Abstract
Lenalidomide with dexamethasone is a standard induction treatment regimen for newly diagnosed myeloma (although a Federal Drug Administration indication is still absent). In the context of the Phase 3 clinical trial E4A03 (lenalidomide plus dexamethasone in low or high doses), we queried whether a fluorescence in situ hybridization (FISH)-based genetic classification into high risk (HR) and standard risk (SR) multiple myeloma (MM) would remain clinically significant. Of 445 E4A03 patients, 126 had FISH analysis; 21 were classified HR with t(4;14), t(14;16), or 17p13 deletions. Median survival follow-up approached 3 years. Patients with FISH data tended to be younger and healthier compared to the rest of the study population and, consequently, had superior overall survival (OS) results. Within the FISH cohort, shorter OS in the HR versus SR group (P=0.004) corresponded to a hazard ratio of 3.48 [95% confidence interval: (1.42–8.53)], an effect also observed in multivariate analysis. Two-year OS rates were 91% for SR MM and 76% for HR MM. There was also evidence of interaction between risk status and treatment (p=0.026). HR patients were less likely to attain good partial response (SR 46% and HR 30%, Odds Ratio=2.0 [0.7–5.6]), but overall response rates were not different. FISH-based risk classification retained prognostic significance in patients receiving lenalidomide-based induction.
Keywords: myeloma, lenalidomide, dexamethasone, bortezomib, FISH
INTRODUCTION
The presence of specific genetic abnormalities contributes to the heterogeneity in outcome among patients with multiple myeloma (MM) (Fonseca et al, 2003; Avet-Loiseau et al, 2007). Translocations involving the immunoglobulin heavy chain locus on chromosome 14 are among the most common abnormalities seen in myeloma, with nearly one half of patients having a balanced or unbalanced translocation between chromosome 14 and chromosomes 4, 6, 11,16 or 20 (Fonseca et al, 2003; Avet-Loiseau et al, 2007). Among this group of patients, the presence of t(4;14), t(14;16) or t(14;20) typically signifies a poor outcome (Fonseca et al, 2003). In addition, abnormalities, such as 17p deletions, can be found in 5–10% of patients at diagnosis, and increase in prevalence with disease progression, comprising a group of patients with particularly poor outcome (Avet-Loiseau et al, 2007; Chng et al, 2007; Drach et al, 1998). Use of these genetic markers along with other traditional laboratory parameters, such as beta-2 microglobulin, or more modern techniques, such as gene expression profiling, have been used in various combinations to develop risk stratification systems (Decaux et al, 2008; Shaughnessy et al, 2007).
It is important to understand how these risk factors affect patient response in the context of new therapies, so that treatment decisions can be individualized on the basis of risk stratification (Kapoor et al, 2010). Subgroup analyses connected with recent clinical trials have demonstrated that high-risk patients can overcome, albeit only partially, poor prognosis with certain treatments, in particular bortezomib, and only for certain subsets (San Miguel et al, 2008; Jagannath et al, 2007; Avet-Loiseau et al, 2010a). Other research has suggested that treatment with lenalidomide and dexamethasone does not have the ability to alter patient outcomes in the high risk group, as particularly seen in two trials of relapse, refractory MM (Reece et al, 2009; Avet-Loiseau et al, 2010b) and in our own pilot study of initial therapy with lenalidomide and dexamethasone (Kapoor et al, 2009). We have therefore examined this question in a larger cohort of patients included in a Phase 3 randomized trial (Rajkumar et al, 2010).
PATIENTS and METHODS
Patients
Patients enrolled on the E4A03 randomized Phase 3 clinical trial, the results of which have been previously published (Rajkumar et al, 2010), were included in the current study. On E4A03, newly diagnosed patients were randomized to lenalidomide plus high-dose dexamethasone (RD) versus lenalidomide plus low-dose dexamethasone (Rd). Patients on both arms received oral lenalidomide 25 mg/day on days 1–21. Patients on RD received oral dexamethasone 40 mg/day on days 1–4, 9–12, and 17–20 of each 28-day cycle while patients on Rd received oral dexamethasone 40 mg/day on days 1, 8, 15 and 22 of each 28-day cycle. After the first 4 cycles of therapy, patients could discontinue therapy to pursue stem cell transplantation (SCT) or other treatments or continue therapy on study until disease progression. All patients provided written informed consent before entering the trial, in accordance with the Declaration of Helsinki. The Institutional Review Boards in the participating Eastern Cooperative Oncology Group (ECOG) institutions approved the study. Patients with adequate bone marrow sample from study entry were assessed, 126 patients of which contributed fluorescence in situ hybridization (FISH) data. The reason we only have data on 126 patients is that research sample submission was voluntary and this was the yield for such collections.
FISH Studies
Aspirate samples were enriched for mononuclear cells using the Ficoll method and cytospin slides were stored for future use at −70°C. FISH analysis was performed as previously described and results published for chromosome13 deletion, 17p13 deletion, t(4;14)(p16;q32) and t(14;16)(q32;q23) (Fonseca et al, 2003). In our study, high-risk (HR) myeloma was defined by the presence of t(4;14), t(14;16) or 17p13 deletion by FISH with the remaining patients considered standard risk (SR) (Dispenzieri et al, 2007).
Statistical Design and Analysis
The association of FISH risk status with overall survival (OS) was of primary interest but additional endpoints evaluated were progression-free survival (PFS), grade three or higher non-haematological toxicity and best overall response (OR rate: ≥partial response). OS and PFS were estimated using the Kaplan-Meier (KM) method and the log-rank test was used for comparisons of survival distributions between groups. PFS was defined as the time from randomization to disease progression or death due to any cause. Patients were censored at the time of alternative therapy. Cox proportional hazards (PH) regression was used to examine the relationship between baseline factors and survival outcome in univariate and adjusted models, using stepwise selection. The effect of treatment within subgroups was also examined using Cox regression models. The response criteria used were standard European Group for Blood and Bone Marrow Transplant (Blade et al, 1998) except that responses were confirmed 4 weeks apart (instead of 6 weeks). In addition, patients were classified as having a very good partial response (VGPR) based on the International Myeloma Working Group response criteria (Durie et al, 2006). A category of immunofixation-negative complete response (CR) was defined as confirmed disappearance of the monoclonal protein in the serum and urine by immunofixation studies without the requirement for bone marrow studies. Fisher’s exact test was used to compare response and toxicity rates. A p-value of 0.05 was considered statistically significant.
RESULTS
Between November 2004 and April 2006, 445 patients were enrolled on the E4A03 treatment trial from participating institutions. Only 54% of patients voluntarily submitted any baseline samples on E4A03. Of these, interphase FISH was performed on 126 patients (28%) with adequate bone marrow samples available. Based on data as of November 2008, the median follow up for the 126 FISH patients was 36 months from registration, the same as the remaining 319 patients (non-FISH cohort) enrolled on the trial.
It should be noted that in the primary study (Rajkumar et al 2010), evidence of an OS advantage on the Rd arm at the first pre-planned interim analysis (n=45 events) in March 2007 prompted the Data Monitoring Committee (DMC) to release the study data and recommend crossover of patients. At this time 79 patients were still on-treatment and, as such, longer term OS analyses by treatment are confounded. Around 3 years, the OS curves by treatment cross, yielding a non-significant p-value. Median OS was not reached for the FISH cohort nor the original study cohort.
Baseline characteristics of the FISH and non-FISH cohorts along with SR and HR subgroups are shown in Table 1. Twenty-one patients (17%) were classified as high risk. Of these 21 patients, 14 had t(4;14), 2 had t(14;16), and 6 had 17p13 deletion present. The FISH cohort was significantly younger and healthier than the remaining non-FISH cohort as evidenced by a median age of 62 versus 66 years along with a lower proportion of International Staging system (ISS) Stage III and ECOG performance score (PS)>0 patients. There were no such substantial differences in the HR versus SR comparison. RD treatment assignment was sufficiently balanced. Notably, HR patients tended to have elevated creatinine and lower haemoglobin levels. Only 9% (n=11) of patients were still on treatment at the time of the current analysis. The median (95% confidence interval [CI]) treatment duration was similar between these groups: 4.5 (3.5–6.6) months for SR and 4.4 (3.5–6.2) months for HR, P=0.46. The proportion of patients that underwent SCT was also similar between groups (SR=41% and HR=38%) and slightly higher than the remaining study cohort (31%). There were no differences between the groups in terms of the reasons for treatment discontinuation. There were 422 patients in the primary analysis of response, representing the eligible population (Table 2). The OR rate for the overall FISH cohort was 77%, with no differences between risk groups (SR=77% and HR=75%) or the non-FISH cohort (76%). The depth and quality of responses were superior amongst SR patients who were more likely to achieve a VGPR or better (SR 46% and HR 30%, OR=2.0 [0.7–5.6]). Response rates by treatment within risk groups are provided in Table 3; due to small numbers the confidence intervals are wide and interpretation is descriptive. OR and VGPR rates by treatment within SR patients generally mirrored the overall cohort results. It appeared especially difficult for HR patients treated with RD or Rd to attain VGPR; however, Rd HR patients seemed to fare just as well as Rd SR patients in terms of partial response. The significant difference in OR by treatment as seen in the primary study was not apparent in the HR subgroup, with OR rates for RD HR patients below the overall cohort. Although interpretation is limited by small numbers, it appears the significant difference in OR by treatment as seen in the primary study (odds ratio RD/Rd 1.85) was not seen in the HR subgroup, with OR rates for RD HR patients below the overall cohort. (Rajkumar et al 2010)
Table 1.
Baseline characteristics
| Variable | Category* | Non-FISH | FISH | p- value | Standard Risk | High Risk | p- value |
|---|---|---|---|---|---|---|---|
| Total patients (n) | 319 | 126 | -- | 105 | 21 | -- | |
| Treatment | RD | 164 (51.4) | 59 (46.8) | 0.401 | 47 (44.8) | 12 (57.1) | 0.345 |
| Age (years) | Median (Q1, Q3) | 66 (59, 73) | 62 (55, 69) | 0.001 | 62 (55, 69) | 63 (56, 67) | 0.878 |
| Gender | Male | 182 (57.1) | 71 (56.3) | 0.916 | 59 (56.2) | 12 (57.1) | 1.000 |
| Race | White | 267 (83.7) | 114 (90.5) | 0.073 | 96 (91.4) | 18 (85.7) | 0.420 |
| ISS | Stage I | 85 (28.5) | 53 (44.2) | 0.004 | 46 (46.0) | 7 (35.0) | 0.663 |
| Stage II | 127 (42.6) | 46 (38.3) | 37 (37.0) | 9 (45.0) | |||
| Stage III | 86 (28.9) | 21 (17.5) | 17 (17.0) | 4 (20.0) | |||
| Unknown/Missing | 21 | 6 | 5 | 1 | |||
| ECOG PS | 1 or 2 | 178 (55.8) | 58 (46.0) | 0.073 | 47 (44.8) | 11 (52.4) | 0.633 |
| Bone Disease | Yes | 197 (61.8) | 79 (62.7) | 0.914 | 68 (64.8) | 11 (52.4) | 0.327 |
| Beta-2 Microglobulin (mg/l) | Median (Q1, Q3) | 3.9 (2.6, 6.1) | 3.3 (2.5, 4.6) | 0.010 | 3.2 (2.4, 4.6) | 3.8 (2.7, 4.8) | 0.287 |
| Freq. of Missing | 8 | 1 | 0 | 1 | |||
| C-reactive protein (mg/l) | Median (Q1, Q3) | 6 (3, 17) | 5 (2, 11) | 0.019 | 5 (2, 11) | 5 (2, 9) | 0.736 |
| Freq. of Missing | 44 | 5 | 1 | 4 | |||
| Serum M Spike (g/l) | Median (Q1, Q3) | 31 (18, 44) | 32 (21, 48) | 0.161 | 31 (19, 45) | 46 (30, 52) | 0.030 |
| Freq. of Missing | 26 | 7 | 7 | 0 | |||
| Albumin (g/l) | Median (Q1, Q3) | 35 (30, 40) | 37 (33, 40) | 0.038 | 37 (35, 40) | 32 (28, 39) | 0.006 |
| Freq. of Missing | 13 | 5 | 5 | 0 | |||
| Lactate dehydrogenase (u/l) | Median (Q1, Q3) | 159 (128, 208) | 154 (119, 209) | 0.455 | Median (Q1, Q3) | 159 (128, 208) | 0217 |
| Freq. of Missing | 24 | 4 | 3 | 1 | |||
| Calcium (mmol/l) | Median (Q1, Q3) | 2.33 (2.2, 2.45) | 2.3 (2.23,2.4) | 0.439 | 2.3 (2.23,2.4) | 2.25 (2.2,2.38) | 0.265 |
| Freq. of Missing | 1 | 0 | 0 | 0 | |||
| Creatinine (μmol/l) | Median (Q1, Q3) | 972 (796, 1238) | 884 (796, 1061) | 0.017 | 884 (707, 1061) | 972 (884, 1326) | 0.025 |
| Haemoglobin (g/l) | Median (Q1, Q3) | 108 (95, 122) | 115 (102, 125) | 0.003 | 116 (105, 126) | 102 (99, 118) | 0.019 |
Data are number (%) unless otherwise noted. Missing values are excluded from calculations of percentages or medians
ISS, International Staging System; ECOG PS, Eastern Cooperative Oncology Group performance status.
Table 2.
Best Overall Response
| Non-FISH (%) N=298 |
FISH (%) N=124 |
Standard Risk (%) N=104 |
High Risk (%) N=20 |
|
|---|---|---|---|---|
| Complete Response | 14 (4.7) | 5 (4.0) | 4 (3.9) | 1 (5.0) |
| ≥ Immunofix negative | 46 (15.4) | 21 (16.9) | 19 (18.3) | 2 (10.0) |
| ≥ Very Good Partial Response | 138 (46.3) | 54 (43.6) | 48 (46.2) | 6 (30.0) |
| ≥ Partial Response | 225 (75.5) | 95 (76.6) | 80 (76.9) | 15 (75.0) |
| Minor Response | 26 (8.7) | 11 (8.9) | 10 (9.6) | 1 (5.0) |
| No Response/Stable Disease | 18 (6.0) | 8 (6.5) | 6 (5.8) | 2 (10.0) |
| Progressive Disease | 8 (2.7) | 5 (4.0) | 4 (3.9) | 1 (5.0) |
| Not evaluable | 21 (7.1) | 5 (4.0) | 4 (3.9) | 1 (5.0) |
Table 3.
Best Overall Response by Treatment
| RD: Prop [freq/n] (95%CI) | Rd: Prop [freq/n] (95%CI) | Abs. Diff. (95%CI) | Odds Ratio (95%CI) | |
|---|---|---|---|---|
| ≥ Very Good Partial Response | ||||
| Overall Cohort | 50.5% [108/214] (43.8%,57.2%) | 40.4% [84/208] (33.7%,47.1%) | 10.1% (0.6%,19.5%) | 1.50 (1.02,2.21) |
| High Risk | 36.4% [4/11] (7.9%,64.8%) | 22.2% [2/9] (0.0%,49.4%) | 14.1% (−25.2%,53.5%) | 2.00 (0.27,14.70) |
| Standard Risk | 50.0% [23/46] (35.6%,64.4%) | 43.1% [25/58] (30.4%,55.8%) | 6.9% (−12.4%,26.2%) | 1.32 (0.61,2.87) |
| ≥ Partial Response | ||||
| Overall Cohort | 81.3% [174/214] (76.1%,86.5%) | 70.2% [146/208] (64.0%,76.4%) | 11.1% (3.0%,19.2%) | 1.85 (1.17,2.91) |
| High Risk | 72.7% [8/11] (46.4%,99.0%) | 77.8% [7/9] (50.6%,100.0%) | −5.1% (−42.9%,32.8%) | 0.76 (0.10,5.96) |
| Standard Risk | 82.6% [38/46] (71.7%,93.6%) | 72.4% [42/58] (60.9%,83.9%) | 10.2% (−5.7%,26.1%) | 1.81 (0.70,4.70) |
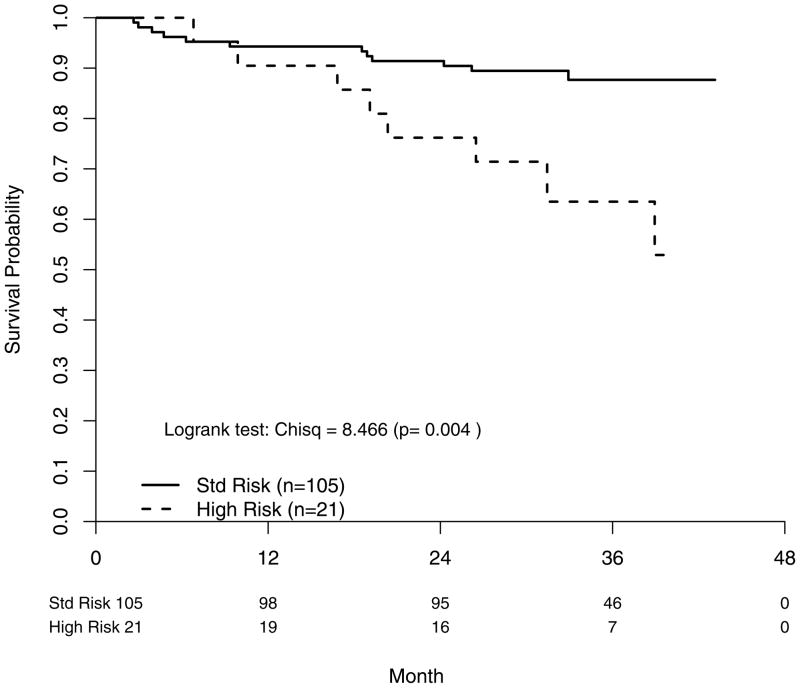
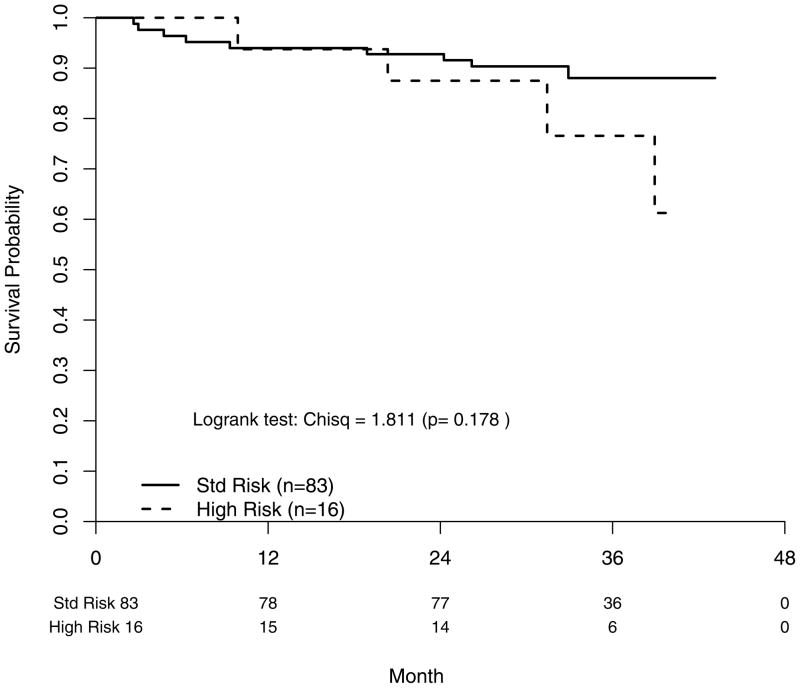
Twenty patients in the FISH cohort had died, representing about one-fifth of all deaths on study as of the last data pull in December 2008. OS for the FISH cohort compared to the remaining subgroup was significantly better (p=0.010) with a widening difference over time (Table 4). Within the FISH cohort, OS was shorter in the HR group (P=0.004) corresponding to a FISH risk status hazard ratio of 3.48 [95% CI: (1.42–8.53)] (Figure 1). The 2-year OS rates were 91% for SR MM and 76% for HR MM (Table 4)
Table 4.
Overall survival, intention-to-treat and progression-free survival Probabilities
| t-year | Overall Cohort | Non-FISH | FISH | Std Risk | High Risk | Difference |
|---|---|---|---|---|---|---|
| N | 445 | 319 | 126 | 105 | 21 | |
| Overall Survival ITT Probabilities | ||||||
| 1 | 0.91 (0.89–0.94) | 0.91 (0.87–0.94) | 0.94 (0.88–0.97) | 0.94 (0.90, 0.99) | 0.90 (0.79, 1.00) | 0.04 (−0.10, 0.17) |
| 2 | 0.83 (0.80–0.86) | 0.81 (0.76–0.85) | 0.89 (0.82–0.93) | 0.91 (0.86, 0.97) | 0.76 (0.60, 0.97) | 0.15 (−0.04, 0.34) |
| 3 | 0.74 (0.70–0.79) | 0.71 (0.65–0.76) | 0.84 (0.75–0.89) | 0.88 (0.81, 0.95) | 0.63 (0.44, 0.91) | 0.24 (0.01, 0.48) |
| Progression-Free Survival Probabilities | ||||||
| 1 | 0.66 (0.60–0.71) | 0.65 (0.59–0.71) | 0.68 (0.57–0.76) | 0.71 (0.62, 0.82) | 0.48 (0.27, 0.84) | 0.23 (−0.06, 0.52) |
| 2 | 0.50 (0.43–0.56) | 0.48 (0.41–0.56) | 0.54 (0.42–0.65) | 0.59 (0.47, 0.73) | 0.24 (0.08, 0.75) | 0.35 (0.05, 0.65) |
| 3 | 0.42 (0.35–0.48) | 0.41 (0.33–0.49) | 0.43 (0.30–0.56) | 0.46 (0.33, 0.64) | 0.24 (0.08, 0.75) | 0.22 (−0.10, 0.53) |
ITT, intention-to-treat
Figure 1.
Survival probability (overall) according to FISH-determined risk status. Std Risk; standard risk.
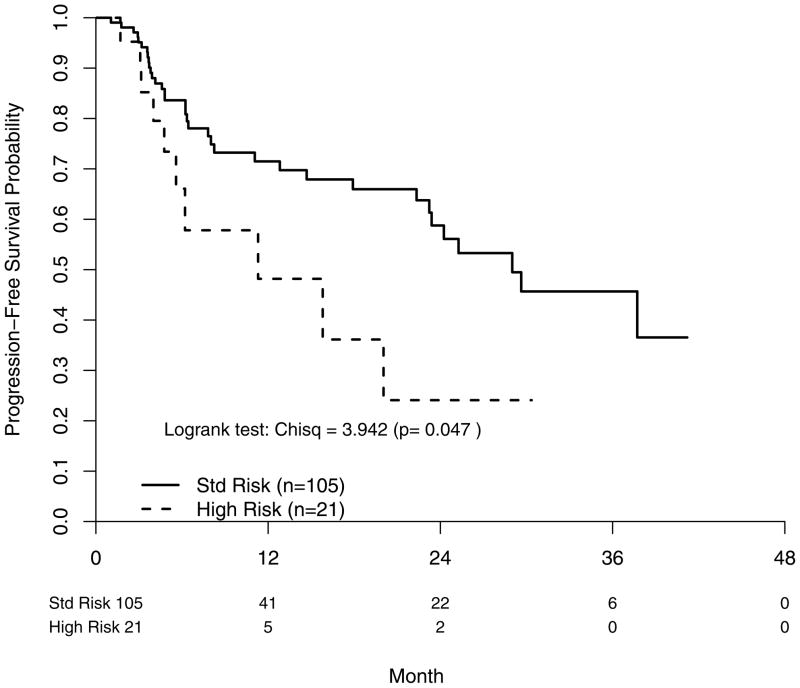
28% of all PFS events on study occurred in the FISH subgroup (45/161). The PFS distributions for the FISH and non-FISH cohorts were overlapping. Median PFS was 29 months (SR) and 11 months (HR) P=0.047, with a corresponding FISH risk status hazard ratio of 2.02 [95% CI: (0.99–4.11)] (Figure 2). 2-year PFS rates were 59% (SR) and 24% (HR) (Table 4).
Figure 2.
Progression free survival according to FISH-determined risk status. Std Risk; standard risk.
As expected, toxicity was not different between risk groups. The rate of grade 3 or higher non-haematological toxicities was 54% (SR) and 57% (SR).
Cox post-hazard regression univariate models were run for the following variables: risk (HR vs. SR), SCT as time-varying, ISS (II/III vs. I), haemoglobin (>110 vs. ≤110 g/l), Rd vs. RD, creatinine (>132.6 vs. ≤132.6 μmol/l), age (≥70 vs. <70 years), ECOG PS (1/2 vs. 0), race (white vs. non-white) and bone disease (present vs. absent). Given the low number of events in the FISH cohort (OS n=20 events and PFS n=45 events), there is power to discern only large effects so non-significance is difficult to interpret. On univariate analyses, only ISS Stage and risk were statistically significant for the OS model. It should be noted that in the non-FISH study cohort, age, ECOG PS, haemoglobin and SCT as a time-varying covariate were significantly associated with OS. Both factors were retained in a stepwise selection model that included all variables.
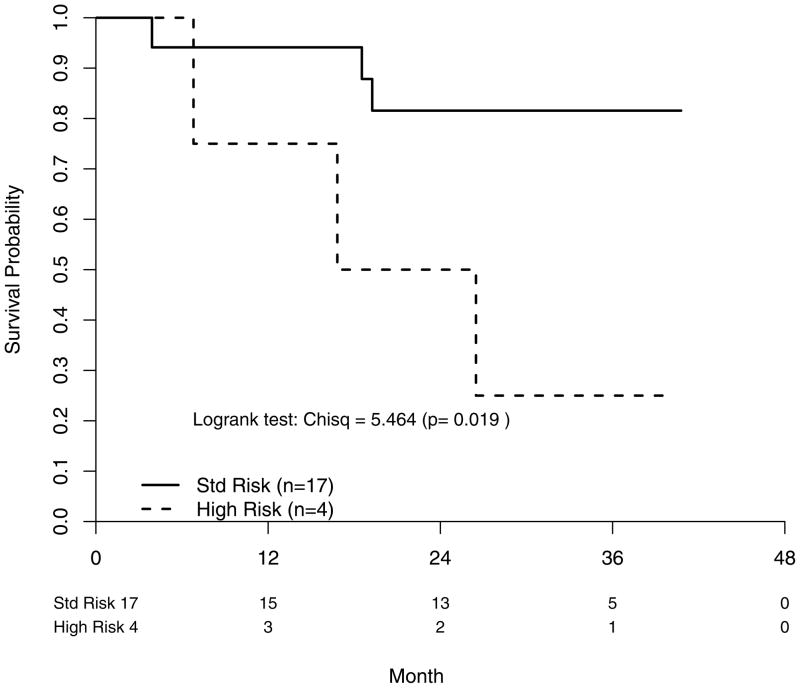
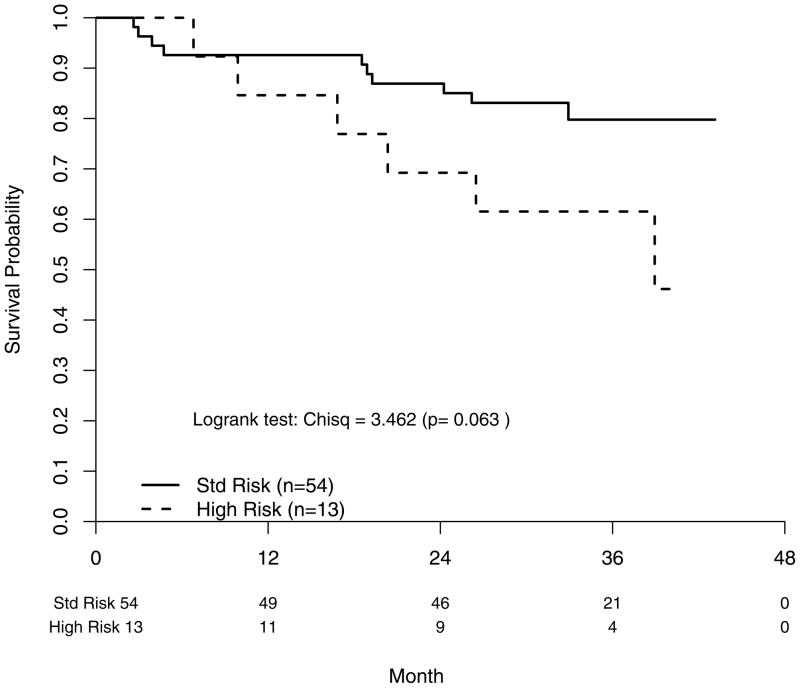
The adjusted FISH risk status and ISS Stage OS hazard ratios were 4.30 [95% CI: (1.25–14.8)] (P=0.021) and 2.63 [95% CI: (1.03–6.72)] (P=0.043), respectively. From our exploratory analyses of the OS outcomes by FISH risk status within ISS subgroups, it appears FISH risk status is further discerning in the worse-off patients. If only ISS III patients are included, HR patients were found to have shorter OS (P=0.019) (Figure 3a). This remained true when we expand the subgroup to ISS II/III patients with a risk status hazard ratio of 2.54 [95% CI: (0.92–7.04)] (P=0.063) (Figure 3b). By contrast, it is not evident that HR patients in the ISS I/II subgroup had comparatively inferior OS with a hazard ratio of 2.20 [95% CI: (0.68–7.17)] (P=0.190) (Figure 3c). For the PFS analyses, only treatment and FISH risk status were associated with PFS in univariate Cox post-hazard regression analyses. In an adjusted Cox post-hazard model, risk status and treatment both remained marginally significant, P<0.08.
Figure 3.
Fig. 3a: Overall survival segregating by FISH-determined risk status in patients with ISS Stage III.
Fig. 3b: Overall survival segregating by FISH-determined risk status in patients with ISS Stages II and III.
Fig 3c: Overall survival segregating by FISH-determined risk status in patients with ISS Stages I and II.
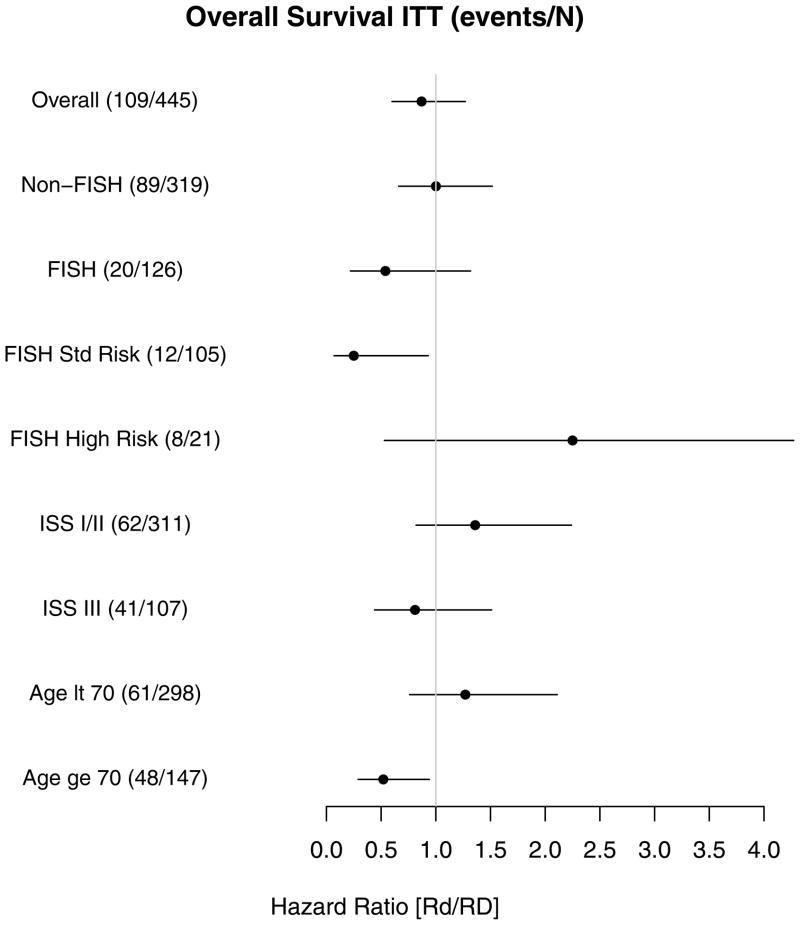
Analyses of the treatment effect on OS within prognostic subgroups were performed as exploratory due to the small number of events. As seen in Figure 4, Rd treatment was strongly favored in the SR and elderly subgroups. The unadjusted treatment hazard ratio (Rd/RD) for OS in SR patients was [0.25 95% CI:(0.07–0.93), P=0.038)] versus HR patients [2.25 95%CI:(0.53, 9.52), P=0.270], leading to significant interaction (P=0.026).
Figure 4.
Forest plot showing the effect of the shown variables on overall survival. Positioning of the hazard ratio to the right indicates an unfavourable prognostic variable, while positioning to the right indicates a favourable prognostic variable. Numbers in parentheses indicate death/at risk. ITT, intention-to-treat. Age lt, age lower than; Age ge, Age greater than or equal to.
DISCUSSION
This study explored whether established adverse genetic markers detected by FISH remain relevant for the prognosis of patients with newly diagnosed MM treated with lenalidomide and dexamethasone. We also evaluated the extent of the difference in clinical outcome depending on treatment. The studies leading to the identification of molecular classes of MM were mostly carried out using older series of patients treated with alkylators, both at standard and high doses with stem cell support (Fonseca et al, 2003) and very limited information exists regarding the validity of these factors when applied to MM patients treated with newer immunomodulatory agents (Kapoor et al, 2010; Reece et al, 2009; Avet-Loiseau et al, 2010b; Kapoor et al, 2009).
We tested patients for three of the most commonly used poor risk genetic prognostic factors: the t(4;14)(p16;q32), t(14;16)(q32;q23) and 17p13 deletion (Fonseca et al, 2009). The first two are clearly biological classifiers of the disease that have known prognostic significance. Irrespective of treatment provided, patients with these two translocations have a propensity to display other features of aggressive disease than those without these markers. In most large series, these individuals have greater tumour burden, circulating plasma cells, proliferative markers, and all of this ultimately translates to inferior outcomes. Multiple studies have previously shown that the t(4;14)(p16;q32) carries negative implications for disease-free survival and OS (Fonseca et al, 2003; Avet-Loiseau et al, 2007; Avet-Loiseau et al, 2010a). Likewise there are several studies showing that t(14;16)(q32;q23) and other MAF translocations are also associated with more aggressive disease features at baseline (Fonseca et al, 2003; Ross et al, 2010; Zhan et al, 2006), even though a single large study failed to confirm inferior long term outcomes (Avet-Loiseau et al, 2011). In this last study, other features of aggressiveness were seen though including a large number of circulating cells at the time of diagnosis (Avet-Loiseau et al, 2011). Deletions of 17p13, most if not all of them involving the TP53 locus, are believed to be the most common progression event in MM, and are always associated with aggressive disease (Fonseca et al, 2003; Avet-Loiseau et al, 2007; Drach et al, 1998). All series that have addressed deletions as a marker have confirmed the inferior outcome of patients with deletions.
To the best of our knowledge, only one other study, performed by the Mayo group and not associated with a clinical trial, showed persistence of prognostic impact of high risk genetic markers for recently diagnosed patients (Kapoor et al, 2009). Two studies have shown that, for patients with relapsed and refractory MM, these markers still identify dissimilar outcomes (Reece et al, 2009; Avet-Loiseau et al 2010b). This is the first study of these markers in the context of a prospective clinical trial showing the discriminating ability of the HR MM FISH classification.
Initial observations with bortezomib were touted as showing that this medication could, at least partially overcome the negative prognostic implications of t(4;14)(p16;q32) (San Miguel et al, 2008) or other markers of high risk, such as del13, by karyotype (Jagannath et al, 2007). Yet in another study, bortezomib treatment was shown to improve outcomes for t(4;14)(p16;q32) patients but not 17p deletion patients (Avet-Loiseau et al, 2010a). Results, however, were limited by the low power intrinsic to the small size of subgroups and limited duration of follow up. With subsequent follow up it has become apparent that the adverse prognostic impact of t(4;14)(p16;q32) is diminished but not totally eliminated (Avet-Loiseau et al, 2010a; Mateos et al 2010). These collective observations have lead to the current practice of favouring bortezomib-based induction regimens for newly diagnosed patients with HR features (Dispenzieri et al, 2007). Conversely, there has been an impression that lenalidomide in combination with dexamethasone is insufficient induction therapy for HR MM. Rd, however, has not been compared directly with bortezomib within HR or SR patient subsets. Some would argue that the two drugs in combination could eliminate the need for risk stratification but this carries an increased cost and no proven long-term benefit. In fact, other combinations, such as bortezomib, cyclophosphamide and dexamethasone, may be superior to lenalidomide, bortezomib and dexamethasone (Reeder et al, 2009; Reeder et al, 2010). Furthermore, the long-term outcome reported in the context of this study for patients who receive induction with Rd/D followed by autologous stem cell transplant are remarkable (Rajkumar et al, 2010). It is currently unknown what the effect on long-term outcomes will be for more active combinations such as VTD (bortezomib, thalidomide, dexamethasone, CVRD (cyclophosphamide, bortezomib, lenalidomide, dexamethasone) and others, but such combinatorial strategies hold the promise of being even more effective in high risk situations (Cavo et al, 2010, Kumar et al, 2010, Richardson et al, 2010). In the Bologna study the addition of bortezomib eliminated the negative effect of t(4;14)(p16;q32) as opposed to those patients only treated with thalidomide and dexamethasone (Cavo et al 2010).
Our analysis of patients with available FISH data imply that in the context of lenalidomide treatment, the presence of adverse genetic features still can identify patients with shorter survival, even after adjustment for ISS Stage. We also observed patients with absent traditional adverse genetic factors treated with Rd had very positive 1- and 2-year OS and PFS outcomes. In the confines of small numbers, we observed that HR patients treated with Rd, however, did not do especially well. An alternative hypothesis is that the more pronounced dichotomy shown for HR MM patients treated with Rd could be a function of a much-improved outcome for this agent in patients with SR MM. This is supported in the study reported by by Kapoor et al (2009), with the PFS outcome of HR patients treated with lenalidomide and dexamethasone (median HR 18.5 vs SR 36.5 months) similar to that of HR patients treated under the VISTA trail (both at approximately 19 months) (San Miguel et al, 2008; Kapoor et al, 2009). We recognize that our study is limited due to small sample size and our analyses are exploratory in nature.
While many efforts aim to improve the accuracy of prognostication of MM using genomics at the RNA- and DNA-based level (Decaux et al, 2008; Shaughnessy et al, 2007; Avet-Loiseau et al, 2009), FISH-based prognostication remains the standard of care, and has great application as a prognostic tool (Fonseca et al, 2009). Further research needs to focus on the differences in risk prediction among platforms and their incremental value over traditional clinical measures. Until more information becomes available, MM patients identified as HR by FISH should be considered at greater risk of relapse and death. While there is much hope in that MM may become a “chronic disease” this promise is not fully substantiated for HR patients. The level of evidence to support the adoption of bortezomib-based regimens for HR MM patients and/or lenalidomide-based regimens for SR MM patients needs to be enhanced. Specifically, prospective randomized trials within risk-stratified populations are needed to establish optimal treatment selection.
Acknowledgments
This study was coordinated by the Eastern Cooperative Oncology Group (Robert L. Comis, M.D., Chair) and supported in part by Public Health Service Grants CA23318, CA66636, CA21115, CA13650 and from the National Cancer Institute, National Institutes of Health and the Department of Health and Human Services. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
Footnotes
Disclosures: Relevant to this work Dr. Fonseca has received a patent for the prognostication of MM based on genetic categorization of the disease. Non relevant; he has received consulting fees from Medtronic, Otsuka, Celgene, Genzyme, BMS and AMGEN. He also has sponsored research from Cylene and Onyx.
AUTHOR CONTRIBUTIONS, ACKNOWLEDGEMENTS AND DISCLOSURES
Contributions: A=performed research, B=designed the research study, C=contributed essential reagents or tools, D=analysed the data, E=wrote the paper.
| S. J. Jacobus | No disclosures Contributions: ABCDE |
| S. Kumar | Clinical Trial Support - Celgene, Millennium, Genzyme, Novartis, Cephalon; Consultant Merck Contributions: ABCDE |
| S.A. Van Wier | No disclosures Contributions: AC |
| G.J. Ahmann | No disclosures Contributions: AC |
| K.J. Henderson | No disclosures Contributions: AC |
| N.S. Callander | No disclosures Contributions: ABCD |
| M.E. Williams | Celgene clinical trial research funding; consultant and Member of a Celgene clinical trial Data Safety Committee (total compensation <$10,000). Cephalon, Genentech, Millennium, Novartis: clinical trial research funding. Contributions: A |
| D.S. Siegel | Celgene Honoraria/Speakers Bureau/Research Support Millennium Honoraria/Speakers Bureau; Onyx-Honoraria Contributions: A |
| P.R. Greipp | No disclosures Contributions: ABCDE |
| S.V. Rajkumar | No disclosures Contributions: ABCDE |
| H. Uno | No disclosures Contributions: ABCDE |
| R. Fonseca | Patent for prognostication of MM based on genetic categorization of the disease. Consulting fees from Medtronic, Otsuka, Celgene, Genzyme, BMS and Amgen. Research funding from Cylene, Onyx. Contributions: ABCDE |
References
- Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, Leyvraz S, Michallet M, Yakoub-Agha I, Garderet L, Marit G, Michaux L, Voillat L, Renaud M, Grosbois B, Guillerm G, Benboubker L, Monocunduit M, Thieblemont C, Casassus P, Caillot D, Stoppa AM, Sotto JJ, Wetterwald M, Dumontet C, Fuzibet JG, Azais I, Dorvaux V, Zandecki M, Bataille R, Minvielle S, Harousseau JL, Facon T, Mathiot C. Genetic abnormailities in multiple myeloma: the experience of the Intergroupe Francophone du Myélome. Blood. 2007;109:3489–95. doi: 10.1182/blood-2006-08-040410. [DOI] [PubMed] [Google Scholar]
- Avet-Loiseau H, Li C, Magrangeas F, Gouraud W, Charbonnel C, Harousseau JL, Attal M, Marit G, Mathiot C, Facon T, Moreau P, Anderson KC, Campion L, Munshi NC, Minvielle S. Prognostic significance of copy-number alterations in multiple myeloma. Journal of Clinical Oncology. 2009;27:4585–90. doi: 10.1200/JCO.2008.20.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avet-Loiseau H, Leleu X, Roussel M, Moreau P, Guerin-Charbonnel C, Caillot D, Marit G, Benboubker L, Voillat L, Mathiot C, Kolb B, Macro M, Campion L, Wetterwald M, Stoppa AM, Hulin C, Facon T, Attal M, Minvielle S, Harousseau JL. Bortezomib plus dexamethasone induction improves outcome of patients with t(4;14) myeloma but not outcome of patients with del(17p) Journal of Clinical Oncology. 2010a;28:4630–4. doi: 10.1200/JCO.2010.28.3945. [DOI] [PubMed] [Google Scholar]
- Avet-Loiseau H, Soulier J, Fermand JP, Yakoub-Agha I, Attal M, Hulin C, Garderet L, Belhadj K, Dorvaux V, Minvielle S, Moreau P IFM and MAG groups. Impact of high-risk cytogenetics and prior therapy on outcomes in patients with advanced relapsed or refractory multiple myeloma treated with lenalidomide plus dexaméthasone. Leukemia. 2010b;24:623–8. doi: 10.1038/leu.2009.273. [DOI] [PubMed] [Google Scholar]
- Avet-Loiseau H, Malard F, Campion L, Magrangeas F, Sebban C, Lioure B, Decaux O, Lamy T, Legros L, Fuzibet JG, Michallet M, Corront B, Lenain P, Hulin C, Mathiot C, Attal M, Facon T, Harousseau JL, Minvielle S, Moreau P Intergroupe Francophone du Myélome. Translocation t(14;16) and multiple myeloma: is it really an independent prognostic factor? Blood. 2011;117:2009–11. doi: 10.1182/blood-2010-07-295105. [DOI] [PubMed] [Google Scholar]
- Bladé J, Samson D, Reece D, Apperley J, Björkstrand B, Gahrton G, Gertz M, Giralt S, Jagannath S, Vesole D. Criteria for evaluating disease response and progression in patients with multiple myeloma treated by high-dose therapy and haemopoietic stem cell transplantation. Myeloma Subcommittee of the EBMT. European Group for Blood and Marrow Transplant. British Journal of Haematology. 1998;102:1115–23. doi: 10.1046/j.1365-2141.1998.00930.x. [DOI] [PubMed] [Google Scholar]
- Cavo M, Tacchetti P, Patriarca F, Petrucci MT, Pantani L, Galli M, Di Raimondo F, Crippa C, Zamagni E, Palumbo A, Offidani M, Corradini P, Narni F, Spadano A, Pescosta N, Deliliers GL, Ledda A, Cellini C, Caravita T, Tosi P, Baccarani M GIMEMA Italian Myeloma Network. Bortezomib with thalidomide plus dexamethasone compared with thalidomide plus dexamethasone as induction therapy before, and consolidation therapy after, double autologous stem-cell transplantation in newly diagnosed multiple myeloma: a randomized phase 3 study. Lancet. 2010;376:2075–85. doi: 10.1016/S0140-6736(10)61424-9. [DOI] [PubMed] [Google Scholar]
- Chng WJ, Price-Troska T, Gonzalez-Paz N, Van Wier S, Jacobus S, Blood E, Henderson K, Oken M, Van Ness B, Greipp P, Rajkumar SV, Fonseca R. Clinical significance of TP53 mutation in myeloma. Leukemia. 2007;21:582–4. doi: 10.1038/sj.leu.2404524. [DOI] [PubMed] [Google Scholar]
- Decaux O, Lodé L, Magrangeas F, Charbonnel C, Gouraud W, Jézéquel P, Attal M, Harousseau JL, Moreau P, Bataille R, Campion L, Avet-Loiseau H, Minivielle S Intergroupe Francophone du Myélome. Prediction of survival in multiple myeloma based on gene expression profiles reveals cell cycle and chromosomal instability signatures in high-risk patients and hyperdiploid signatures in low-risk patients: a study of the Intergroupe Francophone du Myélome. Journal of Clinical Oncology. 2008;26:4798–805. doi: 10.1200/JCO.2007.13.8545. [DOI] [PubMed] [Google Scholar]
- Dispenzieri A, Rajkumar SV, Gertz MA, Fonseca R, Lacy MQ, Bergsagel PL, Kyle RA, Greipp PR, Witzig TE, Reeder CB, Lust JA, Russell SJ, Hayman SR, Roy V, Kumar S, Zeldenrust SR, Dalton RJ, Stewart AK. Treatment of newly diagnosed multiple myeloma based on Mayo Stratification of Myeloma and Risk-adapted Therapy (mSMART): consensus statement. Mayo Clinic Proceedings. 2007;82:323–41. doi: 10.4065/82.3.323. [DOI] [PubMed] [Google Scholar]
- Drach J, Ackerman J, Fritz E, Krömer E, Schuster R, Gisslinger H, DeSantis M, Zojer N, Fiegl M, Roka S, Schuster J, Heinz R, Ludwig H, Huber H. Presence of a p53 gene deletion in patients with multiple myeloma predicts for short survival after conventional-dose chemotherapy. Blood. 1998;92:802–9. [PubMed] [Google Scholar]
- Durie BG, Harousseau JL, Miguel JS, Bladé J, Barlogie B, Anderson K, Gertz M, Dimopoulos M, Westin J, Sonneveld P, Ludwig H, Gahrton G, Beksac M, Crowley J, Belch A, Boccadaro M, Cavo M, Turesson I, Joshua D, Vesole D, Kyle R, Alexanian R, Tricot G, Attal M, Merlini G, Powles R, Richardson P, Shimizu K, Tosi P, Morgan G, Rajkumar SV International Myeloma Working Group. International uniform response criteria for multiple myeloma. Leukemia. 2006;20:1467–73. doi: 10.1038/sj.leu.2404284. [DOI] [PubMed] [Google Scholar]
- Fonseca R, Blood E, Rue M, Harrington D, Oken MM, Kyle RA, Dewald GW, Van Ness B, Van Wier SA, Henderson KJ, Bailey RJ, Greipp PR. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;101:4569–75. doi: 10.1182/blood-2002-10-3017. [DOI] [PubMed] [Google Scholar]
- Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, Morgan G, Van Ness B, Chesi M, Minvielle S, Neri A, Barlogie B, Kuehl WM, Liebisch P, Davies F, Chen-Kiang S, Durie BG, Carrasco R, Sezer O, Reiman T, Pilarski L, Avet-Loiseau H International Myeloma Working Group. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23:2210–21. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannath S, Richardson PG, Sonneveld P, Schuster MW, Irwin D, Stadtmauer EA, Facon T, Harousseau JL, Cowan JM, Anderson KC. Bortezomib appears to overcome the poor prognosis conferred by chromosome 13 deletion in phase 2 and 3 trials. Leukemia. 2007;21:151–7. doi: 10.1038/sj.leu.2404442. [DOI] [PubMed] [Google Scholar]
- Kapoor P, Kumar S, Fonseca R, Lacy MQ, Witzig TE, Hayman SR, Dispenzieri A, Buadi F, Bergsagel PL, Gertz MA, Dalton RJ, Mikhael JR, Dingli D, Reeder CB, Lust JA, Russell SJ, Roy V, Zeldenrust SR, Stewart AK, Kyle RA, Greipp PR, Rajkumar SV. Impact of risk stratification on outcome among patients with multiple myeloma receiving initial therapy with lenalidomide and dexamethasone. Blood. 2009;114:518–21. doi: 10.1182/blood-2009-01-202010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor P, Fonseca R, Rajkumar SV, Sinha S, Gertz MA, Stewart AK, Bergsagel PL, Lacy MQ, Dingli DD, Ketterling RP, Buadi F, Kyle RA, Witzig TE, Greipp PR, Dispenzieri A, Kumar S. Evidence for cytogenetic and fluorescence in situ hybridization risk stratification of newly diagnosed multiple myeloma in the era of novel therapies. Mayo Clinic Proceedings. 2010;85:532–7. doi: 10.4065/mcp.2009.0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SK, Flinn I, Noga SJ, Hari P, Rifkin R, Callander N, Bhandari M, Wolf JL, Gasparetto C, Krishnan A, Grosman D, Glass J, Sahovic EA, Shi H, Webb IJ, Richardson PG, Rajkumar SV. Bortezomib, dexamethasone, cyclophosphamide and lenalidomide combination for newly diagnosed multiple myeloma: phase 1 results from the multicenter EVOLUTION study. Leukemia. 2010;24:1350–6. doi: 10.1038/leu.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos M-V, Gutierrez NC, Paiva B, Oriol A, Martinez-Lopez J, Teruel AI, de Paz R, Garcia-Larana J, Bengoechea E, Martin A, Diaz-Mediavilla J, Palomera L, Gonzalez Y, Hernandez JM, Sureda A, Bello JL, Bargay J, Penalver F-J, Ribera J-M, Martin-Mateos M-L, Garcia-Sanz R, Cibeira MT, Martin-Ramos M-L, Vidriales M-B, Montalban M-A, Lahuerta J-J, Blade J, San Miguel JF., Sr Clinical Outcome According to Both Cytogenetic Abnormalities (CA) Detected by Fluorescence In Situ Hibridization (FISH) and Hyperdiploidy Assessed by Flow Cytometry (FCM) In Elderly Newly Diagnosed Myeloma Patients Treated with A Bortezomib-Based Combination. Blood (ASH Annual Meeting Abstracts) 2010;116:309. [Google Scholar]
- Reece D, Song KW, Fu T, Roland B, Chang H, Horsman DE, Mansoor A, Chen C, Masih-Khan E, Trieu Y, Bruyere H, Stewart DA, Bahlis NJ. Influence of cytogenetics in patients with relapsed or refractory multiple myeloma treated with lenalidomide plus dexamethasone: adverse effect of deletion. Blood. 2009;114:522–5. doi: 10.1182/blood-2008-12-193458. [DOI] [PubMed] [Google Scholar]
- Reeder CB, Reece DE, Kukreti V, Chen C, Trudel S, Hentz J, Noble B, Pirooz NA, Spong JE, Piza JG, Zepeda VH, Mikhael JR, Leis JF, Bergsagel PL, Fonseca R, Stewart AK. Cyclophosphamide, bortezomib and dexamethasone induction for newly diagnosed multiple myeloma: high response rates in a phase III clinical trial. Leukemia. 2009;23:1337–41. doi: 10.1038/leu.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder CB, Reece DE, Kukreti V, Chen C, Trudel S, Laumann K, Hentz J, Pirooz NA, Piza JG, Tiedemann R, Mikhael JR, Bergsagel PL, Leis JF, Fonseca R, Stewart AK. Once-versus twice-weekly bortezomib induction therapy with CyBorD in newly diagnosed multiple myeloma. Blood. 2010;115:3416–7. doi: 10.1182/blood-2010-02-271676. [DOI] [PubMed] [Google Scholar]
- Rajkumar SV, Jacobus S, Callander NS, Fonseca R, Vesole DH, Williams ME, Abonour S, Siegel DS, Katz M, Greipp PR Eastern Cooperative Oncology Group. Lenalidomide plus high-dose dexamethasone versus lenalidomide plus low-dose dexamethasone as initial therapy for newly diagnosed multiple myeloma: an open-label randomised controlled trial. Lancet Oncology. 2010;11:29–37. doi: 10.1016/S1470-2045(09)70284-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson PG, Weller E, Lonial S, Jakubowiak AJ, Jagannath S, Raje NS, Avigan DE, Xie W, Ghobrial IM, Schlossman RL, Mazumder A, Munshi NC, Vesole DH, Joyce R, Kaufman JL, Doss D, Warren DL, Lunde LE, Kaster S, Delaney C, Hideshima T, Mitsiades CS, Knight R, Esseltine DL, Anderson KC. Lenalidomide, bortezomib, and dexamethasone combination therapy in patients with newly diagnosed multiple myeloma. Blood. 2010;116:679–86. doi: 10.1182/blood-2010-02-268862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross FM, Chiecchio L, Dagrada G, Protheroe RK, Stockley DM, Harrison CJ, Cross NC, Szubert AJ, Drayson MT, Morgan GJ UK Myeloma Forum. The t(14;20) is a poor prognostic factor in myeloma but is associated with long-term stable disease in monoclonal gammopathies of undetermined significance. Haematologica. 2010;95:1221–5. doi: 10.3324/haematol.2009.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Miguel JF, Schlag R, Khuageva NK, Dimopoulos MA, Shpilberg O, Kropff M, Spicka I, Petrucci MT, Palumbo A, Samoilova OS, Dmosynska A, Abdulkadyrov KM, Schots R, Jiang B, Mateos MV, Anderson KC, Essentine DL, Liu K, Cakana A, vande Velde H, Richardson PG VISTA Trial Investigators. Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. New England Journal of Medicine. 2008;259:906–17. doi: 10.1056/NEJMoa0801479. [DOI] [PubMed] [Google Scholar]
- Shaughnessy JD, Jr, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, Stewart JP, Kordsmeier B, Randolph C, Williams DR, Xiao Y, Xu H, Epstein J, Anaissie E, Krishna SG, Cottler-Fox M, Hollmig K, Mohiuddin A, Pineda-Roman M, Tricot G, van Rhee F, Sawyer J, Alsayed Y, Walker R, Zangari M, Crowley J, Barlogie B. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109:2276–84. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, Epstein J, Yaccoby S, Sawyer J, Burington B, Anaissie E, Hollmig K, Pineda-Roman M, Tricot G, van Rhee F, Walker R, Zangari M, Crowley J, Barlogie B, Shaughnessy JD., Jr The molecular classification of multiple myeloma. Blood. 2006;108:2020–28. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]