Abstract
Immunological memory is thought to depend upon a stem cell-like, self-renewing population of lymphocytes capable of differentiating into effector cells in response to antigen re-exposure. Here we describe a long-lived human memory T-cell population that displays enhanced self-renewal and multipotent capacity to derive central memory, effector memory and effector T cells. These cells, specific for multiple viral and self-tumor antigens, were found within a CD45RO−, CCR7+, CD45RA+, CD62L+, CD27+, CD28+ and IL-7Rα+ T-cell compartment characteristic of naïve T cells. However, they expressed increased levels of CD95, IL-2Rβ, CXCR3, and LFA-1, and exhibited numerous functional attributes distinctive of memory cells. Compared to known memory populations, these lymphocytes displayed increased proliferative capacity, more efficiently reconstituted immunodeficient hosts and mediated superior anti-tumor responses in a humanized mouse model. The identification of a human stem cell-like memory T-cell population is of direct relevance to the design of vaccines and T-cell therapies.
Long-lived self-renewing memory lymphocytes are a hallmark feature of the adaptive immune system in response to pathogens and tumors1–3. Analogous to organ systems in which non-replicating, terminally-differentiated cells are continually replenished by the progeny of less differentiated stem cells, it has been suggested that memory cells might contain stem cell-like cells4,5. Indeed, several characteristics of stem cells can be found to certain degrees in memory B and T cells, including selective transcriptional profiles6, the capacity to self-renewal and the multipotency to differentiate into progeny with diverse fates4,5.
The memory T-cell compartment is heterogeneous and has been conventionally divided into two subsets based on the expression of the lymph node homing molecules CD62L and CCR77. Central memory T cells (TCM) express high levels of CD62L and CCR7 and were thought to be the stem cell-like memory subset, whereas CD62L− CCR7− effector memory T cells (TEM) are considered committed progenitor cells that undergo terminal differentiation after a limited number of divisions4,5. The identification in mice of a novel population of memory T cells with enhanced stem cell-like qualities compared to conventional TCM cells adds complexity to this dichotomous view8,9. These memory T cells, which were designated memory stem cells (TSCM), exhibit a CD44low CD62Lhigh phenotype like naïve T cells (TN), but co-express stem cell antigen–1 (Sca-1) and high levels of the antiapoptotic molecule B cell lymphoma 2 (Bcl-2), the β chain of the IL-2 and IL-15 receptor (IL2-Rβ), and the chemokine (C-X-C motif) receptor CXCR38,9. Whether a similar memory T-cell population exists in human is currently under intensive investigation10.
A human CD8+ memory T-cell population has been described that shares phenotypic and functional characteristics with hematopoietic stem cells including the expression of the stem cell marker C-KIT and the ability to efflux cellular toxins through the ATP-binding cassette (ABC)–superfamily multidrug efflux protein ABCB111. However, recent data revealed that these cells are predominantly Vα7.2+ mucosal associated invariant T cells (MAIT)12. More recently, Schenkel et al.13 speculated that CD4dimCD8bright T cells expressing high levels of β-catenin, a molecule associated with the generation of mouse TSCM9,14, represent human TSCM, but the definitive identification of human TSCM remains to be accomplished.
Identification of human T memory stem cells
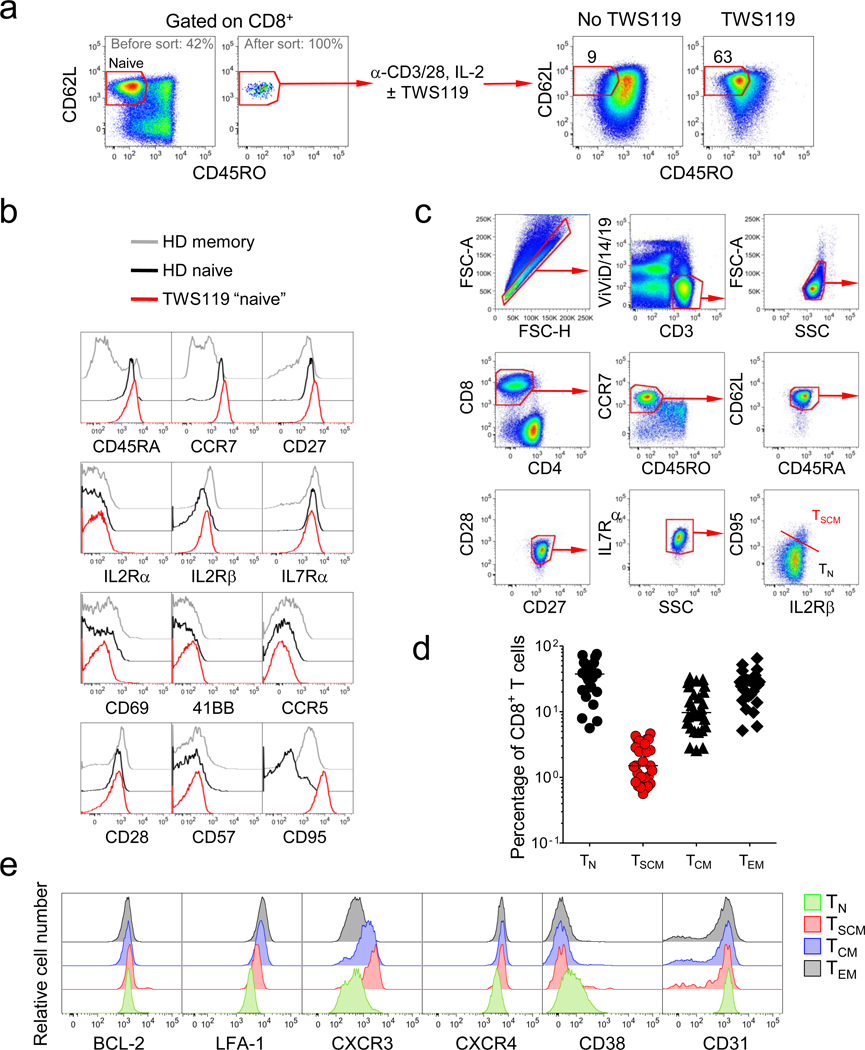
We previously found that mouse TSCM can be generated effectively in vitro by triggering Wnt signaling during T cell priming using Wnt3A or inhibitors of glycogen synthase kinase-3β (GSK-3β)9,14,15. We sought to employ the same strategy to generate candidate human TSCM by activating CD45RO−CD62L+ naïve CD8+ T cells in the presence of the GSK-3β inhibitor TWS119 (Fig. 1a). After two weeks, the majority of T cells cultured with TWS119 retained a CD45RO−CD62L+ naïve-like phenotype, whereas in the absence of GSK-3β inhibition, T cells uniformly upregulated the memory marker CD45RO (Fig. 1a). To determine whether the CD45RO−CD62L+ T cells generated in the presence of TWS119 were truly naïve cells or had acquired memory traits, we performed an extensive phenotypic analysis using established markers of T-cell activation and differentiation (Fig. 1b)16. The vast majority of molecules (CD45RA, CCR7, CD27, IL-2Rα, IL-7Rα, CD69, 41BB, CCR5 and CD57) showed a similar expression pattern between TN and TWS119-generated naïve-like T cells (Fig. 1b). However, the naïve-like T cells expressed levels of CD95 and IL-2Rβ similar to conventional memory T cells (Fig. 1b). Thus, we hypothesized that the expression of CD95 and IL-2Rβ on otherwise phenotypically naïve T cells could identify human TSCM cells.
Figure 1. Identification of TSCM cells in human blood.
a, Flow cytometry analysis of sorted human CD45RO−CD62L+ naïve CD8+ T cells prior to and 14 days after stimulation with α-CD3/CD2/CD28-coated beads and IL-2 in the presence or absence of 5µM TWS119. Numbers indicate the percentage of cells in the CD45RO−CD62L+ gate. b, Flow cytometry analysis of TWS119-generated CD45RO−CD62L+ naïve-like CD8+ T cells overlaid with CD45RO−CD62L+ naïve and memory (non-CD45RO− CD62L+) cells from a healthy donor (HD). c, Flow cytometry analysis of PBMC from a healthy donor. Dot plots show the gating strategy to identify CD95+, IL2Rβ+ TSCM cells. d, Percentages of circulating CD8+ T-cell subsets in 29 healthy donors. e, Flow cytometry analysis of PBMC from a representative healthy donor. Overlaid histogram plots show expression levels of a given molecule in different CD8+ T-cell subsets. CD8+ T-cell subsets were defined as follows: TN cells, CD3+CD8+CD45RO−CCR7+CD45RA+CD62L+CD27+CD28+IL7Rα+CD95−; TSCM cells, CD3+CD8+CD45RO−CCR7+CD45RA+CD62L+CD27+CD28+IL7Rα+CD95+; TCM cells, CD3+CD8+CD45RO+CD45RA− CCR7+CD62L+; TEM cells, CD3+CD8+CD45RO+CD45RA−CCR7−CD62L−.
To determine if candidate TSCM cells occur naturally we used polychromatic flow cytometry (PFC)17. Based on previous data17, we employed a highly stringent criterion of 7 markers to accurately define TN. Strikingly, a CD95+IL-2Rβ+ subset could be found in CD45RO−CCR7+CD45RA+CD62L+CD27+CD28+IL-7Rα+ naïve-like CD8+ (Fig. 1c) and CD4+ (Supplementary Fig. 1a) T cells. In 29 healthy donors, these cells, referred to hereafter as TSCM cells, represented about 2–3% of all circulating CD8+ and CD4+ T lymphocytes (Fig. 1d and Supplementary Fig. 1b). Further phenotypic analysis of T-cell differentiation markers revealed that TSCM also expressed higher levels of BCL-2, LFA-1, CXCR3, CXCR4, and lower levels of CD38 and CD31 compared to TN cells (Fig. 1e and Supplementary Fig. 1c). Notably, TSCM cells expressed all of the core phenotypic markers (IL-2Rβ, BCL-2 and CXCR3) of their mouse counterparts8,9, except SCA-1 which does not have a human ortholog. These T cells were phenotypically different from those described by Turtle et al.11 and were not MAIT12 (Supplementary Fig. 2). Similar to conventional memory, TSCM cells were detected at low frequencies (<1%) in cord blood (Supplementary Fig. 3). The phenotype of TSCM cells indicates a tropism for lymphatic tissues, but full anatomical characterization of TSCM-cell niches remains to be addressed.
TSCM possess attributes of conventional memory T cells
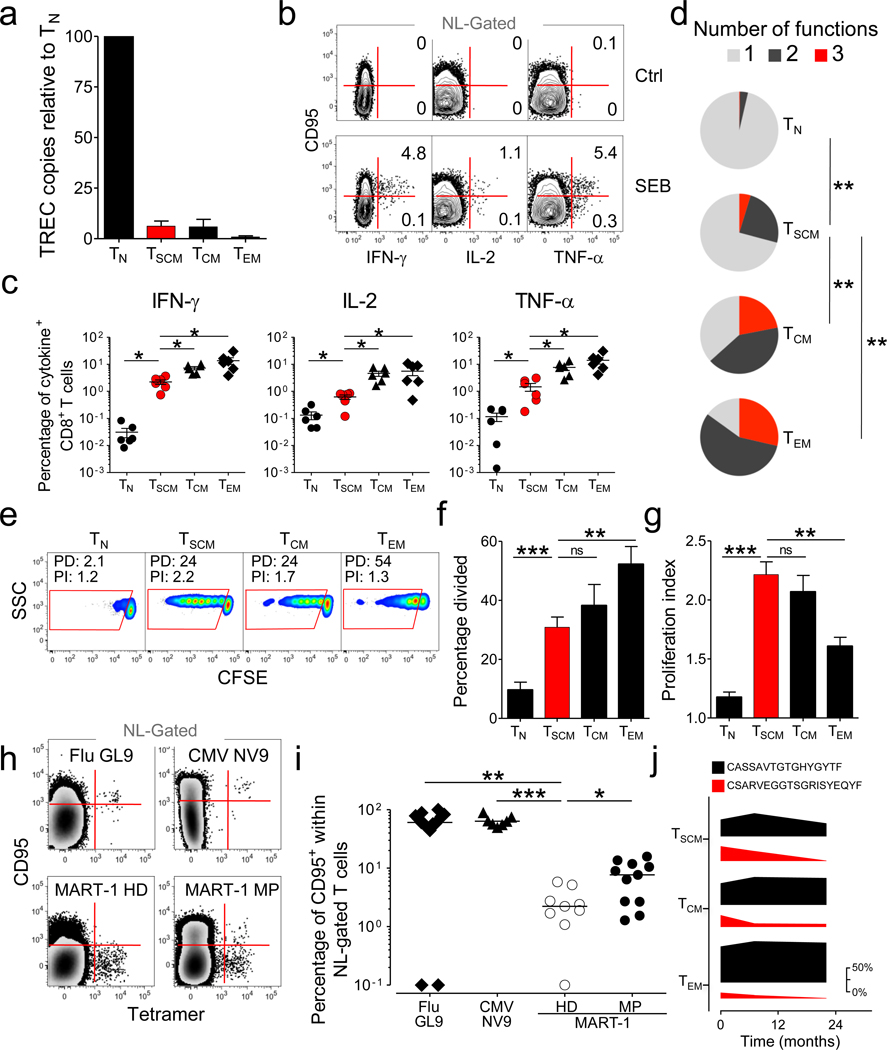
Due to the concomitant expression of numerous markers of naiveté as well as key molecules of memory differentiation, it remained unclear whether TSCM cells were functionally naïve or memory T cells. Naive T cells have high levels of TCR rearrangement excision circles (TREC), which are diluted during clonal proliferation18. Like TCM and TEM cells, we found that TSCM cells had low levels of TREC, indicating that they had undergone several rounds of division (Fig. 2a).
Figure 2. TSCM cells possess attributes of conventional memory T cells.
a, TCR excision circle (TREC) copy number in sorted CD8+ T-cell subsets relative to TN cells. Data are represented as mean ± s.e.m. of 4 donors. b, Intracellular cytokine staining of PBMC from a representative healthy donor after stimulation with SEB. Graphs show naïve-like (NL) gated T cells. NL, CD45RO−CCR7+CD45RA+CD27+CD28+. Numbers represent the percentage of CD95+ (TSCM) and CD95− (TN) cells producing a single cytokine. c, Percentages of CD8+ T-cell subsets producing IFN-γ, IL-2 and TNF-α in 6 healthy donors (obtained as described in panel b). d, Pie charts depicting the quality of the cytokine response in CD8+ T-cell subsets in 6 healthy donors as determined by the Boolean combination of gates identifying IFN-β+, IL-2+ and TNF-α+ cells. e, CFSE dilution in sorted CD8+ T-cell subsets after stimulation with 25 ng ml−1 of IL-15 for 10 days. Data are shown after gating on CD8+ cells. PD: percentage divided; PI: proliferation index. f, Percentage divided cells and g, Proliferation index of different CD8+ T-cell subsets after stimulation as in panel e. Data are represented as means ± s.e.m of 9 donors. h, Flow cytometry analysis of PBMC from HLA-A2+ donors. Graphs show tetramer-binding cells vs. CD95 expression in the NL (CD45RO−CCR7+CD45RA+CD27+IL7Rα+) gate. i, Percentage of tetramer-binding cells expressing CD95 in the NL gate determined as in panel h. Data represent the donors tested for tetramer specificity. HD: healthy donor; MP: melanoma patient. j, Frequency of two immunodominant CMV-specific TCRβ clonotypes relative to all CMV-specific TCRβ clonotypes in pp65 -specific T-cell subsets isolated over a period of 22 months from a representative donor. The figure legend shows the CDR3β amino acid sequences. Changes in the frequencies of immunodominat clonotypes are depicted as the thickness of the bars, with the magnitude scale shown on the right. * = P < 0.05; ** = P < 0.01; *** = P < 0.001; ns= not significant (t test, c,f,g,I and χ2 permutation test, d).
Memory T cells can also be distinguished from TN by their ability to rapidly acquire effector functions upon antigen rechallenge19. We found that within 4h after exposure to Staphylococcus Enterotoxin B (SEB), a significant fraction of CD95+ naïve-like CD8+ T cells produced IFN-γ, IL-2 and TNF-αs while TN cells remained relatively quiescent (Fig. 2b,c). Thus, TSCM cells rapidly acquired effector functions following superantigen stimulation like conventional memory T cells (Supplementary Fig. 4). Interestingly, the fraction of responding cells, as well as T-cell polyfunctionality, progressively increased from TN cells →TSCM cells →TCM cells →TEM cells (Fig. 2c,d), consistent with the hypothesis that TSCM cells are the least differentiated subset. Similar findings were observed for CD4+ T cells (Supplementary Fig. 1d and e). The rapid responsiveness of TSCM cells was also observed after polyclonal stimulation with α-CD3/CD2/CD28 beads (Supplementary Fig. 5). Consistent with the intracellular cytokine staining result, sorted TSCM cells, but not TN cells, secreted IFN-γ, IL-2 and TNF-α in response to α-CD3/CD2/CD28 stimulation (Supplementary Fig. 6). Thus, TSCM possess the memory capability of rapid acquisition of effector functions following TCR stimulation.
Unlike TN, memory T cells undergo robust proliferation in the presence of the homeostatic cytokines IL-15 and IL-720–22. We found that, similar to conventional CD8+ memory T cells, TSCM cells divided extensively in response to IL-15 (Fig. 2e). While the majority of TEM cells proliferated (Fig. 2f), they underwent fewer divisions, revealing a lower proliferative potential compared to other memory subsets (Fig. 2g). By contrast, TSCM cells underwent numerous cell divisions (Fig. 2g), although a greater fraction of these cells remained undivided (Fig. 2f). This behavior is reminiscent of stem cells, which are quiescent but can give rise to progeny capable of extensive proliferation and differentiation. Similar findings were observed in the CD4+ T-cell compartment in response to IL-7 (Supplementary Fig. 1f–h). Thus, TSCM have the replicative history and ability to respond rapidly to antigenic and homeostatic stimuli, characteristics of memory T cells.
The frequency of naïve CD8+ T-cell precursors for a given epitope has been estimated to be between 6 × 10−7 and 5 × 10−6, a range below the limit of peptide-MHC class I (pMHCI) tetramer detection23. We reasoned that if we could measure tetramer-binding, naïve-like T cells, they would be enriched in the CD95+ TSCM-cell compartment. In donors with detectable naïve-like CD8+ T cells specific for influenza or cytomegalovirus (CMV) epitopes, the vast majority of tetramer-binding cells expressed high levels of CD95 (Fig. 2h,i and Supplementary Fig. 7). By contrast, virtually all MART-1-specific naïve-like T cells in healthy donors did not express CD95, indicating that these cells were truly naïve (Fig. 2h,i and Supplementary Fig. 7). Notably, a significant fraction of MART-1-specific CD8+ T cells displayed a CD95+ phenotype in 7/11 patients with metastatic melanoma (Fig. 2h,i and Supplementary Fig. 7). Thus, tetramer-binding T cells found in the “naive-like” T-cell compartment could be derived from either increased thymic output (CD95−), as reported for MART-1 in healthy donors24, or from antigenic encounter, expansion and differentiation (CD95+). These experiments also revealed that TSCM represented a substantial fraction of the corresponding total antigen-specific CD8+ T-cell memory responses, averaging 0.6% for CMV pp65495–503, 4.2% for influenza M158–66 and 7.6% for MART-126–35, and that their frequency tended to correlate with that of conventional memory T cells (Supplementary Fig. 8).
To determine whether TSCM clonotypes represent a long-lived population or merely recently activated cells transitioning from a naïve to a conventional memory state, we analyzed TCRβ sequences of CMV-specific T-cell subsets from the same donor spanning a time period of 22 months. Like conventional memory T cells, we found dominant persisting clonotypes in TSCM cells, indicating that they represent a stable memory T-cell population (Fig. 2j and Supplementary Table 1). These findings demonstrate that TSCM cells are long-lived memory T cells with multiple viral and self–tumor specificities.
TSCM cells represent the least differentiated T-cell memory subset
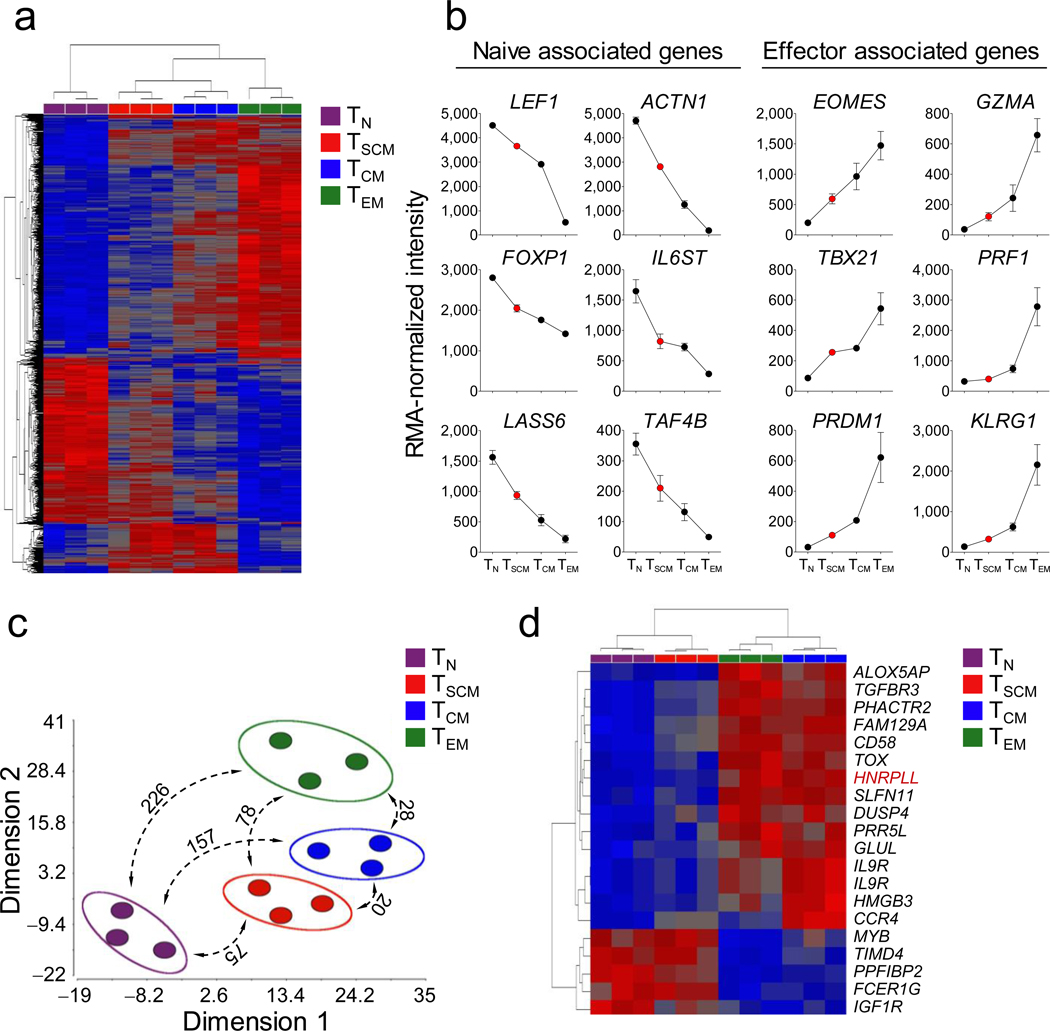
We sought to compare the transcriptome of TSCM cells with naive and conventional memory T-cell subsets and validate key findings with PFC (Fig. 3 and Supplementary Fig. 9). Nine hundred genes were differentially expressed among the four CD8+ T-cell subsets (P < 0.01, FDR < 5%) (Supplementary Table 2). Unsupervised hierarchical clustering revealed that TSCM cells had a distinct gene expression profile more closely related to conventional memory T cells than TN cells, further corroborating that TSCM cells are a unique T-cell memory subset (Fig. 3a). Consistent with the data reported by Willinger et al.25, the majority of genes (565/900) progressively increased (effector-associated genes) or decreased (naïve-associated genes) in the exact order: TN cells →TSCM cells →TCM cells →TEM cells (Supplementary Table 3). For example, transcripts of key regulators of effector differentiation and senescence, such as Eomesodermin26, T-box 2127 and PR Domain Containing 1, with ZNF Domain28, as well as genes encoding for cytotoxic molecules (e.g. Granzyme A and Perforin) and markers of T-cell senescence (e.g. Killer Cell Lectin-like Receptor Subfamily G, Member 1 (KLRG1)27), were increasingly expressed from TN cells to TEM cells (Fig. 3b). Conversely, the expression of transcription factors that inhibit T-cell activation and differentiation such as Lymphoid Enhancer-binding Factor 19 and Forkhead Box P129 and LAG1 Homolog, Ceramide Synthase 6, which promotes cellular quiescence by regulating intracellular ceramide levels30, progressively decreased from TN cells to TEM cells (Fig. 3b). These data are consistent with a linear model of T-cell differentiation, in which TSCM are the least differentiated memory T-cell subset.
Figure 3. TSCM cells represent a distinct, less differentiated T-cell memory subset.
a, Heat map of differentially expressed genes (P < 0.01 One-Way Repeated Measures ANOVA, FDR < 5% Benjamini-Hochberg's method) among CD8+ T-cell subsets. Red and blue colors indicate increased and decreased expression respectively. b, Robust Multichip Analysis (RMA)-normalized intensity of selected display of genes progressively downregulated (naïve associated genes) or upregulated (effector associated genes) from TN cells →TSCM cells →TCM cells →TEM cells. Data are represented as means ± s.e.m of 3 donors. c) Multidimensional scaling (MDS) analysis of differentially expressed genes (P < 0.01, FDR < 5%). Numbers represent the differentially regulated genes among each CD8+ T-cell subset (P < 0.01 (t test) and > 2 fold change in expression level). d, Heat map of differentially-expressed genes among TSCM and TCM cells (P < 0.01 (t test) and > 2 fold change in expression level). Red and blue colors indicate increased and decreased expression, respectively. Full gene names are listed in the Supplementary Information.
Multidimensional scaling (MDS) analysis31 confirmed that TSCM cells was the memory T-cell subset most similar to TN (Fig. 3c). Indeed, only 75 genes were differentially expressed between TN and TSCM (P < 0.01 and > 2-fold change in expression) compared to 157 and 226 for TCM and TEM cells, respectively (Fig. 3c, and Supplementary Tables 4–6). TSCM and TCM cells were the most closely related T-cell subsets, with 20 differentially expressed genes (Fig. 3c,d and Supplementary Tables 4–9). Among these genes, TSCM cells, like TN cells, expressed low levels of Heterogeneous Nuclear Ribonucleoprotein L-like, a key regulator of the alternative splicing of the CD45 pre-mRNA required for efficient CD45RO expression32, confirming the purity of the sorting. When considering this subset of 20 genes, TSCM cells have a pattern of expression similar to TN cells, while TCM cells clustered with TEM cells (Fig. 3d), further underscoring that TSCM cells are less differentiated than TCM cells.
Enhanced self-renewal and multipotency of TSCM cells
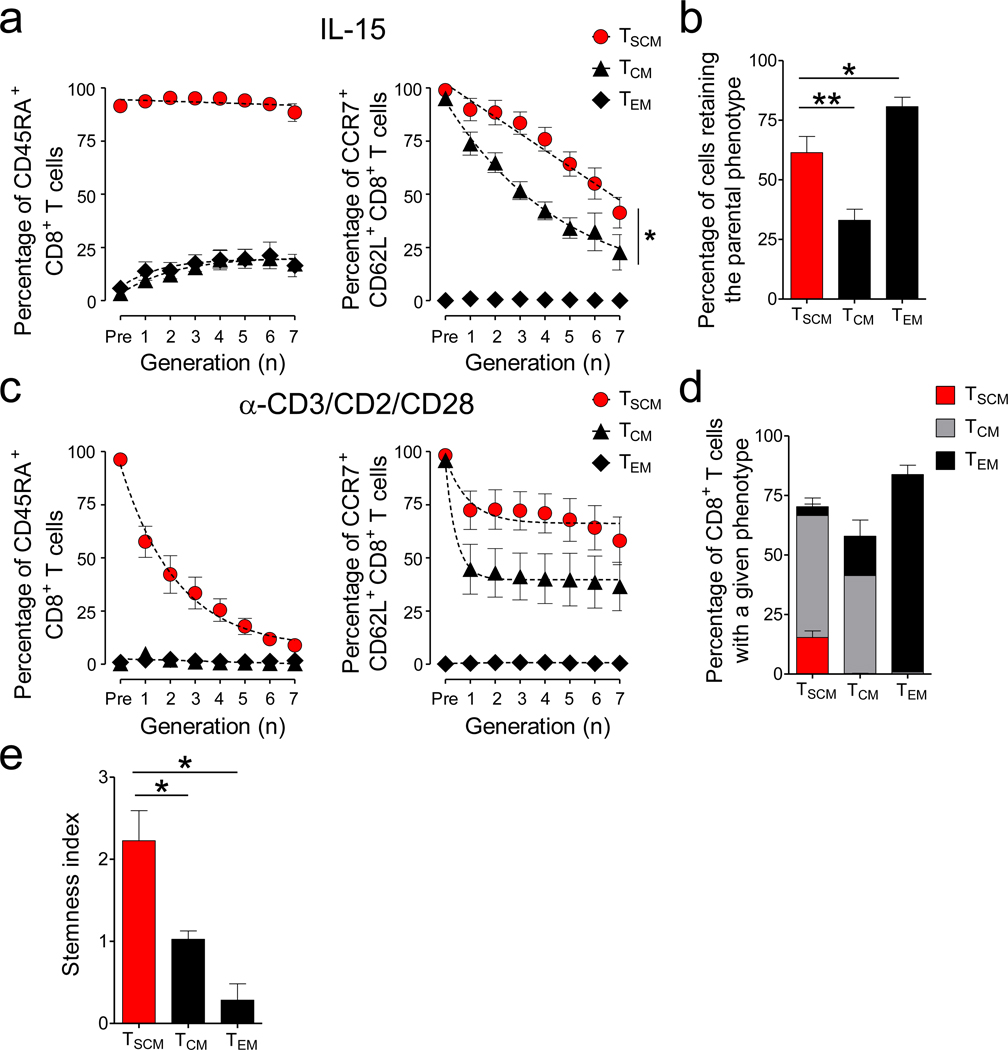
The abilities to self-renew and differentiate into specialized cell types are defining qualities of stem cells. To determine whether TSCM have these stem cell-like properties, we evaluated their capacity to self-renew with homeostatic signals as well as their multipotency after TCR activation. After exposure to IL-15, the vast majority of TSCM cells maintained CD45RA+, and retained significantly higher levels of CD62L and CCR7 than TCM cells (Fig. 4a and Supplementary Fig. 10). At the end of stimulation about 60% of cells derived from TSCM maintained their phenotypic identity (CCR7+CD62L+CD45RA+CD45RO−) while only 30% of TCM cells retained their input phenotype (CCR7+CD62L+CD45RA−CD45RO+) (Fig. 4b). TSCM cells also displayed greater self-renewal capacity compared to TCM cells following a secondary exposure to IL-15 (Supplementary Fig. 11).
Figure 4. Enhanced self-renewal and multipotency of TSCM cells.
a, Percentage of CD8+ T cells expressing CCR7 and CD62L (right panel) and CD45RA (left panel) relative to cell division after exposure to 25ng ml−1 of IL-15 for 10 days. Slopes (S) were compared using a Wilcoxon rank test, *= P = 0.0391. where g = generation number and p = percentage of CD62L+CCR7+ cells (n = 8). b, Percentage of CFSE-diluted CD8+ T cells that retained the parental phenotype following stimulation with 25ng ml−1 of IL-15 for 10 days. c, Percentage of CD8+ T cells expressing CCR7 and CD62L (right panel) and CD45RA (left panel) relative to cell division after stimulation with α-CD3/CD2/CD28-coated beads for 6 days. d, Percentage of CFSE-diluted CD8+ T cells with a given phenotype following stimulation with α-CD3/CD2/CD28-coated beads for 6 days. e, Stemness index of CD8+ memory T-cell subsets. Stemness index was calculated by multiplying self-renewal (SI) and multipotency (MI) indexes. SI was calculated as follows: SI= 2PIPRP, PI=Proliferation index, PRP= Percentage of cells retaining the input phenotype and MI was calculated as the net entropy of the progeny T-cell subsets where p = percentage of a given T-cell subset generated following α-CD3/CD2/CD28 stimulation. Data are represented as mean ± s.e.m of 4 donors; * = P < 0.05 (t test) (n = 4).
Following α-CD3/CD2/CD28 stimulation, however, TSCM cells gradually upregulated CD45RO over several cell divisions while acutely downregulating CD62L and CCR7 (Fig. 4c and Supplementary Fig. 10). These dynamic changes in phenotype resulted in a diverse progeny comprising about 50% of TCM cells and 4% of TEM cells (Fig. 4d). Most importantly, 15% of TSCM-derived cells maintained a CCR7+CD62L+CD45RA+CD45RO− phenotype even after this potent stimulus, indicating that TSCM cells have the multipotent capacity to derive all memory T-cell subsets (Fig. 4d). By contrast, TCM cells retained a central memory phenotype or differentiated into TEM cells, but did not generate TSCM (Fig. 4d). Consistent with their advanced differentiation state, TEM cells did not reacquire CD62L or CCR7 and did not dedifferentiate into TCM or TSCM cells after either IL-15 or α-CD3/CD2/CD28 stimulation (Fig. 4a–d). Taken together, these findings indicate that TSCM cells have the stem cell-like properties of self-renewal and multipotency in vitro (Fig. 4e).
Increased proliferative capacity, survival and anti-tumor activity of TSCM cells
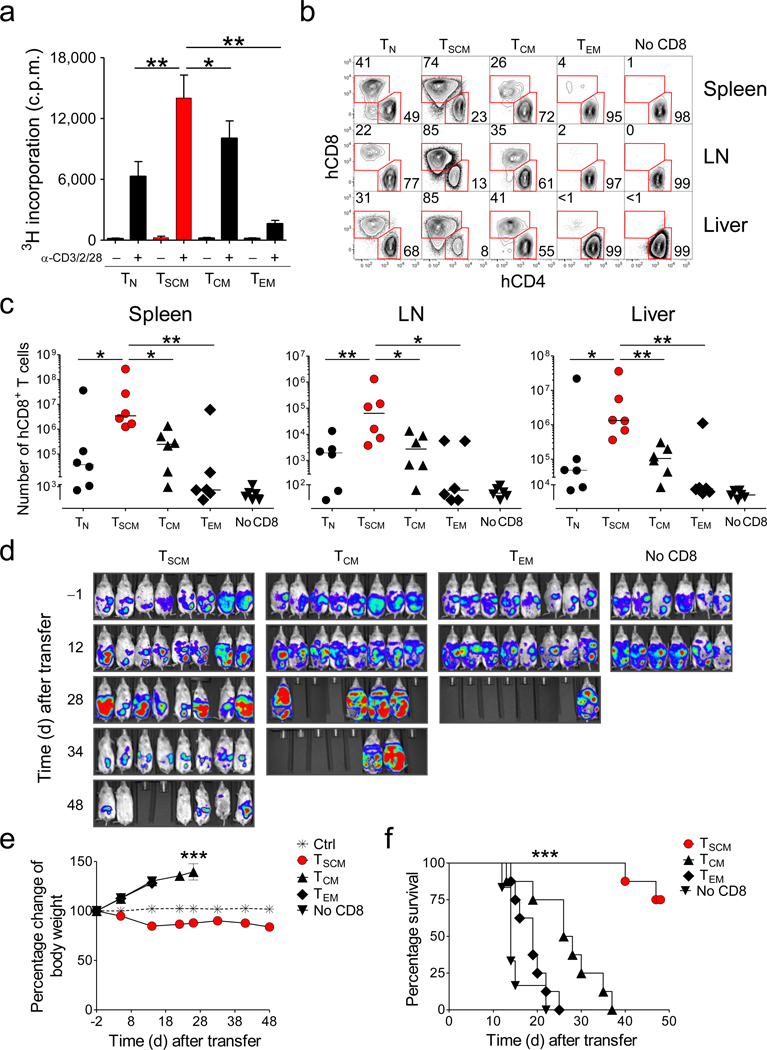
We previously found that mouse TSCM cells have enhanced proliferative and survival capacities compared with TCM and TEM cells9. To evaluate the replicative responses of TSCM cells we measured the levels of 3H-thymidine incorporation after TCR stimulation. As previously reported33, TCM and TN cells displayed increased proliferative responses compared to TEM cells, but they were outpaced by TSCM cells (Fig. 5a). Although assessment of telomerase activity was uninformative (Supplementary Fig. 12), we sought to ascertain the long-term replicative and survival capacities of TSCM. We adoptively transferred CD8+ T-cell subsets into highly immunodeficient NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice and evaluated T-cell engraftment one month after transfer. We co-transferred CD8-depleted peripheral blood mononuclear cells (PBMC) to provide a source of human cytokines and costimulatory molecules34. Strikingly, we found that TSCM engrafted with 10- to 100-fold more progeny than TCM or TN cells in both lymphoid and non-lymphoid tissues (Fig. 5b,c and Supplementary Fig. 13). Notably, TEM cells, which are used in clinical trials for adoptive immunotherapy35,36, had a poor proliferative and survival capability with engraftment comparable to the contaminating CD8+ T-cell population from the co-transferred CD8-depleted PBMC. This humanized mouse model, however, is inadequate to test T-cell self-renewal in vivo because all CD8+ T-cell subsets uniformly differentiated into effector cells (Supplementary Fig. 14) likely as a result of encounter homeostatic cytokines and with xenogeneic major histocompatibility antigens. Nonetheless, adoptive transfer in NSG mice prove that TSCM cells have enhanced replicative and survival capabilities compared to naïve and conventional memory subsets.
Figure 5. Increased proliferative capacity, survival and antitumor activity of TSCM cells.
a, 3H-thymidine incorporation by sorted CD8+ T-cell subsets after stimulation with α-CD3/CD2/CD28-coated beads. Data are represented as means ± s.e.m of 10 donors. Results are normalized to the number of seeded cells, as different cell numbers were obtained from different sorts. * = P < 0.05; ** = P < 0.01; *** = P < 0.001 (t test) b, Flow cytometry analysis of human T cells in the spleen, lymph nodes (LN) and liver of a representative NSG mouse at 4 weeks following adoptive transfer of CD4+ T cells (5 × 106) with or without sorted CD8+ T-cell subsets (106). Graphs show T cells after gating on human CD45+ cells. Numbers indicate the percentage of cells in the CD4+CD8− or CD4−CD8+ gates. c, Total human CD8+ T-cell recovery in the spleens, LN and livers from 6 NSG mice 4 weeks following adoptive transfer of CD4+ T cells with or without sorted CD8+ T-cell subsets. A total of 6 mice per T-cell subset from two independent experiments (3 replicate mice per T-cell subset per experiment) are shown. Horizontal bars indicate median values. * = P < 0.05; ** = P < 0.01 (t test) d–f, In vivo bioluminescent imaging (d), percentage change of body weight (e), and survival of NSG mice (f) bearing M108-luciferase mesothelioma after adoptive transfer of CD4+ T cells (106) with or without sorted CD8+ T-cell subsets (3 × 106) expressing a mesothelin-specific chimeric antigen receptor. *** = P < 0.001 One-Way Repeated Measures ANOVA (e) and Log-rank (Mantel-Cox) Test (f).
T-cell proliferative and survival capacities correlate with the anti-tumor efficacy of adoptively transferred T cells35–39. T cell receptor (TCR) or chimeric antigen receptor (CAR) gene engineering are currently used in the clinic to redirect the specificity of circulating T cells toward the desired target36,40,41. We exploited this approach coupled to the pharmacological activation of Wnt signaling to generate high numbers of mesothelin-specific ex vivo-generated T-cell memory subsets (Supplementary Fig. 15) to test in a xenograft tumor model that we recently established34. Mesothelin-specific TSCM, TCM or TEM cells were co-transferred with mesothelin-specific CD4+ T cells into NSG mice bearing luciferase-expressing M108 mesothelioma established for 3 months in the peritoneum. To generate a treatment window, we administered 3×106 CD8+ T cells and 106 CD4+ T cells, 10-fold fewer cells than previously published34. TEM mediated poor anti-tumor responses as indicated by the increased intensity of the bioluminescent signal in the abdomen (Fig 5d) and the ascites-dependent weight gain (Fig. 5e). Furthermore, transfer of TEM did not significantly extend the survival of the animals compared to CD4+ T cells alone (Fig. 5f). TCM cells were more effective than TEM cells and improved survival, although all mice died from tumor progression within 40 days after treatment (Fig. 5d–f). In stark contrast, TSCM cells triggered tumor regression and cure in mice that otherwise died within two to three weeks (Fig. 5d–f). Late mortality of mice receiving TSCM cells was ascribed to the development of xenogeneic graft-versus-host disease as manifested by loss of body weight (Fig. 5d,e). Thus, adoptively transferred TSCM cells have enhanced anti-tumor activity and are more therapeutically effective than conventional TCM and TEM cells.
Discussion
We identified a long-lived human memory T-cell subset found within the naïve-like T-cell compartment. Once thought to be homogenous, the so-called “naïve T cell subset” is emerging as a complex amalgamation of cell types, including recent thymic emigrants42,43, “super-naïve” CD44very low T cells44, and CXCR3+ or CCR4+ early memory cells45. Evidence presented here indicates that TSCM are a clonally expanded primordial memory subset arising after antigenic stimulation with increased proliferative and reconstitutive capacities. TSCM cells have enhanced self-renewal, and the multipotency to generate all memory and effector T-cell subsets in vitro. These qualities are all consistent with stem cell-like behavior, but formal proof of “stemness” in vivo will have to await clinical trials involving the adoptive transfer of TSCM and assessment of long-term self-renewal and repopulating potential. Nevertheless, the qualities of human TSCM cells that we have measured are consistent with those of mouse TSCM cells that displayed enhanced self-renewal and multipotency in serial transplantation experiments8,9.
Given the young age at which the thymus involutes in humans relative to life expectancy, the presence of such a long-lived less differentiated memory T-cell population might ensure protection against pathogens throughout life. Indeed, the vast majority of naïve-like T cells in centenarians express high levels of CD9546, a phenotype consistent with the TSCM described here.
Expression of CD95 by long-lived memory T cells might seem perplexing because of its well established pro-apoptotic function. However, the outcome of FAS ligand-CD95 interaction might be dependent on the differentiation state of the cellular substrate, as observed in other organ systems47. CD95 triggers apoptosis in terminally differentiated neurons through the formation of a death-inducing complex, but promotes cell proliferation in neural progenitors and cancer stem cells by inducing TCF/β-catenin signaling47. Considering the emerging role of TCF/β-catenin signaling in the formation and maintenance of memory T cells9,14,48,49, CD95 might not only demarcate memory T cells but also be functionally important for their self-renewal and persistence.
The finding that TSCM cells have enhanced proliferative capacity and can sustain the generation of all memory and effector T-cell subsets has considerable implications for the design of T cell-based vaccines to target intracellular pathogens and cancer. Heterologous prime-boost approaches have been used to increase the frequency of memory T cells but they have the undesired effect of driving T cells toward a state of terminal differentiation3,50,51, which compromises their ability to clear systemic infections52,53 and eradicate tumors37. The use of small molecules targeting the mTOR 54,55 and Wnt14,56 signaling pathways might improve vaccines by modulating T-cell differentiation and enriching for TSCM and TCM cells. Finally, by coupling TCR or CAR gene-engineering technology36,40,41 with the pharmacologic modulation of T-cell differentiation56, we have demonstrated a feasible and translatable strategy for the in vitro generation of highly effective TSCM-like cells to use in adoptive T-cell therapy trials for the treatment of cancer and infectious diseases.
METHODS
Antibodies, flow cytometry and cell sorting
We obtained all human samples from healthy donors or patients enrolled in clinical trials approved by the NCI Institutional Review Board. We conjugated unlabeled antibodies (BD Biosciences) in our laboratory reported at http://www.drmr.com/abcon. We purchased fluorescently-conjugated antibodies from BD Biosciences, eBioscience, Biolegend, Invitrogen and Beckman Coulter. Except for MART-126–35 (27L) (Beckman Coulter), we produced recombinant pMHCI tetramers as described previously57. We performed surface and intracellular staining as described previously22. We measured cytokine release using the cytometric bead array/human Th1/Th2/Th17 cytokine kit (BD Biosciences). We performed flow cytometry acquisition on a modified LSR II, equipped to detect 18 fluorescencet parameters. We compensated and analyzed data with FlowJo software (Treestar Inc.). We sorted T cell subsets using a modified FACSAria (BD Biosciences).
In vitro generation of TSCM-like cells
We stimulated CD45RO−CD62L+ cells with α-CD3/CD28 beads (Invitrogen) at 1:1 bead-to-cell ratio and 300 IU ml−1 IL-2 (Chiron) in the presence of 5 µM TWS119 (Cayman Chemical) for two weeks.
Animal experiments
We conducted all animal experiments with the approval of the NCI or University of Pennsylvania Institutional Animal Use and Care Committees. We used NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ mice (NSG) (The Jackson Laboratory) as recipients for adoptive transfer experiments.
Generation of mesothelin-specific T cells
We stimulated T cells with α-CD3/CD28 beads at 1:3 cell to bead ratio and 20 IU ml−1IL-2. We transduced T cells with lentiviral vectors encoding for the anti-mesothelin chimeric receptor SS1:BB:TCR-ζ34 24h after activation and added 5µM TWS119 12h after transduction. Cells were fed every other day with TWS119 and IL-2 for two weeks. We purified CD8+ T memory subsets using MACS positive selection Multisort kits (Miltenyi Biotec). Approximately 2 × 106 CD8+ T cell subsets were mixed with 106 unsorted CD4+ T cells and injected i.v. into tumor-bearing NSG mice.
Mouse xenograft studies
We engineered the M108-km1 human mesothelioma cell line34 with a lentiviral vector to express firefly luciferase, yielding the M108-Luc cell line. Animals were injected i.p. with 8 × 106 M108-Luc cells. We measured tumor burden by bioluminescent imaging and body weight.
Bioluminescence imaging
We injected mice i.p. with 150 mg kg−1 body weight D-luciferin (Caliper Life Sciences) and imaged 10–12 minutes later using a Xenogen Spectrum system and Living Image v3.2 software (Caliper Life Sciences).
Determination of T cell replicative history
We determined the replicative history of sorted subsets by quantifying T cell receptor rearrangement excision circles (TREC) using real time qPCR as described previously18.
Clonotypic analysis of antigen-specific CD8+ T cell populations
We amplified TRB gene products of sorted NV9-specific CD8+ T cell subsets using a template-switch anchored RT-PCR and performed subcloning, sequencing and analysis as described previously57. TCR nomenclature was translated directly from the IMGT database (The ImmunoGeneTics information system® http://imgt.cines.fr) using web-based alignment of molecular TRB transcripts.
Whole genome gene expression analysis
Total RNA from sorted CD8+ T cell subsets was isolated using an RNEasy Micro kit (Qiagen), processed using a WT expression kit (Ambion), fragmented and labeled using a WT Terminal Labeling Kit (Affymetrix), hybridized to WT Human Gene 1.0 ST arrays (Affymetrix) and stained on a Genechip Fluidics Station 450 (Affymetrix). We scanned arrays on a GeneChip Scanner 3000 7G (Affymetrix). We imported the raw data from. CEL files into the Partek Genomincs Suite using the RMA method. We identified differentially expressed genes (DEGs) by One-Way Repeated Measures ANOVA (P < 0.01) corrected by Benjamini-Hochberg's False Discovery Rate method (P < 0.05). For pair-wise comparisons, we further filtered genes for between-group alpha levels of P < 0.01 and a fold change criterion of > 2.0. We deposited array data at the Gene Expression Omnibus (GEO) public depository under the accession number GSE23321.
Analysis of T cell proliferation
We determined cell proliferation by 3H-thymidine incorporation and by 5-(and 6)-carboxyfluorescein diacetate succinimidyl ester (CFSE) dilution. In the former case, we stimulated cells for 24 hours with α-CD3/CD2/CD28-coated beads (Miltenyi Biotec), then pulsed with 3H-thymidine (1 µCi; Perkin Elmer) for an additional 16 hours. We harvested supernatants and determined counts per minute with a β-scintillation counter (Perkin Elmer). In the latter, we labeled cells with 2 µM CFSE for 7 minutes at 37°C and stimulated them with αCD3/CD2/CD28-coated beads (Miltenyi Biotec) for 6 days or with rhIL-7 or rhIL-15 (both 25 ng ml−1; Peprotech) for 14d and 10d, respectively. We determined proliferation index, and percentage of divided cells with FlowJo.
Statistical analysis
We performed statistical analyses using Prism (GraphPad Software) and Spice software. For most of the comparison we used a non-parametric Wilcoxon rank test to compare two groups. We employed One ANOVA to compare three or more groups and χ2 permutation test for pie-chart comparison58. We used non-parametric Spearman’s rank correlation test to measure statistical correlation between two groups and Kaplan-Meier method to analyze survival.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Programs of the US National Institutes of Health, National Cancer Institute, Center for Cancer Research and National Institute of Allergy and Infectious Diseases. We thank S.A. Rosenberg and J.R. Wunderlich for providing samples from HLA-A*0201 patients with melanoma, P. Scheinberg for providing HLA-A*0201 samples, Marianna Sabatino for coordinating phereses, B.J. Hill for assistance with the TREC assay, S.P. Perfetto, R. Nguyen and D.A. Ambrozak for help with cell sorting and P.K. Chattopadhyay, J. Yu for antibody conjugation and R.A. Seder and C.A. Klebanoff for critical review of the manuscript.
Footnotes
Author Contributions
L.G., E.L., Y.J., Z.P., C.M.P., J.R.A, Z.Y. and C.C. performed experiments, L.G., E.L., Y.J., Z.P., C.M.P. and J.R.A analyzed experiments, L.G., E.L., C.M.P., E.W., D.C.D., D.A.P., C.H.J., F.M.M., M.R., and N.P.R. designed experiments, E.G., M.F.Q and D.A.P. contributed reagents, E.L and M.R. edited the manuscript, L.G and N.P.R. wrote the manuscript.
Reference List
- 1.Wakim LM, Bevan MJ. From the thymus to longevity in the periphery. Curr. Opin. Immunol. 2010;22:274–278. doi: 10.1016/j.coi.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim PS, Ahmed R. Features of responding T cells in cancer and chronic infection. Curr. Opin. Immunol. 2010;22:223–230. doi: 10.1016/j.coi.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klebanoff CA, Gattinoni L, Restifo NP. CD8+ T-cell memory in tumor immunology and immunotherapy. Immunol. Rev. 2006;211:214–224. doi: 10.1111/j.0105-2896.2006.00391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fearon DT, Manders P, Wagner SD. Arrested differentiation, the self-renewing memory lymphocyte, and vaccination. Science. 2001;293:248–250. doi: 10.1126/science.1062589. [DOI] [PubMed] [Google Scholar]
- 5.Stemberger C, et al. Stem cell-like plasticity of naive and distinct memory CD8+ T cell subsets. Semin. Immunol. 2009;21:62–68. doi: 10.1016/j.smim.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Luckey CJ, et al. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3304–3309. doi: 10.1073/pnas.0511137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Joe G, Hexner E, Zhu J, Emerson SG. Host-reactive CD8+ memory stem cells in graft-versus-host disease. Nat. Med. 2005;11:1299–1305. doi: 10.1038/nm1326. [DOI] [PubMed] [Google Scholar]
- 9.Gattinoni L, et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat. Med. 2009;15:808–813. doi: 10.1038/nm.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neuenhahn M, Busch DH. The quest for CD8+ memory stem cells. Immunity. 2009;31:702–704. doi: 10.1016/j.immuni.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Turtle CJ, Swanson HM, Fujii N, Estey EH, Riddell SR. A distinct subset of self-renewing human memory CD8+ T cells survives cytotoxic chemotherapy. Immunity. 2009;31:834–844. doi: 10.1016/j.immuni.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dusseaux M, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 2011;117:1250–1259. doi: 10.1182/blood-2010-08-303339. [DOI] [PubMed] [Google Scholar]
- 13.Schenkel JM, Zloza A, Li W, Narasipura SD, Al-Harthi L. Beta-catenin signaling mediates CD4 expression on mature CD8+ T cells. J. Immunol. 2010;185:2013–2019. doi: 10.4049/jimmunol.0902572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gattinoni L, Ji Y, Restifo NP. Wnt/{beta}-catenin signaling in T-cell immunity and cancer immunotherapy. Clin. Cancer Res. 2010;16:4695–4701. doi: 10.1158/1078-0432.CCR-10-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gattinoni L, Ji Y, Restifo NP. beta-catenin does not regulate memory T cell phenotype Reply. Nature Medicine. 2010;16:514–515. doi: 10.1038/nm0510-513. [DOI] [PubMed] [Google Scholar]
- 16.Appay V, van Lier RA, Sallusto F, Roederer M. Phenotype and function of human T lymphocyte subsets: consensus and issues. Cytometry A. 2008;73:975–983. doi: 10.1002/cyto.a.20643. [DOI] [PubMed] [Google Scholar]
- 17.De Rosa SC, Herzenberg LA, Herzenberg LA, Roederer M. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat. Med. 2001;7:245–248. doi: 10.1038/84701. [DOI] [PubMed] [Google Scholar]
- 18.Douek DC, et al. Changes in thymic function with age and during the treatment of HIV infection. Nature. 1998;396:690–695. doi: 10.1038/25374. [DOI] [PubMed] [Google Scholar]
- 19.Kambayashi T, Assarsson E, Lukacher AE, Ljunggren HG, Jensen PE. Memory CD8+ T cells provide an early source of IFN-gamma. J. Immunol. 2003;170:2399–2408. doi: 10.4049/jimmunol.170.5.2399. [DOI] [PubMed] [Google Scholar]
- 20.Surh CD, Sprent J. Homeostatic T cell proliferation: how far can T cells be activated to self-ligands? J. Exp. Med. 2000;192:F9–F14. doi: 10.1084/jem.192.4.f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prlic M, Lefrancois L, Jameson SC. Multiple choices: regulation of memory CD8 T cell generation and homeostasis by interleukin (IL)-7 and IL-15. J. Exp. Med. 2002;195:F49–F52. doi: 10.1084/jem.20020767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lugli E, et al. Transient and persistent effects of IL-15 on lymphocyte homeostasis in nonhuman primates. Blood. 2010;116:3238–3248. doi: 10.1182/blood-2010-03-275438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alanio C, Lemaitre F, Law HK, Hasan M, Albert ML. Enumeration of human antigen-specific naive CD8+ T cells reveals conserved precursor frequencies. Blood. 2010;115:3718–3725. doi: 10.1182/blood-2009-10-251124. [DOI] [PubMed] [Google Scholar]
- 24.Zippelius A, et al. Thymic selection generates a large T cell pool recognizing a self-peptide in humans. J. Exp. Med. 2002;195:485–494. doi: 10.1084/jem.20011658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willinger T, Freeman T, Hasegawa H, McMichael AJ, Callan MF. Molecular signatures distinguish human central memory from effector memory CD8 T cell subsets. J. Immunol. 2005;175:5895–5903. doi: 10.4049/jimmunol.175.9.5895. [DOI] [PubMed] [Google Scholar]
- 26.Pearce EL, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 27.Joshi NS, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutishauser RL, et al. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng X, et al. Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nat. Immunol. 2011;12:544–550. doi: 10.1038/ni.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogretmen B, Hannun YA. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat. Rev. Cancer. 2004;4:604–616. doi: 10.1038/nrc1411. [DOI] [PubMed] [Google Scholar]
- 31.Khan J, et al. Gene expression profiling of alveolar rhabdomyosarcoma with cDNA microarrays. Cancer Res. 1998;58:5009–5013. [PubMed] [Google Scholar]
- 32.Oberdoerffer S, et al. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321:686–691. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinrichs CS, et al. Human effector CD8+ T cells derived from naive rather than memory subsets possess superior traits for adoptive immunotherapy. Blood. 2011;117:808–814. doi: 10.1182/blood-2010-05-286286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carpenito C, et al. Control of large, established tumor xenografts with genetically retargeted human T cells containing CD28 and CD137 domains. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3360–3365. doi: 10.1073/pnas.0813101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gattinoni L, Powell DJ, Jr, Rosenberg SA, Restifo NP. Adoptive immunotherapy for cancer: building on success. Nat. Rev. Immunol. 2006;6:383–393. doi: 10.1038/nri1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.June CH. Adoptive T cell therapy for cancer in the clinic. J. Clin. Invest. 2007;117:1466–1476. doi: 10.1172/JCI32446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gattinoni L, et al. Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8+ T cells. J. Clin. Invest. 2005;115:1616–1626. doi: 10.1172/JCI24480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klebanoff CA, et al. Central memory self/tumor-reactive CD8+ T cells confer superior antitumor immunity compared with effector memory T cells. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9571–9576. doi: 10.1073/pnas.0503726102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hinrichs CS, et al. Adoptively transferred effector cells derived from naive rather than central memory CD8+ T cells mediate superior antitumor immunity. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17469–17474. doi: 10.1073/pnas.0907448106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgan RA, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pule MA, et al. Virus-specific T cells engineered to coexpress tumor-specific receptors: persistence and antitumor activity in individuals with neuroblastoma. Nat. Med. 2008;14:1264–1270. doi: 10.1038/nm.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kimmig S, et al. Two subsets of naive T helper cells with distinct T cell receptor excision circle content in human adult peripheral blood. J. Exp. Med. 2002;195:789–794. doi: 10.1084/jem.20011756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boursalian TE, Golob J, Soper DM, Cooper CJ, Fink PJ. Continued maturation of thymic emigrants in the periphery. Nat. Immunol. 2004;5:418–425. doi: 10.1038/ni1049. [DOI] [PubMed] [Google Scholar]
- 44.Zhao C, Davies JD. A peripheral CD4+ T cell precursor for naive, memory, and regulatory T cells. J. Exp. Med. 2010;207:2883–2894. doi: 10.1084/jem.20100598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song K, et al. Characterization of subsets of CD4+ memory T cells reveals early branched pathways of T cell differentiation in humans. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7916–7921. doi: 10.1073/pnas.0409720102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lugli E, et al. Subject classification obtained by cluster analysis and principal component analysis applied to flow cytometric data. Cytometry A. 2007;71:334–344. doi: 10.1002/cyto.a.20387. [DOI] [PubMed] [Google Scholar]
- 47.Beier CP, Schulz JB. CD95/Fas in the brain--not just a killer. Cell Stem Cell. 2009;5:128–130. doi: 10.1016/j.stem.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 48.Jeannet G, et al. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9777–9782. doi: 10.1073/pnas.0914127107. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou X, et al. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity. 2010;33:229–240. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wirth TC, et al. Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8(+) T cell differentiation. Immunity. 2010;33:128–140. doi: 10.1016/j.immuni.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sallusto F, Lanzavecchia A, Araki K, Ahmed R. From vaccines to memory and back. Immunity. 2010;33:451–463. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wherry EJ, et al. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 2003;4:225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 53.Appay V, Douek DC, Price DA. CD8+ T cell efficacy in vaccination and disease. Nat. Med. 2008;14:623–628. doi: 10.1038/nm.f.1774. [DOI] [PubMed] [Google Scholar]
- 54.Araki K, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pearce EL, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gattinoni L, Klebanoff CA, Restifo NP. Pharmacologic induction of CD8+ T cell memory: better living through chemistry. Sci. Transl. Med. 2009;1:11ps12. doi: 10.1126/scitranslmed.3000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price DA, et al. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J. Exp. Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roederer M, Nozzi JL, Nason MC. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011;79:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.