Abstract
12-Oxophytodienoate reductase 3 (OPR3), one of the enzymes involved in the biosynthesis of the plant hormone jasmonic acid (JA), catalyzes the reduction of the cyclopentenone ring of (9S,13S)-12-oxophytodienoate [(9S,13S)-OPDA]. However, there has been no structural information about the interaction between OPRs and the physiologically relevant (9S,13S)-OPDA. Here we report the crystal structure of Arabidopsis thaliana OPR3 in complex with 8-iso prostaglandin A1 (8-iso PGA1) which has the same stereochemistry in the cyclopentenone ring as in the physiologically relevant 9S,13S-OPDA. This structure reveals a new binding mode for substrate that likely contributes to the relaxed stereospecificity observed for AtOPR3.
Keywords: 12-oxophytodienoate reductase isoform 3, 8-iso prostaglandin A1, jasmonic acid pathway, Arabidopsis thaliana
INTRODUCTION
Jasmonic acid (JA) is a plant secondary messenger that contributes to coordinated responses to many biotic and abiotic stresses, as well as other diverse processes such as pollen maturation and wound responses.1–2 Biosynthesis of JA originates from linolenic acid in chloroplast membranes, and a number of enzymes are involved in the conversion of this polyunsaturated fatty acid into cyclopentane containing compound JA.3 12-oxophytodienoate reductase 3 (OPR3), one of the enzymes involved in the biosynthesis of JA, uniquely catalyzes the reduction of the cyclopentenone ring of (9S,13S)-12-oxophytodienoate [(9S,13S)-OPDA].4 The JA-deficient phenotype of OPR3 loss-of-function mutants in Arabidopsis thaliana indicates that other OPR isoforms cannot substitute for OPR3 in JA biosynthesis.5
The physiologically relevant (9S,13S)-enantiomer of OPDA is not a substrate for the majority of OPRs.4 Thus the unique function of OPR3 in the biosynthesis of JA is a result of its relaxed stereoselectivity with respect to OPDA stereoisomers. While Arabidopsis thaliana OPR3 (AtOPR3) and Lycopersicon esculentum OPR3 (LeOPR3) reduce all four stereoisomers of OPDA, the large majority of OPR isoforms seem to resemble LeOPR1 and AtOPR1/2 in their substrate specificity.6–7 The biological roles of OPR1 and OPR2 have not been established, however, these enzymes efficiently reduce 9R,13R-OPDA but fail to react with the enantiomeric JA precursor 9S,13S-OPDA.7–9
To date, the several ligand- and cofactor-bound structures of OPR1 and OPR3 have been reported.7,10–13 However, there has been no structural information about the interaction between OPRs and the physiologically relevant (9S,13S)-OPDA. Here we report the crystal structure of AtOPR3 in complex with 8-iso prostaglandin A1 (8-iso PGA1) which has the same α,β-unsaturated carbonyl group and stereochemistry in the cyclopentenone ring as in the physiologically relevant 9S,13S-OPDA. Our structure shows that hydrogen bonds and van der Waals’ interactions between AtOPR3 and 8-iso PGA1 stabilize the substrate molecule for reduction of the alkenyl group within the cyclopentenone ring. The positioning of 8-iso PGA1 reveals a new binding orientation for substrate in the active site that likely contributes to the relaxed stereospecificity observed for AtOPR3 relative to other OPR’s.
MATERIALS AND METHODS
Cloning, protein expression, purification, and crystallization
Cloning, expression, purification and crystallization of AtOPR312 were as previously described. The calculated molar absorptivity was 43320 M−1 cm−1. 8-iso PGA1 was from Cayman Chemical (Ann Arbor, MI). HPLC grade methanol and NADPH were from Sigma-Aldrich (St. Louis, MO).
X-ray data collection, structure determination, and model evaluation
X-ray diffraction data were collected using synchrotron beam line 23-ID at the Advanced Photon Source at the Argonne National Laboratory GM/CA-CAT. The diffraction images were integrated and scaled using HKL2000.14 The structure of the 8-iso PGA1 bound AtOPR3 was determined by molecular replacement using AtOPR3 structure in complex with FMN (PDB 1q45) as a template. Cross-rotational search followed by translational search was performed utilizing the program using MolRep from CCP4.15 Subsequent manual model building and refinement were carried out using Coot and Refmac from CCP4 program suite.15 The stereochemical quality of the refined model was assessed using MolProbity.16 Structure figures were generated using PyMOL (DeLano Scientific LLC).
Sedimentation equilibrium studies
Sedimentation equilibrium studies were performed with a Beckman Optima XL-A analytical ultracentrifuge in the Biophysics Instrumentation Facility at the University of Wisconsin-Madison. AtOPR1 was dissolved in 5 mM Tris-Hcl at pH 8, containing 50 mM NaCl, while AtOPR3 was dissolved in 5 mM MES-NaOH at pH 6, containing 50 mM NaCl. From a single stock of each protein, dilutions were made to obtain three samples with UV absorbance readings of 0.2 (4.6 μM), 0.5 (11.5 μM) and 0.8 (18.5 μM) at 280 nm. Solvent densities measured using an Anton Paar DMA5000 at 4°C were 1.00241 and 1.00257 g mL−1, respectively. The partial specific volumes for AtOPR1 and AtOPR3 based on their sequences, neglecting contributions from flavin, were the same, 0.732 mL g−1. For each protein, ~100 μL of each of the three dilutions were placed in 1.2 cm pathlength double-sector, charcoal-filled, epon centerpieces with the appropriate buffers as references. Samples were spun to equilibrium at various speeds at 4°C. Superimposed gradients recorded at 278 nm 2–3 h apart were taken to indicate equilibrium. Data were collected at 278 nm and in most cases at 461 nm and/or 378 nm. A high-speed depletion of protein material from the cells was used as a measure of non-sedimented baseline material. For a given protein, data from all speeds and/or wavelengths were fit to various models including a fixed contribution from the non-sedimented material.
Enzyme reaction
Reactions of AtOPR1, AtOPR2 and AtOPR3 with 8-iso PGA1 and NADPH were monitored by optical spectrometry. A blank was obtained with buffer containing NADPH (150 μM), substrate was added, and the reaction was initiated by addition of enzyme (~1 μM). The oxidation of NADPH resulted in a decrease in absorbance around 340 nm, while reduction of the substrate caused a decrease in the absorption band at ~220 nm. Substrate and product were separated using a Shimadzu VP Series HPLC equipped with a photodiode array spectrophotometer and an ACE C18 column (100 × 4.6 mm, 3 micron particle size) using 70% methanol and 30% water (70:30 v/v) pumped isocratically at 1 mL/min. The substrate and product were monitored and quantified using UV absorption at 218 nm (8-iso PGA1, ε218 = 8200 M−1 cm−1) and 220 nm (reduced product, ε220 = 1700 M−1 cm−1).
The masses of 8-iso PGA1 and the reduced product were determined using a Shimadzu LCMS-2010 with an HPLC inlet, an electrospray ionizer, and a quadrupole analyzer at the Paul Bender Chemical Instrument Center at the Department of Chemistry, University of Wisconsin-Madison.
Data deposition
Atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank (accession code 2G5W).
RESULTS AND DISCUSSION
In order to study the molecular properties that underlie the distinct substrate preferences of OPR3, we analyzed the structure AtOPR3 in complex with 8-iso PGA1 which has the same α,β-unsaturated carbonyl group and stereochemistry in the cyclopentenone ring as in the physiologically relevant (9S,13S)-OPDA (Fig. 1a and 1b). We soaked the AtOPR3 crystals in a crystallization solution containing 5 mM 8-iso PGA1 and the crystal structure of 8-iso PGA1 bound AtOPR3 was solved to 2.6 Å by molecular replacement using AtOPR3 structure as a phasing model (PDB ID: 1q45).12 The final model includes two AtOPR3 monomers per asymmetric unit, and each AtOPR3 monomer contains one FMN and one 8-iso PGA1 molecule. The R and Rfree factors were 20.2% and 26.4%, respectively (Table I). The N-terminal (7 residues in chain A; 8 residues in chain B) and C-terminal regions (3 residues in chain A; 5 residues in chain B) of the protein, as well as the L6 loop region (Try285—Glu301 in chain A; Gly289—Asp299 in chain B) in the crystal have not been included in the refined model. The two chains in the asymmetric unit are highly similar to each other, with r.m.s. deviation of 0.25 Å. When compared with the ligand-free AtOPR3 structure,12 the structure of 8-iso PGA1 bound AtOPR3 has no significant conformational change (Fig. 1c). The previously suspected substrate binding loop region (residues Ile133 to Tyr150), which corresponds to the Lβ3 substrate-binding loop of OPR1, was found to neither move nor interact with 8-iso PGA1. Likewise, comparison of the ligand-free and (9R,13R)-OPDA-bound LeOPR110 showed no significant conformational changes.
Figure 1.
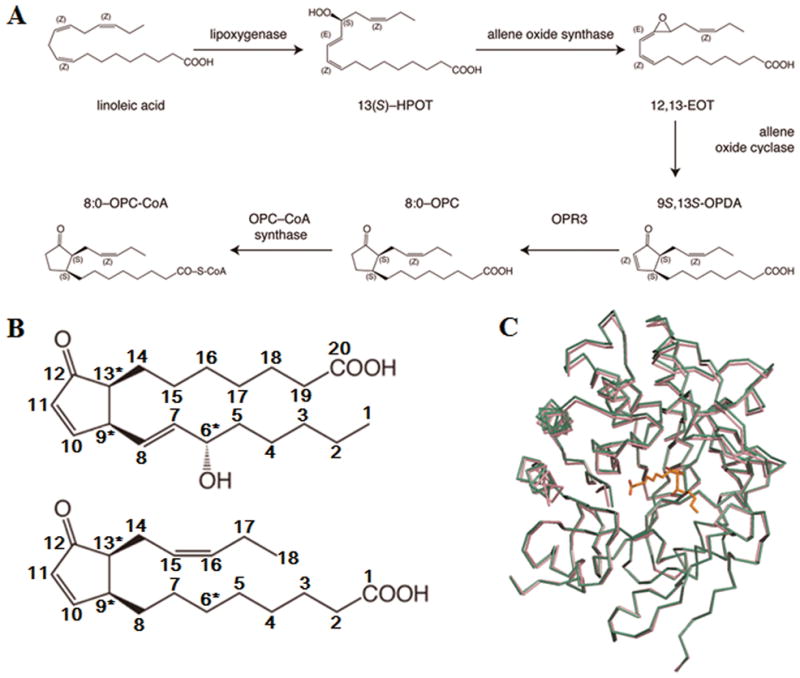
(A) Schematic representation of jasmonic acid biosynthesis.
(B) Structures of 8-iso-PGA1 (top) and (9S,13S)-OPDA (bottom).
(C) Aligned structures of FMN-bound AtOPR3 (colored in pink) and 8-iso-PGA1-FMN-AtOPR3 complex (colored in green). The bound PGA1 is shown in sticks.
Table I.
Statistics for data collection and model refinement
| Data collectiona | AtOPR3 in complex with 8-iso PGA1 |
|---|---|
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 79.70, 86.61, 123.58 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å)b | 50.00–2.58 (2.67-2.58) |
| No. reflections | 22750 |
| Rmerge (%)c | 15.7 (36.6) |
| I/σId | 9.55 (3.42) |
| Completeness (%) | 82 (50) |
| Redundancy | 6.1 (3.9) |
| Refinement e,f | |
| Resolution (Å) | 48.83 - 2.58 (2.64-2.58) |
| No. reflections | 22704 (927) |
| Rwork / Rfree | 0.202 / 0.264 (0.290 / 0.430) |
| No. of atoms | |
| Protein | 5627g |
| Ligand/ion | 110h |
| Water | 108 |
| Mean B-value (overall) | 39.82 |
| Ramachandran analysis | |
| Most favored regions | 273 (89.2%) |
| Additional allowed regions | 33 (10.8%) |
| Disallowed regions | 0 (0%) |
| R.m.s deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 0.977 |
Data collected at Sector 32 of the Advanced Photon Source.
Numbers in parentheses indicate the highest resolution shell of 20.
Rmerge = Σh Σi |I(h)i − <I(h)>| / Σh Σi I(h)i, where I(h) is the observed intensity of relection h, and < I(h) > is the average intensity obtained from multiple measurements.
The root-mean-squared value of the intensity measurements divided by their estimated standard deviation.
Rwork = Σ | | Fo | − | Fc | | / Σ | Fo |, where | Fo | is the observed structure factor amplitude and | Fc | is the calculated structure factor amplitude.
Rfree = R-factor based on 4.96% of the data excluded from refinement.
Number of non-hydrogen protein atoms included in refinement.
These include FMN and 8-iso PGA1.
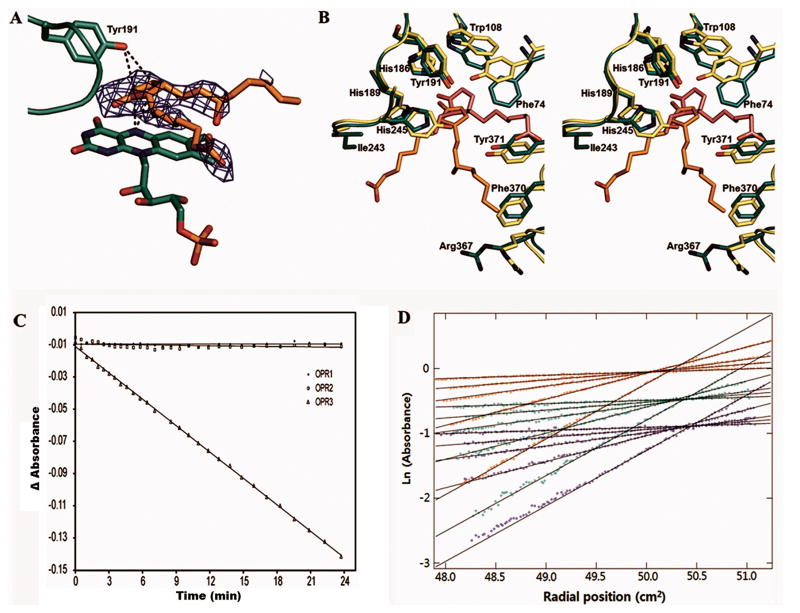
The cyclopentenone ring of 8-iso PGA1, which is structurally comparable to the corresponding cyclopentenone ring of OPDA, forms stacking interactions with the isoalloxazine ring of the FMN (Fig. 2a), and its carbonyl oxygen is in close contact with the two catalytic histidine residues His186 and His189 (Fig. 2b). The double bond formed by C10 and C11 carbon atoms of the cyclopentenone ring of the bound 8-iso-PGA1 is located closely to both hydride (FMN-N5) and proton (Tyr191-OH) donors, suggesting that the cyclopentenone ring in 8-iso-PGA1 has been effectively oriented for redox chemistry in this enzyme-substrate complex (Fig. 2a). While the gatekeeper residues Tyr78 and Tyr246 of AtOPR1 form a narrow entrance into the cavity above the FMN, the corresponding amino acids of AtOPR3, Phe74 and His245, turn away from the active site, giving rise to a widening of the cavity in OPR3 (Fig. 2b). The importance of this position in stereoselectivity is also supported by substitution of the corresponding phenylalanine and histidine residues in LeOPR3 with tyrosine residues, which shifted the specificity of the mutated enzymes toward (9R,13R)-OPDA over (9S,13S)-OPDA.13 These geometrical differences most likely account for the high degree of substrate stereoselectivity of OPR1 and the more relaxed specificity of OPR3 as proposed earlier.13
Figure 2.
(A) Orientation of the bound 8-iso-PGA1 relative to the FMN and the general acid Tyr191. The Fo − Fc electron density of the bound 8-iso-PGA1 is contoured at 2.5 σ. The interactions of the double bond-forming carbon atoms of 8-iso-PGA1 with N5 nitrogen atom of FMN and the general acid Tyr191 are indicated by dotted lines.
(B) Stereo view of the substrate binding site of the 8-iso-PGA1-FMN-AtOPR3 complex structure (green) overlaid with the structure of (9R,13R)-OPDA-FMN-LeOPR1 complex (light green) after structural alignment. Residues are numbered according to the AtOPR3 sequence. The FMN molecules of the enzymes superimpose perfectly with each other and are left out for clarity.
(C) Enzymatic assay results of three A. thaliana OPR isoforms using 8-iso-PGA1 as a substrate. Time-dependent change in A340 observed from AtOPR1, AtOPR2, and AtOPR3 are indicated by circles, rectangles, and triangle, respectively.
(D) Sedimentation equilibrium data for three concentrations of AtOPR3 (blue, 4,6 μM; green, 11.5 μM; red, 18.5 μM) and 5 speeds (4.2k, 6.6k, 8.6k, 12k, 17.5k) detected at 278 nm. Global curve fitting was consistent with the presence of a single species at all concentrations observed. The average molecular weight from the global fit of data to the 278 nm data was 44,101 ± 41 Da for AtOPR3. These results are in good agreement with the molecular weight from sequence of 42,694 Da. Data obtained at other wavelengths (378 nm or 461 nm, corresponding to FAD absorption peaks) were consistent with this analysis and their inclusion in global fits minimally impacted the calculated weight average molecular weights.
Compared to (9R,13R)-OPDA bound to LeOPR1,10 the binding mode of the cyclopentenone ring of 8-iso-PGA1 differs in the distance of the double bond and its angle relative to FMN-N5 (Fig. 2a). The small differences in the positions of the cyclopentenone rings propagate into much larger differences in the positions of the alkyl chains as they emerge from the interior of the active site (Fig. 2b). These discrepancies imply that contacts of the alkyl chains with distal residues may have important contributions to the differences in substrate specificity observed for OPR isoforms. As a result, the 8S tail of 8-iso-PGA1 has van der Waals interactions with the side chains of His189 and Ile243 (Fig. 2b). These interactions potentially mimic hydrophobic interactions with the 13S pentenyl side chain of the natural substrate. The methyl end of the 12S tail of the bound 8-iso-PGA1 locates within 4 Å of the aliphatic part of the Arg367 side chain and within 6 Å of the guanidinium group. Since the methyl group of the 12S tail of 8-iso-PGA1 is replaced by a carboxyl group in the natural substrate (9S,13S)-OPDA, (Fig. 2b), a hydrogen bonding interaction between the natural substrate and the guanidinium group of Arg367 may stabilize the natural substrate-enzyme complex. In addition, the strong hydrogen bond between the PGA1 carboxyl group and the main chain nitrogen of Arg284 that might pull the substrate more out of the cavity than (9S,13S)-OPDA and that leads to very weak hydrogen bonds of the carbonyl oxygen to the two activating histidines.
In order to confirm that 8-iso-PGA1 was an effective substrate for AtOPR3, we performed an enzymatic assay using 8-iso-PGA1 as a substrate (Fig. 2c). The result indicated that 8-iso-PGA1 was an active substrate for AtOPR3 with apparent kcat and KM values of 0.23 s−1 and 22 μM, respectively. Of the three Arabidopsis OPR isoforms tested, only AtOPR3 reduced 8-iso-PGA1, while AtOPR1 and AtOPR2 did not (Fig. 2c). This result is consistent with the stereochemical preferences previously established for the JA pathway and with the known reactions of the AtOPR isoforms with the different OPDA stereoisomers.17
While most OPRs have been described as monomers, LeOPR crystallizes as a self-inhibited dimer, in which the substrate binding pocket is occupied by loop L6 of the neighboring protomer.7,13 In contrast to the self-inhibited dimeric structure of LeOPR3, the structure of substrate-free and 8-iso-PGA1-bound structures of OPR3 showed that the protein exists as a monomer in the crystal. Incorporation of 8-iso-PGA1 into the active site was not affected by crystal packing while soaking the OPDA substrate into LeOPR3 crystals or cocrystallization trials were unsuccessful. Furthermore, sedimentation equilibrium measurements on AtOPR1 and AtOPR3 showed that both proteins were monomers at concentrations up to ~30 μM, at which LeOPR3 forms a self-inhibited dimer (Fig. 2d).13
In the crystal structure of LeOPR3, the sulfate is located at the dimer interface, close to Tyr364, and is positioned perfectly to mimic a phosphorylated tyrosine residue.7 The bound sulfate ion directly interacts with the protruding loop L6 from the protomer, stabilizing the dimer formation of the protein. Thus, it has been suggested that dimerization and hence OPR3 activity may be regulated by the reversible phosphorylation of Tyr364 in vivo.7 In the structures of substrate-free and 8-iso-PGA1 bound AtOPR3 the two monomers observed in the asymmetric unit of AtOPR3 crystals can be superimposed with the corresponding protomers of the LeOPR3 dimer, suggesting a similar dimeric structure for AtOPR3. Although the L6 loop of AtOPR3 is poorly defined in the refined structure, it clearly protrudes toward the active site of the molecular neighbor in a similar fashion as in LeOPR3. Considering that the amino acid sequence of the loop L6 is highly conserved among the OPR3-like enzymes, it is still intriguing that OPR3 activity is regulated by self-inhibitory dimerization, which might be controlled by reversible phosphorylation of the protein, although it awaits further characterizations to confirm this hypothesis.
Acknowledgments
Grant sponsor: NIH; Grant number: GM064598, GM074901 (Protein Structure Initiative), and T32 GM08349 (Institutional Biotechnology Pre-Doctoral Training Grant);
Grant sponsor: NSF; Grant number: MCB-0316232;
Grant Sponsor: Korea Ministry of Education, Science and Technology; Grant number: 2011-0004305, 2011-0027449, and NRF-M1AXA002-2010-0029770 (Basic Science Research Program and the Global Frontier through the National Research Foundation);
Grant sponsor: Korea Ministry of Health and Welfare (Korea Healthcare Technology R&D Project); Grant number: A092006;
Grant sponsor: Seoul National University (Research Settlement Fund for the new faculty).
The authors thank all members of the Center for Eukaryotic Structural Genomics for various important contributions. Beamline 23-ID at the Argonne National Laboratory GM/CA-CAT is supported by the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). The Advanced Photon Source is supported by the U.S. Department of Energy, Basic Energy Sciences.
Abbreviations
- OPR3
12-oxophytodienoate reductase isoform 3
- PGA1
prostaglandin A1
- JA
jasmonic acid
References
- 1.Liechti R, Farmer EE. The jasmonate pathway. Science. 2002;296(5573):1649–1650. doi: 10.1126/science.1071547. [DOI] [PubMed] [Google Scholar]
- 2.Strassner J, Schaller F, Frick UB, Howe GA, Weiler EW, Amrhein N, Macheroux P, Schaller A. Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J. 2002;32(4):585–601. doi: 10.1046/j.1365-313x.2002.01449.x. [DOI] [PubMed] [Google Scholar]
- 3.Vick BA, Zimmermann DC. Distribution of a Fatty Acid cyclase enzyme system in plants. Plant Physiol. 1979;64(2):203–205. doi: 10.1104/pp.64.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaller A, Stintzi A. Enzymes in jasmonate biosynthesis - structure, function, regulation. Phytochemistry. 2009;70(13–14):1532–1538. doi: 10.1016/j.phytochem.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 5.Stintzi A, Browse J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci U S A. 2000;97(19):10625–10630. doi: 10.1073/pnas.190264497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaller F, Biesgen C, Mussig C, Altmann T, Weiler EW. 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta. 2000;210(6):979–984. doi: 10.1007/s004250050706. [DOI] [PubMed] [Google Scholar]
- 7.Breithaupt C, Kurzbauer R, Lilie H, Schaller A, Strassner J, Huber R, Macheroux P, Clausen T. Crystal structure of 12-oxophytodienoate reductase 3 from tomato: self-inhibition by dimerization. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(39):14337–14342. doi: 10.1073/pnas.0606603103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaller F, Hennig P, Weiler EW. 12-Oxophytodienoate-10,11-reductase: occurrence of two isoenzymes of different specificity against stereoisomers of 12-oxophytodienoic acid. Plant physiology. 1998;118(4):1345–1351. doi: 10.1104/pp.118.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biesgen C, Weiler EW. Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10,11-reductases from Arabidopsis thaliana. Planta. 1999;208(2):155–165. doi: 10.1007/s004250050545. [DOI] [PubMed] [Google Scholar]
- 10.Breithaupt C, Strassner J, Breitinger U, Huber R, Macheroux P, Schaller A, Clausen T. X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE. Structure. 2001;9(5):419–429. doi: 10.1016/s0969-2126(01)00602-5. [DOI] [PubMed] [Google Scholar]
- 11.Fox BG, Malone TE, Johnson KA, Madson SE, Aceti D, Bingman CA, Blommel PG, Buchan B, Burns B, Cao J, Cornilescu C, Doreleijers J, Ellefson J, Frederick R, Geetha H, Hruby D, Jeon WB, Kimball T, Kunert J, Markley JL, Newman C, Olson A, Peterson FC, Phillips GN, Jr, Primm J, Ramirez B, Rosenberg NS, Runnels M, Seder K, Shaw J, Smith DW, Sreenath H, Song J, Sussman MR, Thao S, Troestler D, Tyler E, Tyler R, Ulrich E, Vinarov D, Vojtik F, Volkman BF, Wesenberg G, Wrobel RL, Zhang J, Zhao Q, Zolnai Z. X-ray structure of Arabidopsis At1g77680, 12-oxophytodienoate reductase isoform 1. Proteins. 2005;61(1):206–208. doi: 10.1002/prot.20533. [DOI] [PubMed] [Google Scholar]
- 12.Malone TE, Madson SE, Wrobel RL, Jeon WB, Rosenberg NS, Johnson KA, Bingman CA, Smith DW, Phillips GN, Jr, Markley JL, Fox BG. X-ray structure of Arabidopsis At2g06050, 12-oxophytodienoate reductase isoform 3. Proteins. 2005;58(1):243–245. doi: 10.1002/prot.20162. [DOI] [PubMed] [Google Scholar]
- 13.Breithaupt C, Kurzbauer R, Schaller F, Stintzi A, Schaller A, Huber R, Macheroux P, Clausen T. Structural basis of substrate specificity of plant 12-oxophytodienoate reductases. J Mol Biol. 2009;392(5):1266–1277. doi: 10.1016/j.jmb.2009.07.087. [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 15.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 16.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50(3):437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 17.Schaller F. Enzymes of the biosynthesis of octadecanoid-derived signalling molecules. J Exp Bot. 2001;52(354):11–23. [PubMed] [Google Scholar]