Abstract
CsrA is a widely distributed RNA binding protein that regulates translation initiation and/or mRNA stability of target transcripts. CsrA activity is antagonized by sRNA(s) containing multiple CsrA binding sites in several Gram-negative bacterial species. Here we discover FliW, the first protein antagonist of CsrA activity that constitutes a partner switching mechanism to control flagellin synthesis in the Gram-positive organism Bacillus subtilis. Following the flagellar assembly checkpoint of hook completion, secretion of flagellin (Hag) releases FliW protein from a FliW-Hag complex. FliW then binds to CsrA and relieves CsrA-mediated translational repression of hag for flagellin synthesis concurrent with filament assembly. Thus, flagellin homeostatically restricts its own translation. Homeostatic autoregulation may be a general mechanism to precisely control structural subunits required at specific times and in finite amounts such as those involved in the assembly of flagella, type III secretion machines, and pili. Finally, phylogenetic analysis suggests that CsrA, a highly pleiotropic virulence regulator in many bacterial pathogens, had an ancestral role in flagellar assembly and evolved to co-regulate various cellular processes with motility.
Keywords: Flagellin, CsrA, homeostasis, FliW, partner-switching
INTRODUCTION
Bacteria swim by rotating helical propeller-like flagella. Flagella are assembled from over 30 proteins built from the inside-out in three architectural domains (Chevance and Hughes, 2008). The first domain to be assembled is the basal body that anchors the flagellum to the cell membrane, provides the power for rotation, and the apparatus for secretion of the more distal components. The next domain is the hook, a flexible universal joint that changes the angle of flagellar rotation. The final and most distal domain is the filament that polymerizes into a helix made of a single protein flagellin, which when synthesized is among the most abundant proteins made by the cell. Hook and filament proteins are secreted through the basal body and a substrate specificity switch ensures that hook proteins are secreted first and flagellin proteins are secreted second (Chevance and Hughes, 2008). Regulatory checkpoints are presumed to control the ordered expression, synthesis, and assembly of flagellum components.
The best understood morphogenic checkpoint in flagellar assembly inhibits transcription of late class flagellar genes until basal body assembly is complete. Transcription of late class flagellar genes requires RNA polymerase and the alternative sigma factor σ28 (σD) which is inhibited by direct interaction with the anti-sigma factor FlgM (Daughdrill et al., 1997, and Bertero et al., 1999). When the basal body is fully assembled, it becomes proficient for FlgM secretion, which relieves inhibition on σ28 to permit transcription of genes related to the hook and the filament (Hughes et al., 1993, and Barilla et al., 1994). Flagellin synthesis is also regulated at the post-transcriptional level in a variety of bacterial species. In general, post-transcriptional regulation requires the untranslated leader region of the flagellin transcript and an RNA binding protein that is inferred to control either transcript stability or ribosome access (Anderson et al., 2000; Yamamoto et al., 2006; Yakhnin et al., 2007). In some cases, post-transcriptional regulators repress the accumulation of flagellin when cells are mutated for flagellar hook assembly (Anderson et al., 1997; Sal et al., 2008). If and how post-transcriptional regulation is coupled to the assembly state of the flagellum remains unclear.
Here we elucidate the mechanism of a novel assembly checkpoint which prevents flagellin translation in Bacillus subtilis prior to hook completion. The checkpoint is governed by a homeostatic mechanism in which flagellin itself is a critical regulator. Flagellin (Hag) participates in a partner switching mechanism with FliW, a protein of previously unknown function, and CsrA, an RNA binding protein that represses hag translation until Hag protein is secreted. As sRNA(s) were the only known CsrA antagonists (Babitzke and Romeo, 2007), B. subtilis FliW is the first identified protein regulator of CsrA activity in any organism. Translational regulation is the most immediate means to control protein levels, and homeostatic autoinhibition may be a primary mechanism for restricting synthesis to periods of assembly for structural proteins in general. Finally, CsrA is a well-known and highly-studied protein required for virulence in pathogens but may have an ancestral function as a morphogenic checkpoint protein in flagellar assembly.
RESULTS
During secretion of the flagellar hook and filament, each secreted protein typically has one chaperone usher, but the highly abundant flagellin protein Hag in B. subtilis appears to have two: the broadly conserved chaperone FliS and a broadly conserved protein of unknown function FliW (Auvray et al., 2001; Titz et al., 2006; Bange et al., 2010). We wondered why B. subtilis flagellin required a second chaperone for filament assembly. To explore the function of FliW, we generated an in-frame marker-less deletion of the fliW gene in the undomesticated strain 3610 of B. subtilis that exhibits robust swarming motility (Kearns and Losick, 2003). Mutation of fliW abolished migration across the surface of the swarm agar plate, and swarming was restored when the fliW gene was ectopically expressed from an IPTG-inducible promoter (Fig. 1A, 1B). To test if the fliW mutant was defective in flagella assembly, a functional allele of the Hag flagellin (encoded by the hag gene) that can be fluorescently labeled with a cysteine-reactive maleimide stain was ectopically introduced (amyE::Phag-hagT209C) (Blair et al., 2008). Flagellar staining and fluorescence microscopy revealed that the flagella of the fliW mutant were shorter and less numerous than the wild type (Fig. 2A). We conclude that the non-motile fliW phenotype was due to a defect in filament assembly.
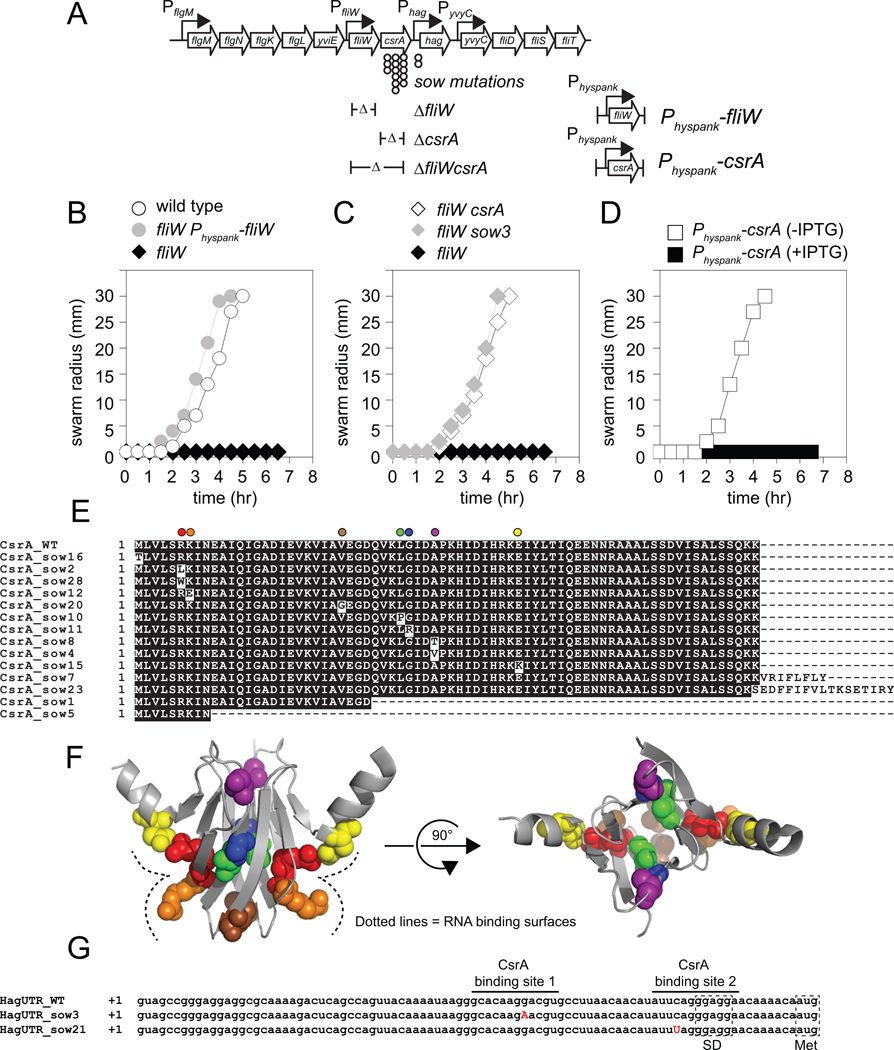
Figure 1. CsrA inhibits motility in the absence of FliW.
A) Genetic neighborhood containing the fliW, csrA, and hag genes. Large open arrows represent open reading frames (lengths not to scale). Bent arrows indicate promoters. Circles indicate the location of sow suppressor mutations. Δ indicates the boundaries of the in-frame deletion mutations. B–D) Quantitative swarm expansion assays for strains: wild type (open circles, 3610), fliW (black diamonds, DS6245), fliW Physpank-fliW in the presence of 1 mM IPTG (grey circles, DS7022), fliW csrA (open diamonds, DS6189), fliW sow3 (gray triangles, DS6530), and Physpank-csrA in the presence (closed squares, DS4940), and absence (open squares, DS4940) of 1 mM IPTG. Each point is the average of three replicas. E) A multiple sequence alignment of the predicted CsrA primary sequences of the twenty-eight suppressor of fliW (sow) alleles as compared to the wild type (WT). All missense mutations were unique, but frameshifts like sow7 and sow23 were common and only one representative is shown. F) Each residue mutated by a sow allele is highlighted on the structure of the homologous protein RsmA from Pseudomonas aeruginosa (Schubert et al., 2007). Each residue is color coded to match the color coded position from Panel A. The RNA binding surface is indicated by a dotted line. G) The complete sequence of the hag leader transcript for the wild type (hagUTR_WT), the sow3 (hagUTR_sow3), and sow21 (hagUTR_sow21) suppressor mutations. Dotted boxes indicate the location of the hag SD sequence (RBS) and start codon (Met). The two CsrA binding sites are indicated by horizontal lines. The location of the suppressor mutations are indicated by bold, capital, red letters.
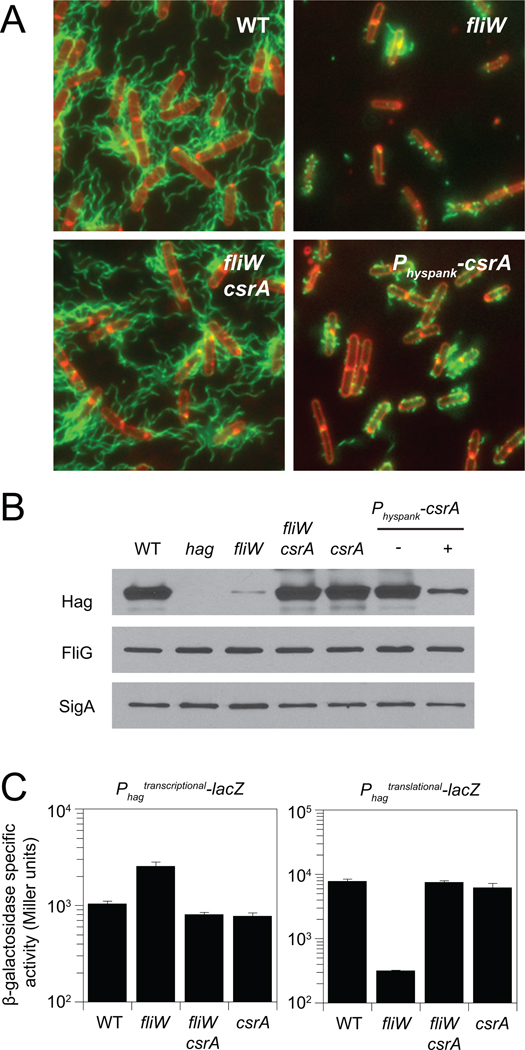
Figure 2. CsrA inhibits flagellin translation in the absence of FliW.
A) Fluorescence micrograph overlays of WT (DS1916), fliW (DS6774), fliW csrA (DS6773), and Physpank-csrA (DS6821) + 1 mM IPTG, membrane stained with FM4–64 (false colored red) and flagella stained (false colored green). B) Western blot analysis of B. subtilis cell lysates of WT (3610), hag (DS1677), fliW (DS6245), fliW csrA (DS6189), csrA (DS6188), and Physpank-csrA (DS4940 −/+ 1mM IPTG), separately probed with anti-Hag, anti-FliG, and anti-SigA primary antibodies. C) β-galactosidase assays of hag-lacZ transcriptional and hag'-'lacZ translational fusions for strains of the indicated genotype (Raw data presented in table S3). Error bars are the standard deviations of three replicates.
It was previously shown that mutation of fliW resulted in a reduced level of flagellin accumulation (Titz et al., 2006). To confirm that the level of flagellin protein was specifically reduced in our fliW mutant, Western blot analysis was conducted on cell lysates using antibodies specific for Hag, the flagellar basal body protein FliG, and the vegetative sigma factor SigA (to serve as a loading control). Levels of Hag were specifically reduced in the fliW mutant relative to the wild type, whereas levels of FliG were unaffected (Fig. 2B). To determine why Hag levels were low in the fliW mutant, hag gene expression was measured using Phag-lacZ transcriptional (Phagtranscriptional-lacZ) and Phag-hag'-'lacZ translational (Phagtranslational-lacZ) fusions. Both fusions were under the control of the hag promoter, but the translational fusion was also under the control of the hag leader region, Shine-Dalgarno (SD) sequence, and start codon. Mutation of fliW resulted in a two-fold increase in expression of the transcriptional fusion but a 24-fold decrease in the expression of the translational fusion (Fig. 2C). We conclude that in the absence of FliW, flagellin protein levels were low due to a reduction in translation of the hag transcript.
To determine the mechanism by which FliW promoted hag translation, thirty independent spontaneous suppressors of fliW (sow) alleles that bypassed the fliW motility defect were clonally isolated. Twenty-eight suppressors contained mutations in the csrA gene located immediately downstream of, and co-oriented with, fliW (Fig. 1A, 1E). Mutation of csrA was epistatic to fliW for all phenotypes tested as an in-frame deletion in csrA not only restored motility (Fig. 1C), but also restored hag translation (Fig. 2C), Hag protein levels (Fig. 2B), and full-length filament assembly to the fliW mutant (Fig. 2A). Furthermore, expression of csrA from the IPTG-inducible Physpank promoter integrated at the chromosomal amyE locus of otherwise wild type cells was sufficient to inhibit motility (Fig. 1D), restrict filament assembly (Fig. 2A), and reduce Hag protein levels (Fig. 2B). We conclude that CsrA is necessary and sufficient to inhibit flagellin synthesis in the absence of FliW.
CsrA is known to inhibit flagellin synthesis in B. subtilis by binding to two sites in the 5’ leader of the hag transcript, one of which overlaps the hag SD sequence (Dubey et al., 2005; Yakhnin et al., 2007). Thus, CsrA inhibits hag translation by competing with ribosome binding. Consistent with this model, three missense mutations in CsrA (R6L, R6W, and K7E) altered residues essential for RNA binding (Fig. 1E, 1F) (Mercante et al., 2006; Mercante et al., 2009). Only two suppressors of fliW were external to csrA. One suppressor (sow21) was located in the CsrA low affinity binding site immediately adjacent to the hag SD sequence (Fig. 1G). The other suppressor (sow3) was located within the highly conserved GGA motif of the high affinity CsrA binding site located ~25 nt upstream of the hag SD sequence (Fig. 1C, 1G) (Yakhnin et al., 2007). Quantitative RNA electrophoretic mobility shift assays (EMSA) conducted with wild type or sow3 hag leader transcripts demonstrated that the sow3 allele reduced the affinity of CsrA binding approximately 25-fold (Fig. 3A, 3B). We conclude that the defect in motility found in the fliW mutant is exclusively due to the repression of hag translation mediated by uncontrolled CsrA.
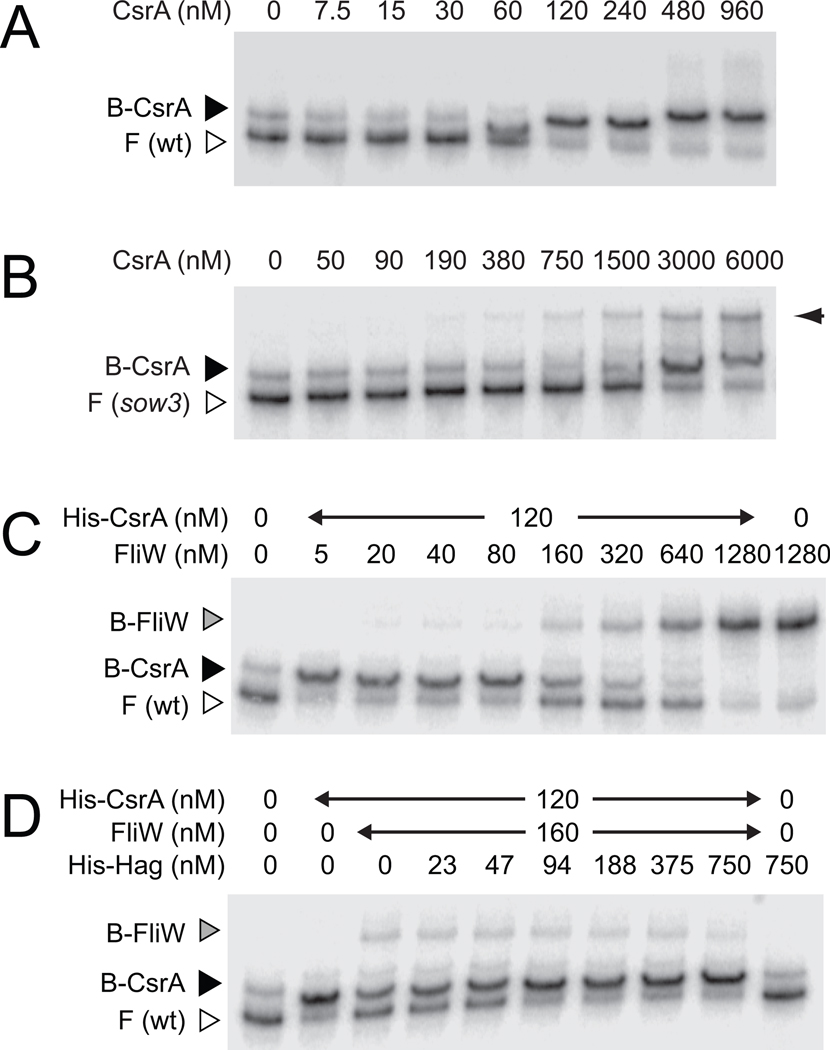
Figure 3. FliW antagonizes CsrA binding to hag RNA.
RNA electrophoretic mobility shift assay (EMSA) using wild type hag 5’UTR RNA (panels A,C, and D) or the sow3 mutant hag leader RNA (panel B) and the indicated amount of His-CsrA, FliW, and His-Hag protein. F indicates free probe, B-CsrA indicates probe bound by CsrA, and B-FliW indicates probe bound by FliW. Right hand arrow in panel B indicates CsrA simultaneously bound to one wild type binding site on two different transcripts (Dubey et al., 2005; Yakhnin et al., 2007).
One way in which FliW might antagonize CsrA is by direct interaction between the two proteins. To determine if FliW binds to CsrA, various concentrations of FliW were incubated with glutathione sepharose beads bound to 5 µM of a GST-CsrA fusion protein. The beads were centrifuged and the supernatant and pellet fractions were separately resolved by SDS polyacrylamide gel electrophoresis (PAGE), then stained with Coomassie Brilliant Blue. FliW was retained in the pellet fraction when both FliW and CsrA were at roughly equimolar concentrations (Fig. 4A). When the FliW concentration was increased above that of CsrA, FliW was retained in the pellet but also accumulated in the supernatant. FliW was poorly retained in the pellet when mixed with glutathione sepharose beads alone or glutathione sepharose beads bound to GST (Fig. S1A, S1B). We conclude that FliW and CsrA directly interacted in vitro.
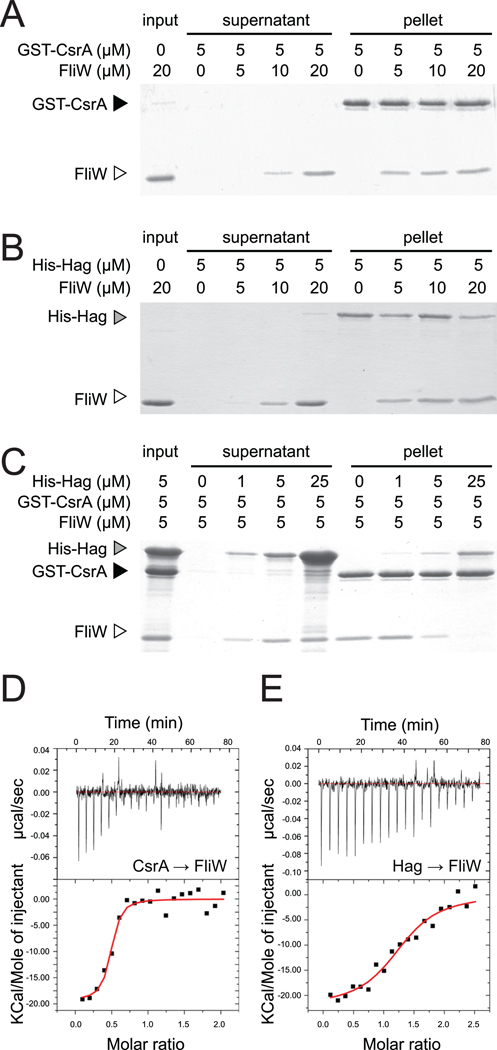
Figure 4. Hag antagonizes binding of FliW to CsrA.
A–C) Protein pull-down assays using the indicated amounts of His-Hag (gray caret), GST-CsrA (black caret), and FliW proteins (open caret). Gels were stained with Coomassie Brilliant Blue. “Input” indicates the starting amount of protein in the assays, “supernatant” indicates the proteins that failed to bind to the beads, and “pellet” indicates the proteins that remained bound to the beads following a series of washes. D–E) ITC titrations of CsrA and Hag into FliW. Top panels are the raw thermograms while the bottom panels are the integrated data. The line in the integrated data panels represents the fit of the titration results to a single-site binding model. (D) Twenty µM of CsrA was titrated into 2 µM FliW at 37°C. (E) 18.5 µM of Hag titrated into 1.5 µM of FliW at 37°C.
To determine the consequences of the FliW-CsrA interaction on the binding of CsrA to hag RNA, various concentrations of FliW were added to an RNA EMSA experiment using the hag leader transcript. FliW competed with CsrA-hag RNA interaction, and free probe began to accumulate when the FliW concentration (160 nM) exceeded that of CsrA (120 nM) (Fig. 3C). We note that a FliW-dependent band with slower mobility also appeared on the gel that was CsrA-independent (B-FliW far right lane, Fig. 3C). Thus, FliW bound to hag RNA but at concentrations higher than necessary to antagonize CsrA. We conclude that the direct protein-protein interaction between FliW and CsrA displaced CsrA from the hag transcript, thereby permitting hag translation.
FliW has also been reported to interact with the Hag protein by two-hybrid and Far Western analyses (Titz et al., 2006). To confirm the Hag–FliW interaction, various concentrations of FliW were incubated with nickel-NTA agarose beads bound to 5 µM of a His-Hag fusion protein. The beads were centrifuged and the supernatant and pellet fractions were resolved by SDS-PAGE, and then stained with Coomassie Brilliant Blue. FliW was retained in the pellet fraction when both FliW and Hag were at roughly equimolar concentrations (Fig. 4B). The retention of FliW in the pellet was dependent on the presence of Hag as FliW was poorly retained in the pellet when mixed with nickel-NTA agarose beads alone (Fig. S1C). We conclude that FliW and Hag directly interacted in vitro.
We next determined the consequences of Hag protein on the FliW-CsrA interaction. Various concentrations of His-Hag were incubated with glutathione sepharose beads to which GST-CsrA and FliW proteins were bound. As the concentration of His-Hag protein increased, FliW proportionately eluted from GST-CsrA into the supernatant (Fig. 4C). We note that a fraction of His-Hag protein was retained in the pellet non-specifically, even in the absence of CsrA (Fig. 4C, Fig. S1B). Furthermore, addition of 94 nM of His-Hag restored CsrA-hag RNA interaction in an EMSA when 160 nM of FliW was present (Fig. 3D). We conclude that Hag competes with CsrA for FliW binding and relieves antagonism on CsrA.
Protein pulldown experiments indicated that both CsrA and Hag interacted with FliW at roughly equimolar concentrations. Isothermal titration calorimetry (ITC) was performed to measure the stoichiometry of each protein interaction. The titration of CsrA into FliW displayed the sigmoidal behavior indicative of exothermic binding (Fig. 4D). Dilution of CsrA into buffer data not shown) revealed that the heat of dilution was minimal and constant, indicating the signal in the CsrA-FliW titration was due to a protein-protein interaction. Fitting the integrated injection enthalpies for CsrA binding to FliW gave a Kd = 20 ± 10 nM. The binding stoichiometry (n) obtained from the fit of the injection enthalpies was n = 0.46 ± 0.02, consistent with two FliW molecules binding to one CsrA dimer (Table 1). Titration of Hag protein into FliW also displayed a thermogram indicative of strong binding between the two proteins and the heat of dilution of Hag was minimal and consistent (Fig. 4E). Fitting of the injection enthalpies for Hag-FliW binding gave a Kd = 140 ± 50 nM. The stoichiometry obtained for Hag-FliW binding can be assigned as 1:1 with n = 1.29 ± 0.05 (Table 1). Thus, two molecules of FliW bound to each CsrA dimer, while FliW bound to Hag in a 1:1 ratio. Furthermore, CsrA bound to FliW with a higher affinity than Hag, and as shown above, both proteins could not bind to FliW simultaneously.
Table 1.
Isothermal calorimetry dataa
| Titration | Kd (nM) |
n | ΔH (kcal mol−1) |
ΔS (cal−1 mol−1 K−1) |
TΔS (kcal mol−1) |
|---|---|---|---|---|---|
| CsrA into FliW Hag into FliW |
20 ± 10 140 ± 50 |
0.46 ± 0.02 1.29 ± 0.05 |
−15.4 ± 0.9 −22 ± 1 |
−15 ± 3 −40 ± 4 |
−4.7 ± 0.9 −12 ± 1 |
errors are from model fitting.
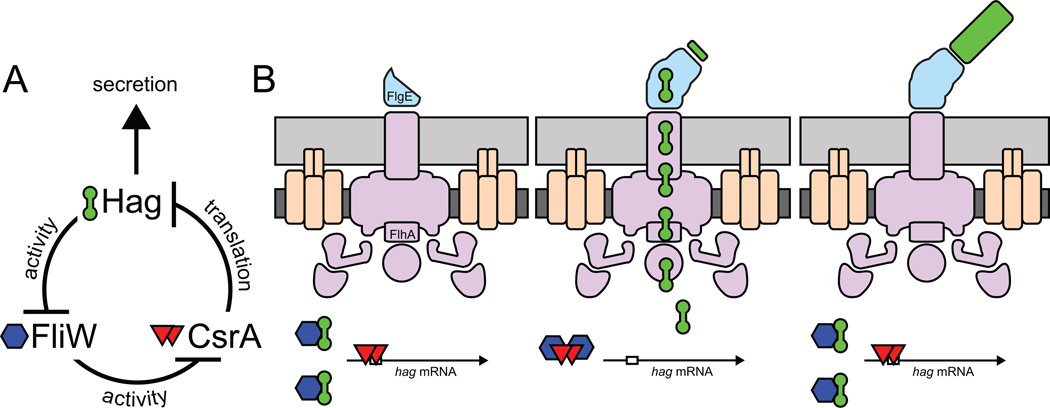
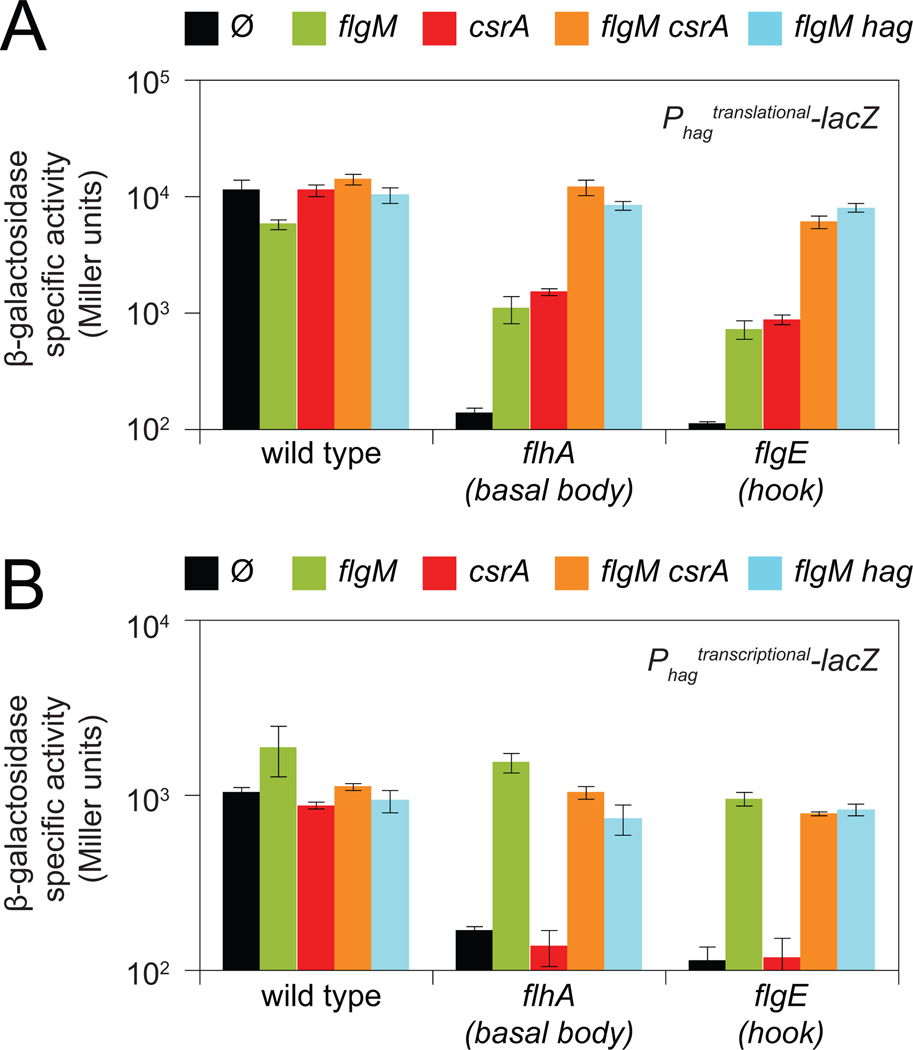
In sum, Hag (flagellin) triggers a partner-switching event with FliW that releases CsrA to repress hag translation (Fig. 5). Thus, Hag acts as an autoinhibitory regulator that is part of a homeostatic mechanism to ensure that Hag concentration is low in the cytoplasm. We hypothesize that when Hag is depleted by secretion, FliW switches partners and binds to CsrA. CsrA in turn releases hag RNA to maximize hag translation, restore cytoplasmic Hag levels, and compensate for rapid efflux. To test the role of secretion in flagellin translation, we mutated the gene flhA, encoding FlhA, a part of the basal body and flagellar secretion apparatus (Bange et al., 2010; Minamino et al., 2010). Cells mutated for flhA reduced expression of the hag translational reporter by 100-fold (Fig 6A). Mutation of the basal body checkpoint protein FlgM in the flhA mutant background increased expression from the hag translational reporter 10-fold due to a comparable increase in hag transcription (Fig. 6A, 6B). Mutation of CsrA in the flhA mutant background also increased expression of the hag translational reporter 10-fold that was transcription independent (Fig 6A, 6B). Simultaneous mutation of flgM and csrA restored full expression of the hag translational reporter to the flhA mutant (Fig 6A). Note that in the flhA mutant background, restoration of full expression of the hag transcriptional reporter by the flgM and csrA mutations was solely due to the flgM mutation (Fig. 6B). We conclude that when flhA is mutated and secretion is blocked, hag expression is reduced by the combined contribution of FlgM, that inhibits hag transcription, and CsrA, that inhibits hag translation.
Figure 5. Model for how Hag-FliW-CsrA interaction governs flagellin homeostasis.
A) The genetic and biochemical relationships of FliW, CsrA, and Hag proteins. T-bars indicate inhibition. B) The partner-switching model of the flagellin homeostasis assembly checkpoint. Hag (barbells), FliW (hexagons), CsrA (triangles), SD sequence (open box in mRNA), basal body (purple), hook (blue), and filament (green), cell membrane (dark gray), and peptidoglycan (light gray).
Figure 6. Hag and CsrA inhibit Hag translation in basal body and hook mutants.
β-galactosidase assays of hag'-'lacZ translational (A) or hag-lacZ transcriptional (B) fusions for background genotypes indicated on the X-axis containing either no additional mutations (Ø) or the mutations indicated at the top of each panel. Error bars are the standard deviations of three replicates. Raw data and strain numbers are provided in Table S4.
Flagellin is not secreted until hook assembly is complete (Minomino et al., 2000; Erhardt et al., 2011). To test the role of hook assembly on flagellin translation, we mutated the gene flgE, encoding FlgE, a structural component of the hook (Kubori et al., 1997; Samatey et al., 2004). Cells mutated for flgE reduced expression of the hag translational reporter more than 100-fold (Fig. 6A). Similar to the flhA mutant background, mutation of either FlgM or CsrA in the flgE mutant background each increased the expression of the hag translational reporter 10-fold due to relief of transcriptional and translational inhibition respectively (Fig. 6). Simultaneous mutation of flgE, flgM, and csrA restored expression of the hag translational reporter to wild type levels, as was the case for the flhA, flgM and csrA triple mutant combination, suggesting that FlgM and CsrA act synergistically prior to the secretion of Hag.
Finally, we tested the autoregulatory role of Hag protein on hag expression. Mutation of hag increased hag translation 8-fold and 11-fold in the flhA flgM and flgE flgM backgrounds respectively, phenocopying a mutation in csrA (Fig 6A). Thus, Hag serves both structural and regulatory roles (Fig 5). We conclude that basal body completion increases hag transcription by antagonizing FlgM, and hag translation is repressed by CsrA until hook assembly is complete and Hag is secreted (Fig. 5B). We further conclude that the CsrA-FliW regulatory module represents a checkpoint mechanism in flagellar assembly that is governed by completion of the hook.
DISCUSSION
Here we report the mechanism of a novel checkpoint on bacterial flagellar assembly. We show that the structural protein flagellin acts as an autoinhibitory regulator to homeostatically restrict its translation and maintain a low level of flagellin in the cytoplasm. Homeostatic equilibrium is perturbed when hook completion permits flagellin secretion and depletes flagellin in the cytoplasm. Accordingly, low levels of flagellin relieve inhibition on flagellin translation to support the massive number of subunits required for extracellular filament assembly. Conversely, cytoplasmic flagellin levels would accumulate when secretion ceases due to completion of the filament, and translational repression would re-engage to restrict flagellin synthesis (Fig. 5B). Translational regulation is a more immediate mechanism for controlling protein levels than transcriptional regulation, and homeostatic feedback ensures that synthesis of flagellin is restricted to the time between the morphogenic events of hook and filament completion.
Flagellin (Hag) homeostasis is mediated by a partner-switching mechanism between the broadly conserved FliW and CsrA proteins. When flagellin (encoded by hag) exceeds a threshold cytoplasmic concentration, it binds to FliW and releases CsrA. Free CsrA then inhibits hag translation by binding to two sites in the leader region of the hag transcript, thereby occluding ribosome access to the SD sequence (Yakhinin et al., 2007). When Hag falls below a threshold concentration, FliW switches partners and binds to CsrA such that CsrA-mediated translational repression of hag is relieved. This mechanism is in sharp contrast to the mechanism by which CsrA regulates motility in E. coli. In E. coli, CsrA activates motility by binding to and stabilizing the flhDC operon transcript (Wei et al., 2001), which encodes a DNA binding activator protein (FlhD4C2) for the motility and chemotaxis cascade (Wang et al., 2006). Furthermore, in Gram-negative bacteria CsrA activity is regulated by sRNAs that contain several CsrA binding sites. Thus the sRNAs competitively inhibit the ability of CsrA to bind to the mRNAs it would normally regulate (Babitzke and Romeo, 2007). Here we discover the first example of CsrA activity being regulated by direct interaction with a protein, FliW, and we identify a direct stimulus for CsrA control, the cytoplasmic level of flagellin.
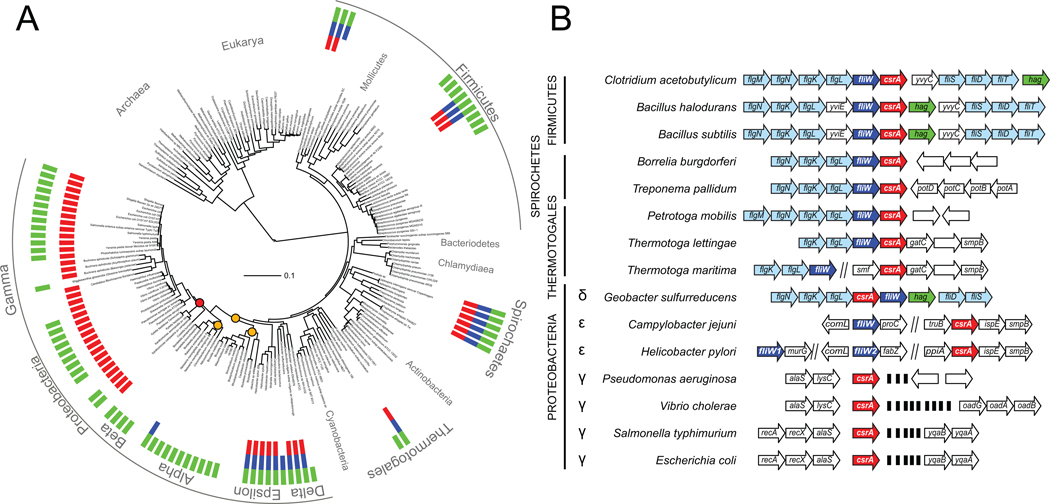
The consequences of CsrA regulation are often highly pleiotropic, and mutation or overexpression of CsrA has been shown to affect biofilm formation, EPS biosynthesis, antibiotic production, glycogen metabolism, motility, and virulence (reviewed in Babitzke and Romeo, 2007; Romeo and Babitzke, 2010; Timmermans and Melderan, 2010). Here we demonstrate that B. subtilis CsrA has a specific function to couple gene expression to the structural state of flagellar hook completion. We questioned whether CsrA in other organisms might also be directly related to flagellar motility by exploring the phylogenetic distribution of CsrA, FliW, and flagellin (Fig. 7A). Microorganisms that encode CsrA represent a subset of those that also encode flagellin, and we note that FliW is an ancestral regulatory component that is often encoded adjacent to CsrA and hook proteins (Fig. 7A, 7B). Furthermore, CsrA was lost from a subset of the proteobacteria but reappears in the γ (gamma) family adjacent to tRNA genes commonly implicated as sites of horizontal gene transfer (Fig. 7A, 7B) (Campbell et al., 2003). Reacquisition of CsrA by the gamma-proteobacteria may have promoted extensive pleiotropy and the use of regulatory sRNAs in place of FliW. Thus it appears that CsrA has a conserved ancestral function in the regulation of flagellar assembly, and evolved other physiological targets that are selectively advantageous when co-regulated with motility.
Figure 7. The gene encoding CsrA is found in a subset of genomes that also encode Hag, and is often linked to genes encoding FliW and flagellar hook proteins.
A) A maximum likelihood phylogeny with genomes that encode flagellin (green), CsrA (red), and FliW (blue) indicated. Orange dots indicate nodes at which CsrA may have been lost, and the red dot indicates the node at which CsrA may have been regained. See Fig. S2 for an enlarged view of the tree. B) The genetic neighborhoods surrounding either the csrA gene (red) and/or the fliW gene (dark blue) of the following species: Clostridium acetobutylicum (ATCC 824), Bacillus halodurans (C-125), Bacillus subtilis (NCIB 3610), Borrelia burgdorferi (B31), Treponema pallidum (SS14), Petrotoga mobilis (SJ95), Termotoga lettingae (TMO), Thermotoga maritima (MSB8), Geobacter sulfurreducens (PCA), Campylobacter jejuni (NCTC 11168), Helicobacter pylori (J99), Pseudomonas aeruginosa (PAO1), Vibrio cholerae (O1 El Tor N16961), Salmonella typhimurium (LT2), and Escherichia coli (MG1655). Genes that encode proteins are indicated by open arrows. Genes that encode tRNAs are indicated by black bars. Genes that encode flagellin are colored green, and genes that encode hook and filament-associated proteins are colored in pale blue. Genetic neighborhoods were generated using information obtained from MicrobesOnline (http://www.microbesonline.org/).
Homeostatic feedback is a simple and elegant mechanism to prevent cytoplasmic accumulation of highly abundant proteins like flagellin, and mechanisms of flagellin homeostasis besides the CsrA-FliW module may exist in other bacteria. We note that Caulobacter crescentus, and other α (alpha)-proteobacteria, do not encode a CsrA homolog (Fig. 7A). Rather, these organisms make FlbT, an RNA-binding protein (like CsrA), and FlaF, a FlbT-antagonist (like FliW), that are often encoded adjacent to hook or flagellin genes. Also similar to CsrA, FlbT represses flagellin translation when hook assembly is disrupted and appears to do so by a convergent mechanism (Llewellyn et al., 2005, and Anderson and Gober, 2000). Finally, we note that other structural components such as those involved in the assembly of the flagellar hook, type III secretion systems, and pili are post-transcriptionally regulated (Chen and Anderson, 2011; Lee and Hughes, 2006; Bonifield et al., 2000; Xie et al., 2004). We speculate that homeostasis may be a general mechanism to govern the synthesis of these and other structural subunits that are required at specific times and in finite amounts.
METHODS
Strains and growth conditions
B. subtilis strains were grown in Luria-Bertani (LB) (10 g tryptone, 5 g yeast extract, 5 g NaCl per L) broth or on LB plates fortified with 1.5% Bacto agar at 37°C. When appropriate, antibiotics were included at the following concentrations: 10 µg/ml tetracycline, 100 µg/ml spectinomycin, 5 µg/ml chloramphenicol, 5 µg/ml kanamycin, and 1 µg/ml erythromycin plus 25 µg/ml lincomycin (mls). Isopropyl β-D-thiogalactopyranoside (IPTG, Sigma) was added to the medium at the indicated concentration when appropriate.
Swarm expansion assay
Cells were grown to mid-log phase at 37°C in LB broth and resuspended to 10 OD600 in pH 8.0 PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4) containing 0.5% India ink (Higgins). Freshly prepared LB containing 0.7% Bacto agar (25 ml/plate) was dried for 20 min in a laminar flow hood, centrally inoculated with 10 µl of the cell suspension, dried for another 10 min, and incubated at 37°C. The India ink demarks the origin of the colony and the swarm radius was measured relative to the origin. For consistency, an axis was drawn on the back of the plate and swarm radii measurements were taken along this transect. For experiments including IPTG, cells were propagated in broth in the presence of IPTG, and IPTG was included in the swarm agar plates.
Western blotting
B. subtilis strains were grown in LB to OD600 ~1.0, 1 ml was harvested by centrifugation, and resuspended to 10 OD600 in Lysis buffer (20 mM Tris-HCl [pH 7.0], 10 mM EDTA, 1 mg/ml lysozyme, 10 µg/ml DNAse I, 100 µg/ml RNAse I and 1 mM PMSF) and incubated for 30 min at 37°C. Ten µl of lysate was mixed with 2 µl 6x SDS loading dye. Samples were separated by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were electroblotted onto nitrocellulose and developed with a 1:80,000 dilution of anti-SigA primary antibody (generous gift of Masaya Fujita, University of Houston), a 1:80,000 diluation of anti-Hag primary antibody, a 1:20,000 dilution of primary antibody with anti-FliG, and a 1:10,000 dilution secondary antibody (horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G). Immunoblot was developed using the Immun-Star HRP developer kit (Bio-Rad).
Microscopy
Fluorescence microscopy was performed with a Nikon 80i microscope with a phase contrast objective Nikon Plan Apo 100X and an Excite 120 metal halide lamp. FM4–64 was visualized with a C-FL HYQ Texas Red Filter Cube (excitation filter 532–587 nm, barrier filter >590 nm).
For fluorescent microscopy of flagella, 0.5 ml of broth culture was harvested at 0.5–2.0 OD600, and washed once in 1.0 ml of PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4). The suspension was pelleted, resuspended in 50 µl of PBS buffer containing 5 µg/ml Alexa Fluor 488 C5 maleimide (Molecular Probes), and incubated for 5 min at room temperature (Blair et al., 2008). Cells were then washed twice with 500 µl PBS buffer. When appropriate, membranes were stained by resuspension in 50 µl of PBS buffer containing 5 µg/ml FM4–64 (Molecular Probes) and incubated for 10 min at room temperature. Three microliters of suspension were placed on a microscope slide and immobilized with a poly-L-lysine-treated coverslip.
Strain construction
All constructs were first introduced into the domesticated strain PY79 by natural competence and then transferred to the undomesticated 3610 background using SPP1-mediated generalized phage transduction (Yasbin and Young, 1974; Canosi et al., 1982). All strains used in this study are listed in Table 2. All plasmids used in this study are listed in Supplemental Table S1. All primers used in this study are listed in Supplemental Table S2.
Table 2.
Strains
| Strain | Genotype | |
|---|---|---|
| 3610 | Wild type | |
| DS278 | amyE::Phagtranslational-lacZ spec | |
| DS793 | amyE::Phagtranscriptional-lacZ cat | (Kearns and Losick, 2005) |
| DS1916 | amyE::Phag-hagT209C spec | (Blair et al., 2008) |
| DS4940 | amyE::Physpank-csrA spec | |
| DS5823 | ΔflgE amyE::Phagtranscriptional-lacZ cat | |
| DS6188 | ΔcsrA | |
| DS6189 | ΔfliWcsrA | |
| DS6245 | ΔfliW | |
| DS6528 | ΔfliW sow1 CsrAQ29STOP | |
| DS6529 | ΔfliW sow2 CsrAR6L | |
| DS6530 | ΔfliW sow3 (hag leader) | |
| DS6604 | ΔfliW sow4 CsrAA36V | |
| DS6605 | ΔfliW sow5 CsrAE10STOP | |
| DS6607 | ΔfliW sow7 CsrAframeshift@K74 | |
| DS6608 | ΔfliW sow8 CsrAA36T | |
| DS6632 | ΔfliW sow10 CsrAL32P | |
| DS6633 | ΔfliW sow11 CsrAG33R | |
| DS6634 | ΔfliW sow12 CsrAK7E | |
| DS6772 | ΔcsrA amyE::Phag-hagT209C spec | |
| DS6773 | ΔfliWcsrA amyE::Phag-hagT209C spec | |
| DS6774 | ΔfliW amyE::Phag-hagT209C spec | |
| DS6784 | ΔflhA amyE::Phagtranscriptional-lacZ cat | |
| DS6821 | amyE::Physpank-csrA spec lacA::Phag-hagT209C mls | |
| DS7022 | ΔfliW amyE::Physpank-fliW spec | |
| DS7596 | ΔcsrA amyE::Phagtranslational-lacZ spec | |
| DS7597 | ΔfliWcsrA amyE::Phagtranslational-lacZ spec | |
| DS7598 | ΔfliW amyE::Phagtranslational-lacZ spec | |
| DS7625 | ΔflgE amyE::Phagtranslational-lacZ spec | |
| DS8143 | ΔflhA amyE::Phagtranslational-lacZ spec | |
| DS8296 | ΔflhA flgM::tet csrA::kan amyE::Phagtranslational-lacZ spec | |
| DS8297 | ΔflhA flgM::tet csrA::kan amyE::Phagtranscriptional-lacZ cat | |
| DS8298 | ΔflhA flgM::tet amyE::Phagtranslational-lacZ spec | |
| DS8299 | ΔflhA flgM::tet amyE::Phagtranscriptional-lacZ cat | |
| DS8323 | ΔcsrA amyE::Phagtranscriptional-lacZ cat | |
| DS8324 | ΔfliWcsrA amyE::Phagtranscriptional-lacZ cat | |
| DS8325 | ΔfliW amyE::Phagtranscriptional-lacZ cat | |
| DS8336 | csrA::kan amyE::Phagtranslational-lacZ spec | |
| DS8337 | flgM::tet csrA::kan amyE::Phagtranslational-lacZ spec | |
| DS8338 | ΔflgE csrA::kan amyE::Phagtranslational-lacZ spec | |
| DS8340 | flgM::tet amyE::Phagtranslational-lacZ spec | |
| DS8341 | csrA::kan amyE::Phagtranscriptional-lacZ cat | |
| DS8342 | flgM::tet csrA::kan amyE::Phagtranscriptional-lacZ cat | |
| DS8343 | ΔflgE csrA::kan amyE::Phagtranscriptional-lacZ cat | |
| DS8345 | flgM::tet amyE::Phagtranscriptional-lacZ cat | |
| DS8371 | ΔflhA csrA::kan amyE::Phagtranslational-lacZ spec | |
| DS8373 | ΔflhA csrA::kan amyE::Phagtranscriptional-lacZ cat | |
| DS8433 | ΔflhA flgM::tet Δhag amyE::Phagtranscriptional-lacZ cat | |
| DS8434 | ΔflhA flgM::tet Δhag amyE::Phagtranslational-lacZ spec | |
| DS8435 | ΔflgE flgM::tet Δhag amyE::Phagtranscriptional-lacZ cat | |
| DS8436 | ΔflhA flgM::tet Δhag amyE::Phagtranslational-lacZ spec | |
| DS8442 | flgM::tet Δhag amyE::Phagtranslational-lacZ spec | |
| DS8443 | flgM::tet Δhag amyE::Phagtranscriptional-lacZ cat | |
| DS8579 | ΔfliW sow15 CsrAE46K | |
| DS8580 | ΔfliW sow16 CsrAM1T | |
| DS8584 | ΔfliW sow20 CsrAV25G | |
| DS8587 | ΔfliW sow23 CsrAframeshift@K73 | |
| DS8592 | ΔfliW sow28 CsrAR6W | |
| DS8806 | ΔflgE flgM::tet amyE::Phagtranscriptional-lacZ cat | |
| DS8924 | ΔflgE flgM::tet csrA::kan amyE::Phagtranscriptional-lacZ cat | |
| DS8927 | ΔflgE flgM::tet amyE::Phagtranslational-lacZ spec | |
| DS8978 | ΔflgE flgM::tet csrA::kan amyE::Phagtranslational-lacZ spec |
In-frame deletions
To generate the ΔfliW in frame marker-less deletion construct, the region upstream of fliW was PCR amplified using the primer pair 1871/1905 and digested with EcoRI and BamHI, and the region downstream of fliW was PCR amplified using the primer pair 1906/1907 and digested with BamHI and SalI. The two fragments were then simultaneously ligated into the EcoRI and SalI sites of pMiniMAD2 which carries a temperature sensitive origin of replication and an erythromycin resistance cassette to generate pJP87 (Patrick and Kearns, 2008). The plasmid pJP87 was introduced to PY79 by single cross-over integration by transformation at the restrictive temperature for plasmid replication (37°C) using mls resistance as a selection. The integrated plasmid was then transduced into 3610. To evict the plasmid, the strain was incubated in 3 ml LB broth at a permissive temperature for plasmid replication (22°C) for 14 hours, diluted 30-fold in fresh LB broth, and incubated at 22°C for another 8 hours. Dilution and outgrowth was repeated 2 more times. Cells were then serially diluted and plated on LB agar at 37°C. Individual colonies were patched on LB plates and LB plates containing mls to identify mls sensitive colonies that had evicted the plasmid. Chromosomal DNA from colonies that had excised the plasmid was purified and screened by PCR using primers 1871/1907 to determine which isolate had retained the ΔfliW allele.
The ΔcsrA construct was built in a way similar to ΔfliW using the primer pairs 1875/1876 and 1877/1878 to generate pSG35. The primer pairs 1871/1872 and 1877/1878 were used for the ΔfliWcsrA deletion construct pSG36.
ΔcsrA::kan
The ΔcsrA::kan insertion deletion allele was generated by long flanking homology PCR (using primers 2704 and 2707, 2708 and 2709), and DNA containing a kanamycin drug resistance gene (pDG780) was used as a template for marker replacement (Guérout-Fleury et al., 1995, and Wach, 1996).
ΔflgM::tet
The ΔflgM::tet insertion deletion allele was generated by long flanking homology PCR (using primers 140 and 141, 142 and 143), and DNA containing a tetracycline drug resistance gene (pDG1515) was used as a template for marker replacement (Guérout-Fleury et al., 1995, and Wach, 1996).
Inducible constructs
To generate the inducible amyE::Physpank-fliW spec construct pLC108, a PCR product containing fliW was amplified from B. subtilis 3610 chromosomal DNA using the 1541/1542 primer pair, digested with NheI and SphI, and cloned into the NheI and SphI sites of pDR111 containing a spectinomycin resistance cassette, a polylinker downstream of the Physpank promoter, and the gene encoding the LacI repressor between the two arms of the amyE gene (Ben-Yehuda et al., 2003). Similarly, the inducible amyE::Physpank-csrA spec construct pLC109 was built using the primer pair 1543/1544.
LacZ reporter constructs
To generate the translational fusion of Hag and β-galactosidase (lacZ) reporter construct pDP42, a PCR product containing the hag promoter, the hag 5’ UTR and the 5’ end of the hag open reading frame was amplified from B. subtilis 3610 chromosomal DNA using the primers 131 and 132. The PCR product was digested with BamHI and SalI and cloned into the BamHI and SalI sites of plasmid pDG1728, which carries a spectinomycin-resistance marker and a polylinker upstream of the lacZ gene between two arms of the amyE gene (Guérout-Fleury et al., 1996).
Fluorescent filament lacA::Phag-hagT209C construct
The Phag-hagT209C allele was excised from plasmid pNE4 (Blair et al., 2008) with BamHI and SalI and ligated into the BamHI/SalI sites of plasmid pDR183 which contains a polylinker and an erythromycin resistance marker between the arms of the lacA gene (Doan et al., 2005) to generate plasmid pKB142.
SUMO-FliG fusion protein expression vector
To generate the translational fusion of FliG to the His-SUMO tag, a fragment containing fliG was amplified using 3610 as a template and the primer pair 882/883 and was digested with SapI and XhoI. The fragment was ligated into the SapI and XhoI sites of pTB146 containing ampicillin resistance cassette (Bendezu et al., 2009) to create pKB43.
SUMO-FliW fusion protein expression vector
To generate the translational fusion of FliW to the His-SUMO tag, a fragment containing fliW was amplified using 3610 as a template and the primer pair 2230/2231 and was digested with SapI and XhoI. The fragment was ligated into the SapI and XhoI sites of pTB146 containing ampicillin resistance cassette (Bendezu et al., 2009) to create pSM12.
6His-Hag fusion protein expression vector
To generate the translational fusion of Hag to the 6His tag, a fragment containing hag was amplified using 3610 as a template and the primer pair 663/664 and was digested with NheI and XhoI. The fragment was ligated into the NheI and XhoI sites of pET28A containing kanamycin resistance cassette to create pJP4.
GST-CSrA fusion protein expression vector
To generate the translational fusion of CsrA to the GST tag, a fragment containing csrA was amplified using 3610 as a template and the primer pair 2140/2141 and was digested with EcoRI and BamHI. The fragment was ligated into the EcoRI and BamHI sites of pGEX-2TK containing containing ampicillin resistance cassette to create pSM6.
Hag antibody preparation
One mg of purified Hag protein was sent to Cocalico Biologicals Inc. for serial injection into a rabbit host for antibody generation. Anti-Hag serum was mixed with Hag-conjugated Affigel-10 beads and incubated overnight at 4°C. Beads were packed onto a 1 cm column (BioRad) and then washed with 100 mM glycine (pH 2.5) to release the antibody and immediately neutralized with 2 M Tris base. Purification of the antibody was verified by SDS-PAGE. Purified anti-Hag antibody was dialized into 1× PBS supplemented with 50% glycerol and stored at −80°C.
Sequencing sows
The region around fliW was amplified using the primer pair 2093/848 and sequenced with primer 1565.
SPP1 phage transduction
To 0.2 ml of dense culture grown in TY broth (LB broth supplemented after autoclaving with 10 mM MgSO4 and 100 µM MnSO4), serial dilutions of SPP1 phage stock were added and statically incubated for 15 min at 37°C. To each mixture, 3 ml TYSA (molten TY supplemented with 0.5% agar) was added, poured atop fresh TY plates, and incubated overnight at 37°C. Top agar from the plate containing near confluent plaques was harvested by scraping into a 50 ml conical tube, vortexed, and centrifuged at 5,000 × g for 10 min. The supernatant was treated with 25 µg/ml DNase I final concentration before being passed through a 0.45 µm syringe filter and stored at 4°C.
Recipient cells were grown to stationary phase in 2 ml TY broth at 37°C. 0.9 ml cells were mixed with 5 µl of SPP1 donor phage stock. Nine ml of TY broth was added to the mixture and allowed to stand for 30 min at 37°C. The transduction mixture was then centrifuged at 5,000 × g for 10 min, the supernatant was discarded and the pellet was resuspended in the remaining volume. Cell suspension (100 µl) was then plated on TY fortified with 1.5% agar, the appropriate antibiotic, and 10 mM sodium citrate.
β-galactosidase assay
One millilitre of cells were harvested from a mid-log-phase (OD600 ~0.5) culture grown in LB broth shaken at 37°C, harvested and re-suspended in an equal volume of Z buffer (40 mM NaH2PO4, 60 mM Na2HPO4, 1 mM MgSO4, 10 mM KCl and 38 mM β-mercaptoethanol). To each sample, lysozyme was added to a final concentration of 0.2 mg/ml and incubated for 15 min at 30°C. Each sample was diluted appropriately to 500 µl in Z buffer and the reaction was started with 100 µl of 4 mg/ml O-nitrophenyl β-D-galactopyranoside (in Z buffer) and stopped with 250 µl of 1 M Na2CO3. The OD420 of the reaction mixtures was recorded and the β-galactosidase-specific activity was calculated according to the equation: [OD420/(time × OD600)] × dilution factor ×1000.
FliW protein purification
The FliW protein expression vector pSM12 was transformed into Rosetta gami Escherichia coli, grown to ~0.7 OD600 in 500 ml of Terrific Broth, induced with 1 mM IPTG and grown overnight at 16°C. Cells were pelleted and resuspended in Lysis Buffer (50 mM Na2HPO4, 300 mM NaCl and 10 mM Imidazole), treated with lysozyme and lysed by sonication. Lysed cells were ultracentrifuged at 9000 rpm for 30 min. Cleared supernatant was combined with Ni-NTA resin (Novagen) and incubated for 1 h at 4°C. The bead/lysate mixture was poured onto a 1 cm separation column (Bio-Rad), the resin were allowed to pack and was washed with Wash Buffer (50 mM Na2HPO4, 300 mM NaCl and 30 mM Imidazole). His–SUMO-FliW bound to the resin was then eluted using a stepwise elution of Wash Buffer with 50–500 mM Imidazole. Elutions were separated by SDS-PAGE and stained with Coomassie Brilliant Blue to verify purification of the His-SUMO-FliW fusion. Purified His–SUMO-FliW was combined with Ubiquitin Ligase (protease) and Cleavage Buffer and incubated overnight at 4°C to cleave the SUMO tag from the FliW protein (Butt et al., 2005). The cleavage reaction was combined with Ni-NTA beads, incubated for 2 h at 4°C and centrifuged to pellet the resin. Supernatant was removed and dialized in to 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 10% Glycerol and 1 mM DTT and stored at −20°C. Removal of the SUMO tag was verified by SDS-PAGE and staining with Coomassie Brilliant Blue.
6His-Hag protein purification
The Hag protein expression vector pJP4 was transformed into Rosetta gami E. coli, grown to ~0.5 OD600 in 500 ml of Terrific Broth, induced with 1 mM IPTG and grown overnight at 16°C. Cells were pelleted and resuspended in Lysis Buffer (50 mM Na2HPO4, 300 mM NaCl and 10 mM Imidazole) and lysed by treatment with BugBuster for 1 h at room temperature. Lysed cells were ultracentrifuged at 35,000 rpm. Cleared supernatant was combined with Ni-NTA resin (Novagen) and incubated for 1 h at 4°C. The bead/lysate mixture was poured onto a 1 cm separation column (BioRad), the resin was allowed to pack and was washed with Wash Buffer (50 mM Na2HPO4, 300 mM NaCl and 30 mM Imidazole). His-Hag bound to the resin was then eluted using a stepwise elution of Wash Buffer with 50 mM – 500 mM Imidazole. Elutions were separated by SDS-PAGE and stained with Coomassie Brilliant Blue to verify purificaton of the His-Hag fusion.
GST-CsrA protein purification
The GST-CsrA protein expression vector pSM6 was transformed into Rosetta gami E. coli, grown to ~0.7 OD600 in 500 ml of Luria-Bertani Broth, induced with 1 mM IPTG and grown for 3 h at 37°C. Cells were pelleted and resuspended in Lysis Buffer (25 mM Tris-HCl [pH 8.0], 1 mM DTT, 1 mM EDTA, 0.1% Triton X-100, 150 mM NaCl, 250 µg/ml lysozyme, 0.5 mM PMSF and 5 µg/ml leupeptin), and frozen at −80°C for overnight. The frozen cell pellet was thawed and lysed by sonication. Lysed cells were ultracentrifuged at 12000 rpm for 30 min at 4°C. Cleared supernatant was combined with Glutathione-Sepharose (GE Healthcare) and incubated for 3 h at 4°C. The bead/lysate mixture was poured onto a 1 cm separation column (Bio-Rad), the resin was allowed to pack and was washed with Wash Buffer (25 mM Tris-HCl [pH 8.0], 1 mM DTT, 1 mM EDTA, 0.1% NP-40, 250 mM NaCl, 10% glycerol, 0.5 mM PMSF and 5 µg/ml leupeptin). GST-CsrA bound to the resin was then eluted using GST-elution buffer (25 mM Tris-HCl [pH 8.5], 20 mM Glutathione, 1 mM DTT, 1 mM EDTA, 250 mM NaCl, 10% glycerol, 0.5 mM PMSF and 5 µg/ml leupeptin). Elutions were separated by SDS-PAGE and stained with Coomassie Brilliant Blue to verify purification of the GST-CsrA fusion. GST-CsrA was dialyzed into 25 mM Tris-HCl (pH 8.0), 1 mM DTT, 1 mM EDTA, 250 mM NaCl, 10% glycerol, 20% Sucrose, 0.5 mM PMSF and 5 µg/ml leupeptin.
His-CsrA protein purification
CsrA containing six His residues at the C-terminus was purified as described previously (Yakhnin et al., 2007). BL21 E. coli cells containing pCSB9 were grown at 37°C in LB broth and 100 µg/ml of ampicillin to an OD600 of 0.6, at which time 1 mM IPTG was added to the culture and incubation was continued for 5 h. Cells were harvested by centrifugation and the cell pellet (10 g) was suspended in lysis buffer containing 100 mM Tris-HCl (pH 8.0) and 400 mM NaCl (5 ml of buffer per gram of cells). Cell lysate was prepared using a French press, followed by centrifugation at 20,000 × g for 30 min at 4°C. The resulting supernatant was diluted with an equal volume of water and loaded onto a 50 ml DEAE 52 (Whatman) column that had been pre-equilibrated with 50 mM Tris-HCl (pH 8.0) and 200 mM NaCl. The flowthrough from the DEAE column was mixed with 2 ml of prewashed Ni-NTA resin (Qiagen) for 1 hr at 4°C and packed into a column. The column was washed successively with buffer A (50 mM Tris-HCl, pH 8.0, 200 mM NaCl and 10% Glycerol), buffer A containing 5 mM imidazole, and buffer A containing 15 mM imidazole. His-CsrA was eluted with buffer A containing 500 mM imadazole. Elutions were separated by SDS-PAGE and stained with Coomassie Brilliant Blue to verify purification of the His-CsrA fusion. Fractions containing pure His-CsrA were combined and dialyzed against 20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 10% Glycerol and 1 mM EDTA and stored ay −80°C.
FliG protein purification
The FliG protein expression vector pKB43 was transformed into Rosetta gami E. coli, grown to ~0.5 OD600 in 500 ml of Terrific Broth, induced with 1 mM IPTG and grown overnight at 16°C. Cells were pelleted and resuspended in Lysis Buffer (50 mM Na2HPO4, 300 mM NaCl and 10 mM Imidazole) and lysed by treatment with BugBuster for 1 h at room temperature. Lysed cells were ultracentrifuged at 35,000 rpm. Cleared supernatant was combined with Ni-NTA resin (Novagen) and incubated for 1 h at 4°C. The bead/lysate mixture was poured onto a 1 cm separation column (BioRad), the resin was allowed to pack and was washed with Wash Buffer (50 mM Na2HPO4, 300 mM NaCl and 30 mM Imidazole). SUMO-FliG bound to the resin was then eluted using a stepwise elution of Wash Buffer with 50 mM – 500 mM Imidazole. Elutions were separated by SDS-PAGE and stained with Coomassie Brilliant Blue to verify purificaton of the SUMO-FliG fusion. Purified SUMO-FliG was combined with Ubiquitin Ligase (protease) and Cleavage Buffer and incubated overnight at 4°C to cleave the SUMO tag from the FliG protein (Bendezu et al., 2009). The cleavage reaction was combined with Ni-NTA beads, incubated for 2 h at 4°C, and centrifuged to pellet the resin. Supernatant was removed and dialized in to 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 10% Glycerol and 1 mM DTT and stored at −20°C. Removal of the SUMO tag was verified by SDS-PAGE and stained with Coomassie Brilliant Blue. Pooled fractions were dialyzed in to 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 10% Glycerol and 1 mM DTT and stored at −20°C.
FliG antibody preparation
1.7 mg of purified FliG protein was cut from an SDS-PAGE gel and sent to Cocalico Biologicals Inc. for serial injection into a rabbit host for antibody generation. Anti-FliG serum was mixed with FliG-conjugated Affigel-10 beads and incubated overnight at 4°C. Beads were packed onto a 1 cm column (BioRad) and then washed with 100 mM glycine (pH 2.5) to release the antibody and immediately neutralized with 2 M Tris base. Purification of the antibody was verified by SDS-PAGE. Purified anti-FliG antibody was dialized into 1× PBS supplemented with 50% glycerol and stored at −80°C.
Protein-Protein interaction assay
a) GST-CsrA and FliW interaction
Glutathione-Sepharose beads were washed with T(0.1) buffer [25 mM Tris-HCl (pH 8.0), 20% glycerol, 100 mM NaCl, 1 mM DTT and 1X Protease inhibitor cocktail (from Roche)]. Thirty µl of washed beads was mixed with 30 µl of 5 µM GST-CsrA protein and rotated on labquake for 2 h at 4°C. Next the beads bound to GST-CsrA protein were centrifuged at 1000 rpm for 2 min and the pellet was washed twice with T(1.0) 25 mM Tris-HCl (pH 8.0), 20% glycerol, 1M NaCl, 1 mM DTT and 1X Protease inhibitor cocktail (Roche) and again twice with T(0.1). Then 30 µl of increasing concentrations of FliW (0, 5, 10, and 20 µM) were added to the washed beads bound to GST-CsrA and rotated on labquake for 2 h at 4°C. The samples were centrifuged at 1000 rpm for 2 min and the supernatant was saved. The pellet was washed 4 times with T(0.1). The supernatant and pellet fractions were subjected to SDS-PAGE analysis and stained with Coomassie Brilliant Blue.
b) His-Hag and FliW interaction
Ni-NTA beads were washed with T(0.1) buffer [25mM Tris-HCl (pH 8.0), 20% glycerol, 100 mM NaCl, 1 mM DTT and 1X Protease inhibitor cocktail] containing 10 mM Imidazole. Thirty µl of washed beads was mixed with 30 µl of 5 µM His-Hag protein and rotated on labquake at 4°C for 2 h. Next the beads bound to His-Hag protein were centrifuged at 1000 rpm for 2 min and the pellet was washed twice with T(1.0) [25 mM Tris-HCl (pH 8.0), 20% glycerol, 1 M NaCl, 1 mM DTT and 1X Protease inhibitor cocktail] containing 10 mM Imidazole and again twice with T(0.1) containing 10 mM Imidazole. Then 30 µl of increasing concentrations of FliW (0, 5, 10, and 20 µM) were added to the washed beads bound to His-Hag and rotated on labquake at 4°C for 2 h. The samples were centrifuged at 1000 rpm for 2 min and the supernatant was saved. The pellet was washed 4 times with T(0.1) containing 10 mM Imidazole. The supernatant and pellet fractions were subjected to SDS-PAGE analysis and stained with Coomassie Brilliant Blue.
c) GST-CsrA-FliW-Hag interaction
Glutathione-Sepharose beads were washed with T(0.1) buffer [25 mM Tris-HCl (pH 8.0), 20% glycerol, 100 mM NaCl, 1 mM DTT and 1X Protease inhibitor cocktail]. Thirty µl of washed beads was mixed with 30 µl of 5 µM GST-CsrA protein and rotated on labquake at 4°C for 2 hours. Next the beads bound to GST-CsrA protein were centrifuged at 1000 rpm for 2 min and the pellet was washed twice with T(1.0) [25mM Tris-HCl (pH 8.0), 20% glycerol, 1 M NaCl, 1 mM DTT and 1X Protease inhibitor cocktail] and again twice with T(0.1). Then 30 µl of a mixture of 5 µM of FliW and increasing concentrations of Hag (0, 1, 5, 25 µM) were added to the washed beads bound to GST-CsrA and rotated on labquake at 4°C for 2 h. The samples were centrifuged at 1000 rpm for 2 min and the supernatant was saved. The pellet was washed 4 times with T(0.1). The supernatant and pellet fractions were subjected to SDS-PAGE analysis and stained with Coomassie Brilliant Blue.
Isothermal Titration Calorimetry (ITC)
His-CsrA, FliW, and His-Hag proteins were purified as described above and exhaustively dialyzed against the same batch of buffer containing 10 mM HEPES (pH 7.5), 10 mM MgCl2, 100 mM KCl, 1 mM β-mercaptoethanol, and 7.5% glycerol. The concentrations of protein were determined by a Bio-Rad assay using BSA as a standard (Bio-Rad Laboratories, Hercules CA). All experiments were performed on a MicroCal AutoiTC200 (MicroCal, Inc. Northampton, MA) with both cell and syringes samples loaded into a 96-well plate. In all titrations, FliW was in the sample cell with 19 injections (2 µl each) of CsrA or Hag. Dilution control runs were done with CsrA or Hag titrated into pure buffer. The titrations were carried out at 37°C with the standard 5 µcal/s reference power. The data was analyzed and fit to a single site binding model using the Origin AutoITC software (OriginLab Corporation, Northampton MA). This single site binding model allows the macromolecule to have several identical binding sites. The last five injection heats for each titration were averaged and subtracted from the data to correct for protein dilution.
Electrohoretic Mobility Shift Assay (EMSA)
Quantitative electrophoretic mobility shift assays used to examine CsrA-hag RNA followed a published procedure (Dubey et al., 2005). DNA templates for in vitro transcription were synthesized by PCR using chromosomal DNA from B. subtilis strains 3610 (wild type) and DS6530 (sow3). RNA was synthesized in vitro with the Stratagene RNA-Maxx transcription kit. Each template contained a T7 RNA polymerase promoter and hag-specific sequences (+1 to +151 relative to the start of hag transcription).
Phylogenetic analysis
The maximum likelihood phylogeny was constructed from a concatenated alignment of 31 conserved genes and was annotated to indicate the occurrence of COGs 1551 (csrA), 1699 (fliW), and 1344 (flagellin and related hook-associated proteins) in sequenced bacterial genomes as identified by the Integrated Microbial Genomes platform (Ciccarelli et al., 2006, and Markowitz et al., 2010). Tree annotation and display used the Interactive Tree of Life website tools (Letunic and Bork, 2011).
Supplementary Material
ACKNOWLEDGEMENTS
We thank K. Blair, C. Courtney, L. Cozy, S. Mukhopadhyay, and J. Patrick, for reagents, constructs, and technical assistance. We thank Y. Brun and C. Fuqua for critical reading of the manuscript. Isothermal titration calorimetry was supported by the Penn State Automated Biological Calorimetry Facility and grant NSF MRI 0922974. This work was supported by NIH grants GM055969 (to P.B.) and GM093030 (to D.B.K.).
REFERENCES
- Anderson DK, Newton A. Posttranscriptional regulation of Caulobacter flagellin genes by a late flagellum assembly checkpoint. J Bacteriol. 1997;179:2281–2288. doi: 10.1128/jb.179.7.2281-2288.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson PE, Gober JW. FlbT, the post-transcriptional regulator of flagellin synthesis in Caulobacter crescentus, interacts with the 5′ untranslated region of flagellin mRNA. Mol Microbiol. 2000;38:41–52. doi: 10.1046/j.1365-2958.2000.02108.x. [DOI] [PubMed] [Google Scholar]
- Auvray F, Thomas J, Fraser GM, Hughes C. Flagellin polymerization control by a cytosolic export chaperone. J Mol Biol. 2001;308:221–229. doi: 10.1006/jmbi.2001.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Bange G, Kummerer N, Engel C, Bozkurt G, Wild K, Sinning I. FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc Natl Acad Sci USA. 2010;107:11295–11300. doi: 10.1073/pnas.1001383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barilla D, Caramori T, Galizzi A. Coupling of flagellin gene transcription to flagellar assembly in Bacillus subtilis. J Bacteriol. 1994;176:4558–4564. doi: 10.1128/jb.176.15.4558-4564.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 2009;28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science. 2003;299:532–536. doi: 10.1126/science.1079914. [DOI] [PubMed] [Google Scholar]
- Bertero MG, Gonzales B, Tarricone C, Ceciliani F, Galizzi A. Overproduction and characterization of the Bacillus subtilis anti-sigma factor FlgM. J Biol Chem. 1999;274:12103–12107. doi: 10.1074/jbc.274.17.12103. [DOI] [PubMed] [Google Scholar]
- Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science. 2008;320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- Bonifield HR, Yamaguchi S, Hughes KT. The flagellar hook protein, FlgE, of Salmonella enterica Serovar Typhimurium is posttranscriptionally regulated in response to the stage of flagellar assembly. J Bacteriol. 2000;182:4044–4050. doi: 10.1128/jb.182.14.4044-4050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt TR, Edavettal SC, Hall JP, Mattern MR. SUMO fusion technology for difficult to express proteins. Protein Expr Purif. 2005;43:1–9. doi: 10.1016/j.pep.2005.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A. Prophage insertion sites. Res Microbiol. 2003;154:277–282. doi: 10.1016/S0923-2508(03)00071-8. [DOI] [PubMed] [Google Scholar]
- Canosi U, Luder G, Trautner TA. SPP1-mediated plasmid transduction. J Virol. 1982;44:431–436. doi: 10.1128/jvi.44.2.431-436.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Anderson DM. Expression hierarchy in the Yersinia type III secretion system established through YopD recognition of RNA. Mol Microbiol. 2011;80:966–980. doi: 10.1111/j.1365-2958.2011.07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FFV, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Daughdrill GW, Chadsey MS, Karlinsey JE, Hughes KT, Dahlquist FW. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, σ28. Nat Struct Biol. 1997;4:285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- Doan T, Marquis KA, Rudner DZ. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol. 2005;55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- Dubey AK, Baker CS, Romeo T, Babitzke P. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA. 2005;11:1579–1587. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt M, Singer HM, Wee DH, Keener JP, Hughes KT. An infrequent molecular ruler controls flagellar hook length in Salmonella enterica. EMBO J. 2011:1–14. doi: 10.1038/emboj.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Guérout-Fleury AM, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Losick R. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol. 2003;49:581–590. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- Kubori T, Okumura M, Kobayashi N, Nakamura D, Iwakura M, Aizawa SI. Purification and characterization of the flagellar hook-basal body complex of Bacillus subtilis. Mol Microbiol. 1997;24:399–410. doi: 10.1046/j.1365-2958.1997.3341714.x. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Hughes KT. Posttranscriptional control of the Salmonella enterica flagellar hook protein FlgE. J Bacteriol. 2006;188:3308–3316. doi: 10.1128/JB.188.9.3308-3316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn M, Dutton RJ, Easter J, O’Donnol D, Gober JW. The conserved flaF gene has a critical role in coupling flagellin translation and assembly in Caulobacter crescentus. Mol Microbiol. 2005;57:1127–1142. doi: 10.1111/j.1365-2958.2005.04745.x. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Chen IA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Anderson I, Lykidis A, Mavromatis K, Ivanova NN, Kyrpides NC. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 2010;38:D382–D390. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercante J, Edwards AN, Dubey AK, Babitzke P, Romeo T. Molecular geometry of CsrA (RsmA) binding to RNA and its implications for regulated expression. J Mol Biol. 2009;392:511–528. doi: 10.1016/j.jmb.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercante J, Suzuki K, Cheng X, Babitzke P, Romeo T. Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. J. Biol. Chem. 2006;281:31832–31842. doi: 10.1074/jbc.M606057200. [DOI] [PubMed] [Google Scholar]
- Minamino T, Macnab RM. Domain structure of FlhB, a flagellar export component responsible for substrate specificity switching. J. Bacteriol. 2000;182:4906–4914. doi: 10.1128/jb.182.17.4906-4914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Shimada M, Okabe M, Saijo-Hamano Y, Imada K, Kihara M, Namba K. Role of the C-terminal domain of FlhA in bacterial flagellar type III protein export. J. Bacteriol. 2010;192:1929–1936. doi: 10.1128/JB.01328-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick JE, Kearns DB. MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol Microbiol. 2008;70:1166–1179. doi: 10.1111/j.1365-2958.2008.06469.x. [DOI] [PubMed] [Google Scholar]
- Romeo T, Babitzke P. The Second Messenger Cyclic Di-GMP. Washington DC: ASM Press; 2010. Csr (Rsm) system and its overlap and interplat with cyclic di-GMP regulatory systems; pp. 201–214. [Google Scholar]
- Sal MS, Chunhao L, Motalab MA, Shibata S, Aizawa S, Charon NW. Borrelia burgdorferi uniquely regulates its motility genes and has an intricate flagellar hook-basal body structure. J Bacteriol. 2008;190:1912–1921. doi: 10.1128/JB.01421-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samatey FA, Matusnami H, Imada K, Nagashima S, Shaikh TR, Thomas DR, Chen JZ, DeRosier DJ, Kitao A, Namba K. Structure of the bacterial flagellar hook and implications for the molecular universal joint mechanism. Nature. 2004;431:1062–1068. doi: 10.1038/nature02997. [DOI] [PubMed] [Google Scholar]
- Schubert M, Lapouge K, Duss O, Oberstrass FC, Jelesarov I, Haas D, Allain FH. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Mol Biol. 2007;14:807–813. doi: 10.1038/nsmb1285. [DOI] [PubMed] [Google Scholar]
- Timmermans J, Melderan LV. Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci. 2010;67:2897–2908. doi: 10.1007/s00018-010-0381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titz B, Rajagopala SV, Ester C, Häuser R, Uetz P. Novel Conserved Assembly Factor of the Bacterial Flagellum. J Bacteriol. 2006;188:7700–7706. doi: 10.1128/JB.00820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Wang S, Fleming RT, Westbrook EM, Matsumura P, McKay DB. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J Mol Biol. 2006;355:798–808. doi: 10.1016/j.jmb.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Wei BL, Brun-Zinkernagel A-M, Simecka JW, Prüβ BM, Babitzke P, Romeo T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol. 2001;40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- Xie H, Kozlova N, Lamont RJ. Porphyromonas gingivalis genes involved in fimA regulation. Infect Immun. 2004;72:651–658. doi: 10.1128/IAI.72.2.651-658.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin H, Pandit P, Petty TJ, Baker CS, Romeo T, Babitzke P. CsrA of Bacillus subtilis regulates translation initiation of the gene encoding the flagellin protein (hag) by blocking ribosome binding. Mol Microbiol. 2007;64:1605–1620. doi: 10.1111/j.1365-2958.2007.05765.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Kutsukake K. FljA-mediated posttranscriptional control of Phase 1 flagellin expression in flagellar phase variation of Salmonella enterica Serovar Typhimurium. J Bacteriol. 2006;188:958–967. doi: 10.1128/JB.188.3.958-967.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasbin RE, Young FE. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol. 1974;14:1343–1348. doi: 10.1128/jvi.14.6.1343-1348.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.