytosol of Dictyostelium. J. Biol. Chem. 1996;271:12024–12035. doi: 10.1074/jbc.271.20.12024. [DOI] [PubMed] [Google Scholar]
- 22.Yamanaka A, Yada M, Imaki H, Koga M, Ohshima Y, Nakayama K. Multiple Skp1-related proteins in Caenorhabditis elegans: diverse patterns of interaction with Cullins and F-box proteins. Curr. Biol. 2002;12:267–275. doi: 10.1016/s0960-9822(02)00657-7. [DOI] [PubMed] [Google Scholar]
- 23.Mohanty S, Lee S, Yadava N, Dealy MJ, Johnson RS, Firtel RA. Regulated protein degradation controls PKA function and cell-type differentiation in Dictyostelium. Genes Dev. 2001;15:1435–1148. doi: 10.1101/gad.871101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koivunen P, Hirsilä M, Kivirikko KI, Myllyharju J. The length of peptide substrates has a marked effect on hydroxylation by the hypoxia-inducible factor prolyl 4-hydroxylases. J. Biol. Chem. 2006;281:28712–28720. doi: 10.1074/jbc.M604628200. [DOI] [PubMed] [Google Scholar]
- 25.Hurd RE. Gradient-enhanced spectroscopy. J. Magn. Reson. 1990;87:422–428. doi: 10.1016/j.jmr.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 26.Cai M, Huang Y, Liu J, Krishnamoorthi R. Solution conformations of proline rings in proteins studied by NMR spectroscopy. J. Biomolecular NMR. 1995;6:123–128. doi: 10.1007/BF00211775. [DOI] [PubMed] [Google Scholar]
- 27.Taylor CM, Hardre R, Edwards PJB. The impact of pyrrolidine hydroxylation on the conformation of proline-containing peptides. J. Org. Chem. 2005;70:1306–1305. doi: 10.1021/jo0490043. [DOI] [PubMed] [Google Scholar]
- 28.Taylor CM, Hardre R, Edwards PJB, Park JH. Factors affecting conformation in proline-containing peptides. Org. Lett. 2003;5:4413–4416. doi: 10.1021/ol035711r. [DOI] [PubMed] [Google Scholar]
- 29.Henzl MT, Thalmann I, Thalmann R. OCP2 exists as a dimer in the organ of Corti. Hear. Res. 1998;126:37–46. doi: 10.1016/s0378-5955(98)00148-8. [DOI] [PubMed] [Google Scholar]
- 30.Teng-umnuay P, Morris HR, Dell A, Panico M, Paxton T, West CM. The cytoplasmic F-box binding protein SKP1 contains a novel pentasaccharide linked to hydroxyproline in Dictyostelium. J. Biol. Chem. 1998;273:18242–18249. doi: 10.1074/jbc.273.29.18242. [DOI] [PubMed] [Google Scholar]
- 31.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 32.Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 33.West CM, van der Wel H, Sassi S, Gaucher EA. Cytoplasmic glycosylation of protein-hydroxyproline and its relationship to other glycosylation pathways. Biochim. Biophys. Acta. 2004;1673:29–44. doi: 10.1016/j.bbagen.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 35.Ivan M, Haberberger T, Gervasi DC, Michelson KS, Günzler V, Kondo K, Yang H, Sorokina I, Conaway RC, Conaway JW, Kaelin WG., Jr Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc. Natl. Acad. Sci. USA. 2002;99:13459–13464. doi: 10.1073/pnas.192342099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li T, Pavletich NP, Schulman BA, Zheng N. High-Level Expression and Purification of recombinant SCF ubiquitin ligases. Meth. Enzymol. 2005;398:125–142. doi: 10.1016/S0076-6879(05)98012-9. [DOI] [PubMed] [Google Scholar]
- 37.Nelson MK, Clark A, Abe T, Nomura A, Yadava N, Funair CJ, Jermyn KA, Mohanty S, Firtel RA, Williams JG. An F-Box/WD40 repeat-containing protein important for Dictyostelium cell-type proportioning, slug behaviour, and culmination. Dev. Biol. 2000;224:42–59. doi: 10.1006/dbio.2000.9793. [DOI] [PubMed] [Google Scholar]
- 38.Chowdhury R, McDonough MA, Mecinović J, Loenarz C, Flashman E, Hewitson KS, Domene C, Schofield CJ. Structural basis for binding of hypoxia-inducible factor to the oxygen-sensing prolyl hydroxylases. Structure. 2009;17:981–989. doi: 10.1016/j.str.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Ketcham C, Wang F, Fisher SZ, Ercan A, van der Wel H, Locke RD, Doulah kS, Matta KL, West CM. Specificity of a soluble UDP-galactose:fucoside α1,3galactosyltransferase that modifies the cytoplasmic glycoprotein Skp1 in Dictyostelium. J. Biol. Chem. 2004;279:29050–29059. doi: 10.1074/jbc.M313858200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.