Abstract
Objective
Responses to therapies, either with regards to toxicities or efficacy, are expected to involve complex relationships of gene products within the same molecular pathway or functional gene set. Therefore, pathways or gene sets, as opposed to single genes, may better reflect the true underlying biology and may be more appropriate units for analysis of pharmacogenomic studies. Application of such methods to pharmacogenomic studies may enable the detection of more subtle effects of multiple genes in the same pathway that may be missed by assessing each gene individually.
Methods
A gene set analysis of 3,821 gene sets is presented assessing the association between basal mRNA expression and drug cytotoxicity using ethnically defined human lymphoblastoid cell lines for two classes of drugs: pyrimidines (dFdC and AraC) and purines (6-TG and 6-MP).
Results
The gene set nucleoside-diphosphatase activity was found to be significantly associated with both dFdC and AraC, while gene set gamma-aminobutyric acid catabolic process was associated with dFdC and 6-TG. These gene sets were significantly associated with the phenotype even after adjusting for multiple testing. In addition, five associated gene sets were found in common between the pyrimidines and two gene sets for the purines (3′,5′-cyclic-AMP phosphodiesterase activity and gamma-aminobutyric acid catabolic process) with p < 0.0001. Functional validation was attempted with 4 genes each in gene sets for thiopurine and pyrimidine anti-metabolites. All four genes selected from the pyrimidine gene sets (PSME3, CANT1, ENTPD6, ADRM1) were validated, but only one (PDE4D) was validated for the thiopurine gene sets.
Conclusions
In summary, results from the gene set analysis of pyrimidine and purine therapies, used often in the treatment of various cancers, provide novel insight into the relationship between genomic variation and drug response.
Keywords: gene set enrichment analysis, mRNA expression, cytotoxicity, pharmacogenomics, bioinformatics
1. INTRODUCTION
Many drug-related phenotypes are expected to involve complex relationships of gene products within the same molecular pathway or functional gene set. Therefore, pathways or gene sets, as opposed to single genes, may better reflect the true underlying biology and be more appropriate units for analysis of pharmacogenomic studies[1]. Pathway or gene set methods for analysis of expression data incorporate prior biological knowledge into the statistical analysis by evaluating the overall evidence of association of a phenotype with expression of all genes in a given pathway or gene set[2,3]. Application of such methods may enable the detection of more subtle effects of multiple genes in the same pathway that may be missed by assessing each gene individually. Moreover, incorporation of biological knowledge in the statistical analysis may aid researchers in the interpretation of results.
In the field of pharmacogenomics, use of gene set analysis or gene set enrichment analysis have been utilized in the past, as responses to therapies are thought to be the results of a complex relationship of multiple genes within the same pathway or gene set. For example, an enrichment analysis identified cellular pathways associated with resistance to chemotherapy[4], while another study report that variants associated with response to chemotherapeutic drugs are enriched in expression quantitative trait loci (eQTLs)[5]. Therefore, analysis was completed to determine if expression patterns in pre-defined gene sets were associated with response to two classes of drugs: pyrimidines (gemcitabine (dFdC) and cytosine arabinoside (AraC)) and purines (6-mercaptopurine (6-MP), 6-thioguanine (6-TG)).
The drugs dFdC and AraC have been used to treat many cancers [6,7], and they share similar chemical structures, metabolic pathways, and mechanisms of actions [8,9,10]. Both are prodrugs that must be transported into the cell and activated by kinases to form active di- and tri-phosphorylated metabolites. Although gemcitabine and AraC are both a type of pyrimidine (cytidine analogues), gemcitabine is used to treat solid tumors, [7,11]; while AraC is a major agent for the treatment of acute myelogenous leukemia (AML) [7,9,12]. Clinical response to both drugs varies widely [8,13,14,15]. Nearly all previous studies of gemcitabine and AraC have focused on variation in the expression of genes within their known metabolism and activation pathway [16,17,18]. However, very little information is available with regard to genes outside of that pathway.
In contrast to these two pyrimidines, the purine drugs of 6-mercaptopurine (6-MP) and 6-thioguanine (6-TG) are used to treat acute lymphoblastic leukemia (ALL) of childhood and autoimmune disorders[19]. Several genes within the known “thiopurine pathway” can contribute to individual variation in thiopurine response. For example, inherited variation in thiopurine S-methyltransferase (TPMT) activity is a major factor responsible for individual variation in 6-TGN concentrations and thiopurine response.[20] Patients homozygous for TPMT alleles associated with very low enzyme activity are at greatly increased risk for the development of life-threatening bone-marrow suppression.[20] However, level of TPMT activity does not account for all of the variation observed during thiopurine therapy.[21] Thus, assessment of variation across the genome, in the form of gene set analysis, might provide novel candidate genes and pathways that could help explain individual response to thiopurine drugs.
To determine if variation in basal gene expression within pre-defined gene sets is associated with response to chemotherapeutic agents, a lymphoblastoid cell line model system, consisting of cell lines from the Coriell Human Variation Panel, was used. This model system consists of ethnically-defined cell lines for which gene expression data using Affymetrix U133 Plus 2.0 GeneChips and drug-related cytotoxicity phenotypes has been obtained, as previously described[22,23]. In this study, two pyrimidines, gemcitabine (dFdC) and cytosine arabinoside (AraC), and two purine drugs, 6-mercaptopurine (6-MP) and 6-thioguanine (6-TG), were used to test whether individual variation in basal gene expression for known pathways and gene sets might influence drug sensitivity. Gene set analysis of these four drugs will provide novel gene sets implicated in the response to the individuals drugs or possibly, the class of drugs.
2. METHODS AND MATERIALS
2.1. Cell Lines, Drug and Cell proliferation Assays
The study is based on 175 EBV-transformed B lymphoblastoid cells derived from Caucasian-American (CA), African-American (AA) and Han Chinese-American (HCA) were purchased from the Coriell Institute (Camden, NJ). The dFdC was provided by Eli Lilly (Indianapolis, IN). AraC and the thiopurines, 6-TG and 6-MP, were purchased from Sigma Aldrich (St Louis, MO). dFdC and AraC were dissolved in PBS and frozen at −80°C, while 6-MP and 6-TG were prepared in DMSO immediately before use and then further diluted with media. Cells were plated at a density of 5 × 104 cells per well in triplicate in 96-well plates (Corning, NY). One hour after plating, cells were treated with dFdC, AraC, 6-MP or 6-TG. The CellTiter96 Aqueous Non-Radioactive Cell Proliferation Assays (Promega, Madison, WI) were performed as described by the manufacturer after 72 h incubations. Plates were read in a Safire2 microplate reader (Tecan AG, Switzerland), with subsequent cytotoxicity measurements recorded at various doses of dFdC, AraC, 6-MP and 6-TG for the cell lines. For the pyrimidine studies, drug dosages of 10 μl at concentrations ranging from 0.1 nM to 1 mM where used cytotoxicity studies involving dFdC and 1 nM to 10 mM for cytotoxicity studies involving AraC, as previously described[22]. The doses used in the purine studies were 0.01, 0.1, 0.5, 1, 2.5, 5, 10 and 100 μM for 6-MP and 0.005, 0.05, 0.25, 0.5, 1, 2.5, 5, and 50 μM for 6-TG.
To determine IC50 and AUC values for each drug, a logistic function (was fitted to the cytotoxicity data using the R package drc (http://cran.r-project.org/)[24]. Let Rj represent the response for a cell line at dose j (Dj) of the drug, j =1,…, 8 with Rj ~ N(ηj, σ2) and , β1, β2 > 0. The responses at infinite and zero concentration are represented by β1i and β2i, respectively. The parameter β3i represents the log(IC50) and β4 represents the slope of the dose-response curve[25,26]. From the fitted logistic dose-response curve for each subject, the IC50 phenotype (effective dose that kills 50% of the cells) was estimated based on the estimate of β3i and AUC phenotype (area under the dose-response curve) was determined by numerically computing the area under the estimated dose-response curve. This procedure for estimating the IC50 and AUC for each subject was completed for each of the four drugs. Cytotoxicity data for gene set analysis of dFdC, AraC, 6-TG and 6-MP was available for 172, 174, 171 and 171 cell lines, respectively.
2.2. Genome-wide mRNA Expression Data
Whole Genome expression data for cell lines was obtained with Affymetrix U133 plus 2.0 expression array chip. The RNA extraction and the expression array assays were performed following the Affymetrix GeneChip® expression technical manual (Affymetrix, Inc., Santa Clara, CA). Before the assay, RNA quality was tested using an Agilent 2100 Bioanalyzer. The Affymetrix GeneChip® contains over 54,000 probe sets whose design is based on build 34 of the Human Genome Project. The mRNA expression array data were normalized on the log2 scale using GCRMA methodology[27,28]. The mRNA expression data for the entire set of cell lines (96 CA, 96 AA and 96 HCA) has been assigned a GEO number GSE23120 is available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23120.
2.3. Gene Sets Definitions and Analysis
Gene sets were determined based on four categories: Gene Ontology (GO) Biological Process, GO Cellular Component, GO Molecular Function and known pathways (e.g., KEGG, PharmGKB). The mapping of expression array probe sets to gene sets was provided by Affymetrix, resulting in 6,849 gene sets. All probe sets were included in the analysis, regardless of whether or not multiple probe sets occurred for a given gene. The distribution of the gene set sizes was highly skewed with the mean (median) number of probe sets within a gene set be 47.34 (6.00) (Minimum = 1.00, first quartile = 2.00, third quartile = 15.00, maximum = 13500). For the gene set analysis of the four drugs, analyses was restricted to gene sets with 5 to 5,000 genes, resulting in 3,821 gene sets for analysis (removed 7 gene sets with more than 5,000 genes with the remaining 3,021 gene sets removed having 4 or less genes).
To assess association of gene set expression values with drug IC50, the self-contained gene set method, termed the “global model”, of Goeman et al[29] was utilized,. Based on previous research, the global model has been shown to be more powerful than alternative approaches for conducting gene set analysis for expression studies for a variety of situations (e.g., size of gene set, correlation between mRNA values within a gene set, number and size of individual mRNA effects on the phenotypes)[30]. The global modeling approach is based on a linear random effects model in which the continuous drug phenotype IC50 (Y) is modeled as a function of the expression values for the genes within the gene-set of interest. That is, Y is modeled as Y|X ~ N(Xβ, σ2) with the gene expression effects, β, having a common distribution with mean 0 and variance τ2. Under the null hypothesis of no gene expression effects on the phenotype, the variance of the expression effects is zero (τ2 = 0), which can be tested with a score test [29]. Instead of relying on asymptotic determine p-values for association, empirical p-values based on 500,000 permutations were computed.
Prior to analysis, the normalized expression data were then regressed on gender and race. Residuals from this regression were then standardized by subtracting the mean residual for individual probe sets and dividing by the standard deviation to derive a “standardized adjusted expression value”, as previously described[22,31]. The phenotypes IC50 and AUC values were log transformed for dFdC and AraC and rank transformed (i.e., Van der Waerden transformation) for 6-TG and 6-MP and adjusted in a fashion similar to that described for the expression data. All gene set analyses were based on adjusted standardized expression array data and the IC50 and AUC phenotypes using the R library “globaltest” available for download in Bioconductor (http://bioconductor.org/packages/2.6/bioc/html/globaltest.html).
2.4. Functional validation studies
Human pancreatic cancer SU86 and human glioma U251 cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and were cultured in RPMI 1640 and DMEM medium containing 10% FBS, respectively. Pools of 4 specific siRNAs for the candidate genes and negative non-targeting control siRNA pools were purchased from Dharmacon (Chicago, IL). Reverse transfection with siRNA was performed in 96-well plates with SU86 (pyrimidines) and U251 (thiopurines) cells, using 0.2 μL of lipofectamine RNAi-MAX reagent (Invitrogen, Carlsbad, CA) and siRNA pools at final concentrations of 30 nM for 24 hours. The cells were then treated with a series of drug concentrations. After 72 hours, cytotoxicity assays were performed using the CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay kit (Promega Corporation, Madison, WI), followed by absorbance measurements at 490 nm in a Safire2 microplate reader (Tecan AG, Switzerland).
Total RNA was isolated from the cultured cells using the Quick-RNA™ MiniPrep kit (Zymo Research, Orange, CA), followed by qRT-PCR performed with the Power SYBR® Green RNA-to-CTTM 1-Step Kit (AB Foster CA). Specifically, primers purchased from QIAGEN were used to perform qRT-PCR using the Stratagene Mx3005P Real-Time PCR detection system (Agilent Technologies, Santa Clara, CA). All experiments were corrected by using beta-actin as an internal control.
Cytotoxicity for AraC and dFdC in SU86 cell lines was compared between cells treated with negative control siRNA and gene-specific siRNAs, and similar comparisons were made for 6-TG using U251 cells.
3. RESULTS
3.1. Gene Set Analysis
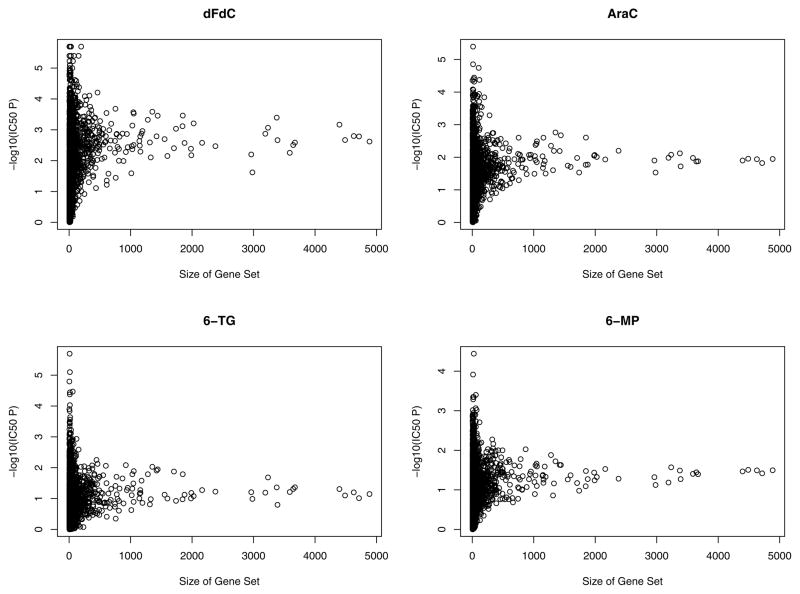
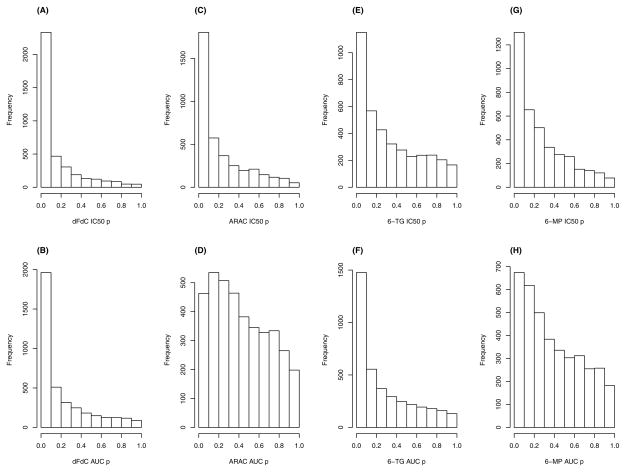
The correlation between IC50 and AUC for dFdC, AraC, 6-MP and 6-TG was 0.20, 0.68, 0.39, and 0.49, respectively. Statistically significant gene sets, at a Bonferroni adjusted significance level (0.05/3821), for the four drugs and two drug related phenotypes (IC50 and AUC) are presented in Tables 1 and 2. Figure 1 displays the relationship between gene set size and significance of the gene set. As the figure illustrates, the gene sets with more genes had larger p-values, indicating that these gene sets may contain a diverse set of genes (probe sets) that “waters-down” the association. The drug dFdC had the most significant gene sets associated with the phenotypes IC50 (36) and AUC (13), while AraC only had a single significant gene set associated with IC50 (nucleoside-diphosphatase activity). In contrast, the purine 6-MP had no gene sets significantly associated with either IC50 or AUC while 6-TG had three gene sets associated with IC50 and four associated with AUC. Figure 2 shows the histograms of the p-values for the various drug/phenotype combinations. As Figure 2 illustrates, for each drug/phenotype combination, there appears to be deviation from the null hypothesis of no association of gene sets with the phenotype, with the most striking results for the drug dFdC and IC50.
Table 1.
Statistical significant gene sets associated with IC50.
| Drug | IC50 p | Gene Set Size | Gene Set |
|---|---|---|---|
| dFdC | 1.2E-05 | 14 | Wnt receptor activity |
| 1.2E-05 | 21 | proton-transporting ATP synthase complex, catalytic core F(1) | |
| 1.2E-05 | 7 | negative regulation of Ras protein signal transduction | |
| <2.0E-06 | 13 | regulation of mitochondrial membrane permeability | |
| 2.0E-06 | 5 | antigen process/presentation, exogenous lipid antigen via MHC class Ib | |
| 2.0E-06 | 5 | positive regulation of NK T cell differentiation | |
| 1.0E-05 | 14 | regulation of mitochondrial membrane potential | |
| <2.0E-06 | 27 | diacylglycerol kinase activity | |
| 1.0E-05 | 52 | protein kinase inhibitor activity | |
| 4.0E-06 | 81 | small GTPase regulator activity | |
| <2.0E-06 | 17 | FK506 binding | |
| 4.0E-06 | 9 | laminin-5 complex | |
| 8.0E-06 | 15 | caveola | |
| 8.0E-06 | 11 | mitochondrial electron transport, ubiquinol to cytochrome c | |
| <2.0E-06 | 8 | DNA ligation | |
| 2.0E-06 | 8 | eye pigment biosynthetic process | |
| 2.0E-06 | 196 | protein kinase cascade | |
| 2.0E-06 | 12 | phosphatidylinositol transporter activity | |
| <2.0E-06 | 6 | proteasome activator complex | |
| <2.0E-06 | 7 | proteasome activator activity | |
| <2.0E-06 | 12 | negative regulation of survival gene product activity | |
| 2.0E-06 | 32 | dorsal/ventral pattern formation | |
| <2.0E-06 | 15 | ATP hydrolysis coupled proton transport | |
| 4.0E-06 | 5 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity | |
| <2.0E-06 | 37 | protein processing | |
| 8.0E-06 | 12 | cyclin-dependent protein kinase regulator activity | |
| 4.0E-06 | 9 | flotillin complex | |
| 1.2E-05 | 12 | nucleoside-diphosphatase activity | |
| 4.0E-06 | 30 | stress fiber | |
| 6.0E-06 | 7 | L-ascorbic acid metabolic process | |
| 4.0E-06 | 154 | diacylglycerol binding | |
| 2.0E-06 | 30 | secretory granule | |
| 1.2E-05 | 20 | estrogen receptor activity | |
| <2.0E-06 | 45 | estrogen receptor signaling pathway | |
| 1.0E-05 | 8 | blastocyst development | |
| 6.0E-06 | 63 | forebrain development | |
| AraC | 4.0E-06 | 12 | nucleoside-diphosphatase activity |
| 6-TG | <2.0E-06 | 26 | 3′,5′-cyclic-AMP phosphodiesterase activity |
| 8.0E-06 | 14 | O-methyltransferase activity | |
| 2.0E-06 | 8 | acetylserotonin O-methyltransferase activity | |
| 2.0E-06 | 8 | melatonin biosynthetic process |
Table 2.
Statistical significant gene sets associated with AUC.
| Drug | AUC p | Gene Set Size | Gene Set |
|---|---|---|---|
| dFdC | 6.0E-06 | 15 | neurotransmitter catabolic process |
| 1.2E-05 | 31 | Gemcitabine Pathway | |
| <2.0E-06 | 12 | peptide-aspartate beta-dioxygenase activity | |
| 6.0E-06 | 5 | ribonucleoside-diphosphate reductase activity | |
| 1.0E-05 | 8 | thymidine kinase activity | |
| <2.0E-06 | 160 | nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | |
| <2.0E-06 | 19 | mRNA catabolic process | |
| 6.0E-06 | 24 | mitotic chromosome condensation | |
| 8.0E-06 | 39 | condensed chromosome | |
| 4.0E-06 | 54 | transaminase activity | |
| 2.0E-06 | 5 | gamma-aminobutyric acid catabolic process | |
| <2.0E-06 | 9 | peptidyl-amino acid modification | |
| 2.0E-06 | 70 | heterogeneous nuclear ribonucleoprotein complex | |
| 6-TG | 6.0E-06 | 5 | gamma-aminobutyric acid catabolic process |
| <2.0E-06 | 10 | choline transmembrane transporter activity | |
| <2.0E-06 | 10 | choline transport |
Figure 1.
Scatterplot of the size of the genes sets versus the −log10(p-values) for association with IC50 for the 4 drugs.
Figure 2.
Histogram of association p-values from gene set analysis for the four drugs and two phenotypes. (A) dFdC & IC50, (B) dFdC & AUC, (C) AraC & IC50, (D) AraC & AUC, (E) 6-TG & IC50, (F) 6-TG & AUC, (G) 6-MP & IC50, (H) 6-MP & AUC.
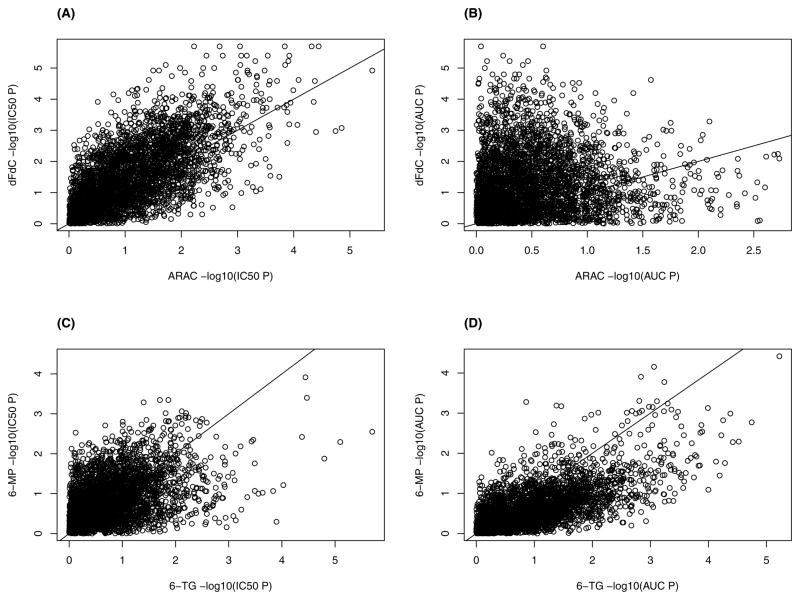
For the statistically significant gene sets, there was some overlap in findings between the various drugs. The gene set nucleoside-diphosphatase activity was found to be associated with both dFdC and AraC IC50, while gene set gamma-aminobutyric acid catabolic process was associated with dFdC and 6-TG AUC. Next, for gene sets with p-values < 0.0001, trends between the two drug classes were investigated. The relationship between the −log10(p-values) between the pyrimidines and the purines is displayed in Figure 3. For the pyrimidines (dFdC and AraC), five gene sets were found to overlap for the phenotype IC50: regulation of cell shape, dorsal/ventral pattern formation, nucleoside-diphosphatase activity, secretory granule, and regulation of axon extension. However, for the phenotype AUC, no gene sets were found to be in common between dFdC and AraC. For the purines (6-TG and 6-MP), one associated gene set was detected for both IC50 and AUC; gene set 3′,5′-cyclic-AMP phosphodiesterase activity and gamma-aminobutyric acid catabolic process for IC50 and AUC, respectively. No other gene sets detected were in common between the two drug classes.
Figure 3.
Scatterplot of the relationship between the −log10(p-values) for the pyrimidines (dFdC, AraC) and purines (6-TG, 6-MP). (A) dFdC & IC50 &. AraC & IC50, (B) dFdC & AUC & AraC & AUC, (C) 6-TG & IC50 & 6-MP & IC50, (D) 6-TG & AUC & 6-MP & AUC
Finally, the amount of overlap between the significant gene sets of potential pharmacogenomic relevance for the pyramides and purines was assessed. For the statistically significant gene sets detected for 6-TG, gene sets melatonin biosynthetic process and acetylserotonin O-methyltransferase activity contained the same set of genes and both gene sets were completely contained within gene set O-methyltransferase activity. In addition to this overlap in gene sets, gene sets choline transmembrane transporter activity and choline transport contained the same set of genes. No other overlap in the top gene sets for the two thiopurine drugs was observed. In contrast, for the statistically significant gene sets for dFdC and AraC, there was limited overlap in the gene sets with only pair of gene sets sharing one gene.
3.2. Functional Validation Studies
Since statistical significance does not always lead to functional relevance (e.g., false positive associations), using the gene set results shown in Tables 1 to 4 we selected gene sets based on both statistical significance and biological relevance to attempt functional validation. For dFdC and AraC, we selected four novel genes (CANT1, ENTPD6, ADRM1, PSME3) within the nucleoside-diphosphatase activity, proteasome activator activity and proteasome activator complex gene sets for functional validation (Table 3). Unlike dFdC and AraC, which shared common genes of interest, only 6-TG showed statistically significant gene sets for the thiopurine drugs. Therefore, we selected four genes (ABAT, ASMTL, PDE4D, and SLC44A1) for functional validation based on the 6-TG results in the gene sets 3′-5′-cyclic-AMP phosphodiesterase activity, choline transport (which overlaps with gene set choline transmembrane transporter activity) and O-methyltransferase activity (Table 4).
Table 4.
Gene level results in statistical and biologically relevant GSs for 6-TG. Bolded genes are genes selected for functional studies. For genes not selected for functional studies, we present only the results for one probe set for the gene (most significant) with the number of probe sets for the gene represented in parentheses.
| IC50 | AUC | |||||
|---|---|---|---|---|---|---|
| Probe set ID | Gene | Gene Set* | p-value | r | p-value | r |
| 208818_s_at | COMT (3) | O-methyltransferase activity | 6.35E-01 | 0.04 | 6.65E-01 | −0.03 |
| 226870_at | COMTD1 | O-methyltransferase activity | 9.10E-01 | 0.01 | 7.35E-01 | −0.03 |
| 223515_s_at | COQ3 (2) | O-methyltransferase activity | 3.30E-01 | −0.07 | 7.03E-01 | −0.03 |
| 206779_s_at | ASMT (2) | O-methyltransferase activity | 6.15E-02 | 0.14 | 5.44E-01 | 0.05 |
| 209394_at | ASMTL | O-methyltransferase activity | 1.29E-06 | 0.36 | 1.54E-03 | 0.24 |
| 36553_at | ASMTL | O-methyltransferase activity | 9.43E-06 | 0.33 | 5.24E-03 | 0.21 |
| 36554_at | ASMTL | O-methyltransferase activity | 3.71E-06 | 0.35 | 9.10E-03 | 0.20 |
| 209394_at | ASMTL | MBP/OMA/acetylserotonin OMA | 1.29E-06 | 0.36 | 1.54E-03 | 0.24 |
| 36554_at | ASMTL | MBP/OMA/acetylserotonin OMA | 3.71E-06 | 0.35 | 9.10E-03 | 0.20 |
| 36553_at | ASMTL | MBP/OMA/acetylserotonin OMA | 9.43E-06 | 0.33 | 5.24E-03 | 0.21 |
| 228447_at | SFRS17A (3) | MBP/OMA/acetylserotonin OMA | 2.15E-01 | 0.10 | 8.35E-02 | 0.13 |
| 211447_s_at | PDE4A (5) | 3′-5′-cyclic-AMP phosphodiesterase activity | 1.02E-02 | 0.20 | 1.60E-01 | 0.11 |
| 211302_s_at | PDE4B (3) | 3′-5′-cyclic-AMP phosphodiesterase activity | 2.03E-02 | −0.18 | 4.25E-02 | −0.16 |
| 206791_s_at | PDE4C (4) | 3′-5′-cyclic-AMP phosphodiesterase activity | 4.67E-03 | 0.22 | 5.65E-01 | −0.04 |
| 210837_s_at | PDE4D | 3′-5′-cyclic-AMP phosphodiesterase activity | 2.42E-06 | −0.35 | 8.61E-01 | −0.01 |
| 204491_at | PDE4D | 3′-5′-cyclic-AMP phosphodiesterase activity | 3.83E-04 | −0.27 | 2.84E-01 | −0.08 |
| 210836_x_at | PDE4D | 3′-5′-cyclic-AMP phosphodiesterase activity | 1.02E-03 | −0.25 | 2.96E-01 | −0.08 |
| 228962_at | PDE4D | 3′-5′-cyclic-AMP phosphodiesterase activity | 1.09E-03 | −0.25 | 3.43E-01 | −0.07 |
| 1554717_a_at | PDE4D | 3′-5′-cyclic-AMP phosphodiesterase activity | 5.41E-02 | −0.15 | 8.87E-01 | −0.01 |
| 211840_s_at | PDE4D | 3′-5′-cyclic-AMP phosphodiesterase activity | 9.56E-01 | 0.00 | 2.86E-01 | −0.08 |
| 230500_at | PDE7A (5) | 3′-5′-cyclic-AMP phosphodiesterase activity | 8.74E-02 | −0.13 | 1.14E-03 | −0.25 |
| 220343_at | PDE7B (3) | 3′-5′-cyclic-AMP phosphodiesterase activity | 4.67E-01 | 0.06 | 2.94E-01 | −0.08 |
| 203608_at | ALDH5A1 (2) | gamma-aminobutyric acid catabolic process | 9.81E-03 | 0.20 | 1.22E-04 | 0.29 |
| 209459_s_at | ABAT | gamma-aminobutyric acid catabolic process | 1.49E-02 | 0.19 | 2.21E-06 | 0.35 |
| 209460_at | ABAT | gamma-aminobutyric acid catabolic process | 7.38E-02 | 0.14 | 1.15E-03 | 0.25 |
| 206527_at | ABAT | gamma-aminobutyric acid catabolic process | 7.09E-01 | 0.03 | 5.17E-01 | 0.05 |
| 222364_at | SLC44A1 | choline transport/CTTA | 1.62E-04 | −0.28 | 1.96E-05 | −0.32 |
| 224596_at | SLC44A1 | choline transport/CTTA | 2.34E-04 | −0.28 | 4.93E-05 | −0.31 |
| 228485_s_at | SLC44A1 | choline transport/CTTA | 3.38E-04 | −0.27 | 1.38E-05 | −0.33 |
| 228486_at | SLC44A1 | choline transport/CTTA | 5.10E-04 | −0.26 | 1.54E-06 | −0.36 |
| 227620_at | SLC44A1 | choline transport/CTTA | 5.59E-04 | −0.26 | 7.78E-07 | −0.37 |
| 224595_at | SLC44A1 | choline transport/CTTA | 2.84E-03 | −0.23 | 4.26E-04 | −0.27 |
| 220722_s_at | SLC5A7 (2) | choline transport/CTTA | 5.49E-01 | 0.05 | 9.01E-01 | −0.01 |
| 224609_at | SLC44A2 (2) | choline transport/CTTA | 9.04E-01 | 0.01 | 7.72E-02 | −0.14 |
CTTA = choline transmemberane transporter activity; MBP = melatonin biosynthetic process; OMA = O-methyltransferase activity
Table 3.
Gene level results in statistical and biologically relevant GSs for dFdC and AraC. Bolded genes are genes selected for functional studies.
| dFdC IC50 | AraC IC50 | |||||
|---|---|---|---|---|---|---|
| Probe set ID | Gene | Gene Set* | p-value | r | p-value | r |
| 46323_at | CANT1 | nucleoside-diphosphatase activity | 8.45E-04 | −0.25 | 1.70E-04 | −0.28 |
| 221732_at | CANT1 | nucleoside-diphosphatase activity | 7.21E-04 | −0.26 | 4.74E-04 | −0.26 |
| 1554327_a_at | CANT1 | nucleoside-diphosphatase activity | 1.50E-03 | −0.24 | 1.27E-02 | −0.19 |
| 206191_at | ENTPD3 | nucleoside-diphosphatase activity | 6.14E-01 | 0.04 | 7.25E-01 | −0.03 |
| 1555118_at | ENTPD3 | nucleoside-diphosphatase activity | 3.30E-01 | −0.07 | 4.81E-01 | −0.05 |
| 204076_at | ENTPD4 | nucleoside-diphosphatase activity | 4.98E-01 | −0.05 | 2.20E-03 | −0.23 |
| 1555358_a_at | ENTPD4 | nucleoside-diphosphatase activity | 3.83E-01 | −0.07 | 3.47E-01 | −0.07 |
| 204077_x_at | ENTPD4 | nucleoside-diphosphatase activity | 1.89E-01 | −0.10 | 6.05E-01 | −0.04 |
| 205757_at | ENTPD5 | nucleoside-diphosphatase activity | 4.01E-03 | −0.22 | 4.17E-02 | −0.15 |
| 1554094_at | ENTPD5 | nucleoside-diphosphatase activity | 8.33E-01 | 0.02 | 8.51E-01 | −0.01 |
| 201704_at | ENTPD6 | nucleoside-diphosphatase activity | 3.58E-04 | −0.27 | 2.66E-03 | −0.23 |
| 234946_at | ENTPD6 | nucleoside-diphosphatase activity | 5.33E-02 | 0.15 | 9.68E-03 | 0.20 |
| 201281_at | ADRM1 | proteasome activator activity | 4.53E-04 | −0.26 | 6.30E-02 | −0.14 |
| 200814_at | PSME1 | proteasome activator complex and PAA | 9.20E-01 | 0.01 | 9.86E-01 | 0.00 |
| 201762_s_at | PSME2 | proteasome activator complex and PAA | 1.77E-01 | −0.10 | 5.45E-01 | −0.05 |
| 200988_s_at | PSME3 | proteasome activator complex and PAA | 2.60E-06 | −0.35 | 2.66E-04 | −0.27 |
| 209852_x_at | PSME3 | proteasome activator complex and PAA | 1.60E-05 | − 0.32 | 4.41E-04 | −0.26 |
| 200987_x_at | PSME3 | proteasome activator complex and PAA | 4.10E-05 | −0.31 | 4.54E-02 | −0.15 |
| 209853_s_at | PSME3 | proteasome activator complex and PAA | 5.58E-07 | −0.37 | 2.09E-02 | −0.18 |
PAA = proteasome activator activity
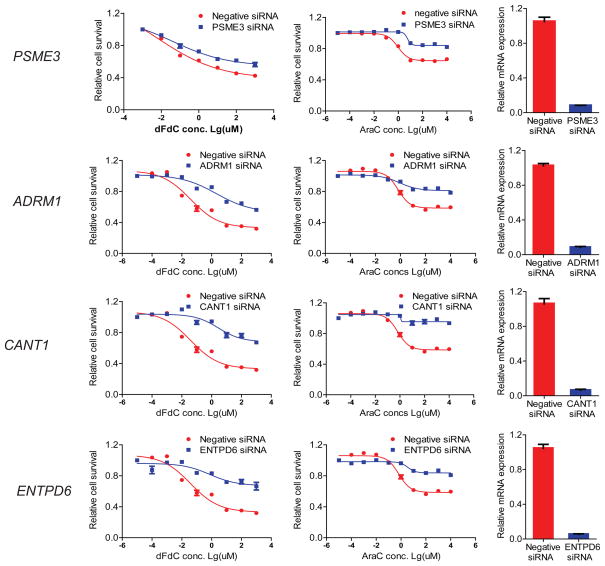
The human pancreatic cancer SU86 cell line was chosen to test dFdC and AraC because dFdC is first-line therapy for pancreatic cancer, and the genes of interest were expressed in that cell line. Knockdown of all four genes (PSME3, CANT1, ENTPD6, ADRM1) in SU86 cells significantly desensitized the cells to dFdC and AraC (Figure 4), confirming the results of the gene set analysis. Knockdown efficiency of specific siRNAs against the four genes was confirmed using real-time QRT-PCR assays (Figure 4).
Figure 4.
dFdC and AraC functional validation in human SU86 cells. siRNA knockdown was performed, followed by drug cytotoxicity (left panels) and real-time QRT-PCR assays (right panels). SU86 cells were transfected with control siRNA or specific siRNA for PSME3, ADRM1, CANT1, and ENTPD6. 24 h after siRNA transfection, cells were treated with either dFdC or AraC for an additional 72 h.
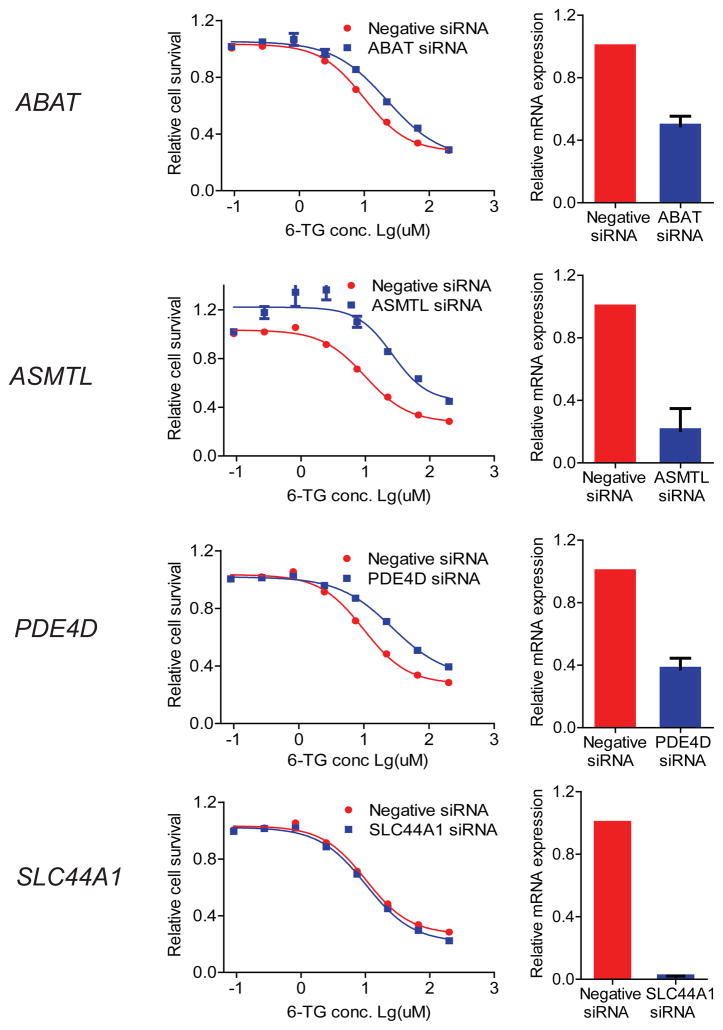
For 6-TG, 4 candidate genes were tested in the U251 cell line. That cell line expressed all four of these genes. Knockdown of PDE4D, ABAT and ASMTL resulted in a shift in the cytotoxicity curves (Figure 5), but only PDE4D showed a shift compatible with the direction of drug effect observed during the initial gene expression studies (Table 4). Therefore, in the case of 6-TG, only the PDE4D results among the four genes tested were validated.
Figure 5.
6-TG functional validation in U251 cells. siRNA knockdown was performed, followed by drug cytotoxicity (left panels) and real-time QRT-PCR assays (right panels). U251 cells were transfected with control siRNA or specific siRNA for ABAT, ASMTL, PDE4D, and SLC44A1. 24 h after siRNA transfection, cells were treated with 6-TG for an additional 72 h.
4. DISCUSSION
With the advent of high-throughput technologies, such as microarrays, complete genome-wide studies of genomic predictors for response to therapies are becoming commonplace. Responses to therapies, either with regard to toxicities or efficacy, are expected to involve complex relationships of gene products within the same molecular pathway or functional gene set. Therefore, pathways or gene sets, as opposed to single genes, may better reflect the true underlying biology and be more appropriate units for analysis for pharmacogenomic studies. Application of such methods to pharmacogenomic studies may enable the detection of more subtle effects of multiple genes in the same pathway that may be missed by assessing each gene individually. Moreover, the incorporation of biological knowledge in the statistical analysis may aid researchers in the interpretation of results from genome-wide pharmacogenomic studies. However, results from our gene set analysis are limited to the current definition of gene sets and pathways, realizing that knowledge about the genome and definitions of GSs are evolving and that no single definition of a GS exists. In addition, Gene Ontology (GO) uses a hierarchical ontology that classifies genes and therefore a level of the hierarchy (or specificity) must be specified for defining gene sets. For this analysis, we chose level 4 to determine genes sets as a compromise between specificity and sensitivity. In this manuscript, a gene set analysis is presented to assess the impact of mRNA expression on drug cytotoxicity for two classes of drugs: pyrimidines (dFdC, AraC) and purines (6-TG and 6-MP).
Gene set analysis of the drug dFdC detected the gene set of the gemcitabine metabolic pathway to be significantly associated with dFdC AUC (p = 1.2 × 10−5), which confirmed the confidence of the gene set methods for incorporating the known biological knowledge into the statistical analysis. The gene set of nucleoside-diphosphatase activity was also found to be associated with both dFdC (p = 1.2 × 10−5) and AraC (p = 4 × 10−6) IC50. It is well-known that these two cytidine analogues share similar chemical structures, metabolic pathways, and mechanisms of action. Both are prodrugs that must be transported into cells, activated by kinases to form active diphosphorylated and triphosphorylated metabolites, and inactivated by dephosphorylation[6,7]. One of nucleotidase family members, NT5C3, that catalyzes the dephosphorylation of nucleoside monophosphates, has been previously demonstrated to be associated with sensitivity to dFdC and AraC[22], with these results indicating the possible involvement of other genes in the nucleoside-diphosphatase activity gene set in the inactivation of these cytidine analogues. In particular, we hypothesize that nucleoside diphosphatases might convert nucleoside diphosphate to nucleoside monophosphate, and consequently to decreased bioactive metabolite levels of the cytidine analogues, dFdC- or AraC- triphosphorate. Therefore, it is plausible that genes in this gene set might be associated with the clinical response to these two drugs. Functional validation by siRNA knockdown of CANT1 and ENTPD6, followed by dFdC and AraC cytotoxicity assay in SU86 cells, validated our gene set results (lowest p = 7.21E-04 and r = − 0.26 for CANT1, while lowest p = 3.58E-04 and r = − 0.27 for ENTPD6). These results suggest that CANT1 and ENTPD6 might be involved in the mechanism of response to dFdC and AraC in pancreatic cancer cells.
Additionally, two of the gene sets statistically associated with dFdC IC50 were proteasome activator activity (p<2.0E-06) and proteasome activator complex (p<2.0E-06). The survival genes are very likely to contribute to tumor cell proliferation and chemotherapy, as well as the variation in drug response. The ubiquitin-proteasome system is involved in degradation of many intracellular proteins and is necessary for proper functioning of the cell under normal conditions and its survival under stress conditions[32]. The genes within the gene set of survival genes product activity might be negatively regulated and consequently dysfunction through the proteasome pathway. Some part of survival genes were components of the 20S and 26S proteasome complexes[33]. It is possible to hypothesize that those genes involved in proteasome activator complex and activity might also play roles in the regulation of the drug response to these two cytidine analogues. In follow-up functional validation studies, we demonstrated that deceased expression of PSME3 and ADRM1 in SU86 cells resulted in resistance to dFdC and AraC. These observations confirmed the gene set results (lowest p = 5.58E-07 and r = − 0.37 for PSME3, while p = 4.53E-04 and r = − 0.26 for ADRM1), suggesting possible roles for PSME3 and ADRM1 in response to dFdC and AraC.
In contrast to the results from the gene set analysis of the pyrimidines, gene set analysis for the two purine antimetabolites, 6-MP and 6-TG, identified gene sets 3′, 5′-cyclic-AMP phosphodiesterase activity and gamma-aminobutyric acid catabolic process in common between 6-TG and 6-MP for IC50 and AUC phenotypes, respectively. In addition, the analysis identified 7 gene sets that are uniquely associated with 6-TG cytotoxic phenotypes (4 for IC50 and 3 for AUC). The similarity and uniqueness of gene sets detected for the two drugs was not surprising, as 6-MP and 6-TG are anticancer purine analogues that shares similar structure and metabolomic pathways but different mechanism of actions to inhibit the growth of cancer cells. Specifically, after metabolomic activation by hypoxanthine-guanine phosphoribosyl transferase (HGPRT), the active metabolites of 6-MP and 6-TG (TIMP and TGMP) will function as inhibitor for de novo purine biosynthesis and inhibitors for DNA/RNA synthesis, respectively[34]. The gene set for 3′, 5′-cyclic-AMP phosphodiesterase activity includes 26 genes that may influence intracellular level of cAMP. A previous study has demonstrated that efflux of nucleotide (especially cyclic nucleotides, e.g. cAMP) through nucleotide efflux transporters that alters the concentration of intracellular cyclic nucleotide and their signaling and could confer resistance to cytotoxic thiopurine nucleotides[35]. These findings provide further evidence on the importance of intracellular level of cAMP in chemosensitivity of the cells to purine analogues. Only one of the 6-TG related genes, PDE4D, could be functionally validated (Figure 5). PDE4D encodes a cAMP-specific 3′, 5′-cyclic phosphodiesterase and is involved in purine metabolism. By selectively degrading cGMP and cAMP to the corresponding nucleotide monophosphates[36,37], PDE4D regulates their cellular concentrations, localization and duration of action. Therefore, PDE4D is a biologically plausible candidate to modify 6-TG response.
One-carbon metabolism that includes the Methionine and the Folate cycles, is a major resource of one carbon unit for many essential cellular events, including de novo biosynthesis of purines and thymidylates[38]. The Methionine and the Folate cycles are coupled through remethylation of homocysteine that also generates methionine. S-Adenosylmethionine (SAM) that is converted from methionine is a high-energy methyl group donor and is required for most SAM-dependent methyltransferases for metabolisms of many neurotransmitters. A recent study by Ross et al. demonstrated that genetic variation in two SAM-dependent methyltransferase, TPMT and catechol O-methyltransferase (COMT) are significantly associated with cisplatin-induced hearing loss in children[39], linking the essential one-carbon metabolism to cytotoxicity of chemotherapeutic agents. Of interest, gene set analyses discovered several gene sets associated with 6-TG cytotoxic profiles (both IC50 and AUC) that could relate to the One-carbon metabolism process. Gene sets for o-methyltransferase activity could influence the balance of the Methionine cycle by regulating level of SAM, and thus its precursor, methionine, as well as the remethylation process of methionine, therefore, indirectly influence the Folate cycle and further, the purine biosynthesis. Since choline is converted to betaine, which is the substrate of the betaine-homocysteine methyltransferase, the primary enzyme catalyzes the homocysteine remethylation in liver; gene sets for choline transport could also indirectly affect the folate pathway and the purine synthesis through the homocysteine remethylation step. However, knockdown of SLC44A1 and ASMTL were not functionally validated.
In summary, a comprehensive gene set analysis of two commonly used therapeutic drug classes, pyrimidines and purines, used in the treatment of cancer found novel gene sets associated with IC50 and 5 of the 8 genes chosen for functional studies were validated. These findings provide insight into the relationship between genomic variation and drug response. The results of this analysis also illustrate the usefulness of gene set analysis methods for the development of novel hypotheses that can then be followed-up with additional clinical or functional studies. The strengths of this study are: (1) the extensive amount of information available for multiple drugs within the same class of drugs; allowing for the comparison of results between the drugs within the same class and across drug classes; and (2) the functional validation of genes detected from the gene set analysis. Moreover, application of gene set methods to these pharmacogenomic studies detected significant gene sets (and the corresponding genes within these gene sets) that impact drug cytotoxicity.
Acknowledgments
The research was supported by the NIH U01 HG005137 (RMW, LW), U01 GM61388 (RMW, BLF, LL, YJ, LW, GDJ, FL), R21 CA140879 (BLF), R01 GM28157 (RMW, FL), R01 CA132780 (RMW, BLF), K22 CA130828 (LW), R01 CA138461 (LW, LL), Minnesota Partnership for Biotechnology and Medical Genomics grant H9046000431 (BLF, GDJ, AB) and the Mayo Foundation.
Footnotes
Conflict of Interest: none to declare
References
- 1.Bild AH, Potti A, Nevins JR. Linking oncogenic pathways with therapeutic opportunities. Nat Rev Cancer. 2006;6:735–741. doi: 10.1038/nrc1976. [DOI] [PubMed] [Google Scholar]
- 2.Goeman JJ, Buhlmann P. Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics. 2007;23:980–987. doi: 10.1093/bioinformatics/btm051. [DOI] [PubMed] [Google Scholar]
- 3.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 4.Riedel RF, Porrello A, Pontzer E, Chenette EJ, Hsu DS, et al. A genomic approach to identify molecular pathways associated with chemotherapy resistance. Mol Cancer Ther. 2008;7:3141–3149. doi: 10.1158/1535-7163.MCT-08-0642. [DOI] [PubMed] [Google Scholar]
- 5.Gamazon ER, Huang RS, Cox NJ, Dolan ME. Chemotherapeutic drug susceptibility associated SNPs are enriched in expression quantitative trait loci. Proc Natl Acad Sci U S A. 107:9287–9292. doi: 10.1073/pnas.1001827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berlin JD, Catalano P, Thomas JP, Kugler JW, Haller DG, et al. Phase III study of gemcitabine in combination with fluorouracil versus gemcitabine alone in patients with advanced pancreatic carcinoma: Eastern Cooperative Oncology Group Trial E2297. J Clin Oncol. 2002;20:3270–3275. doi: 10.1200/JCO.2002.11.149. [DOI] [PubMed] [Google Scholar]
- 7.Kern W, Estey EH. High-dose cytosine arabinoside in the treatment of acute myeloid leukemia: Review of three randomized trials. Cancer. 2006;107:116–124. doi: 10.1002/cncr.21543. [DOI] [PubMed] [Google Scholar]
- 8.Heinemann V, Hertel LW, Grindey GB, Plunkett W. Comparison of the cellular pharmacokinetics and toxicity of 2′,2′-difluorodeoxycytidine and 1-beta-D-arabinofuranosylcytosine. Cancer Res. 1988;48:4024–4031. [PubMed] [Google Scholar]
- 9.Wiley JS, Taupin J, Jamieson GP, Snook M, Sawyer WH, et al. Cytosine arabinoside transport and metabolism in acute leukemias and T cell lymphoblastic lymphoma. J Clin Invest. 1985;75:632–642. doi: 10.1172/JCI111741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fawcett T. An introduction to ROC analysis. Pattern Recognition Letters. 2006;27:861–874. [Google Scholar]
- 11.Kindler HL. In focus: advanced pancreatic cancer. Clin Adv Hematol Oncol. 2005;3:420–422. [PubMed] [Google Scholar]
- 12.Castaigne S, Tilly H, Sigaux F, Solal Celigny P, Bordessoule D, et al. Treatment of malignant hemopathies with aracytine in low doses. Analysis of 159 cases. Nouv Rev Fr Hematol. 1985;27:377–382. [PubMed] [Google Scholar]
- 13.Braess J, Jahns-Streubel G, Schoch C, Haase D, Haferlach T, et al. Proliferative activity of leukaemic blasts and cytosine arabinoside pharmacodynamics are associated with cytogenetically defined prognostic subgroups in acute myeloid leukaemia. Br J Haematol. 2001;113:975–982. doi: 10.1046/j.1365-2141.2001.02866.x. [DOI] [PubMed] [Google Scholar]
- 14.Schoch C, Haferlach T, Haase D, Fonatsch C, Loffler H, et al. Patients with de novo acute myeloid leukaemia and complex karyotype aberrations show a poor prognosis despite intensive treatment: a study of 90 patients. Br J Haematol. 2001;112:118–126. doi: 10.1046/j.1365-2141.2001.02511.x. [DOI] [PubMed] [Google Scholar]
- 15.Mini E, Nobili S, Caciagli B, Landini I, Mazzei T. Cellular pharmacology of gemcitabine. Ann Oncol. 2006;17 (Suppl 5):v7–v12. doi: 10.1093/annonc/mdj941. [DOI] [PubMed] [Google Scholar]
- 16.Seve P, Mackey JR, Isaac S, Tredan O, Souquet PJ, et al. cN-II expression predicts survival in patients receiving gemcitabine for advanced non-small cell lung cancer. Lung Cancer. 2005;49:363–370. doi: 10.1016/j.lungcan.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Rosell R, Danenberg KD, Alberola V, Bepler G, Sanchez JJ, et al. Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2004;10:1318–1325. doi: 10.1158/1078-0432.ccr-03-0156. [DOI] [PubMed] [Google Scholar]
- 18.Smid K, Bergman AM, Eijk PP, Veerman G, van Haperen VW, et al. Micro-array analysis of resistance for gemcitabine results in increased expression of ribonucleotide reductase subunits. Nucleosides Nucleotides Nucleic Acids. 2006;25:1001–1007. doi: 10.1080/15257770600890269. [DOI] [PubMed] [Google Scholar]
- 19.Lennard L. The clinical pharmacology of 6-mercaptopurine. Eur J Clin Pharmacol. 1992;43:329–339. doi: 10.1007/BF02220605. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Weinshilboum R. Thiopurine S-methyltransferase pharmacogenetics: insights, challenges and future directions. Oncogene. 2006;25:1629–1638. doi: 10.1038/sj.onc.1209372. [DOI] [PubMed] [Google Scholar]
- 21.Gearry RB, Barclay ML, Burt MJ, Collett JA, Chapman BA, et al. Thiopurine S-methyltransferase (TPMT) genotype does not predict adverse drug reactions to thiopurine drugs in patients with inflammatory bowel disease. Aliment Pharmacol Ther. 2003;18:395–400. doi: 10.1046/j.1365-2036.2003.01690.x. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Fridley B, Kalari K, Jenkins G, Batzler A, et al. Gemcitabine and Cytosine Arabinoside Cytotoxicity: Association with Lymphoblastoid Cell Expression. Cancer Res. 2008;68:7050–7058. doi: 10.1158/0008-5472.CAN-08-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F, Fridley BL, Matimba A, Kalari KR, Pelleymounter L, et al. Ecto-5′-nucleotidase and thiopurine cellular circulation: association with cytotoxicity. Drug Metab Dispos. 2010;38:2329–2338. doi: 10.1124/dmd.110.035220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritz C, Streibig JC. Bioassay Analysis using R. Journal of Statistical Software. 2005:12. [Google Scholar]
- 25.Gallant AR. Nonlinear Statistical Models. New York: Wiley; 1987. [Google Scholar]
- 26.Davidian M, Giltinan DM. Nonlinear models for repeated measurement data. New York: Chapman & Hall; 1995. [Google Scholar]
- 27.Wu Z, Irizarry R, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligobucleotide expression arrays. Journal of the American Statistical Association. 2004;99:909–917. [Google Scholar]
- 28.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 29.Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 30.Fridley BL, Jenkins GD, Biernacka JM. Self-contained gene-set analysis of expression data: an evaluation of existing and novel methods. PLoS One. 2010;5:e12693. doi: 10.1371/journal.pone.0012693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, Fridley BL, Kalari K, Jenkins G, Batzler A, et al. Gemcitabine and arabinosylcytosin pharmacogenomics: genome-wide association and drug response biomarkers. PLoS One. 2009;4:e7765. doi: 10.1371/journal.pone.0007765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang L, Chen CH. Proteasome regulators: activators and inhibitors. Curr Med Chem. 2009;16:931–939. doi: 10.2174/092986709787581860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thaker NG, Zhang F, McDonald PR, Shun TY, Lewen MD, et al. Identification of survival genes in human glioblastoma cells by small interfering RNA screening. Mol Pharmacol. 2009;76:1246–1255. doi: 10.1124/mol.109.058024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karran P, Attard N. Thiopurines in current medical practice: molecular mechanisms and contributions to therapy-related cancer. Nat Rev Cancer. 2008;8:24–36. doi: 10.1038/nrc2292. [DOI] [PubMed] [Google Scholar]
- 35.Sampath J, Adachi M, Hatse S, Naesens L, Balzarini J, et al. Role of MRP4 and MRP5 in biology and chemotherapy. AAPS PharmSci. 2002;4:E14. doi: 10.1208/ps040314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huai Q, Colicelli J, Ke H. The crystal structure of AMP-bound PDE4 suggests a mechanism for phosphodiesterase catalysis. Biochemistry. 2003;42:13220–13226. doi: 10.1021/bi034653e. [DOI] [PubMed] [Google Scholar]
- 37.Nemoz G, Zhang R, Sette C, Conti M. Identification of cyclic AMP-phosphodiesterase variants from the PDE4D gene expressed in human peripheral mononuclear cells. FEBS Lett. 1996;384:97–102. doi: 10.1016/0014-5793(96)00300-6. [DOI] [PubMed] [Google Scholar]
- 38.Carmel R, Jacobson DW. Homocyseine in Health and Disease. New York, NY: Cambridge University Press; 2001. [Google Scholar]
- 39.Ross CJ, Katzov-Eckert H, Dube MP, Brooks B, Rassekh SR, et al. Genetic variants in TPMT and COMT are associated with hearing loss in children receiving cisplatin chemotherapy. Nat Genet. 2009;41:1345–1349. doi: 10.1038/ng.478. [DOI] [PubMed] [Google Scholar]