Abstract
The Y chromosome is paternally inherited and therefore serves as an evolutionary marker of patrilineal descent. Worldwide DNA variation within the non-recombining portion of the Y chromosome can be represented as a monophyletic phylogenetic tree in which the branches (haplogroups) are defined by at least one SNP. Previous human population genetics research has produced a wealth of knowledge about the worldwide distribution of Y-SNP haplogroups. Here, we apply previous and very recent knowledge on the Y-SNP phylogeny and Y-haplogroup distribution by introducing two multiplex genotyping assays that allow for the hierarchical detection of 28 Y-SNPs defining the major worldwide Y haplogroups. PCR amplicons were kept small to make the method sensitive and thereby applicable to DNA of limited amount and/or quality such as in forensic settings. These Y-SNP assays thus form a valuable tool for researchers in the fields of forensic genetics and genetic anthropology to infer a man's patrilineal bio-geographic ancestry from DNA.
Keywords: Y chromosome, Y-SNP, Haplogroup, Patrilineal ancestry, Bio-geographic ancestry, Multiplex SNaPshot
Introduction
Knowledge about the bio-geographic ancestry revealed from crime-scene samples can be relevant for investigative intelligence purposes in search for unknown sample donors who usually cannot be identified via conventional forensic STR profiling. DNA-based bio-geographic ancestry inference is also applied in genealogical and anthropological research for various purposes. The human Y chromosome is widely studied as an evolutionary marker of patrilineal descent. A well-established Y-chromosome phylogeny is available [6] and is continuously being expanded as novel SNPs are discovered. A wealth of data has been produced previously on the worldwide distribution and allele frequencies of numerous Y-SNPs and the respective Y haplogroups they define. Here, we take advantage of existing knowledge on the Y-SNP phylogeny and worldwide Y haplogroup distribution and introduce two Y-SNP multiplex assays, based on single-base primer extension (SNaPshot™) technology, for the detection of the major worldwide Y haplogoups. Together with well-known Y-SNPs, we have also included some relatively novel Y-SNPs such as M522 [4], M526 [4], P326 [8] and M412 [9], acknowledging most recent progress in Y-chromosome research.
Materials and methods
DNA samples
A subset of DNA samples from the HapMap 3 reference panel [1], belonging to various Y haplogroups, was obtained from the Coriell Institute for Medical Research (http://www.coriell.org/).
Primer design
Primers were designed using Primer3Plus [11] with a Tm around 60°C for PCR primers and around 55°C for extension primers. Potential interactions between primers in the same multiplex were evaluated with the AutoDimer version 1.0 software [12]. In order to minimize allelic dropouts due to primer mismatches, we avoided as much as possible that primer-annealing sites overlapped with known Y-chromosome polymorphisms. Extension primers were varied in length through the addition of 5′ non-homologous poly(GACT) tails to ensure electrophoretic separation of extended fragments.
PCR amplification
Multiplex PCR amplification was carried out in a reaction volume of 6 μL, containing 1× GeneAmp PCR Gold Buffer (Applied Biosystems, CA, USA), 4.5 mM MgCl2 (Applied Biosystems), 100 μM of each dNTP (Roche, Mannheim, Germany), 0.35 units of AmpliTaq Gold DNA polymerase (Applied Biosystems), 1–2 ng of genomic DNA template, and PCR primers (desalted; Metabion, Martinsried, Germany) in concentrations as specified in Tables 1 and 2. The reactions were performed in a Dual 384-well GeneAmp PCR System 9700 (Applied Biosystems) using the following cycling conditions: 10 min at 95°C, followed by 30 cycles of 94°C for 15 s, 60°C for 45 s, and a final extension at 60°C for 5 min. PCR products were purified by adding 2 μL ExoSAP-IT (USB Corporation, OH, USA) to 6 μL PCR product, followed by incubation at 37°C for 30 min and 80°C for 15 min.
Table 1.
Genotyping details of Y-SNP multiplex 1
| Locus | Mutation | PCR amplification | Single-base extension | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Primer sequences (5′-3′) | Conc. (μM) | Amplicon size (bp) | Primer sequence (5′–3′) (5′ aspecific tail in lowercase italics) | Conc. (μM) | Length (nt) | Orientation | Alleles (dye) | |||
| M91 | ins T | F | CAAAAATCCCCCTACATTGC | 0.600 | 144/143 | g CTACAGTAGTGAACTGATTAAAAAAAA | 0.300 | 28 | R | a (yellow), i (green)a |
| R | GCAGTGCCCTTCCAAATAAA | 0.600 | ||||||||
| M60 | ins T | F | TCTTTACATTTCAAAATGCATGACT | 0.600 | 128/129 | ct(gact) 6 TAACCACTGTGTGCCTGAT | 0.600 | 45 | R | a (yellow), i (green)a |
| R | GAGAAGGTGGGTGGTCAAGA | 0.600 | ||||||||
| M145 | G->A | F | GCATACTTGCCTCCACGACT | 0.200 | 96 | ct(gact) 3 gac TAGGCTAAGGCTGGCTCT | 0.450 | 35 | R | G (yellow), A (red) |
| R | CCTCCCACTCCTTTTTGGAT | 0.200 | ||||||||
| M174 | T->C | F | TCTCCGTCACAGCAAAAATG | 0.450 | 178 | ct(gact) 5 g ATACCTTCTGGAGTGCCC | 0.100 | 41 | F | T (red), C (yellow) |
| R | AGGAGAAGGACAAGACCCATC | 0.450 | ||||||||
| M96 | G->C | F | TGAGCTGTGATGTGTAACTTGG | 0.200 | 117 | act(gact) 10 gac TGGAAAACAGGTCTCTCATAATA | 0.200 | 69 | F | G (blue), C (yellow) |
| R | CACCCACTTTGTTGCTTTGT | 0.200 | ||||||||
| M216 | C->T | F | CCTCAACCAGTTTTTATGAAGCTA | 0.100 | 102 | ct(gact) 6 g CTGCTAGTTATGTATACCTGTTGAAT | 0.075 | 53 | R | C (blue), T (green) |
| R | TTCTAAATCTGAATTCTGACACTGC | 0.100 | ||||||||
| M89 | C->T | F | CAGCTTCCTGGATTCAGCTC | 0.200 | 105 | ct(gact) 13 ga AACTCAGGCAAAGTGAGAGAT | 0.300 | 77 | R | C (blue), T (green) |
| R | CACTTTGGGTCCAGGATCAC | 0.200 | ||||||||
| M282 | A->G | F | TGTGCAACCTCAACTTTGCTT | 0.750 | 106 | t(gact) 15 GAAAGCAAAATCTCAATATGATAA | 1.000 | 85 | F | A (green), G (blue) |
| R | TGTGATCAACTTCTTTCCCTCA | 0.750 | ||||||||
| P257 | G->A | F | ACCCCTCAGTCTCTCCGATT | 0.200 | 71 | (gact) 9 g ATTATCCCACTGCATTTCTG | 0.300 | 57 | F | G (blue), A (green) |
| R | TCATCTCCAACCCCCATCT | 0.200 | ||||||||
| M69 | T->C | F | GGAGGCTGTTTACACTCCTGA | 0.300 | 87 | (gact) 10 g GGCTGTTTACACTCCTGAAA | 0.150 | 61 | F | T (red), C (yellow) |
| R | TCTCCCCTTAGCTCTCCTGTT | 0.300 | ||||||||
| M522 | G->A | F | TCCAATTCCCATGTCCTCTC | 0.100 | 109 | t(gact) 11 CTACTACGCCTCTCTTGTCC | 0.075 | 65 | F | G (blue), A (green) |
| R | CAGTGCAGAAAATCACGGTAGA | 0.100 | ||||||||
| M258 | T->C | F | TTCAGGATTTGTCAAGGATGG | 0.200 | 108 | t(gact) 3 gac GGGATTCCAAGTTCCCA | 0.300 | 33 | R | T (green), C (blue) |
| R | GCTATGACTAAGAGGGATTCCAA | 0.200 | ||||||||
| M304 | A->C | F | TTGTAACAAACAGTATGTGGGATTT | 0.200 | 88 | act(gact) 11 ga TTATACCAAAATATCACCAGTTGT | 0.300 | 73 | R | A (red), C (blue) |
| R | CGTCTTATACCAAAATATCACCAGTT | 0.200 | ||||||||
| M9 | C->G | F | CTGCAAAGAAACGGCCTAAG | 0.100 | 90 | t(gact) 7 g CGGCCTAAGATGGTTGAAT | 0.100 | 49 | F | C (yellow), G (blue) |
| R | AACTAAGTATGTAAGACATTGAACGTTTG | 0.100 | ||||||||
a a ancestral, i insertion
Table 2.
Genotyping details of Y-SNP multiplex 2
| Locus | Mutation | PCR amplification | Single-base extension | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Primer sequences (5′–3′) | Conc. (μM) | Amplicon size (bp) | Primer sequence (5′–3′) (5′ aspecific tail in lowercase italics) | Conc. (μM) | Length (nt) | Orientation | Alleles (dye) | |||
| M9 | C->G | F | CTGCAAAGAAACGGCCTAAG | 0.300 | 90 | t(gact) 7 g CGGCCTAAGATGGTTGAAT | 0.100 | 49 | F | C (yellow), G (blue) |
| R | AACTAAGTATGTAAGACATTGAACGTTTG | 0.300 | ||||||||
| M526 | A->C | F | TAGAGGCAGGGTGTTGCTCT | 0.300 | 100 | ct(gact) 10 ga TGTCATCAGGCTGAATCATAC | 0.450 | 65 | F | A (green), C (yellow) |
| R | TACTTTGGGAGGCTGCTGTT | 0.300 | ||||||||
| M147 | ins T | F | CCTGAATAAGCTGGTGAAAGAAA | 0.500 | 114/115 | ct(gact) 11 ga CCTGTCTCTGAAAGAAAAAAA | 1.000 | 69 | R | a (yellow), i (green)a |
| R | GGAGACCCTGTCTCTGAAAGAA | 0.500 | ||||||||
| P308 | C->T | F | GCTACCAATACCCCCAAAGA | 0.050 | 108 | gactgac GAAATGATTAAGTAAGTGCCTTCT | 0.150 | 31 | R | C (blue), T (green) |
| R | CCTGGAATATGGCACGAAAT | 0.050 | ||||||||
| P79 | T->C | F | TTGCTTAGTATAATGTCTTTCATGCTC | 0.500 | 101 | ct(gact) 4 g TGCTCATTCGCATCTTTG | 1.000 | 37 | F | T (red), C (yellow) |
| R | AAATGAGGCTAATCAATGGAACA | 0.500 | ||||||||
| P261 | G->A | F | TCCTAGAAGGTAACCCACTACCC | 0.500 | 93 | t(gact)7gac TTTTTGTTTTTATTAATGAATGCTA | 1.000 | 57 | R | G (yellow), A (red) |
| R | TGTGCATATGTTATCCACCATGT | 0.500 | ||||||||
| P256 | G->A | F | TCTTGGTTTTCCCATTGACC | 0.200 | 91 | t(gact) 13 ga TGCCCTACACTAGATAGAAAGG | 0.150 | 77 | F | G (blue), A (green) |
| R | CATCTCCCAACTTGTCTGTGC | 0.200 | ||||||||
| M231 | G->A | F | AACAACATTTACTGTTTCTACTGCTTTC | 0.300 | 119 | act(gact) 3 g CGATCTTTCCCCCAATT | 0.450 | 33 | R | G (yellow), A (red) |
| R | TTCACATCATCCAGTACAGCAA | 0.300 | ||||||||
| M175 | 5 bp del | F | CCCAAATCAACTCAACTCCAG | 0.300 | 101/96 | t(gact) 10 CACATGCCTTCTCACTTCTC | 0.600 | 61 | F | a (red), d (green)a |
| R | TTCTACTGATACCTTTGTTTCTGTTCA | 0.300 | ||||||||
| M45 | G->A | F | CATCGGGGTGTGGACTTTA | 0.400 | 109 | act(gact) 6 g AATTGGCAGTGAAAAATTATAGATA | 0.750 | 53 | F | G (blue), A (green) |
| R | CCTCAGAAGGAGCTTTTTGC | 0.400 | ||||||||
| M242 | C->T | F | AAAAAGGTGACCAAGGTGCT | 0.400 | 46 | ct(gact) 6 g CGTTAAGACCAATGCCAA | 0.100 | 45 | R | C (blue), T (green) |
| R | AAAAACACGTTAAGACCAATGC | 0.400 | ||||||||
| M207 | A->G | F | GGGGCAAATGTAAGTCAAGC | 0.300 | 83 | (gact) 14 g AATGTAAGTCAAGCAAGAAATTTA | 0.300 | 81 | F | A (green), G (blue) |
| R | TCACTTCAACCTCTTGTTGGAA | 0.300 | ||||||||
| M412 | G->A | F | GGCACTCCTCCGTCATCTT | 0.300 | 114 | ct(gact) 16 GGGTACAATCTGATGAGGC | 0.300 | 85 | F | G (blue), A (green) |
| R | GGTGAAGTGGACCCTATCCA | 0.300 | ||||||||
| P202 | T->A | F | AAACTTCCCAGTTTGTGGTTC | 0.300 | 125 | ct(gact) 12 ga CCAGTTTGTGGTTCTTTGTTA | 0.300 | 73 | F | T (red), A (green) |
| R | TGATCCCTTAATTAATAGCAAGACC | 0.300 | ||||||||
| P326 | T->C | F | TTCAGATATCAGGCCGCTTT | 0.200 | 61 | t(gact) 3 CCTAAGCAGAGGAAAATAGTACAG | 0.150 | 37 | R | T (green), C (blue) |
| R | GAGCTGTCAGCCTGCCTAAG | 0.200 | ||||||||
a a ancestral, i insertion, d deletion
Single-base extension
Multiplex single-base primer extension was carried out in a reaction volume of 6 μL, containing 1 μL SNaPshot™ Ready Reaction Mix (Applied Biosystems), 1 μL purified PCR product, and extension primers (HPLC-purified; Metabion, Martinsried, Germany) in concentrations as specified in Tables 1 and 2. The reactions were performed in a Dual 384-well GeneAmp PCR System 9700 (Applied Biosystems) using the following cycling conditions: 2 min at 96°C, followed by 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 30 s. The reaction products were purified by adding 1 unit of Shrimp Alkaline Phosphatase (USB Corporation) to 6 μL of extension product, followed by incubation at 37°C for 45 min and 75°C for 15 min.
Capillary electrophoresis
The extended fragments were separated and detected by capillary electrophoresis on a 3130xl Genetic Analyzer (Applied Biosystems) using POP-7 polymer. A mixture of 1 μL purified extension product, 8.7 μL Hi-Di formamide (Applied Biosystems) and 0.3 μL GeneScan-120 LIZ internal size standard (Applied Biosystems) was run with 10 s injection time at 1.2 kV and 500 s run time at 15.0 kV. Results were analysed using GeneMapper version 3.7 software (Applied Biosystems).
Results and discussion
Two genotyping multiplex assays were developed targeting a total of 28 Y-SNPs that define the major worldwide Y-chromosome haplogroups (Fig. 1). During the course of this work, a paper was published that reported a reorganization of the deepest clades of the Y-chromosome phylogeny, one of the consequences being that marker M91 no longer defines a monophyletic haplogroup A, but rather should be placed on the stem leading to the BCDEF (also referred to as BT) clade [5]. We have incorporated this change in our tables and figures to conform with the latest Y-chromosome topology. Furthermore, we took advantage of some recently discovered Y-SNPs (P326, M526, M522 and M412) that, as far as we know, were not included in previous Y genotyping systems [e.g. 2, 3, 10]. Of these novel SNPs, P326 (also known as L298) defines a new branch that joins haplogroups L and T into a single clade now called LT [8]. M526 is located downstream of marker M9 and encompasses haplogroups K1 to K4 as well as M to S [4]; the branch defined by M526 is now referred to as haplogroup K, and the former haplogroup K (defined by M9) is now relabelled as KLT. M522 (also known as L16 or S138) defines a new node within haplogroup F that encompasses haplogroups I, J and KLT [4] and is referred to as haplogroup IJKLT. M412 (also known as L51 or S167) defines a significant subhaplogroup within haplogroup R that is most abundant in western parts of Europe [9].
Fig. 1.
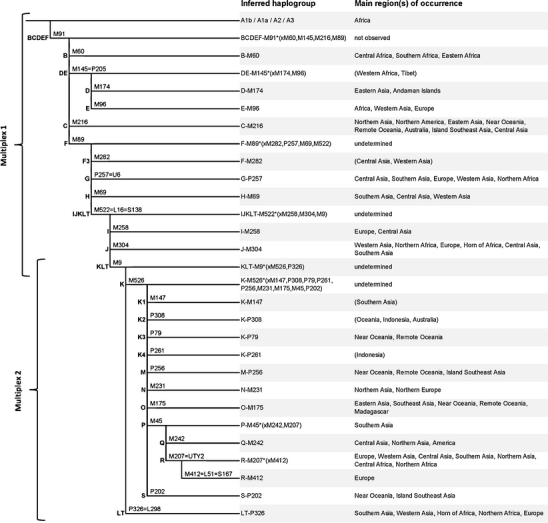
Y-SNP marker phylogeny, inferred haplogroups and their geographic distributions as covered by the two Y-SNP multiplex assays introduced here. The phylogeny shown is a truncation of the entire Y-chromosome tree. Due to the fact that some haplogroups have so far been observed only sporadically, their regions of occurrence are less certain and are therefore shown in parentheses
The 28 Y-SNPs were divided into two multiplexes such as to allow a hierarchical typing strategy. Multiplex 1 covers haplogroups BCDEF, B, C, DE, D, E, F, F3, G, H, IJKLT, I, J and KLT. If a sample is found to belong to the latter, it can subsequently be typed with multiplex 2 which covers haplogroups KLT, K, K1, K2, K3, K4, M, N, O, P, Q, R, R-M412 (also known as R1b1a2a1a), S and LT.
To maintain high sensitivity of the multiplexes, PCR amplicons were kept short with an average length of 103 bp (minimum, 46 bp; maximum, 178 bp). The recommended amount of template DNA for the PCR reactions is 1–2 ng, which gives satisfactory results when the DNA is of reasonable quality (Fig. 2). Although we did not further evaluate the sensitivity of the two multiplex assays, we expect that in many cases, lower amounts of template DNA will still yield informative genotypes.
Fig. 2.
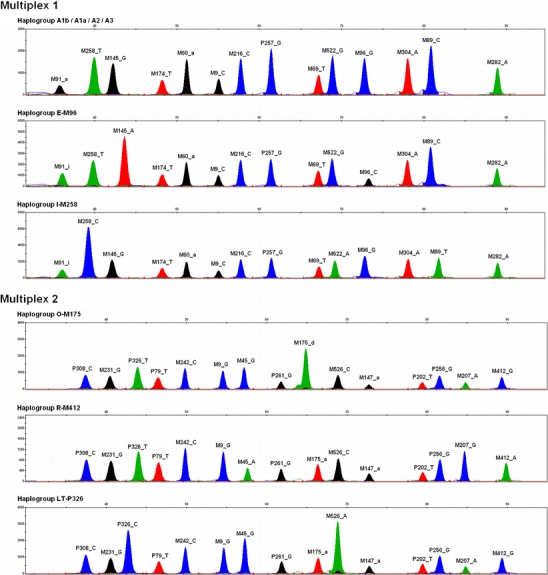
Typical electropherograms obtained with the two Y-SNP multiplex assays introduced here, using DNA samples belonging to a range of Y haplogroups. For each peak, the detected allele is indicated in concordance with Tables 1 and 2. As is convention, the yellow dye is shown as black for better contrast
The multiplexes were optimized on a Genetic Analyzer using POP-7 polymer. We noticed in the past that the type of POP polymer has some influence on the relative electrophoretic mobilities as well as peak intensities of the extended fragments. Therefore, re-adjustment of 5′ tail lengths as well as reaction concentrations of the extension primers might be necessary when employing a POP polymer that is different from the one used here.
Conclusion
The multiplex assays presented here form a convenient tool for detecting the major worldwide Y haplogroups, hence giving a first idea about the patrilineal bio-geographic ancestry of men, being of relevance in forensic investigation and anthropological research. Notably, for most of the haplogroups covered here, more detailed phylogenetic resolution can be obtained by genotyping additional Y-SNPs. Hence, we foresee that additional multiplex assays, targeting more downstream Y-SNPs and dedicated to the dissection of particular (sub)haplogroups, will form useful additions to the global assays presented here. For more complete reconstruction of a person's overall bio-geographic ancestry, we recommend that Y-chromosome markers are combined with ancestry-informative markers from mitochondrial DNA and autosomal DNA, as already achievable with efficient multiplex tools offering resolution on a continental level [e.g. 7, 13].
Acknowledgements
We thank Qasim Ayub and Chris Tyler-Smith for help in providing access to DNA samples belonging to various Y haplogroups. This work was supported in parts by funding provided by the Netherlands Forensic Institute (NFI) and by the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) within the framework of the Forensic Genomics Consortium Netherlands (FGCN).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Altshuler DM, Gibbs RA, Peltonen L, International HapMap 3 Consortium et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berniell-Lee G, Sandoval K, Mendizabal I, Bosch E, Comas D. SNPlexing the human Y-chromosome: a single-assay system for major haplogroup screening. Electrophoresis. 2007;28(18):3201–3206. doi: 10.1002/elps.200700078. [DOI] [PubMed] [Google Scholar]
- 3.Brión M, Sanchez JJ, Balogh K, Thacker C, Blanco-Verea A, Børsting C, Stradmann-Bellinghausen B, Bogus M, Syndercombe-Court D, Schneider PM, Carracedo A, Morling N. Introduction of an single nucleodite polymorphism-based “Major Y-chromosome haplogroup typing kit” suitable for predicting the geographical origin of male lineages. Electrophoresis. 2005;26(23):4411–4420. doi: 10.1002/elps.200500293. [DOI] [PubMed] [Google Scholar]
- 4.Chiaroni J, Underhill PA, Cavalli-Sforza LL. Y chromosome diversity, human expansion, drift, and cultural evolution. Proc Natl Acad Sci USA. 2009;106(48):20174–20179. doi: 10.1073/pnas.0910803106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruciani F, Trombetta B, Massaia A, Destro-Biol G, Sellitto D, Scozzari R. A revised root for the human Y chromosomal phylogenetic tree: the origin of patrilineal diversity in Africa. Am J Hum Genet. 2011;88(6):1–5. doi: 10.1016/j.ajhg.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18(5):830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lao O, Vallone PM, Coble MD, Diegoli TM, van Oven M, van der Gaag KJ, Pijpe J, de Knijff P, Kayser M. Evaluating self-declared ancestry of U.S. Americans with autosomal, Y-chromosomal and mitochondrial DNA. Hum Mutat. 2010;31(12):E1875–E1893. doi: 10.1002/humu.21366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendez FL, Karafet TM, Krahn T, Ostrer H, Soodyall H, Hammer MF. Increased resolution of Y chromosome haplogroup T defines relationships among populations of the Near East, Europe, and Africa. Hum Biol. 2011;83(1):39–53. doi: 10.3378/027.083.0103. [DOI] [PubMed] [Google Scholar]
- 9.Myres NM, Rootsi S, Lin AA, Järve M, King RJ, Kutuev I, Cabrera VM, Khusnutdinova EK, Pshenichnov A, Yunusbayev B, Balanovsky O, Balanovska E, Rudan P, Baldovic M, Herrera RJ, Chiaroni J, Di Cristofaro J, Villems R, Kivisild T, Underhill PA. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur J Hum Genet. 2011;19(1):95–101. doi: 10.1038/ejhg.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onofri V, Alessandrini F, Turchi C, Pesaresi M, Buscemi L, Tagliabracci A. Development of multiplex PCRs for evolutionary and forensic applications of 37 human Y chromosome SNPs. Forensic Sci Int. 2006;157(1):23–35. doi: 10.1016/j.forsciint.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 11.Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vallone PM, Butler JM. AutoDimer: a screening tool for primer-dimer and hairpin structures. Biotechniques. 2004;37(2):226–231. doi: 10.2144/04372ST03. [DOI] [PubMed] [Google Scholar]
- 13.van Oven M, Vermeulen M, Kayser M. Multiplex genotyping system for efficient inference of matrilineal genetic ancestry with continental resolution. Investig Genet. 2011;2:6. doi: 10.1186/2041-2223-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


