Abstract
Nkx2.2 and Arx are essential pancreatic transcription factors. Nkx2.2 is necessary for the appropriate specification of the islet alpha, beta, PP and epsilon cell lineages, whereas Arx is required to form the correct ratio of alpha, beta, delta and PP cells. To begin to understand the cooperative functions of Nkx2.2 and Arx in the development of endocrine cell lineages, we generated progenitor cell-specific deletions of Arx on the Nkx2.2 null background. The analysis of these mutants demonstrates that expansion of the ghrelin cell population in the Nkx2.2 null pancreas is not dependent on Arx; however, Arx is necessary for the upregulation of ghrelin mRNA levels in Nkx2.2 mutant epsilon cells. Alternatively, in the absence of Arx, delta cell numbers are increased and Nkx2.2 becomes essential for the repression of somatostatin gene expression. Interestingly, the dysregulation of ghrelin and somatostatin expression in the Nkx2.2/Arx compound mutant (Nkx2.2null;ArxΔpanc) results in the appearance of ghrelin+/somatostatin+ co-expressing cells. These compound mutants also revealed a genetic interaction between Nkx2.2 and Arx in the regulation of the PP cell lineage; the PP cell population is reduced when Nkx2.2 is deleted but is restored back to wildtype numbers in the Nkx2.2null;ArxΔpanc mutant. Moreover, conditional deletion of Arx in specific pancreatic cell populations established that the functions of Arx are necessary in the Neurog3+ endocrine progenitors. Together, these experiments identify novel genetic interactions between Nkx2.2 and Arx within the endocrine progenitor cells that ensure the correct specification and regulation of endocrine hormone-producing cells.
Keywords: Nkx2.2, Arx, transcriptional regulation, endocrine cell fate, ghrelin, PP, somatostatin
Introduction
At embryonic day (E) 9.5, the region of dorsal foregut endoderm fated to become pancreas evaginates from the gut tube creating the dorsal pancreatic bud. One day later, the ventral pancreatic domain is similarly produced from the ventral foregut endoderm (Jorgensen et al., 2007). The generation of these pancreatic anlagen requires signals from the surrounding tissues, which ultimately permit the patterning of all organs along the length of the primitive gut tube (Hebrok et al., 1998; Kim et al., 1997; Martin et al., 2005; Wells and Melton, 2000; Zorn and Wells, 2009). Cells within this pancreatic domain expressing the transcription factors Pdx1 and Ptf1a mark the pancreatic progenitor cells (Burlison et al., 2008; Kawaguchi et al., 2002; Offield et al., 1996). Once these progenitor cells are specified, a cascade of transcription factors together with signals from the mesenchyme (Ahlgren et al., 1997; Bhushan et al., 2001), peripheral nervous system (Lausier et al., 2010; Nekrep et al., 2008; Olerud et al., 2009; Plank et al., 2011) and developing vasculature (Yoshitomi and Zaret, 2004) are required to specify the three pancreatic lineages: endocrine, exocrine and ductal cells. Cells of the exocrine lineage will function in the production and secretion of digestive enzymes (Macdonald et al., 2010), while the ductal lineage will generate the epithelial cords that compose the ductal network, which runs throughout the pancreas and contains Neurogenin3+ (Neurog3) endocrine progenitor cells (Gradwohl, 2006; Villasenor et al., 2011). Activation of Neurog3, a pro-endocrine bHLH transcription factor, is necessary to specify the endocrine progenitors that will give rise to the five major endocrine subtypes located in the islets of Langerhans, including alpha, beta, delta, PP and epsilon cells, and expressing the hormones glucagon, insulin, somatostatin, pancreatic polypeptide and ghrelin, respectively (Gradwohl et al., 2000; Prado et al., 2004).
A major wave of endocrine differentiation occurs from E12.5 to E15.5, the stage of pancreas development termed the secondary transition (Pictet and Rutter, 1972). This differentiation and subsequent maturation of all hormone-producing endocrine cells is reliant upon the temporal and spatial activation and/or repression of a complex network of transcription factors (Pan and Wright, 2011). Two transcription factors necessary for specific endocrine cell lineage development are NK2 homeobox 2 (Nkx2.2) and Aristaless-related homeobox (Arx). The developing pancreas of the Nkx2.2 null embryo shows a complete absence of insulin-expressing beta cells, a ∼90% reduction of glucagon-expressing alpha cells and a ∼50% reduction in pancreatic polypeptide-expressing PP cells. In place of these cell populations there is an increase in ghrelin-expressing epsilon cells, although the small population of glucagon/ghrelin coexpressing cells that exist in wildtype pancreata is lost (Chao et al., 2007; Prado et al., 2004; Sussel et al., 1998). Similar to the Nkx2.2 null embryos, deletion of Arx results in a significant loss of alpha cells and the glucagon/ghrelin coexpressing cells, but unlike Nkx2.2 mutants, display an increase of the beta and delta cell populations (Collombat et al., 2003). Although Arx does not appear to be necessary for PP cell development, the misexpression of Arx in Pdx1- or Pax6-expressing cells results in an increase of alpha and PP cells (Collombat et al., 2007). The cumulative phenotypic analyses indicate that the primary commonality between the Nkx2.2 and Arx single mutants is the absence of the glucagon-expressing alpha cells, as well as the rare population of glucagon+/ghrelin+ cells (Chao et al., 2007; Heller et al., 2005).
Bihormonal cells have been described in several endocrine organs, including the developing pancreas and stomach. In particular, glucagon+/insulin+, ghrelin+/glucagon+ or ghrelin+/PP+ co-expressing islet cells have been identified in the embryonic pancreatic domain of the mouse, rat and human (De Krijger et al., 1992; Heller et al., 2005; Herrera, 2000; Lukinius et al., 1992; Wierup et al., 2004). In the developing stomach, gastrin and somatostatin co-expressing cells have also been observed (Larsson, 2000). Interestingly, while it is believed that mature gastrin-secreting G cells and somatostatin-secreting D cells derive from the common bihormonal precursor cells in the gastric epithelium (Larsson, 2000; Larsson et al., 1996), the pancreatic glucagon+/insulin+ bihormonal cells do not appear to represent endocrine progenitors; cells expressing insulin do not derive from those previously expressing glucagon, and the reverse is also true (Herrera, 2000). Therefore, hormone coexpression may or may not indicate a lineage relationship between two endocrine subtypes. Similarly, the emergence of bihormonal cell populations in genetically mutant backgrounds could represent either the expansion of rare bipotential cells or single lineage populations with dysregulated hormone genes. Both situations are likely influenced by the presence or absence of specific transcription factors within each lineage. While mutations in single transcription factors can lead to dysregulated lineage decisions and/or hormone expression (Ahlgren et al., 1998; Collombat et al., 2003; Prado et al., 2004; Wang et al., 2008), it is also possible that specific combinations of transcription factors are required to regulate appropriate lineage decisions or hormone gene regulation.
Given the overlapping, and sometimes opposing, roles for Nkx2.2 and Arx in specifying endocrine lineages, we hypothesized Nkx2.2 and Arx genetically interact to regulate endocrine cell differentiation and hormone gene expression in the developing pancreas. In this study we identified Arx+/Nkx2.2+ co-expressing cells in the early pancreatic progenitor domain, and demonstrated that Arx is expressed in wildtype and Nkx2.2-deficient ghrelin+ cells. To investigate the requirement of Arx and Nkx2.2 in regulating islet cell fates, we deleted Arx in the pancreas progenitor cells (Pdx1-cre; (Hingorani et al., 2003)) or endocrine progenitor cells (Neurog3-cre; (Schonhoff et al., 2004)) in the Nkx2.2 null background. These compound mutants revealed a genetic interaction between Arx and Nkx2.2 in the development of the PP cell lineage. Moreover, simultaneous deletion of Arx and Nkx2.2 lead to the dysregulation of ghrelin and somatostatin gene expression. Arx-dependent ghrelin gene regulation and Nkx2.2- dependent somatostatin gene regulation were altered in delta and epsilon cells, respectively, resulting in the expansion of a ghrelin+/somatostatin+ co-expressing cell population. Taken together, these data indicate that Nkx2.2 and Arx genetically interact in the regulation of islet PP cell specification and endocrine hormone gene expression in the ghrelin- and somatostatin-expressing cell populations.
Materials and Methods
Mice
All mice were bred and maintained on an outbred Black Swiss background (NTac:NIHBS, Taconic), according to Columbia University IACUC approved protocols. All strains were previously generated. Cell-type specific Arx null mice were generated by intercrossing Arxtm1Gldn (Arxflox/flox(or Y); (Fulp et al., 2008)) and either Tg(Ipf1-cre)1Tuv (Pdx1-cre; (Hingorani et al., 2003)) or Tg(Neurog3-cre)C1Able (Neurog3-cre; (Schonhoff et al., 2004)) mice. Arxflox/flox(or Y);Pdx1-cre and Arxflox/flox(or Y);Neurog3-cre mice were viable and fertile. The Pdx1-cre deletes Arx in all pancreatic progenitor cells; however, the Pdx1 expression domain also includes the glandular stomach and the duodenum (Larsson et al., 1996; Offield et al., 1996). These mice were then crossed to Nkx2-2tm1Jlr knock-in mice (Sussel et al., 1998) to generate compound heterozygotes. Embryos were collected from timed matings between Nkx2.2+/-;Arxflox/flox(orY);Pdx1-cre or Nkx2.2+/-;Arxflox/flox(or y);Neurog3-cre mice The Nkx2.2LacZ/+ knock-in reporter line was used for expression analysis; the homozygous Nkx2.2LacZ/LacZ mice are phenotypically identical to the Nkx2.2 null allele (Arnes and Sussel, in preparation). Noon on the day of appearance of a vaginal plug was considered embryonic day (E) 0.5. The experimental genotypes of wildtype, Nkx2.2-/- (Nkx2.2null), Arxflox/flox(or Y);Pdx1-cre (ArxΔpanc), Nkx2.2-/-;Arxfiox/flox(or Y);pdx1-cre (Nkx2.2null;ArxΔpanc), Arxflox/flox(or Y);Neurog3-cre (ArxΔendo), and Nkx2.2-/-;Arxflox/flox(or Y);Neurog3-cre (Nkx2.2null;ArxΔendo) were studied. Litters were assessed at E12.5, E15.5, and postnatal day (P) 0. All embryo dissections were carried out in cold PBS, using a dissecting microscope (Leica MZ8). Tail or yolk sac was removed from the embryo, digested with proteinase K, and DNA extracted for genotyping purposes. Genotyping was carried out with standard conditions and primers as previously described (Fulp et al., 2008; Hingorani et al., 2003; Schonhoff et al., 2004; Sussel et al., 1998).
Realtime PCR
Pancreas was dissected from each embryo and stored in RNAlater (Ambion) until RNA was extracted using the NucleoSpin RNAII Kit (Clonetech). Subsequently, cDNA was made with equal amounts of RNA for each sample and oligo(dT) (Superscript III Kit, Invitrogen, CA). Realtime PCR was performed using TaqMan gene expression assays (Applied biosystems) for glucagon (Mm00801712_m1), ghrelin (Mm00445450_m1), somatostatin (Mm00436671_m1), insulin1 (Mm01950294_s1), insulin2 (Mm00731595_gH), pancreatic polypeptide (Mm00435889_m1), Neurod1 (Mm01280117_m1), Pdx1 (Mm00435565_m1), Nkx6.1 (Mm00436671_m1), Hes1 (Mm00468601_m1), Arx (Mm00545903_m1), and Brn4 (Pou3f4; Mm00447171_s1). Nkx2.2 and cyclophilinB, which was used as a control housekeeping gene, were also assayed using probe and primer sets previously described (Chao et al., 2007). A standard two-step realtime PCR program was used for all genes assessed, with an annealing temperature of 61°C and 40 cycles of amplification (CFX96 RealTime System C1000 Thermal Cycler, Biorad). All gene expression values were normalized to the internal control gene, cyclophilinB, and relative quantification was performed using a standard curve from embryonic age-matched wildtype cDNA. A standard two-tailed Student t-test was performed to determine significance.
Immunofluorescence
Immunofluorescence was performed according to standard protocols, on E12.5 and P0 whole embryos that were embedded in OCT, after fixation with 4% PFA and cryopreservation in 30% sucrose. Transverse frozen sections (8 μm) were cut and mounted on glass slides. Sections were stained with rabbit α-ghrelin (1:800; Phoenix Pharmaceuticals, CA), goat α-ghrelin (1:800; Santa Cruz), guinea pig α-glucagon (1:1000; Linco/Millipore, MA), guinea pig α-insulin (1:1000; Millipore), rabbit α-insulin (1:1000; Cell Signaling Technology), rabbit α-somatostatin (1:200; Phoenix Pharmaceuticals), rabbit α-pancreatic polypeptide (1:200; Zymed), rabbit α-amylase (1:1000; Sigma), goat α-carboxypeptidase A (1:800; R&D Systems), rabbit α-Arx (1:500; kind gift from Dr. Kanako Miyabayashi, Kyushu University, Japan; (Kitamura et al., 2002)), rabbit α-Pdx1 (1:1000; Millipore), guinea pig α-Pdx1 (1:1000; C. Wright, Vanderbilt University; supplied by BCBC), rabbit α-Sox9 (1:500; Santa Cruz), rabbit α-Neurog3 (1:500; BCBC Antibody Core), and chicken α-beta-galactosidase (1:250; Abcam). Donkey α-guinea pig-Cy3 or Cy5, α-rabbit-Cy2 or Cy3, α-chicken-Cy3, and α-goat Cy2 or Cy5 secondary antibodies were used (1:400, Jackson ImmunoResearch). DAPI (1:1000; Invitrogen) was applied for 30 minutes following secondary antibody incubation. Images were acquired on either a fluorescent (Leica DM5500) or confocal (Zeiss) microscope. Morphometric analysis was performed by immunostaining every 10th section throughout each embryo (N=3 or 4 for each genotype). For quantification of individual hormone-expressing cells, cell number was assessed versus total pancreas as defined by amylase area, and calculated using ImagePro software.
Results
Arx expression is detected in Nkx2.2null ghrelin cells
Despite the dramatic reduction of the glucagon-expressing alpha cell population in Nkx2.2null pancreata, expression of the alpha cell factor, Arx, was not reduced (Chao et al., 2007) (Figure 1A). To identify which cell populations expressed Arx in the Nkx2.2 mutant pancreas, Arx expression was examined by immunostaining in wildtype and Nkx2.2null pancreas. In addition to the few remaining alpha cells that were positive for Arx (Figure 1B, D; arrows), Arx expression in the Nkx2.2null pancreas was found in the ghrelin-expressing population (Figure 1C,E; arrows). Since the loss of Nkx2.2 leads to an increase in the number of Arx+ ghrelin cells, we hypothesized that Nkx2.2 and Arx genetically interact to control early cell specification events in the pancreas. Consistent with this idea, we determined that Nkx2.2 and Arx were co-expressed in the early E10.5 developing pancreatic domain in both the Nkx2.2 heterozygote (Nkx2.2LacZ/+) and Nkx2.2 mutant (Nkx2.2LacZ/LacZ) mice (Figure 1F-I; Arnes, Sussel, in preparation; see Materials and Methods). We determined that, while cells expressing either Arx or Nkx2.2 expressed glucagon in the early Pdx1+ pancreatic domain (Figure 1F-I, dotted boxes; Figure 1J-M), there was also a population of Arx+/Nkx2.2+ co-expressing cells that did not express hormone (Figure 1F-I, arrows).
Figure 1. Arx is co-expressed in glucagon+, ghrelin+ and Nkx2.2+ cells.

Quantitative realtime PCR demonstrates that glucagon expression is decreased and ghrelin expression is increased in the Nkx2.2null, whereas there is no change in the expression of the alpha cell factor, Arx (A; * p>0.05). Using immunofluorescence and confocal imaging, both glucagon+ and ghrelin+ cells express Arx (arrows), in the wildtype and Nkx2.2null pancreas at E13.5 (B-E; 48×). Using adjacent sections, Arx+/Nkx2.2(β-gal)+ coexpressing cells are observed in the E10.5 Pdx1+ pancreatic domain in both the Nkx2.2 heterozygote (Nkx2.2LacZ/+) and Nkx2.2 mutant (Nkx2.2LacZ/LacZ) (F-I, arrows; 40×). Dotted boxes encircle Arx+ or Nkx2.2 (β-gal)+ cells that are glucagon+. Direct costaining of Arx or Nkx2.2 with glucagon+ alpha cells at E10.5 (J-M; arrows denote the remaining glucagon+ cells in the null; 40×). In B-I, top and right rectangular panels represent a Z projection of 10 stack pictures at the level of intersection of the red/green crosshairs. In C and E, asterisks mark blood cell or apoptotic cell autofluorescence. DAPI marks all nuclei.
To further investigate which cell populations normally produced Arx during early pancreas development, Arx expression was examined in conjunction with islet progenitor markers or islet hormones. As expected, Arx expression was detected in glucagon+ cells from E9.5 through E15.5 (Figure 2A-H, dotted boxes; Figure 1B,D; (Collombat et al., 2003); Supplemental Figure 1). However, a small subset of Pdx1+ progenitor cells in the pancreatic bud, which did not express glucagon, also expressed Arx as early as E9.5; the presence these Arx+/Pdx1+ cells persisted into the secondary transition at E15.5 (Figure 2A-H, arrows and inset panels). A small population of Neurog3+ cells also co-expressed Arx at the beginning of the secondary transition, E12.5 (Figure 2I), and these cells were located in or adjacent to the epithelial cords, which were defined by the expression of the transcription factor Sox9 (Figure 2J). Given the established role for Arx in regulating the formation of the glucagon+/ghrelin+ cell population (Heller et al., 2005), we examined wildtype E15.5 pancreas for glucagon, ghrelin and Arx and confirmed the expression of Arx in glucagon+/ghrelin+ cells (Figure 2K-N, arrows). Interestingly, similar to what was observed in the Nkx2.2null pancreas, we also detected Arx expression in ghrelin-expressing single hormone+ cells (Figure 2K-N; dotted boxes identify a ghrelin+/Arx+ cell), as well as in a population of cells that did not express hormone (Figure 2K-N).
Figure 2. Arx expression during pancreas development.
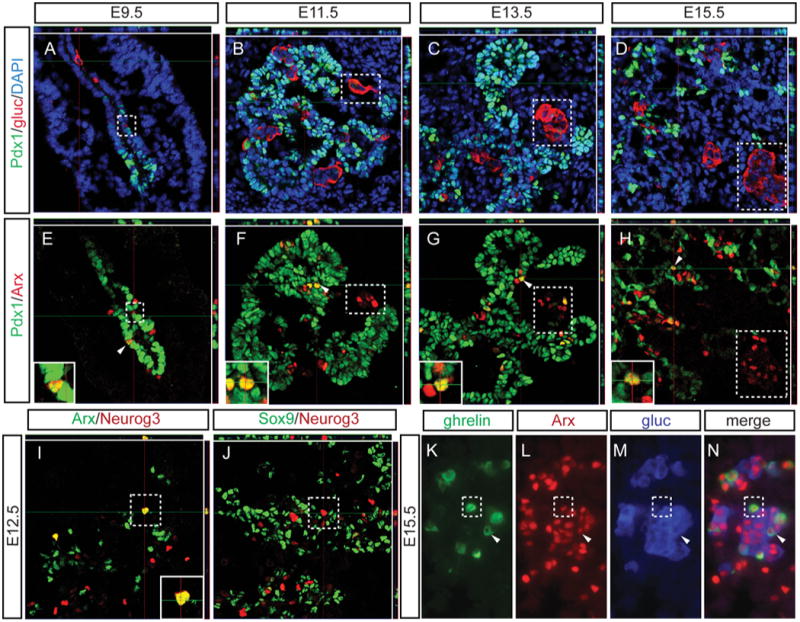
Sections are stained for Pdx1, to identify the pancreatic area, and glucagon to identify alpha cells (A-D; 40× confocal). Adjacent sections are stained for Pdx1 and Arx, and demonstrate Arx+ cells that are glucagon+ (dotted boxes) as well as Pdx1+/Arx+ coexpressing cells that do not express glucagon (arrows); Arx expression is detected in Pdx1+ progenitor cells from the beginning of pancreas development at E9.5 through the secondary transition at E15.5 (E-H, 40× confocal; insets 60×, confocal). Neurog3+ cells co-express Arx at the beginning of the secondary transition (E12.5), and these cells are located in the epithelial cords, defined by the expression of Sox9 (I-J; 40× confocal; inset 60×). In wildtype E15.5 tissue, Arx is expressed in ghrelin+ (dotted box) or glucagons+ single hormone+ cells, glucagon+/ghrelin+ coexpressing cells (arrow), and cells that do not express either hormone (K-M; 40×). In A-J, top and right rectangular panels represent a Z projection of 10 stack pictures at the level of intersection of the red/green crosshairs. DAPI marks all nuclei.
Arx is required for ghrelin expression but not ghrelin cell specification
Based on the observation that Arx was expressed in Nkx2.2-deficient and wildtype ghrelin single positive cells, and was also co-expressed with Nkx2.2 in the early pancreatic progenitors, Arx was removed in the Pdx1+ pancreatic progenitor cells in the Nkx2.2null background to examine the role of Arx in ghrelin cell specification. Using immunofluorescence, the number of ghrelin cells was quantified and compared between the single mutant mice, Nkx2.2null and Arxflox/flox(or Y);Pdx1-cre (from hereon referred to as ArxΔpanc), the compound double mutant, Nkx2.2-/-;Arxflox/flox(or Y);Pdx1-cre (Nkx2.2null;ArxΔpanc), and control littermates. The Nkx2.2null and Nkx2.2null;ArxΔpanc mice displayed an increase in the number of ghrelin-expressing cells compared with wildtype, and there was no significant difference in ghrelin cell number between the Nkx2.2null and Nkx2.2null;ArxΔpanc (Figure 3A; Supplemental Table 1). Using realtime PCR, we confirmed that ghrelin expression was unchanged in the P0 pancreas of the ArxΔpanc, and the Nkx2.2null displayed the expected increase in ghrelin compared with wildtype (Figure 3B); however, surprisingly, a reduced level of ghrelin expression was detected in the Nkx2.2null;ArxΔpanc when compared with the Nkx2.2null (Figure 3B). The observed reduction of ghrelin expression in the Nkx2.2null;ArxΔpanc compared with the Nkx2.2null, without a corresponding reduction in ghrelin cell numbers was confirmed using immunofluorescence analysis, imaging all pancreas sections with identical exposure time. The difference in intensity of ghrelin expression in equivalent numbers of cells was quantified using ImagePro Plus software and determined to be greater in the Nkx2.2null (17637 arbitrary intensity units) compared with Nkx2.2null;ArxApanc (6703 arbitrary intensity units). Therefore as predicted, we did not detect a difference in ghrelin cell numbers between the Nkx2.2null and Nkx2.2null;ArxΔpanc (Figure 3C-F), but the expression of ghrelin per cell was greatly reduced in the Nkx2.2null;ArxΔpanc compared with the Nkx2.2null (Figure 3E-F, inset images). Taken together these results suggest that Arx is not necessary for ghrelin cell specification, but is essential in the regulation of ghrelin gene expression.
Figure 3. Expression of ghrelin in the Nkx2.2null;ArxΔpanc mutants.
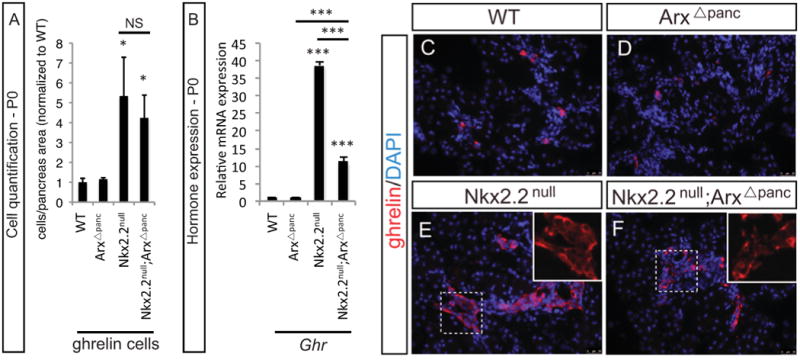
At P0, the number of ghrelin-expressing cells is unchanged between the wildtype (WT) and ArxΔpanc, whereas the number of cells is dramatically increased in the Nkx2.2null and Nkx2.2null;ArxΔpanc (A). Realtime PCR was used to quantitate ghrelin expression at P0. Expression levels match that of cell numbers except for the Nkx2.2null;ArxΔpanc, where ghrelin expression was significantly decreased compared with the Nkx2.2null (B). Representative images display ghrelin cells in the pancreas at P0. When imaged with identical exposure time, the intensity of ghrelin expression is decreased in the Nkx2.2null;ArxΔpanc compared with the Nkx2.2null (C-F; 20×). Each inset image (E-F, 40×; DAPI has been removed) contains 23 ghrelin+ cells; and the intensity of ghrelin expression in these cells was quantified using ImagePro Plus software and determined to be greater in the Nkx2.2null (17637 arbitrary intensity units) compared with Nkx2.2null;ArxΔpanc (6703 arbitrary intensity units). DAPI marks all nuclei.
Nkx2.2 and Arx genetically interact to affect multiple endocrine lineages
Arx and Nkx2.2 are each known to affect the development of several endocrine lineages, therefore we investigated whether these two factors also function together in the development or specification of the other islet endocrine populations. The expected phenotype for glucagon-expressing alpha cells and insulin-expressing beta cells was recapitulated in the ArxΔpanc and Nkx2.2null mutants at P0 (Supplemental Figure 2). In the Nkx2.2null;ArxAΔpanc, the simultaneous deletion of Arx did not affect the absence of beta cells or loss of expression of the insulin genes (Ins1, Ins2), resulting from the loss of Nkx2.2 (Figure 4 A-D; Supplemental Figure 2). Moreover, the transcription factors Neurod1, Pdx1 and Nkx6.1 were unaffected by the deletion of Arx alone, but displayed a decrease in expression when Nkx2.2 was deleted either alone or in combination with Arx in the Nkx2.2null;ArxΔpanc (Supplemental Figure 3). The expected loss of alpha cells in both the ArxΔpanc and Nkx2.2null was also observed in the Nkx2.2null;ArxΔpanc (Figure 4E-H); however, when both Arx and Nkx2.2 were deleted from the pancreas, glucagon (Gcg) expression showed an additive decrease. The augmented decrease was also observed at E15.5, but not earlier in development at E12.5 (Supplemental Figure), suggesting that Nkx2.2 and Arx function together during the secondary transition to regulate the major wave of alpha cell development and differentiation. The alpha cell factor Pou3f4 (Brn4) was also decreased in all mutants, which mimicked the loss of glucagon expression and the observed reduction of alpha cells (Supplemental Figure 3).
Figure 4. Pancreatic phenotype in the Nkx2.2null;ArxΔpanc mutants at P0.
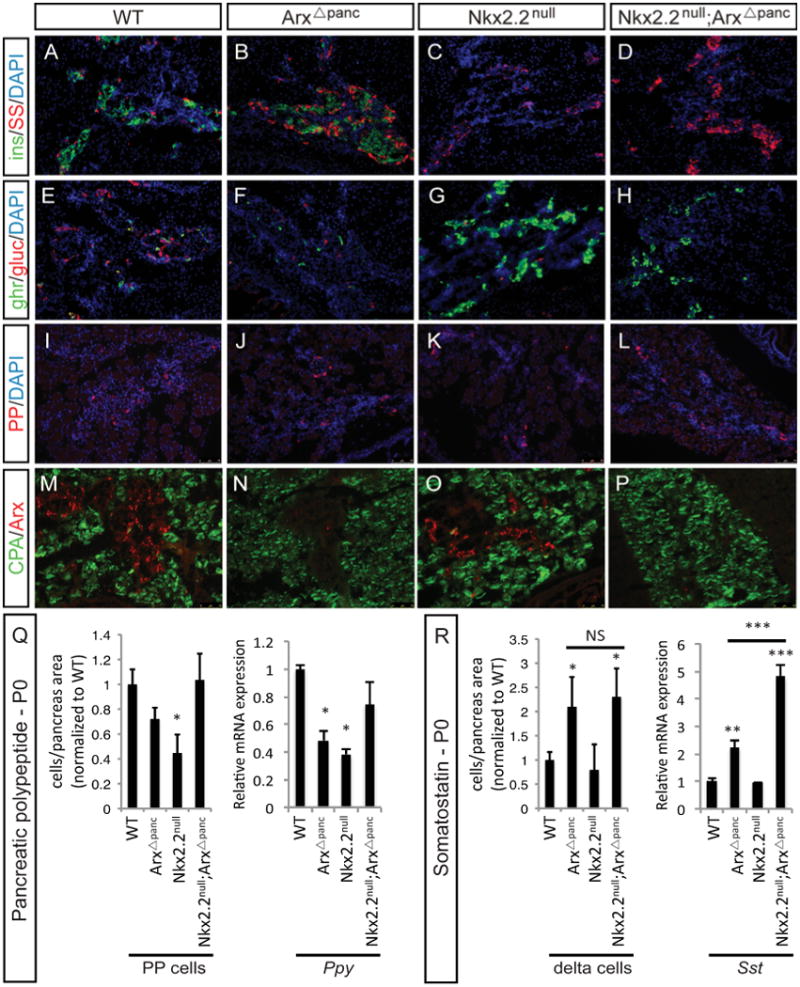
Insulin-expressing beta cells are absent in the Nkx2.2null and Nkx2.2null;ArxΔpanc compared to wildtype (WT) and ArxΔpanc (A-D). Somatostatin-expressing delta cells are increased in both the ArxΔpanc and Nkx2.2null;ArxΔpanc, but unchanged in the Nkx2.2null (A-D). Ghrelin-expressing cells are increased in both the Nkx2.2null and Nkx2.2null;ArxΔpanc compared to wildtype and ArxΔpanc (E-H). Glucagon-expressing alpha cells are reduced in both single mutants and the Nkx2.2null;ArxΔpanc compared to wildtype (E-H). Pancreatic polypeptide-expressing PP cells are reduced in the Nkx2.2null and Nkx2.2null;ArxΔpanc(I-L). Arx expression was used to confirm deletion in the ArxΔpanc single mutant and the Nkx2.2null;ArxΔpanc compound mutant, compared with the wildtype and Nkx2.2null, which maintain Arx expression; carboxypeptidase-A (CPA) marks the exocrine tissue (M-P). Quantitation of cell populations reveals that PP cells are reduced in the Nkx2.2null, but this reduction is restored to wildtype levels in the Nkx2.2null;ArxΔpanc pancreas (Q). Using realtime PCR, the expression of Ppy is reduced in both the ArxΔpanc and Nkx2.2null, and Ppy expression is restored in the Nkx2.2null;ArxΔpanc (Q). Somatostatin cells are increased in the ArxΔpanc, and this increase is maintained in the Nkx2.2null;ArxΔpanc pancreas (R). Expression of Sst is increased in the ArxΔpanc, and Sst expression in the Nkx2.2null;ArxΔpanc is further significantly increased compared with the ArxΔpanc (R). Cell numbers quantified relative to total pancreas area and displayed normalized to wildtype. * p>0.05; ** p>0.01; *** p>0.001. DAPI marks all nuclei. All images are 20×.
Immunostaining analyses also showed that while PP cell numbers were reduced in the Nkx2.2null pancreas, the PP cell population was restored to wildtype levels in the Nkx2.2null;ArxΔpanc (Figure 4I-L, Q; Supplemental Table 1). Pancreatic polypeptide (Ppy) expression reflected the change in cell number, with the restoration of Ppy hormone expression to wildtype levels in the Nkx2.2null;ArxΔpanc (Figure 4Q). These findings revealed an unexpected combined role for Nkx2.2 and Arx in the development of PP cells, and further suggests that these two lineage specification factors are likely to genetically interact to regulate the PP cell population.
The somatostatin-expressing delta cell population was previously shown to be increased in the Arx null mice (Collombat et al., 2005; Collombat et al., 2003), but was unaffected by the loss of Nkx2.2 (Sussel et al., 1998). Consistent with these findings, the number of delta cells was increased in both the ArxΔpanc and Nkx2.2null;ArxΔpanc, but there was no difference in total delta cell number between these two mutants (Figure 4A-D, R; Supplemental Table 1). Interestingly, realtime PCR analysis demonstrated not only an increase in somatostatin (Sst) expression in the ArxΔpanc but also revealed a further increase in Sst in the Nkx2.2null ArxΔpanc at P0 that was significantly higher than in the ArxΔpanc (Figure 4R). These data confirm that Arx, but not Nkx2.2, is necessary for delta cell development; however, the additional deletion of Nkx2.2 in an Arx-deficient pancreas further alters somatostatin transcript expression.
Ghrelin+/Somatostatin+ co-expressing cells are identified in the Nkx2.2null;ArxΔpanc
The unexpected finding in the Nkx2.2null;ArxΔpanc that ghrelin was downregulated by the Arx deletion and somatostatin was upregulated by the Nkx2.2 deletion (Figure 3 and 4), prompted us to further explore the potential interaction of Nkx2.2 and Arx in regulating the expression of these two hormones. Analysis of ghrelin expression at earlier embryonic stages demonstrated that the reduction in ghrelin expression in the Nkx2.2null;ArxΔpanc compared with Nkx2.2null was observed as early as E12.5 and was consistent through gestation (Figure 5A). Interestingly, we also detected a precocious and significant upregulation of somatostatin expression in both the ArxΔpanc and Nkx2.2null;ArxΔpanc at E12.5; this increase in somatostatin expression was increased to significantly higher levels in the Nkx2.2null;ArxΔpanc at E15.5 compared with the ArxΔpanc (Figure 5B). Using immunofluorescence, we stained tissue at E12.5 for ghrelin and somatostatin and discovered an unexpected population of cells in the Nkx2.2null;ArxΔpanc that co-express ghrelin and somatostatin (Figure 5C-F); these co-expressing cells were also present at P0 (Figure 5G-J). While a small number of ghrelin+/somatostatin+ co-expressing cells were detected in the ArxΔpanc at each time point (Figure 5D,H), no such cells were found in the wildtype or Nkx2.2null pancreata (Figure 5C,E,G,I) indicating that the deletion of both Arx and Nkx2.2 results in a significant increase of these bihormonal cells.
Figure 5. Ghrelin+/somatostatin+ co-expressing cells are present in the Nkx2.2;Arx compound mutant.
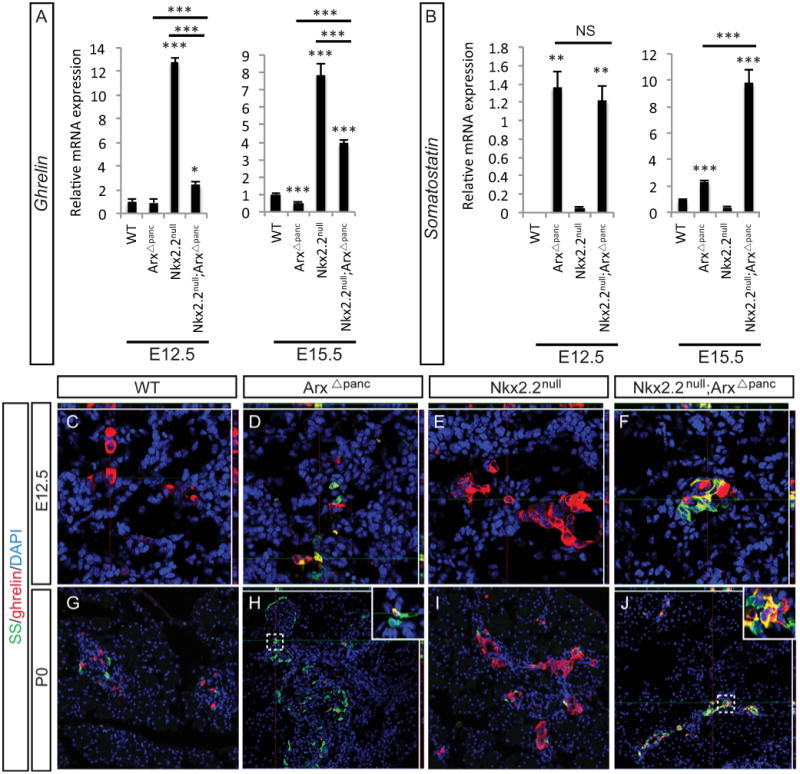
At E12.5 and E15.5 ghrelin expression is reduced in the Nkx2.2null;ArxΔpanc compared to the Nkx2.2null (A). Somatostatin expression is increased in both the ArxΔpanc and Nkx2.2null;ArxΔpanc at E12.5 and E15.5; the increase at E15.5 in the Nkx2.2null;ArxΔpanc is also further significantly upregulated compared with the ArxΔpanc (B). Using immunofluorescence and confocal microscopy, E12.5 (C-F; 48× confocal) and P0 (G-J, 20×; insets 48× confocal) pancreas sections were examined for ghrelin- and somatostatin-expressing cells. At both ages, precocious somatostatin cells are present in the ArxΔpanc (D, H) and increased ghrelin+ cells are present in the Nkx2.2null (E, I). Whereas ghrelin+/somatostatin+ cells are rare or absent in the ArxΔpanc (H inset) or Nkx2.2null, this population of co-expressing cells in greatly increased in the Nkx2.2null;ArxΔpanc (F, J, inset). In C-F, H and J, top and right rectangular panels represent a Z projection of 10 stack pictures at the level of intersection of the red/green crosshairs. DAPI marks all nuclei. * p>0.05; ** p>0.01; *** p>0.001
Endocrine-specific deletion of Arx in the Nkx2.2null background phenocopies the Nkx2.2null;ArxΔpanc
We demonstrated that Arx was expressed in the Pdx1+ pancreatic progenitors and in the Neurog3+ endocrine progenitors (Figure 2). To determine whether Arx function was required early within the pancreas progenitor population to influence the competency of the endocrine progenitor population or whether it is specifically required in the Neurog3+ endocrine progenitor population, we generated an endocrine-specific (Neurog3-cre) deletion of Arx in the Nkx2.2 mutant background. Hormone expression was assessed by immunostaining and realtime PCR at P0 in Nkx2.Z-/-;Arxflox/flox(or Y);Neurog3-cre (Nkx2.2null;ArxΔendo) and associated wildtype, Arxflox/flox(or Y);Neurog3-cre (ArxΔendo), and Nkx2.2null littermates. Specifically, glucagon (Gcg) expression was decreased in all mutants, with an additive decrease in expression in the Nkx2.2nuII;ArxΔendo compared with the single mutants (Figure 6A-D,Q). Interestingly, a significant decrease in glucagon expression was also observed between Nkx2.2null and ArxΔendo; this change was not observed between the Nkx2.2null and ArxΔpanc. The increase in ghrelin (Ghr) expression in the Nkx2.2null was also reduced when Arx was also deleted in the Neurog3+ cells (Figure 6E-H, Q). The expression changes of both somatostatin (Sst) (Figure 6I-L, Q) and pancreatic polypeptide (Ppy) (Figure 6M-P,Q) were identical to the ArxΔpanc and Nkx2.2null;ArxΔpanc mutants. Both insulin1 (Ins1) and insulin2 (Ins2) were absent in the Nkx2.2null and Nkx2.2null;ArxΔendo; the ArxΔendo displayed a trend toward increased insulin1 and unaltered insulin2 expression (Supplemental Figure 4). Altogether, the changes in gene expression were consistent with the Nkx2.2null;ArxΔpanc mutant, suggesting that Arx functions within the Neurog3+ endocrine progenitor cells to affect the development and differentiation of hormone-expressing cells in the embryonic pancreas.
Figure 6. Phenotype of endocrine-specific Nkx2.2;Arx compound and single mutants at P0.
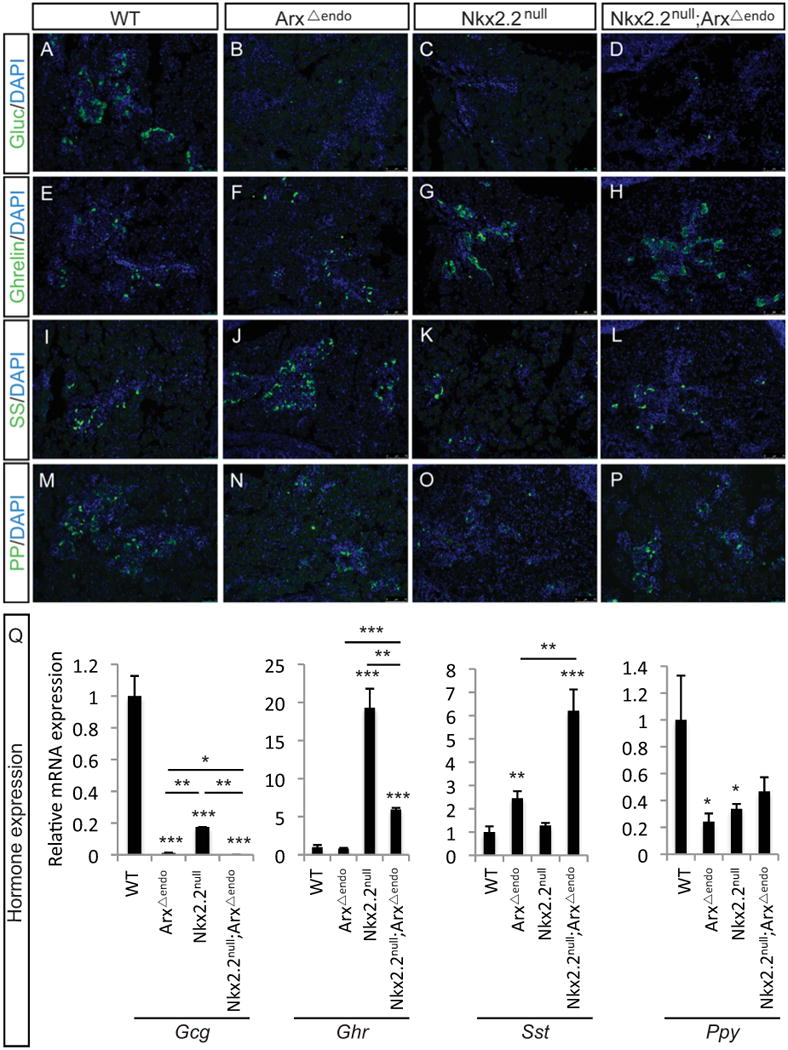
The pancreatic phenotype at P0 for the Nkx2.2null;Arxflox/flox(or Y);Neurog3-cre (Nkx2.2null;ArxΔendo) and associated wildtype (WT), Arxflox/flox(or Y);Neurog3-cre (ArxΔendo), and Nkx2.2null littermates. Using immunofluorescence analysis, glucagon cells are reduced in the single mutants and DKO (A-D). Ghrelin cells are unchanged in the ArxΔendo compared to wildtype, whereas ghrelin cells are increased in both the Nkx2.2null and Nkx2.2null;ArxΔendo (E-H). Somatostatin cells are increased in the ArxΔendo and Nkx2.2null;ArxΔendo (M-P). PP cells are reduced in the Nkx2.2null, but this reduction appears rescued in the Nkx2.2null;ArxΔendo (M-P). Using realtime PCR the hormone expression profile was determined (Q). Glucagon (Gcg) expression is reduced in all mutants, with an additive reduction in the Nkx2.2null;ArxΔendo. Somatostatin (Sst) is increased in the ArxΔendo and further increased in the Nkx2.2null;ArxΔendo. Ghrelin (Ghr) expression is increased in the Nkx2.2null and Nkx2.2null;ArxΔendo compared to wildtype, but the expression in the Nkx2.2null;ArxΔendo is reduced compared with the Nkx2.2null. Pancreatic polypeptide (Ppy) is reduced in both single mutants, and this expression is rescued in the Nkx2.2null;ArxΔendo. Relative mRNA expression was normalized to the housekeeping gene, cyclophilinB. * p>0.05; ** p>0.01; *** p>0.001. DAPI marks all nuclei. All images are 20×.
Discussion
The specification and differentiation of all hormone-expressing endocrine lineages in the pancreas relies on the temporal and spatial activation of a network of transcription factors. Using loss-of-function and gain-of-function mouse models, the contribution of individual transcription factors to these lineage decisions has been well described (Pan and Wright, 2011); however, the relationships among these transcription factors within each endocrine cell type have yet to be identified. In this study, using compound mutants of Nkx2.2 and Arx, we uncover a previously unappreciated genetic interaction between these two transcription factors within the endocrine progenitors, which regulates endocrine cell differentiation and hormone gene expression.
The hallmark of the pancreatic phenotype in the Nkx2.2 mutant mouse is the aberrant increase in ghrelin-expressing cells in place of the beta, alpha and (a subset of) PP cells (Prado et al., 2004). Here, we have determined that Arx is not required for ghrelin cell specification, but functions in the Neurog3+ endocrine progenitor cells to regulate ghrelin gene expression in the absence of Nkx2.2. Interestingly, we performed in silico analysis examining the ghrelin promoter for Arx consensus binding motifs (TAATTA; (Fulp et al., 2008)) and identified two putative Arx binding sites upstream of the transcriptional start site that are conserved between mouse and rat (Supplemental Figure 5), which suggests that Arx may directly regulate ghrelin; however, the ability of Arx to activate ghrelin expression appears to depend on the absence of Nkx2.2.
Previous studies have identified a role for Arx in delta cell specification (Collombat et al., 2005; Collombat et al., 2003), and as expected we demonstrated that delta cells and somatostatin expression were increased when Arx was deleted in the Pdx1+ cells. We observed a similar outcome when Arx was removed in the Neurog3+ cells; these data indicate that Arx is functioning in the endocrine progenitors to regulate the delta cell fate. Interestingly, delta cell numbers do not change between the ArxΔpanc and Nkx2.2null;ArxΔpanc but somatostatin expression is significantly different. Moreover, ghrelin cell numbers do not change between the Nkx2.2null and Nkx2.2null;ArxΔpanc but ghrelin expression is significantly altered. These data suggest that the population of ghrelin+/somatostatin+ co-expressing cells observed in the Nkx2.2null;ArxΔpanc are mutant somatostatin+ cells that misexpress ghrelin and mutant ghrelin+ cells that misexpress somatostatin. Previous reports have identified that deletion of Nkx2.2 directly affects the expression of Neurod1 (Anderson et al., 2009a), which is expressed in all endocrine lineages except delta cells (Anderson et al., 2009b) and is a known repressor of the somatostatin gene (Itkin-Ansari et al., 2005). Therefore we hypothesize that the further increase in somatostatin expression observed in the Nkx2.2null;ArxΔpanc compared with the ArxΔpanc single mutant could be indirectly caused by the reduction of Neurod1, which is associated with the loss of Nkx2.2. Interestingly, a triple deletion of Nkx2.2, Neurod1 (Neurod1tm1Jle; (Miyata et al., 1999)) and Arx in the Pdx1+ cells (Nkx2.2null;Neurod1null;Arxflox/flox;Pdx1-cre) shows a very dramatic increase in somatostatin expression (n=2; data not shown), supporting the idea that an additive increase in somatostatin expression could be caused by the combined deletion of Arx, Nkx2.2 and complete loss of Neurod1.
Our findings demonstrate that Nkx2.2 and Arx are required for ghrelin cell specification and ghrelin gene regulation, respectively. Previous studies have also indicated that Nkx2.2 is necessary for a subset of the PP cell lineage (Chao et al., 2007; Sussel et al., 1998) and spurious expression of Arx induces the PP cell fate; however, no specific alteration in the PP cell population was reported in the Arx null mouse (Collombat et al., 2005; Collombat et al., 2003). Therefore, it was quite unexpected that deletion of Arx in an Nkx2.2null background restored the PP cell population reduced in the Nkx2.2null. While this suggests that Nkx2.2 and Arx genetically interact to regulate the PP cell lineage, it also indicates that additional transcription factors, possibly regulated by Arx, are involved in the PP lineage decision. Interestingly, the PP cell lineage is affected by the deletion of Notch in Pdx1-expressing cells (Apelqvist et al., 1999). The Arx single mutant displays a significant increase in the Notch effector Hes1 and the compound mutant shows a trend toward increased Hes1 expression (data not shown); therefore, it is possible that aberrant notch function as a result of Arx deletion contributes to the restoration of the PP cell lineage in the Nkx2.2;Arx compound mutant.
Similar to studies conducted in transcription factor compound mutants, including Pax4;Pax6 (St-Onge et al., 1997), Nkx6.1;Nkx6.2 (Henseleit et al., 2005), Nkx2.2;Neurod1 (Chao et al., 2007), this study provides further evidence that a specific “transcription factor code” is required to fine tune both cell identify, and appropriate timing and location of gene products in pancreatic endocrine cells. In addition, the genetic interaction we uncovered between Nkx2.2 and Arx may also be necessary for cell fate decisions during the development of other organs, given that both transcription factors are expressed in the brain and central nervous system (Briscoe et al., 1999; Fulp et al., 2008; Poirier et al., 2004), and intestine ((Desai et al., 2008); May CL, unpublished observation). In particular, Nkx2.2 and Arx may cooperate to regulate the ghrelin cell lineage in the stomach, given that many ghrelin+ cells express Arx and Nkx2.2 in the adult stomach (Du and May, Arnes and Sussel, unpublished observation). Ultimately distilling the interactions of transcription factors within specific settings will provide great insight into how these factors function in the larger context of organ development.
Conclusion
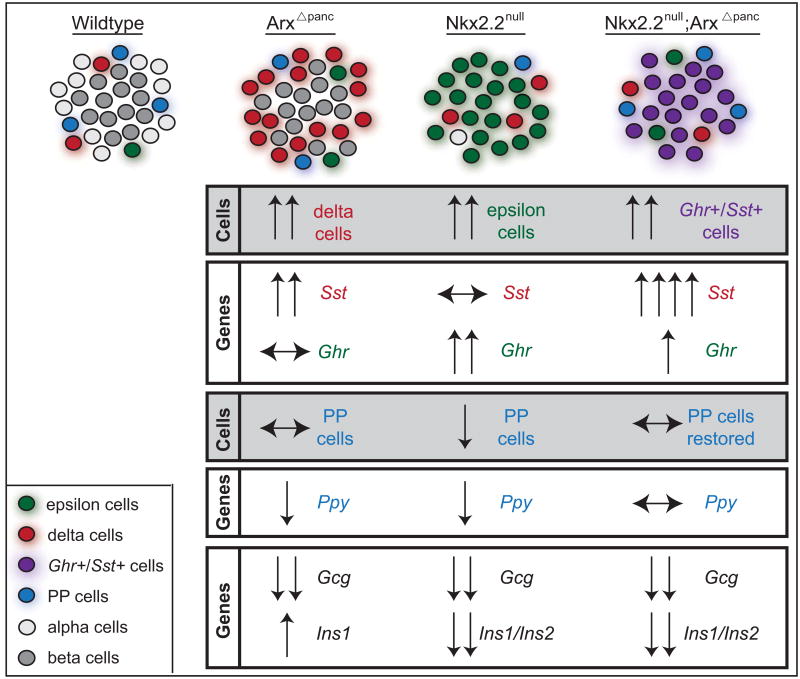
Our study clearly demonstrates that Nkx2.2 and Arx genetically interact in the endocrine progenitor cells to regulate both pancreatic endocrine cell development and hormone gene expression. A proposed model illustrating how these two key transcription factors function in regulating endocrine lineages is summarized in Figure 7. Specifically, increased numbers of delta and ghrelin cells are detected in Arx and Nkx2.2 single mutants, respectively, and the combined deletion of these two transcription factors leads to an increase in mutant ghrelin+/somatostatin+ co-expressing cells in the compound mutant and restoration of the PP cell lineage.
Figure 7. Proposed model for Nkx2.2 and Arx in specifying endocrine cell lineages.
A summary of the novel changes to normal endocrine development when both Nkx2.2 and Arx are deleted in the embryonic pancreas. As expected, somatostatin (Sst) expression and delta cells are increased in the ArxΔpanc. Similarly, in the Nkx2.2null ghrelin (Ghr) expression and ghrelin-expressing cells are increased. However, when Nkx2.2 and Arx are both absent in the pancreas, Sst expression is further increased and Ghr expression is reduced, but remains increased compared to wildtype. In addition, a mutant population of ghrelin+/somatostatin+ cells appears in the Nkx2.2null;ArxΔpanc. Most interestingly, PP cells and pancreatic polypeptide (Ppy) expression are reduced in the Nkx2.2null, and both are restored in the Nkx2.2null;ArxΔpanc. As expected, glucagon (Gcg) expression and alpha cells are reduced in both the ArxΔpanc and Nkx2.2null; the Nkx2.2null;ArxΔpanc compound mutant demonstrates the same phenotype. While insulin1 expression is increased in the ArxΔpanc, insulin1/insulin2 expression and beta cells are absent from both the Nkx2.2null; the Nkx2.2null;ArxΔpanc. The expression changes observed in the ArxΔendo and Nkx2.2null;ArxΔendo mutant mimics those of the ArxΔpanc and Nkx2.2null;ArxΔpanc mutants. Grey boxes represent data from morphometric analyses; white boxes summarize data from gene expression analyses.
Supplementary Material
Costaining of glucagon and Arx identifies Arx+/glucagon+ co-expressing cell populations and Arx+/glucagon- co-expressing cell populations exist normally throughout pancreas development, including E11.5 (C), E13.5 (D) and E15.5 (E) (40×).
At E12.5 the expression of glucagon (Gcg) is reduced in the pancreas of all mutants (A). At E15.5 and P0, glucagon expression is also decreased in the ArxΔpanc, Nkx2.2null and Nkx2.2null;ArxΔpanc mice compared to wildtype; however, there is an additive reduction of glucagon observed in the Nkx2.2null;ArxΔpanc mice (A). At P0, the expression of insulin1 (Ins1) and insulin2 (Ins2) are undetectable in the Nkx2.2null and Nkx2.2null;ArxΔpanc (B). In the ArxΔpanc, insulin1 is increased but insulin2 remains unchanged compared to wildtype (B). Relative mRNA expression was normalized to the housekeeping gene, cyclophilinB. * p>0.05; ** p>0.01; *** p>0.001
Using realtime PCR, transcription factor expression was assessed in P0 pancreata. The alpha cell marker, Brn4 (Pou3f4), is decreased in all mutants. The transcription factors Neurod1, Nkx6.1 and Pdx1 are all reduced in the Nkx2.2null and Nkx2.2null;ArxΔpanc, but unchanged in the ArxΔpanc. Relative mRNA expression was normalized to the housekeeping gene, cyclophilinB. * p>0.05; ** p>0.01; *** p>0.001
Immunofluorescence was used to confirm the presence of Arx expression in the wildtype (WT) and Nkx2.2null, and the deletion of Arx in the ArxΔendo and Nkx2.2null;ArxΔendo; carboxypeptidase-A (CPA) marks the exocrine tissue (A-D). Insulin cells are completely absent in the Nkx2.2null and Nkx2.2null;ArxΔendo (E-H). Realtime PCR was used to determine that the expression of insulin1 (Ins1) and insulin2 (Ins2) is undetectable in the Nkx2.2null and Nkx2.2null;ArxΔendo (I). In the ArxΔendo there is a trend for increased insulin1; insulin2 is not changed (I). Relative mRNA expression was normalized to the housekeeping gene, cyclophilinB. DAPI marks all nuclei. All images are 20×. * p>0.05; ** p>0.01; *** p>0.001
In silico analysis of sequence 20kb upstream of the ghrelin gene transcriptional start site was investigated for Arx binding motifs. Two motifs were identified that matched the published consensus sequence, TAATTA; these are located at -3303 bp and -5354 bp upstream of the transcriptional start site. Both motifs are conserved between mouse and rat. The published region within the ghrelin promoter that contains two Nkx2.2 consensus sequences (between -488 and -714) is also demarked.
Highlights.
Nkx2.2 and Arx cooperate to regulate pancreatic endocrine cell fates
Removal of Arx in the pancreas of Nkx2.2 mutants results in restoration of pancreatic polypeptide-expressing PP cells
Ghrelin+/somatostatin+ co-expressing cells are formed in the Nkx2.2/Arx compound mutant pancreas
Transcriptional regulation of ghrelin and somatostatin gene expression by Arx and Nkx2.2, respectively
Genetic interaction between Nkx2.2 and Arx occurs specifically in Neurog3+ endocrine progenitor cells
Acknowledgments
The authors would like to thank Dr. Aiping Du, Dr. Marcia Manterola and Ruth Singer for technical assistance, Dr. Kanako Miyabayashi for the kind gift of the Arx antibody, and Dr. Angela Christiano for use of the Zeiss confocal microscope. TLM is supported by a JDRF Postdoctoral Fellowship (3-2010-791). JAG is supported by NIH grant R01-NS46616 and NIH IDDRC grant P30-HD-26979. CLM is supported by NIH-DK078606 and JDRF2-2007-703. This study and LS were supported by NIH Beta Cell Biology Consortium (BCBC) grants U01-DK072504 and U01-DK089523, and R01-DK082590. Additional support provided by Columbia University DERC P30-DK63608 and University of Pennsylvania DERC P30-DK19525.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12:1763–8. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlgren U, Pfaff SL, Jessell TM, Edlund T, Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385:257–60. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- Anderson KR, Torres CA, Solomon K, Becker TC, Newgard CB, Wright CV, Hagman J, Sussel L. Nkx2.2 regulates NeuroD1 transcription through two distinct promoter elements. J Biol Chem. 2009a doi: 10.1074/jbc.M109.048694. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KR, White P, Kaestner KH, Sussel L. Nkx2.2 regulates endocrine and exocrine genes during early embryonic pancreas development. BMC Dev Biol. 2009b doi: 10.1186/1471-213X-9-65. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U, Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–81. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Bhushan A, Itoh N, Kato S, Thiery JP, Czernichow P, Bellusci S, Scharfmann R. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development. 2001;128:5109–17. doi: 10.1242/dev.128.24.5109. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Sussel L, Serup P, Hartigan-O'Connor D, Jessell TM, Rubenstein JL, Ericson J. Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature. 1999;398:622–7. doi: 10.1038/19315. [DOI] [PubMed] [Google Scholar]
- Burlison JS, Long Q, Fujitani Y, Wright CV, Magnuson MA. Pdx-1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells. Dev Biol. 2008;316:74–86. doi: 10.1016/j.ydbio.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao CS, Loomis ZL, Lee JE, Sussel L. Genetic identification of a novel NeuroD1 function in the early differentiation of islet alpha, PP and epsilon cells. Dev Biol. 2007;312:523–32. doi: 10.1016/j.ydbio.2007.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Hecksher-Sorensen J, Broccoli V, Krull J, Ponte I, Mundiger T, Smith J, Gruss P, Serup P, Mansouri A. The simultaneous loss of Arx and Pax4 genes promotes a somatostatin-producing cell fate specification at the expense of the alpha- and beta-cell lineages in the mouse endocrine pancreas. Development. 2005;132:2969–80. doi: 10.1242/dev.01870. [DOI] [PubMed] [Google Scholar]
- Collombat P, Hecksher-Sorensen J, Krull J, Berger J, Riedel D, Herrera PL, Serup P, Mansouri A. Embryonic endocrine pancreas and mature beta cells acquire alpha and PP cell phenotypes upon Arx misexpression. J Clin Invest. 2007;117:961–70. doi: 10.1172/JCI29115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Mansouri A, Hecksher-Sorensen J, Serup P, Krull J, Gradwohl G, Gruss P. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev. 2003;17:2591–603. doi: 10.1101/gad.269003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Krijger RR, Aanstoot HJ, Kranenburg G, Reinhard M, Visser WJ, Bruining GJ. The midgestational human fetal pancreas contains cells coexpressing islet hormones. Dev Biol. 1992;153:368–75. doi: 10.1016/0012-1606(92)90121-v. [DOI] [PubMed] [Google Scholar]
- Desai S, Loomis Z, Pugh-Bernard A, Schrunk J, Doyle MJ, Minic A, McCoy E, Sussel L. Nkx2.2 regulates cell fate choice in the enteroendocrine cell lineages of the intestine. Dev Biol. 2008;313:58–66. doi: 10.1016/j.ydbio.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulp CT, Cho G, Marsh ED, Nasrallah IM, Labosky PA, Golden JA. Identification of Arx transcriptional targets in the developing basal forebrain. Hum Mol Genet. 2008;17:3740–60. doi: 10.1093/hmg/ddn271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradwohl G. Development of the endocrine pancreas. Diabetes Metab. 2006;32:532–3. doi: 10.1016/s1262-3636(06)72807-5. [DOI] [PubMed] [Google Scholar]
- Gradwohl G, Dierich A, LeMeur M, Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 2000;97:1607–11. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebrok M, Kim SK, Melton DA. Notochord repression of endodermal Sonic hedgehog permits pancreas development. Genes Dev. 1998;12:1705–13. doi: 10.1101/gad.12.11.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RS, Jenny M, Collombat P, Mansouri A, Tomasetto C, Madsen OD, Mellitzer G, Gradwohl G, Serup P. Genetic determinants of pancreatic epsilon-cell development. Dev Biol. 2005;286:217–24. doi: 10.1016/j.ydbio.2005.06.041. [DOI] [PubMed] [Google Scholar]
- Henseleit KD, Nelson SB, Kuhlbrodt K, Hennings JC, Ericson J, Sander M. NKX6 transcription factor activity is required for alpha- and beta-cell development in the pancreas. Development. 2005;132:3139–49. doi: 10.1242/dev.01875. [DOI] [PubMed] [Google Scholar]
- Herrera PL. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127:2317–22. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–50. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- Itkin-Ansari P, Marcora E, Geron I, Tyrberg B, Demeterco C, Hao E, Padilla C, Ratineau C, Leiter A, Lee JE, Levine F. NeuroD1 in the endocrine pancreas: localization and dual function as an activator and repressor. Dev Dyn. 2005;233:946–53. doi: 10.1002/dvdy.20443. [DOI] [PubMed] [Google Scholar]
- Jorgensen MC, Ahnfelt-Ronne J, Hald J, Madsen OD, Serup P, Hecksher-Sorensen J. An illustrated review of early pancreas development in the mouse. Endocr Rev. 2007;28:685–705. doi: 10.1210/er.2007-0016. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32:128–34. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Kim SK, Hebrok M, Melton DA. Notochord to endoderm signaling is required for pancreas development. Development. 1997;124:4243–52. doi: 10.1242/dev.124.21.4243. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Yanazawa M, Sugiyama N, Miura H, Iizuka-Kogo A, Kusaka M, Omichi K, Suzuki R, Kato-Fukui Y, Kamiirisa K, Matsuo M, Kamijo S, Kasahara M, Yoshioka H, Ogata T, Fukuda T, Kondo I, Kato M, Dobyns WB, Yokoyama M, Morohashi K. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat Genet. 2002;32:359–69. doi: 10.1038/ng1009. [DOI] [PubMed] [Google Scholar]
- Larsson LI. Developmental biology of gastrin and somatostatin cells in the antropyloric mucosa of the stomach. Microsc Res Tech. 2000;48:272–81. doi: 10.1002/(SICI)1097-0029(20000301)48:5<272::AID-JEMT4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Larsson LI, Madsen OD, Serup P, Jonsson J, Edlund H. Pancreatic-duodenal homeobox 1 -role in gastric endocrine patterning. Mech Dev. 1996;60:175–84. doi: 10.1016/s0925-4773(96)00609-0. [DOI] [PubMed] [Google Scholar]
- Lausier J, Diaz WC, Roskens V, LaRock K, Herzer K, Fong CG, Latour MG, Peshavaria M, Jetton TL. Vagal control of pancreatic ss-cell proliferation. Am J Physiol Endocrinol Metab. 2010;299:E786–93. doi: 10.1152/ajpendo.00202.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinius A, Ericsson JL, Grimelius L, Korsgren O. Ultrastructural studies of the ontogeny of fetal human and porcine endocrine pancreas, with special reference to colocalization of the four major islet hormones. Dev Biol. 1992;153:376–85. doi: 10.1016/0012-1606(92)90122-w. [DOI] [PubMed] [Google Scholar]
- Macdonald RJ, Swift GH, Real FX. Transcriptional control of acinar development and homeostasis. Prog Mol Biol Transl Sci. 2010;97:1–40. doi: 10.1016/B978-0-12-385233-5.00001-5. [DOI] [PubMed] [Google Scholar]
- Martin M, Gallego-Llamas J, Ribes V, Kedinger M, Niederreither K, Chambon P, Dolle P, Gradwohl G. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Dev Biol. 2005;284:399–411. doi: 10.1016/j.ydbio.2005.05.035. [DOI] [PubMed] [Google Scholar]
- Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13:1647–52. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrep N, Wang J, Miyatsuka T, German MS. Signals from the neural crest regulate beta-cell mass in the pancreas. Development. 2008;135:2151–60. doi: 10.1242/dev.015859. [DOI] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–95. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Olerud J, Kanaykina N, Vasylovska S, King D, Sandberg M, Jansson L, Kozlova EN. Neural crest stem cells increase beta cell proliferation and improve islet function in co-transplanted murine pancreatic islets. Diabetologia. 2009;52:2594–601. doi: 10.1007/s00125-009-1544-z. [DOI] [PubMed] [Google Scholar]
- Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn. 2011;240:530–65. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- Pictet R, Rutter WJ. Development of the embryonic endocrine pancreas. Williams and Wilkins; Washington, DC: 1972. [Google Scholar]
- Plank JL, Mundell NA, Frist AY, LeGrone AW, Kim T, Musser MA, Walter TJ, Labosky PA. Influence and timing of arrival of murine neural crest on pancreatic beta cell development and maturation. Dev Biol. 2011;349:321–30. doi: 10.1016/j.ydbio.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier K, Van Esch H, Friocourt G, Saillour Y, Bahi N, Backer S, Souil E, Castelnau-Ptakhine L, Beldjord C, Francis F, Bienvenu T, Chelly J. Neuroanatomical distribution of ARX in brain and its localisation in GABAergic neurons. Brain Res Mol Brain Res. 2004;122:35–46. doi: 10.1016/j.molbrainres.2003.11.021. [DOI] [PubMed] [Google Scholar]
- Prado CL, Pugh-Bernard AE, Elghazi L, Sosa-Pineda B, Sussel L. Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc Natl Acad Sci U S A. 2004;101:2924–9. doi: 10.1073/pnas.0308604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonhoff SE, Giel-Moloney M, Leiter AB. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev Biol. 2004;270:443–54. doi: 10.1016/j.ydbio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature. 1997;387:406–9. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- Sussel L, Kalamaras J, Hartigan-O'Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, German MS. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development. 1998;125:2213–21. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- Villasenor A, Chong DC, Henkemeyer M, Cleaver O. Epithelial dynamics of pancreatic branching morphogenesis. Development. 2011;137:4295–305. doi: 10.1242/dev.052993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Elghazi L, Martin S, Martins I, Srinivasan RS, Geng X, Sleeman M, Collombat P, Houghton J, Sosa-Pineda B. Ghrelin is a novel target of Pax4 in endocrine progenitors of the pancreas and duodenum. Dev Dyn. 2008;237:51–61. doi: 10.1002/dvdy.21379. [DOI] [PubMed] [Google Scholar]
- Wells JM, Melton DA. Early mouse endoderm is patterned by soluble factors from adjacent germ layers. Development. 2000;127:1563–72. doi: 10.1242/dev.127.8.1563. [DOI] [PubMed] [Google Scholar]
- Wierup N, Yang S, McEvilly RJ, Mulder H, Sundler F. Ghrelin is expressed in a novel endocrine cell type in developing rat islets and inhibits insulin secretion from INS-1 (832/13) cells. J Histochem Cytochem. 2004;52:301–10. doi: 10.1177/002215540405200301. [DOI] [PubMed] [Google Scholar]
- Yoshitomi H, Zaret KS. Endothelial cell interactions initiate dorsal pancreas development by selectively inducing the transcription factor Ptf1a. Development. 2004;131:807–17. doi: 10.1242/dev.00960. [DOI] [PubMed] [Google Scholar]
- Zorn AM, Wells JM. Vertebrate endoderm development and organ formation. Annu Rev Cell Dev Biol. 2009;25:221–51. doi: 10.1146/annurev.cellbio.042308.113344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Costaining of glucagon and Arx identifies Arx+/glucagon+ co-expressing cell populations and Arx+/glucagon- co-expressing cell populations exist normally throughout pancreas development, including E11.5 (C), E13.5 (D) and E15.5 (E) (40×).
At E12.5 the expression of glucagon (Gcg) is reduced in the pancreas of all mutants (A). At E15.5 and P0, glucagon expression is also decreased in the ArxΔpanc, Nkx2.2null and Nkx2.2null;ArxΔpanc mice compared to wildtype; however, there is an additive reduction of glucagon observed in the Nkx2.2null;ArxΔpanc mice (A). At P0, the expression of insulin1 (Ins1) and insulin2 (Ins2) are undetectable in the Nkx2.2null and Nkx2.2null;ArxΔpanc (B). In the ArxΔpanc, insulin1 is increased but insulin2 remains unchanged compared to wildtype (B). Relative mRNA expression was normalized to the housekeeping gene, cyclophilinB. * p>0.05; ** p>0.01; *** p>0.001
Using realtime PCR, transcription factor expression was assessed in P0 pancreata. The alpha cell marker, Brn4 (Pou3f4), is decreased in all mutants. The transcription factors Neurod1, Nkx6.1 and Pdx1 are all reduced in the Nkx2.2null and Nkx2.2null;ArxΔpanc, but unchanged in the ArxΔpanc. Relative mRNA expression was normalized to the housekeeping gene, cyclophilinB. * p>0.05; ** p>0.01; *** p>0.001
Immunofluorescence was used to confirm the presence of Arx expression in the wildtype (WT) and Nkx2.2null, and the deletion of Arx in the ArxΔendo and Nkx2.2null;ArxΔendo; carboxypeptidase-A (CPA) marks the exocrine tissue (A-D). Insulin cells are completely absent in the Nkx2.2null and Nkx2.2null;ArxΔendo (E-H). Realtime PCR was used to determine that the expression of insulin1 (Ins1) and insulin2 (Ins2) is undetectable in the Nkx2.2null and Nkx2.2null;ArxΔendo (I). In the ArxΔendo there is a trend for increased insulin1; insulin2 is not changed (I). Relative mRNA expression was normalized to the housekeeping gene, cyclophilinB. DAPI marks all nuclei. All images are 20×. * p>0.05; ** p>0.01; *** p>0.001
In silico analysis of sequence 20kb upstream of the ghrelin gene transcriptional start site was investigated for Arx binding motifs. Two motifs were identified that matched the published consensus sequence, TAATTA; these are located at -3303 bp and -5354 bp upstream of the transcriptional start site. Both motifs are conserved between mouse and rat. The published region within the ghrelin promoter that contains two Nkx2.2 consensus sequences (between -488 and -714) is also demarked.