Abstract
The ability to retain DNA labels over time is a property proposed to be associated with adult stem cells. Recently, label retaining cells (LRC) were indentified in cancer. LRC were suggested to be the result of either slow-cycling or asymmetric-cell-division with nonrandom-chromosomal-cosegregation (ACD-NRCC). ACD-NRCC is proposed to segregate the older template DNA strands into daughter stem cells and newly synthesized DNA into daughter cells destined for differentiation. The existence of cells undergoing ACD-NRCC and the stem-like nature of LRC remain controversial. Currently, to detect LRC and ACD-NRCC, cells need to undergo fixation. Therefore, testing the stem-cell nature and other functional traits of LRC and cells undergoing ACD-NRCC has been limited. Here, we show a method for labeling DNA with single and dual-color nucleotides in live human liver cancer cells avoiding the need for fixation. We describe a novel methodology for both the isolation of live LRC and cells undergoing ACD-NRCC via fluorescence-activated cell sorting with confocal microscopy validation. This has the potential to be a powerful adjunct to stem-cell and cancer research.
Introduction
Adult stem cells are defined, at least, by their ability to differentiate into multiple progeny and maintain self-renewal activity [1,2]. Adult stem cells have been identified in a variety of tissues including the liver [3]. Recent studies have suggested the presence of cancer-stem-cells (CSC) in solid organs such as the breast, brain, colon, liver, melanoma, and pancreas. (4] To date, the ability to identify CSC has been limited to various surface markers such as CD133, CD24, CD44, CD90, and the side-population based on the efflux of Hoechst [4,5].
Traditionally, label retaining cells (LRC) are identified by repeatedly exposing cells either in-vivo or in-vitro to nucleotide analogs such as Bromodeoxyuridine (BrdU) or 3H-thymine-deoxyribose. After a prolonged chase period, the DNA labels are diluted with each subsequent cell division and become undetectable. However, there is a population of cells that retain the DNA labels over a prolonged period of time. These cells are called LRCs. LRC were associated with population of cells comprised or highly enriched with adult tissue stem-cells [6–8].
It has been proposed that LRC are the result of either slow-cycling or asymmetric-cell-division with nonrandom-chromosomal-cosegregation (ACD-NRCC). These 2 plausible hypotheses continue to be debated. Several studies have shown that LRC are actively dividing, mitigating the slow-cycling hypothesis [9–12]. However, more research is required to definitively demonstrate that LRC undergo meaningful cell division. The other hypothesis is based on the concept of ACD-NRCC that was introduced by Cairns more than thirty years ago [13]. ACD-NRCC proposes that each chromosome in a stem-cell contains 1 DNA strand that is conserved throughout multiple asymmetric divisions (Fig. 1A). As a result, stem-cells are able to avoid accumulation of mutations from DNA replication errors by preferentially segregating replication-errors into the daughter-cell fated to differentiate and ultimately be eliminated [13]. This principle has been demonstrated in various studies and reviewed by several authors [14–24]. In particular, dual-labeling studies using 1 halogenated thymidine and 1 radionuclide-labeled thymidine analog [16] and dual-labeling studies using 2 halogenated thymidine analogs (eg, antibody to BrdU, CldU or IdU) pioneered by Conboy et al. [25,26]. in murine models both in vivo and in vitro have been successful in demonstrating ACD-NRCC. More recently, ACD-NRCC has been identified in fixed human lung cancer cells [26]. However, several investigators have been unable to confirm the existence of ACD-NRCC or LRC [27,28] If the intention is to label the “older template strands,” detection of these phenomena indicates that at some point, stem-cells should undergo symmetric division otherwise DNA labeling could have not occurred. Alternatively, ACD-NRCC can occur with the labeling occurring in the newer template strands. Subsequently, stem-cells can undergo either symmetric cell division or asymmetric cell division or slow cycling, possibly under control of external cues. Currently, identification of LRC or cells undergoing ACD-NRCC is done after fixation. Thus, there is a need for a technique that will afford us testing of the properties of live LRC and cells undergoing ACD-NRCC.
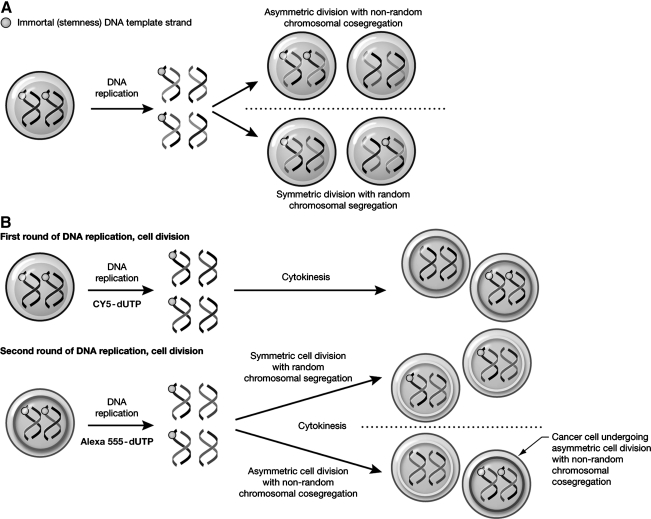
FIG. 1.
The immortal strand hypothesis and detection of ACD-NRCC by dual nucleotide-analogue DNA labeling. (A) The immortal strand hypothesis proposes that each chromosome in a stem-cell has 1 template-DNA strand that can be conserved after numerous asymmetric cell divisions (orange dot). These strands are preferentially cosegregated into daughter stem-cells by ACD-NRCC, whereas chromosomes that do not carry these strands segregate into daughter cells fated for differentiation. (B) To detect ACD-NRCC by dual nucleotide-analog DNA labeling procedure, cells are labeled sequentially for 2 cell cycles with 2 different nucleotides; first Cy5 (pseudo-colored green) then Alexa555 (red). When symmetric division occurs, both nuclei of the daughter cells will incorporate both nucleotides. However, when ACD-NRCC occurs, 1 nucleus will incorporate both Cy5 and Alexa555 (yellow), whereas the other nucleus will incorporate only Alexa555. ACD-NRCC, asymmetric cell division with nonrandom chromosomal cosegregation.
Here, we report on 2 novel techniques for the isolation of live LRC and cells undergoing ACD-NRCC. Using innovative, single-color, and dual-color nucleotide-DNA-labeling techniques, we isolated both live LRC and live cells undergoing ACD-NRCC from human liver cancer cell lines and confirmed the results using confocal microscopy with 3-dimensional (3D) surface rendering. Potentially, these techniques have implications for both cancer-stem-cells and regenerative medicine research.
Methods
Technique for dual-color nucleotide labeling in live cells (ACD-NRCC)
Cells were plated in antibiotic-free serum (AFS) media, washed, and incubated in 0.5 μM carboxyfluorescein diacetate succinimidyl ester (CFSE). After AFS media incubation, cells were microporated to label with 100 μM unlabeled dUTP or Cy5-dUTP. After 1 cell cycle (35 h), the first fluorescence-activated cell sorting (FACS) was performed. The 50%-CFSE+/Cy5+ sorted cells (99% purity) were plated and 22 h later, they were harvested for the second round of dUTP- labeling with Alexa-555-dUTP. The cells were microporated to label with 100 μM unlabeled dUTP or Alexa-555-dUTP. The final FACS sort was performed after completion of the second cell division. Two groups were isolated: 25%-CFSE+/Alexa555+ cells (Alexa555+ purity of 98%) and 25%-CFSE+/Alexa555+/Cy5+ cells (Alexa555+ purity of 97% and a Cy5+ purity of 95%). After the final FACS sort, the cells were prepared for confocal microscopy.
Technique for single-color nucleotide DNA labeling in live cells (LRC)
Cells were labeled with 100 μM Cy5-dUTP by microporation. After the first cell cycle, Cy5+ cells were sorted by FACS. A population of Cy5+ high cells (>55% Cy5+) was sorted to >99% purity. Cells were propagated in culture for 6 and 8 cell cycles. After completion of the sixth and eigth cell cycle, the cells were then sorted again for Cy5+(LRC) and Cy5- control cells. The cells were prepared for confocal microscopy.
Microporation
Extensive experiments for optimization of fluorophore labeled-dUTP incorporation via microporation were conducted using the MicroPorator MP-100 (BTX-Harvard Apparatus). Manufacturing guidelines were followed using both the 10 μL and 100 μL tip kits. Various instrument settings, cell concentrations, and labeled-dUTP concentrations were optimized for the HCC cell line. The selected dUTPs used were unlabeled dUTP (GE Healthcase/Amersham 28406542), Cy5-dUTP (GE Healthcare/Amersham PA55032), and Alexa Fluor 555-dUTP (Invitrogen A32762). Cells were plated in AFS media for 22 h before harvesting for microporation. Before microporation, cells were trypsinized, washed in Dulbecco's phosphate buffered saline (DPBS), and resuspended in R buffer at a concentration of 1.5e5 cells per 10 μL for the 10 μL tips and 5e6 cells per 100 μL for the 100 μL tips. All dUTPs were used at a final concentration of 100 mM. Cells were loaded into 10 μL or 100 μL tips and placed into the microporation chamber containing 3 mL of microporation buffer. The cells were microporated at 1400 V for 20 ms and 2 pulsations. Then, they were immediately plated in AFS media for culture at 37°C.
Fluorescence confocal microscopy
After microporation and FACS sorting, cells were plated in 8-well chamber slides and fixed immediately (Ibidi 80822). Cells were plated at every stage of the experiment to validate the nature of the cells sorted by FACS. Cells were washed with DPBS and fixed with 4% PFA for 15 min at room temperature. The cells were washed with DPBS and incubated at 37°C for 1 h. Several drops of Vectashield/DAPI stain (Vector Laboratories H-1200) were placed in each chamber and then stored at 4°C before confocal images were acquired. Confocal images were sequentially acquired with Zeiss AIM software on a Zeiss LSM 510 Confocal system (Carl Zeiss Inc.) with a Zeiss Axiovert 100M inverted microscope and 50 mW argon UV laser tuned to 364 nm, a 25 mW Argon visible laser tuned to 488 nm, a 1 mW HeNe laser tuned to 543 nm, and a 5 mW HeNe laser tuned to 633 nm. A 63× Plan-Apochromat 1.4 NA oil immersion objective was used at digital zoom settings of 1 or 2. Emission signals after sequential excitation of DAPI (blue), FITC (pseudo-colored white), Alexa-Fluor 555 (red), and Alexa Fluor 568 (pseudo-colored green) by 364 nm, 488 nm, 543 nm, or 633 nm lazer lines were collected with a BP 385-470 filter, BP 505-550 filter, LP 560 filter, or LP 650 filter, respectively, using individual photomultipliers. Z-stacks consisted of 30 to 50 slices at 0.38 μm intervals, and these stacks were used with Bitplane's (Zurich) Imaris software (v6.0) for surface rendering. In some cases, a cutting plane was used to expose internal surfaces or outer surfaces were made transparent. 3D video imaging can be seen for a symmetrically dividing Alexa555+/Cy5+ cell (Supplementary Online Video S1), an asymmetrically dividing Alexa555+/Cy5+ and Alexa555+ cell (Supplementary Online Video S2), and an asymmetrically dividing Alexa555+/Cy5+ and Cy5+ cell (Supplementary Online Video S3).
More detailed methods are described in Supplementary Methods (Supplementary Data, available online at www.liebertonline.com/scd).
Results
Detection of cells undergoing ACD-NRCC: technique for dual-color nucleotide labeling in live cells
Based on work reported by others [10,15,18,19,21,25,26,28], we reasoned that to detect ACD-NRCC it is necessary to follow exactly 2 complete cell cycles (Fig. 1B). In order to guarantee that 2 cell divisions have occurred, measuring the doubling-times of the tested cells is paramount. Following various culture conditions to match our new technique, the doubling-times of the first liver cancer cell line tested (PLC/PRF/5) was approximately 35 h (Supplementary Fig. S1). Cell division was monitored using the cytoplasmic stain CFSE (0.5 μM) and started before the first round of DNA replication. The cytoplasmic dye-protein complexes within the CFSE-labeled cells are retained by the cells throughout mitosis and then passed onto daughter cells after each division at a fixed ratio (Supplementary Fig. S2). We used the CFSE to monitor and follow exactly 2 cell divisions.
To identify ACD-NRCC, cells were labeled sequentially with 2 different fluorophore-labeled nucleotides before each round of replication: Cy5-dUTP (pseudo-colored green) then Alexa-fluor 555-dUTP (red) (Fig. 2). The optimal time for incorporation of nucleotides is when cells are predominately in the G1-S phase that was found to be approximately 22 h after initial plating (Supplementary Fig. S3). Twelve hours after plating, human liver cancer cells (PLC/PRF/5) were stained with 0.5 μM CFSE (100% CFSE, Fig 2B). The cells were given ample time to recover before transfection with the initial fluorophore-labeled dUTP, Cy5-dUTP (100 μM), using microporation (Fig. 2C). After 1 complete cell cycle (35 h), the initial FACS sort was performed (Fig. 2D–F).
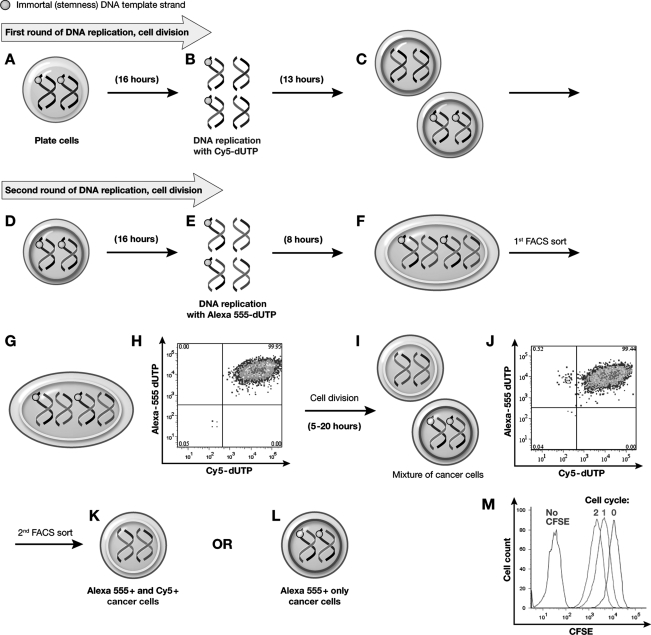
FIG. 2.
Isolation of live cells undergoing ACD-NRCC. After the schema described in Fig. 1B, for the first cell cycle of DNA replication, liver cancer cells were plated (A), stained with CFSE (B), and labeled with Cy5 (C). Subsequently, cells that underwent exactly 1 cell division (50% CFSE; D, E) and were Cy5+ High (F) were sorted by FACS with 99% purity (G, H). For the second round of DNA replication, cells were labeled with Alexa555 (I). After completion of the second cycle of DNA replication (J), cells that underwent exactly 2 cell divisions (25% CFSE) were sorted (K) and were Alexa555+ high and Cy5 high (L, M). Among these cells, 2 populations were identified and sorted: 25% CFSE+/Alexa555+ high cells (N, O) and 25% CFSE+/Alexa555+/Cy5+ high cells (P–R). The 25% CFSE+/Alexa555+ subpopulation represents cells that were generated by ACD-NRCC. The 25% CFSE+/Alexa555+/Cy5+ subpopulation represents the cells in which random chromosomal segregation has occurred (symmetric division). CFSE, carboxyfluorescein diacetate succinimidyl ester; FACS, fluorescence-activated cell sorting.
The aims of the first sort was to isolate cells that have completed exactly 1 cell division (containing 50% CFSE) and were labeled with Cy5-dUTP (Cy5+ high, Fig. 2G, H). To detect ACD-NRCC, it is imperative to start the second cycle of DNA replication with cells that are labeled with Cy5-dUTP (Cy5+ high) and that these cells underwent exactly 1 cell cycle (50% CFSE). Cell-cycle control cells were stained with 0.5 μM CFSE to provide the 100% CFSE controls for the FACS sorting. Cell viability was determined using light scatter properties yielding greater than 90% viability after the initial CFSE/Cy5+ high FACS sort. At all times, we gated, sorted, and counted only viable cells. Cells were then gated based on 50% CFSE-staining (cells that completed 1 cell cycle), which accounted for 94% of the viable cells (Fig. 2D). After Cy5-labeling, 95% of the cells were Cy5+ when compared with the unlabeled-dUTP controls (Fig. 2F); however, to ensure the best post-sort purity and to include only cells that incorporated efficiently Cy5 into their replicating DNA, the gating was set using only Cy5-high cells, those cells which were above the 55% percentile Cy5+ (Fig. 2F). After each sort, we ran the samples again through the FACS machine to ascertain purity. Testing the post sort cells (post-50% CFSE+/Cy5+ high) for purity resulted in >99% purity of Cy5+ high cells that underwent only 1 cell division (Fig. 2H).
Cells from the previous sort that is, cells that underwent only 1 cell division while incorporating Cy5 (post-50% CFSE+/Cy5) and were >99% pure were taken into the second cycle of DNA replication. The aim of the final sort was to isolate cells that completed exactly 2 cell divisions (containing 25% CFSE) and were labeled with Alexa-fluor 555-dUTP only (Alexa555+ high) or labeled with both Cy5-dUTP (Cy5+) and Alexa-fluor 555-dUTP (Alexa555+Cy5+ cells, Fig. 2I–K). Cells that underwent ACD-NRCC are expected to be Alexa555+ high cells, and cells that underwent symmetric division are expected to be Alexa555+/Cy5+ cells. Cell division controls from the initial sort, which were cultured in parallel (100% CFSE-staining initially), were now used as the new controls for the final sort in order to set the gating for 50% CFSE (1 cell division) and 25% CFSE (2 cell divisions). The presort population after the second round of fluorophore-labeling with Alexa-fluor 555-dUTP had viability of greater than 90%. For the final sort, we gated only viable cells. Cells were then gated based on 25% CFSE-staining, which accounted for >95% of the viable cells (Fig. 2J). During the second cycle of DNA replication, we used 3 fluorophores; therefore, we had to adjust the voltages accordingly. This resulted in a shift of the CFSE profile, but ratios were strictly maintained relative to the controls that is, the correct ratio of CFSE as an indicator of cell division was maintained. At all times, only viable cells were sorted and counted. After the Alexa555-labeling and the second cell cycle, >85% of the cells were Alexa555+ high (Fig. 2L) and >65% were also Cy5+ high (Fig. 2M) when compared with the unlabeled-dUTP controls. Only Cy5+ high and Alexa555+ high cells were selected. Two select populations were then sorted to yield cells that underwent ACD-NRCC (25% CFSE/Alexa555+ high, Fig 2N, O) and cells that underwent symmetric cell division (25% CFSE/Alexa555+/Cy5+, Fig. 2P–R). The post-sort analysis for purity demonstrated >98% Alexa555+ high purity in the 25% CFSE/Alexa555+ population, and 95% Cy5+ purity and 97% Alexa555+ purity in the 25% CFSE/Alexa555+/Cy5+ population. Purity was determined after re-running the sorted samples through FACS.
To further ascertain these results, we wanted to use a second method to validate the results just described (Fig. 3). We wanted to test whether cells that are DNA labeled with both Alexa555+ and Cy5+ before the end of the second cycle of DNA replication, before cytokinesis, will complete their cell division into daughter cells that are 25% CFSE/Alexa555+/Cy5+ and daughter cells that are 25% CFSE/Alexa555+ high (Fig. 3). In essence, we wanted to take “yellow cells” (Alexa555+/Cy5+ cells) during mitosis (Fig. 3F) and follow them through completion of mitosis, asking the question whether they will yield “red cells” (Alexa555+ high cells) and “yellow cells” (Alexa555+/Cy5+ cells), indicating that these cells underwent ACD-NRCC. Detecting such cells by this second method will build on the results demonstrated by the first method (described above, Fig. 2) and will lend further credence to the notion that these cells underwent ACD-NRCC in this experimental system. Therefore, we followed a similar labeling scheme as just described (Fig. 2) with modifications (Fig. 3). In brief, cells were labeled with Cy5-dUTP for the first cell cycle (Fig 3A–D), and subsequently with Alexa-555-dUTP for the second cycle of DNA replication (Fig. 3D, E). Then, only Alexa555+/Cy5+ cells were sorted during mitosis and before completion of cytokinesis of the second cell cycle (Fig. 3F). To ascertain that only cells at the beginning of mitosis and before cytokinesis were sorted, we used strict time table based on previous cell cycle studies (Supplementary Figs. S1 and S3). In addition, we sorted only single cells that were 50% CFSE and carefully sorted out the doublets (Fig. 3G, H). The purity of the 50% CFSE/Alexa555+/Cy5+ cells was >99%. These cells were then allowed to complete cytokinesis in culture (Fig. 3I, J). After cytokinesis, we tested the whole cell population for CFSE, Alexa555+, and Cy5+. We found that the >99% pure 50% CFSE/Alexa555+/Cy5+ cells (Fig. 3G, H) yielded cells that were 25% CFSE/Alexa555+ high only (Fig. 3L) and cells that were 25% CFSE/Alexa555+/Cy5+ only (Fig. 3K). The FACS dot plots illustrate that the initial population of cells sorted that is, >99% pure 50% CFSE/Alexa555+/Cy5+ cells before cytokinesis (Fig. 3H) shifted into 2 distinct populations: 25% CFSE/Alexa555+ only cells and 25% CFSE/Alexa555+/Cy5+ only cells indicating that ACD-NRCC did occur (Fig. 3J).
FIG. 3.
Isolation of live cells undergoing ACD-NRCC (second method). For the first cell cycle of DNA replication, liver cancer cells were plated (A), labeled with Cy5 (B), and were let to complete the first cell division in culture (C). Subsequently, cells completing the first round of DNA replication were labeled with Alexa555 (D,E). Before completion of the second round of DNA replication (F), cells that were Alexa555-high/Cy5-high and undergoing mitosis were sorted by FACS and placed in a culture to complete mitosis (G, H). At the completion of mitosis of the second round of DNA replication (I), cultured cells that underwent 2 cell divisions (25% CFSE) were analyzed and subsequently sorted (J). The cultured cells that previously contained only Alexa555-high/Cy5-high cells now generated 2 populations of cells: CFSE-25%/Alexa555-high/Cy5-high cells generated by symmetric division (K), and CFSE25%/Alexa555-high only cells generated by ACD-NRCC (L). CFSE was used to detect contemporaneously cell division status (M).
Detecting 25%-CFSE/Alexa555+ cells and 25%-CFSE/Alexa555+/Cy5+ cells by FACS alone is not sufficient. We wanted to validate that what we see in the dot plots generated by FACS can be also seen in 3D confocal microscopy. Another important aspect of validating the FACS results using 3D confocal microscopy was to confirm that labeling occurred only in the nucleus and not in the cytoplasm. After each stage of the DNA labeling and sorting in parallel with the procedure to detect cells undergoing either symmetric division or ACD-NRCC as shown in Fig. 2, aliquots of cells were sorted into slides, fixed, and examined by confocal microscopy. The post-sort 50% CFSE/Cy5+ population was seen with cytoplasmic CFSE-staining (pseudo-colored white), nuclear DAPI-staining (blue), and Cy5-labeled nucleotides (pseudo-colored green) (Fig. 4A). After the second round of fluorophore-labeling with Alexa555-dUTP and a second cell division, 2 subpopulations were sorted, cells that underwent symmetric division, that is, 25%-CFSE+ (pseudo-colored white)/Cy5+ (pseudo-colored green)/Alexa555+ (red) cells (Fig. 4B) and cells that underwent ACD-NRCC 25%-CFSE+ (pseudo-colored white)/Alexa555+ (red) cells (Fig. 4C). Confocal microscopy demonstrated that all fluoro-nucleotides were incorporated within the nucleus and none in the cytoplasm. To further validate that cells underwent either symmetric cell division or ACD-NRCC, cell couplets were sorted and fixed during the second labeling procedure. As a result, cells arrested at cytokinesis were observed: Cells arrested during symmetric cell division in which 2 nuclei labeled with both fluorophores (pseudo-colored green and red, Fig. 4D) in the same cytoplasmic space, ACD-NRCC in which 1 nucleus containing Alexa555+ (red) only labeled DNA, and the other nucleus containing both labeled-nucleotides Cy5+ (pseudo-colored green)/Alexa555+ (red) (Fig. 4E). Figure 4F illustrates the same cell from Fig. 4E, stripped of the DNA labels, demonstrating 2 nuclei within the same cytoplasmic space, during division, the division furrow is indicated by the white arrow; the yellow arrow shows that the 2 nuclei did not complete full division yet and are still connected by a nuclear bridge (blue DAPI). To further validate these results and ascertain that these nuclei are found within a single cytoplasmic space without intervening membrane, 3D reconstruction movies from Fig. 4D (Supplementary Online Video 1) and Fig. 4E, F (Supplementary Online Video 2). An additional asymmetrically dividing cell with reverse color labeling (Supplementary Online Video 3) demonstrates no intervening cell membrane between the 2 nuclei, thus indicating localization of both nuclei within the same cytoplasmic space during cytokinesis. Statistically, we performed 9 separate experiments and tested for the 2-tailed P value for the exact binomial test. The resulting was P=0.0001, indicating that our observations were statistically significant. For full statistical analysis and actual numbers of cells sorted, please see Supplementary Methods-Statistics.
FIG. 4.
Three-dimensional fluorescent confocal microscopy capturing symmetric and asymmetric cell division in live cells. To confirm the results described in Fig. 2, we tested the products the cell sorting after each stage. (A) shows cells that were isolated by FACS after the first round of DNA replication, [50% CFSE+ (pseudo-colored white)/Cy5+ (pseudo-colored green) cells]. (B, C) shows the 2 populations of cells isolated after the second round of DNA replication: 25% CFSE+ (white)/Cy5+(green)/Alexa555+(red) cells (B) and cells that underwent ACD-NRCC 25% CFSE+(white)/Alexa555+(red) cells (C). (D) Live cells undergoing symmetric cell division containing 2 nuclei labeled with both nucleotides during mitosis (green and red). (E) Live cell undergoing ACD-NRCC is illustrated containing 1 nucleus with the second labeled nucleotides only (red) and the other nucleus containing both nucleotides (green and red). The DAPI (blue) reveals 2 nuclei in the same cytoplasmic space (white) without an intervening membrane. (F) Shows the same cell from E during mitosis where the nucleotide labeling was subtracted to demonstrate 2 nuclei (DAPI-blue) halted at cytokinesis, still connected by a small nuclear bridge (yellow arrow), as division was halted and in the same cytoplasmic space. The white arrow shows the furrow as the cell begins to divide. Three-dimensional reconstruction movies from D (Supplementary Video S1) and E, F (Supplementary Video S2) further demonstrate these phenomena.
Isolation of LRC: technique for single-color nucleotide DNA labeling in live cells
Previous studies [6–9,14–17,19,21,29–31] demonstrated that LRC can be detected after exposure to nucleotide analogues such as BrdU (pulse period) or 3H-thymidine with a subsequent chase period. However, the actual detection of LRC could be performed only on fixed cells. Thus, no direct functional assessment can be performed on these cells. To solve this problem, we explored the methodology just described with modifications utilizing microporation of fluorophore-labeled nucleotides in order to assess label retention in live cells. If cells are initially labeled with Cy5-dUTP during the first cell cycle, then a subpopulation of Cy5+ high labeled cells can be isolated using FACS. In an attempt to simplify the process, we chose to label only 1 strand of the replicating chromosome during 1 cell cycle (Fig. 5). In previous experiments (described above) using a similar method, we have shown that the Cy5+ high DNA labeling is very uniform (Fig. 4). Theoretically, if labeling is uniform and detection is very sensitive, and assuming that after each cell division the Cy5+ labeled DNA is diluted by approximately 50% (although, potentially, it might be more than 50% but unlikely to follow a complete Gaussian distribution), we calculated that potentially after 6 and 8 cell-cycles approximately 1.56% and 0.39% of the cells will be Cy5+ (Supplementary Fig. S4). These calculations do not take into account other biological process that might affect the theoretical considerations just described, and they are based on high degree of uniform label incorporation and highly sensitive detection system. They are used here only to illustrate the potential existence of LRC by normal cell division and not as a confirmation or rejection of the existence of LRC as a stem cell property.
FIG. 5.
Isolation of LRC. Liver cancer cells were plated (A) and microporated using Cy5 (pulse phase) (B). On completion of the first round of DNA replication, we sorted only Cy5+ high cells (C, D). Subsequently, cells that were 100% Cy5+ with 99% purity (E, F) were sorted by FACS and plated (G). For the chase phase, cells were proprogated in culture for 6 to 8 cell generations (H). At the completion of the chase phase, we detected and FACS sorted both LRC and non-LRC (I). LRC (Cy5+ high cells) comprised 5% of the total cell population (J, K). This is a statistically significant observation, P=0.016). We used the ImageStreamX Cytometer (Amnis Corporation) to validate these results (Fig. S6). LRC, live label-retaining-cells.
Human liver cancer cells (PLC/PRF/5) were microporated with unlabeled dUTP (100 μM) as a control, and with the labeled-nucleotide Cy5-dUTP (100 μM), Figure 5A and B. After 1 complete cell cycle (35 h, Fig. 5C and SI Fig. 1) the initial FACS sort was performed. Cell viability was greater than 95%; only viable cells were sorted and counted. In order to ensure the purest subpopulation of Cy5+ high cells, we sorted cells that were above the 50th percentile in terms of Cy5+ fluorescence or Cy5+ high cells (Fig. 5D). To test for purity, we re-ran the cells through FACS and determined that the initial sort of Cy5+ high cells was >99% pure (Fig. 5E, F). This Cy5+ high subpopulation was then propagated in culture for 6 and 8 cell cycles (Fig. 5G, H); FACS was used to sort the Cy5+ LRCs. Cell viability of the sorted LRC-(Cy5+) was greater than 95%. The LRC-(Cy5+) that were detected after 6 and 8 cell generations was 5.0% and 1.54% to 5.0%, respectively (Fig. 5J, K). These represent approximately 3-fold greater than the theoretical calculated values, respectively. Further, 95.35%±0.01% and 88.40%±0.08% of the LRC and the non-LRC expressed the Ki-67 antigen, indicating that LRC are as proliferative as non-LRC cells (Supplementary Fig. S5).
To further validate these results and ascertain that the LRC are indeed retaining DNA labels and that the non-LRC do not retain labels, we sorted both the LRC cells and the non-LRC onto slides and examined the cells using confocal microscopy. Supplementary Fig. S6 demonstrates that our method is highly accurate and that cells sorted as LRC indeed retain labels and cells sorted as non-LRC do not retain labels. Further, after using FACS to sort LRC and non-LRC, we validated the sorting results by testing 15,000 post FACS cells individually, each of LRC and non-LRC. We used Multi-spectral flow cytometry (ImageStreamX Cytometer, Amnis Corp) to examine LRC and non-LRC in real time as they go through the nozzle of the FACS machine. In Supplementary Fig. S7, we show individual cells, LRC all positive for Cy5 and non-LRC and all negative for Cy5 individually. Statistically, we performed 7 experiments in various cancers. We found that 1.54% to 5.0% of the recovered cells were LRCs. We used a 2-tailed Wilcoxon signed rank test with a P=0.016. Therefore, our findings are statistically significant, thus suggesting that the LRC could be the result of ACD-NRCC. For full statistical analysis and actual numbers of cells sorted, please see Supplemental Material-Statistics.
Discussion
LRCs and cells undergoing ACD-NRCCare proposed to be properties related to stem cells. Several investigators hypothesized that LRC represent a sub-population of cells that is highly enriched with adult tissue specific stem cells. However, the existence of LRC and cells undergoing ACD-NRCC as a specific property of stem cells is not fully accepted. This study exploits Cairn's hypothesis by demonstrating ACD-NRCC in human cancer cells [32]. To test the stem cell properties of LRC and cells undergoing ACD-NRCC, there is a need to isolate live LRC and/or live cells that underwent ACD-NRCC. The major obstacle to performing these tests is that thus far, detection of LRC and cells undergoing ACD-NRCC could only be done on fixed cells. These techniques lack the ability to isolate these cells and continue further analysis of the cells. Here we report on 2 new and related methods for the isolation of live LRC and live cells undergoing ACD-NRCC.
These methods involve either single-color (LRC) or dual-color (ACD-NRCC) nucleotide analog DNA labeling using microporation. Isolation of these 2 populations of cells was done using FACS. To validate the nature of the cellular species that were seen on the FACSs dot plots on a subcellular level, we used confocal microscopy. The validation studies have demonstrated that the fluorescent-labeled-nucleotides are localized only within the nucleus in a uniform fashion (other investigators reported patchy BrdU labeling). In this study, we might have underestimated the existence of LRC and cells undergoing ACD-NRCC, because we sorted the “high” fractions. The 2 methods described in this article provide an important new tool to study the stem cell properties of LRC and/or cells undergoing ACD-NRCC.
Identification and isolation of liver cancer cells that have undergone ACD-NRCC and/or LRC has potential implications for the study of putative cancer stem cells and in regenerative medicine. We utilized fundamental characteristics of stem-cells to develop a technique that can be used for the isolation of a live population of cells with functional stem cells traits from human liver cancer cell lines. The method reported here will afford investigators testing the stem-cells properties of LRC and cells undergoing ACD-NRCC. If LRC cells are confirmed to be indeed stem-cells (the stem-cell nature of LRC was tested in xenotransplantation experiments and confirmed by our group recently), it will provide a relatively simple method for the isolation of adult tissue specific stem cells and stem-like cells from various cancers. As we look ahead to future endeavors, we plan to capitalize on these methodologies in order to characterize the properties of these isolated live LRC. The isolation of live cells undergoing ACD-NRCC and/or LRC will enable investigators to study the mechanisms that drive ACD-NRCC. If in the future one will be able to specifically target the mechanism of ACD-NRCC, one could potentially inhibit cancer cells' ability to self-renew. As a result, tumor progression or recurrence might be irreversibly altered and have a significant impact on tumor progression. Our data provide evidence of 2 unique methodologies that support the existence of ACD-NRCC and in live human liver cancer cells and provide a technique to isolate cells undergoing ACD-NRCC. It has the potential to become an important tool both in regenerative medicine and cancer research.
Supplementary Material
Acknowledgments
We thank L. Liam and P. Mannan for the confocal microscopy and surface rendering work. A. Mixon and S. Farid for help with the FACS analysis experiments. This work was supported by an NCI intramural grant (1ZIABC011005-02).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Morrison SJ. Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 2.McCulloch EA. Till JE. Perspectives on the properties of stem cells. Nat Med. 2005;11:1026–1028. doi: 10.1038/nm1005-1026. [DOI] [PubMed] [Google Scholar]
- 3.Strain AJ. Crosby HA. Nijjar S. Kelly DA. Hubscher SG. Human liver-derived stem cells. Semin Liver Dis. 2003;23:373–384. doi: 10.1055/s-2004-815563. [DOI] [PubMed] [Google Scholar]
- 4.Visvader JE. Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 5.Goodell MA. Brose K. Paradis G. Conner AS. Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potten CS. Early and late incorporation of tritiated thymidine into skin cells and the presence of a long-lived G 0-specific precursor pool. J Cell Biol. 1971;51:855–861. doi: 10.1083/jcb.51.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bickenbach JR. Identification and behavior of label-retaining cells in oral mucosa and skin. J Dent Res. 1981;60(Spec No C):1611–1620. doi: 10.1177/002203458106000311011. [DOI] [PubMed] [Google Scholar]
- 8.Morris RJ. Potten CS. Slowly cycling (label-retaining) epidermal cells behave like clonogenic stem cells in vitro. Cell Prolif. 1994;27:279–289. doi: 10.1111/j.1365-2184.1994.tb01425.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhang HB. Ren CP. Yang XY. Wang L. Li H. Zhao M. Yang H. Yao KT. Identification of label-retaining cells in nasopharyngeal epithelia and nasopharyngeal carcinoma tissues. Histochem Cell Biol. 2007;127:347–354. doi: 10.1007/s00418-006-0251-9. [DOI] [PubMed] [Google Scholar]
- 10.Potten CS. Gandara R. Mahida YR. Loeffler M. Wright NA. The stem cells of small intestinal crypts: where are they? Cell Prolif. 2009;42:731–750. doi: 10.1111/j.1365-2184.2009.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F. Lu L. Lu J. Identification and location of label retaining cells in mouse liver. J Gastroenterol. 45:113–121. doi: 10.1007/s00535-009-0139-2. [DOI] [PubMed] [Google Scholar]
- 12.Oliver JA. Klinakis A. Cheema FH. Friedlander J. Sampogna RV. Martens TP. Liu C. Efstratiadis A. Al-Awqati Q. Proliferation and migration of label-retaining cells of the kidney papilla. J Am Soc Nephrol. 2009;20:2315–2327. doi: 10.1681/ASN.2008111203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 14.Lark KG. Consigli RA. Minocha HC. Segregation of sister chromatids in mammalian cells. Science. 1966;154:1202–1205. doi: 10.1126/science.154.3753.1202. [DOI] [PubMed] [Google Scholar]
- 15.Potten CS. Hume WJ. Reid P. Cairns J. The segregation of DNA in epithelial stem cells. Cell. 1978;15:899–906. doi: 10.1016/0092-8674(78)90274-x. [DOI] [PubMed] [Google Scholar]
- 16.Potten CS. Owen G. Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci. 2002;115:2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- 17.Booth BW. Boulanger CA. Smith GH. Selective segregation of DNA strands persists in long-label-retaining mammary cells during pregnancy. Breast Cancer Res. 2008;10:R90. doi: 10.1186/bcr2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merok JR. Lansita JA. Tunstead JR. Sherley JL. Cosegregation of chromosomes containing immortal DNA strands in cells that cycle with asymmetric stem cell kinetics. Cancer Res. 2002;62:6791–6795. [PubMed] [Google Scholar]
- 19.Karpowicz P. Morshead C. Kam A. Jervis E. Ramunas J. Cheng V. van der Kooy D. Support for the immortal strand hypothesis: neural stem cells partition DNA asymmetrically in vitro. J Cell Biol. 2005;170:721–732. doi: 10.1083/jcb.200502073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rambhatla L. Ram-Mohan S. Cheng JJ. Sherley JL. Immortal DNA strand cosegregation requires p53/IMPDH-dependent asymmetric self-renewal associated with adult stem cells. Cancer Res. 2005;65:3155–3161. doi: 10.1158/0008-5472.CAN-04-3161. [DOI] [PubMed] [Google Scholar]
- 21.Shinin V. Gayraud-Morel B. Gomes D. Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 22.Cairns J. Cancer and the immortal strand hypothesis. Genetics. 2006;174:1069–1072. doi: 10.1534/genetics.104.66886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rando TA. Stem cells, ageing and the quest for immortality. Nature. 2006;441:1080–1086. doi: 10.1038/nature04958. [DOI] [PubMed] [Google Scholar]
- 24.Rando TA. The immortal strand hypothesis: segregation and reconstruction. Cell. 2007;129:1239–1243. doi: 10.1016/j.cell.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 25.Conboy MJ. Karasov AO. Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 2007;5:e102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pine SR. Ryan BM. Varticovski L. Robles AI. Harris CC. Microenvironmental modulation of asymmetric cell division in human lung cancer cells. Proc Natl Acad Sci U S A. 107:2195–2200. doi: 10.1073/pnas.0909390107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lansdorp PM. Immortal strands? Give me a break. Cell. 2007;129:1244–1247. doi: 10.1016/j.cell.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 28.Kiel MJ. He S. Ashkenazi R. Gentry SN. Teta M. Kushner JA. Jackson TL. Morrison SJ. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449:238–242. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris RJ. Potten CS. Highly persistent label-retaining cells in the hair follicles of mice and their fate following induction of anagen. J Invest Dermatol. 1999;112:470–475. doi: 10.1046/j.1523-1747.1999.00537.x. [DOI] [PubMed] [Google Scholar]
- 30.Potten CS. Keratinocyte stem cells, label-retaining cells and possible genome protection mechanisms. J Investig Dermatol Symp Proc. 2004;9:183–195. doi: 10.1111/j.1087-0024.2004.09305.x. [DOI] [PubMed] [Google Scholar]
- 31.Bussard KM. Boulanger CA. Kittrell FS. Behbod F. Medina D. Smith GH. Immortalized, premalignant epithelial cell populations contain long-lived, label-retaining cells that asymmetrically divide and retain their template DNA. Breast Cancer Res. 12:R86. doi: 10.1186/bcr2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hart LS. El-Deiry WS. Invincible, but not invisible: imaging approaches toward in vivo detection of cancer stem cells. J Clin Oncol. 2008;26:2901–2910. doi: 10.1200/JCO.2008.16.9573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.