Abstract
Epstein-Barr virus (EBV), a human γ-herpesvirus, establishes lifelong infection by targeting the adaptive immune system of the host through memory B cells. While normally benign, EBV contributes to lymphoid malignancies and lymphoproliferative syndromes in immunocompromised individuals. The viral oncoprotein Latent Membrane Protein 1 (LMP-1) is essential for B lymphocyte immortalization by EBV. The constitutive signaling activity of LMP-1 is dependent on homo-oligomerization of its six-spanning hydrophobic transmembrane domain (TMD). However, the mechanism driving LMP-1 intermolecular interaction is poorly understood. Here we show that the fifth transmembrane helix (TM5) of LMP-1 strongly self-associates, forming a homotrimeric complex mediated by a polar residue embedded in the membrane, D150. Replacement of this aspartic acid residue with alanine disrupts TM5 self-association in detergent micelles and bacterial cell membranes. A full length LMP-1 variant harboring the D150A substitution is deficient in NFκB activation, supporting the key role of the fifth transmembrane helix in constitutive activation of signaling by this oncoprotein.
Keywords: Membrane proteins, NF-kappa B, Peptide chemical synthesis, Protein-protein interactions, CD40, Epstein-Barr virus
INTRODUCTION
The ubiquitous human herpesvirus Epstein-Barr virus (EBV) is commonly recognized as the causal agent of infectious mononucleosis.1,2 EBV is also a human tumor virus linked to a number of human lymphoid malignancies and lymphoproliferative disorders, including Burkitt’s lymphoma and Hodgkin’s disease.3,4 Immunocompromised individuals, such as transplant recipients and AIDS patients, are at high risk for development of EBV-dependent lymphoproliferative diseases and lymphoma.5 Infection of B lymphocytes, the primary target of EBV, triggers cell proliferation through mechanisms dependent upon the expression of a subset of viral gene products.6,7 Latent viral gene products, via molecular mimicry, activate antigen-activated signaling pathways with the goal of promoting viral access to B cell memory, the site of EBV persistence. Tumor formation can occur if virally infected cells escape immune surveillance.5
Latent Membrane Protein 1 (LMP-1) is a viral transforming protein essential for B cell immortalization by EBV.8 LMP-1 is expressed in many EBV-dependent tumors,5,9 playing a key role in EBV-dependent malignancies. LMP-1 constitutively mimics the activity of the Tumor Necrosis Factor Receptor (TNFR), CD40 (Figure 1).10 Both LMP-1 and activated CD40 bind to many of the same signaling proteins (e.g. TRAFs 1, 2, 3 and 5 and JAK3) via their cytoplasmic C-terminal signaling domains and activate the same downstream pathways, including the NFκB and JNK pathways.10,11 The activation of NFκB and JNK pathways triggers B cell activation/differentiation pathways that promote proliferation, survival and differentiation of B cells into memory cells.12–14
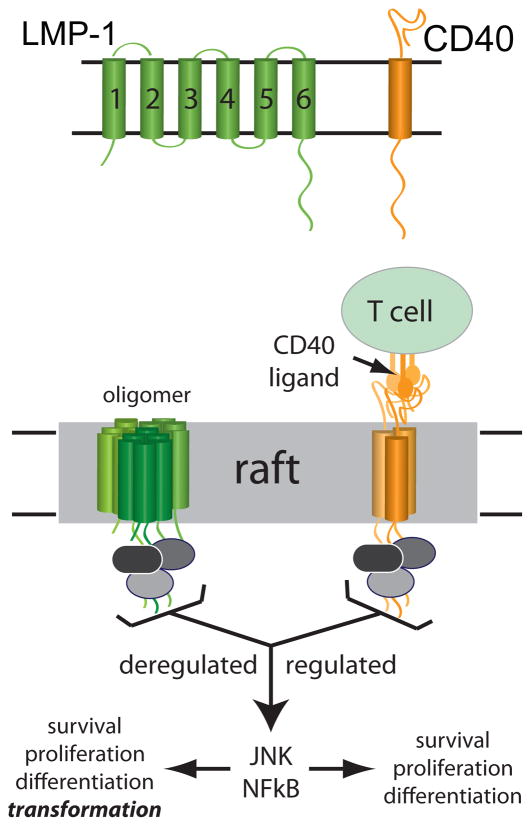
Figure 1.
LMP-1 and CD40 signaling. LMP-1 and CD40 are shown to illustrate their predicted topology in the membrane (TOP). LMP-1 is a six-spanning polytopic membrane protein with very little extracellular domain and a large cytoplasmic signaling domain, whereas CD40 is a single spanning membrane protein with an extra-cellular ligand binding domain and a cytoplasmic signaling domain. LMP-1 and CD40 share no homology in their transmembrane domains. LMP-1 and CD40 are shown hypothetically in oligomeric/raft associated (active) forms (BOTTOM). Membrane-associated CD40 ligand on T cells binds to CD40, CD40 moves to lipid rafts and interacts with intracellular signaling proteins. LMP-1 and activated CD40 interact with many of the same intracellular signaling proteins (TRAFs, for example, grey ovals) and activate the same downstream effectors (NFκB and JNK pathways to regulate B cell survival, proliferation and differentiation). The signal from LMP-1 is potent, deregulated (constitutive) and transforming, whereas the CD40 signal is less potent, regulated (ligand dependent) and non-transforming.
Importantly, while LMP-1 functions as a CD40 mimic, the two proteins share very little sequence homology other than a short consensus sequence for TRAF binding within their C-termini (PxQxT).10 The key role of LMP-1 in signaling activation as well as the lack of homology between LMP-1 and CD40 makes LMP-1 a strategic target for therapeutic intervention against EBV-dependent malignancies. The challenge in understanding and disrupting LMP-1 signaling activation resides in the fact that LMP-1 is a self-associating, multi-pass integral membrane protein. Interestingly, structure/function analyses of LMP-1/CD40 chimeras indicate that the LMP-1 TMD contributes to signaling beyond that of a simple membrane-anchoring domain (Figure 1).15 Without its intact TMD, LMP-1 cannot homo-oligomerize, localize to lipid rafts or activate NFκB.16–20 Thus, the TMD functions as the activation domain of LMP-1 by mediating oligomerization and raft localization.21–25 Nevertheless, the mechanism that drives TMD-dependent activities remains poorly understood, largely due to the lack of probes to study the highly hydrophobic transmembrane region of LMP-1.
In this work we seek to answer specific structural questions in an effort to better understand the LMP-1 mechanism of activation. We utilize a modular approach with synthetic peptides representing individual transmembrane helices to study protein-protein interactions of multi-spanning transmembrane proteins where conventional approaches prove untenable. Synthesis of a single hydrophobic transmembrane helix (26~30 residues) is readily achievable using modern microwave-assisted solid-phase peptide synthesis,26 facilitating the study of each individual pass of a transmembrane domain. While such a modular approach might not be feasible with water-soluble proteins, we have capitalized on the inherent stability of hydrophobic helices. Transmembrane helices have a high level of intrinsic stability due to the energetic requirement to sequester the polar backbone atoms from the hydrophobic environment of the membrane causing them to adopt native-like structures in membrane-mimetic environments.27 Protein-oligomerization hotspots identified through biophysical experiments of individual transmembrane helices were further studied with biological assays using full-length LMP-1.
RESULTS
The fifth transmembrane helix of LMP-1 self-associates
Multi-spanning, integral membrane proteins are difficult to study at the structural level due to their highly hydrophobic nature. LMP-1 is especially challenging in this regard because of the self-aggregating properties of its TMD. To circumvent this problem, we measured the propensity of individual TMs to self-associate. Each of the six LMP-1 TMs was synthesized by solid-state peptide synthesis for analysis using biochemical and biophysical approaches (Figure 2A).
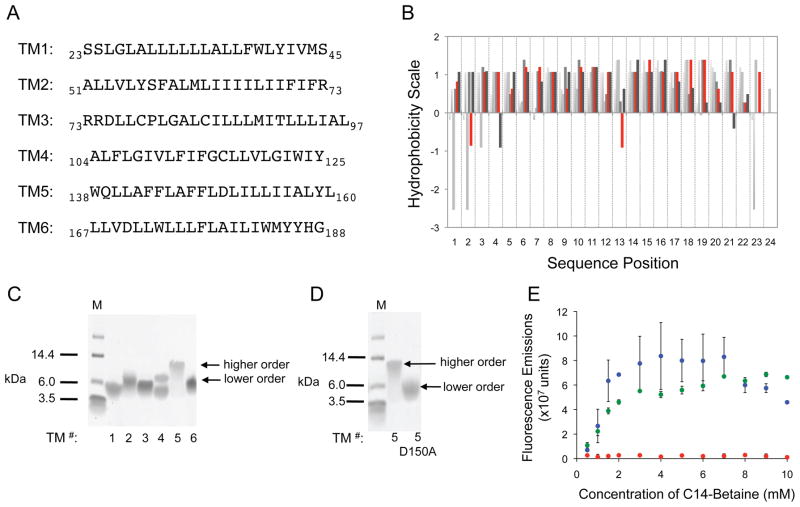
Figure 2.
Identification of D150, in the fifth transmembrane helix of LMP-1, as a key residue driving TM5 self-association. (A) The amino acid sequence for each of the six transmembrane helices of LMP-1. (B) Plot showing the hydrophobicity score for each sequence position in the six TMs of LMP-1. TM5 values shown in red. D150 stands out as it is the only centrally located polar residue, with no obvious hydrogen-bonding partner amongst the other TMs. (C) 12% Bis-Tris SDS-PAGE gel of each of the six transmembrane helices, labeled 1 through 6, of LMP-1. (D) 12% Bis-Tris SDS-PAGE gel comparing wild-type TM5 and TM5 D150A. (E) Fluorescence quenching assay of coumarin-labeled TM1 (blue), TM5 (red), and TM5 D150A (green).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) is commonly used to identify native interactions and tertiary/quaternary structure in membrane proteins because it provides a micellar environment that mimics phospholipid bilayers and does not disrupt non-covalent interactions between hydrophobic peptides.28 Monomeric and oligomeric peptides can, therefore, be distinguished by their rate of migration through SDS-PAGE. As an initial screen for the self-aggregating potential of TM peptides 1–6, 12% Bis-Tris gels were employed (Figure 2C). The migration of TM1 was consistent with its peptide molecular weight whereas the migration of TM5 was much slower than predicted. TMs 2, 3, 4 and 6 migrated more slowly than TM1, with TM4 migrating as a doublet. The unusually slow migration of TM5 was consistent with the formation of a higher-order species. This result suggests that TM5 has a high propensity to oligomerize in a detergent micellar environment. The migration patterns of the TM peptides in 12% Bis-Tris gels were confirmed using a 10% tricine gel (Supporting Information Figure 1A). TM4 migrated as a single band, TM3 migrated as a doublet, and other TM peptides showing similar migration patters to those seen in the 12% Bis-Tris gels. As observed in the 12% Bis-Tris gel system, TM5 migrated as a higher order oligomer. The consistent and unusual migration of TM5 in SDS-PAGE gels prompted further analysis of its oligomeric status.
Although SDS-PAGE analysis is a useful starting point in the determination of oligomerizing species, the assay suffers from a number of drawbacks and a more rigorous biophysical assay is required to confirm peptide self-interaction.29 A fluorescence quenching assay was used to investigate the self-association of the TM5 peptide in solution (Figure 2E). The peptides were labeled with the self-quenching fluorophore, 7-hydroxycoumarin, and fluorescence emissions were monitored to distinguish between monomeric and oligomeric peptide species. 7-Hydroxycoumarin, when compared to other self-quenching fluorophores, has been shown to cause the least amount of perturbation to leucine zipper peptide behavior, such as the stabilization of a dimeric complex or the induction of peptide aggregation.30 TM peptides can be driven to self-associate when dissolved in low detergent concentrations because the micellar peptide concentration is high (high peptide/micelle ratio). Non-specific peptide interactions are disrupted as detergent concentrations increase (low peptide/micelle ratio), while specific interactions persist. The TM1 peptide consistently migrated with the molecular weight of a monomeric species in SDS-PAGE and was used as a monomeric control for this assay. Consistent with SDS-PAGE analysis, TM5 displayed a markedly different dissociation pattern than TM1. The fluorescence emissions from coumarin-labeled TM5 remained low over the entire range of detergent concentrations tested suggesting TM5 self-associates, while coumarin-labeled TM1 rapidly dissociated as evidenced by the increase in fluorescence intensity as detergent concentration increased. Together, these results demonstrate that TM5 has the ability to homo-oligomerize.
Disruptive point mutation interferes with TM5 self-association
TM5 was examined to identify potential sequence motifs that might drive self-association. Self-assembly of transmembrane helices can be driven by small-large-large-small “GXXXG” motifs,31–33 the association of aromatic residues34,35 and knob-into-hole interactions, such as leucine zippers.36,37 Most of the LMP-1 TMs contain multiple aromatic amino acids. Leucine residues comprise nearly 38% of the LMP-1 TMD, yielding multiple potential heptad-repeat motifs that could induce helix associations. However, neither of these motifs is specific to TM5. Interestingly, TM5 contains an aspartic acid, D150, located centrally in the TM helix. Polar residues have been shown to drive self-association of rationally designed transmembrane peptides.38–40 In fact, hydrophilic amino acids are commonly found in the hydrophobic environment of integral membrane proteins.41 However they generally have additional hydrophilic amino acids with whom they can hydrogen bond in order to offset the energetic cost associated with the burial of a polar amino acid in a non-polar environment. But where might such a hydrogen-binding partner be located? Hydrophobicity analysis of the LMP-1 transmembrane helices reveals the unusually high hydrophobic nature of the LMP-1 TMD and the unique placement of the aspartic acid within TM5 (Figure 2B).42 The aspartic acid at position 150 has no apparent internal hydrogen bonding partners in any other LMP-1 TM. The conspicuous absence of other polar residues in the membrane-buried region of LMP-1 suggests that D150 may be involved in an intermolecular hydrogen bond network between TM5 helices on adjacent LMP-1 molecules. To test the impact of D150 on the self-association of TM5, we compared the behavior of TM5 to that of a TM5 peptide with an alanine in place of the aspartic acid (TM5 D150A). Migration of TM5 and TM5 D150A was assessed by SDS-PAGE (Figure 2D and Supporting Information Figure 1B). TM5 D150A migrated faster than wild type TM5, with an apparent molecular weight similar to that of the other LMP-1 TMs. Fluorescence quenching analysis confirmed the altered self-association of TM5 D150A; TM5 D150A rapidly dissociated upon dilution in detergent in a manner analogous to TM1 (Figure 2E). From these data we can conclude that D150 drives the self-association of TM5.
LMP-1 TM5 forms a homo-trimeric complex
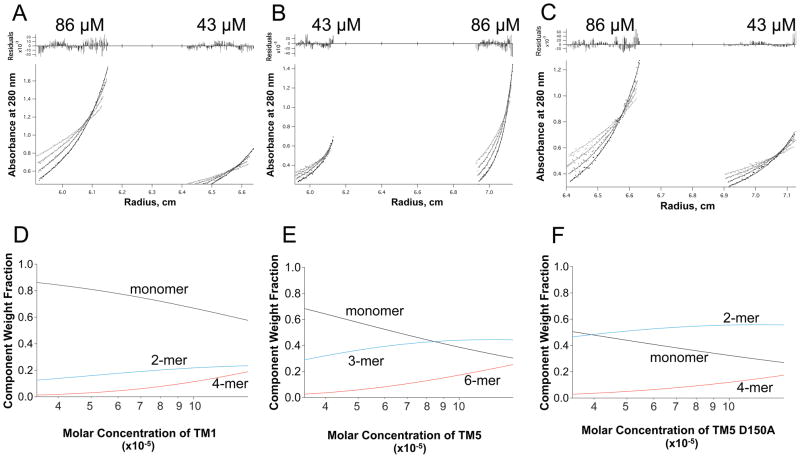
SDS-PAGE analysis (Figure 2C) revealed that TM5 formed an oligomeric species in detergent micelles. SDS-PAGE analysis is useful for evaluating native interactions of transmembrane peptides. Detergent binding to the peptide, however, can retard its progress through the gel, leading to an inaccurate estimation of the mass of the peptide.28 Sedimentation equilibrium analytical ultracentrifugation (SE AUC) is widely used to accurately determine the oligomeric state of macromolecules. The molecular weight and stoichiometry of a molecule in solution can be determined by monitoring the sedimentation profile of the sample in a centrifugal field. SE AUC was used to investigate the stoichiometry of TM1, TM5 and TM5 D150A peptides, solubilized in C14-betaine buffer with 21% deuterium (Figure 3). Results from SE AUC analysis of TM peptides were in agreement with the SDS-PAGE and fluorescence quenching studies described above. The sedimentation profile for TM1 fits a model in which TM1 is primarily monomeric until high local peptide concentrations are reached, after which dimeric and tetrameric species are observed (Figure 3D). The sedimentation profile of TM5 best fits a trimeric model, with some monomeric species observed at low concentrations (Figure 3E). By contrast, TM5 D150A is mostly dimeric with no detectable trimeric species (Figure 3F). Circular dichroism was used to confirm that the TM peptides maintain the native α-helical secondary structure in the C14-betaine solution used for AUC experiments (Supporting Information Figure 2). From these results we conclude that D150 drives TM5 trimerization and the substitution of an alanine for the aspartic acid at position 150 does not disrupt but alters the TM5 oligomeric complex.
Figure 3.
Sedimentation equilibrium analytical ultracentrifugation (SE AUC) results comparing TM1, TM5 and TM5 D150A. (A–C) Sedimentation equilibrium A280-radius profiles and residuals of TM1 in panel (A), TM5 in panel (B), and TM5 D150A in panel (C) at peptide concentrations of 43 μM and 86 μM, solubilized in C14-betaine micelles, spun at 30, 35, 40 and 45K rpm (data point, fit and residuals shown in light gray to black respectively). (D–F) The calculated relative contributions (y-axis) of monomers and oligomers as a function of the total peptide concentration (x-axis). Sedimentation profiles fit as previously described.54 Interestingly, while the TM5 sedimentation profile fits a model of a trimeric complex, the D150A mutation does not disrupt the trimeric complex but instead changes the oligomeric state to that of a dimer.
TM5 oligomerization in cellular membranes
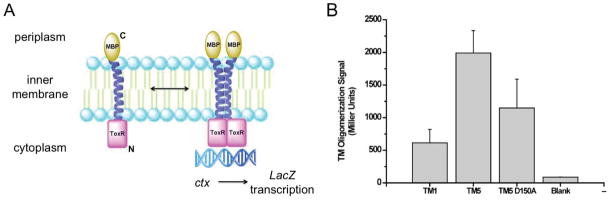
The experiments thus far have been performed with peptides in micellar environments, where orientation is not controlled. To study the oligomerization of TM5 in a natural membrane, a ToxR-based transcriptional reporter assay was used to monitor TM5 oligomerization in the inner membrane of E. coli.43,44 ToxR assays employ a chimeric protein composed of maltose binding protein for localization to the periplasm, the transmembrane region of interest and the N-terminal DNA-binding domain of ToxR, expressed in E. coli. The orientation of the transmembrane helix is controlled by expression between two fusion proteins, thus, in the ToxR assay, only parallel helix-helix interactions can occur. TM-driven oligomerization of the chimeric proteins in the bacterial membrane results in transcriptional activation of the ctx promoter and expression of β-galactosidase. The level of β-galactosidase activity observed corresponds to the strength of TM oligomerization (Figure 4A). Correct membrane insertion was verified by growing transformed cells in minimal media with maltose as the only carbon source (Supporting Information Figure 3A). Only cells expressing the correctly folded chimeric protein with maltose binding protein at the periplasm exhibit correct growth rates. Western blotting against maltose binding protein was used to confirm protein expression (Supporting Information Figure 3B). The oligomerization of TM1, TM5 and TM5 D150A was compared by ToxR assay (Figure 4B). The β-galactosidase activity showed that TM5 strongly oligomerized while TM1 showed limited interaction. TM5 D150A was impaired in its ability to oligomerize in comparison with wild-type TM5. These results demonstrate that TM5 can oligomerize in intact cellular membrane and that D150 plays a key role in this interaction.
Figure 4.
TM5 self-associates in a bacterial cell membrane. (A) Schematic illustration of the ToxR transmembrane oligomerization assay. Homo-oligomerization of TM helices activates ToxR, promoting binding to the ctx promoter and initiation of lacZ transcription. (B) ToxR measurement of relative oligomerization propensities of TM1, TM5 and TM5 D150A of LMP-1. TM5 oligomerized strongly in comparison to TM1. Mutation of TM5 to TM5 D150A significantly decreased oligomerization. Blank represents the signal from cells not transformed with any plasmid.
A model of the TM5 trimeric complex
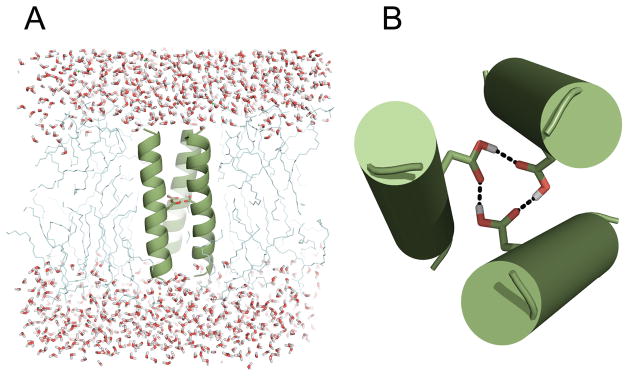
Although nuclear magnetic resonance and X-ray crystallographic analyses of transmembrane proteins are challenging, valuable structural insights can be obtained by combining experimentally derived structural information with computational approaches. Guided by our experimental observations showing that TM5 predominantly oligomerizes as a trimer, the TM5 sequence was first threaded onto a model coiled-coil (PDB id 1AQ5)45 selected by visual inspection to suggest a reasonable seed structure for further optimization. Optimization of the hydrogen-bonding network was carried out using an all-atom Molecular Dynamic (MD) simulation within a phospholipid membrane environment using an explicit solvation model. Energy-optimized structures of the TM5 trimer illustrate how the D150 hydrogen-bonding network could stabilize the oligomer in a trimeric configuration (Figure 5). Such a hydrogen-bonding network is possible because the carboxylic acid groups of the aspartic acid residues remain protonated in a phospholipid bilayer, which is consistent with previous reports that the pKa values of the acid are significantly elevated in a hydrophobic environment.46
Figure 5.
Structural model of TM5 homotrimer. (A) Side view of the molecular dynamics simulation setup showing TM5 trimer in explicit lipid and water environment. (B) Top view of the TM5 trimer showing the hydrogen bonding network formed by D150 residues during MD simulations.
D150 regulates LMP-1 activation in whole cells
Applying insights gained from the analysis of individual TM helices to the study of the intact integral membrane proteins like LMP-1 can be invaluable in relating membrane protein structure to function. To this end, cell-based functional assays were used to evaluate the effect of the TM5 D150A mutation on the function of full-length LMP-1. Expression vectors encoding LMP-1 with an aspartic acid to alanine substitution at position 150 (LMP-1 D150A) were constructed for this purpose.
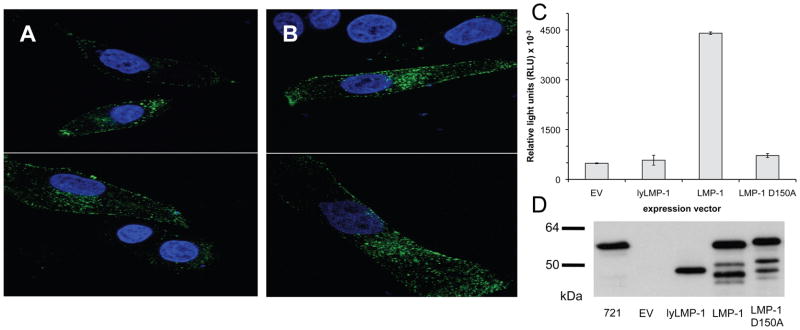
Because amino acid substitutions can perturb protein stability and folding, the subcellular localization of LMP-1 and LMP-1 D150A in HEp2 cells was compared to ensure that the D150A substitution did not affect LMP-1 folding or subcellular localization. As shown previously in HEp2 cells,24 LMP-1 localized to plasma membrane and intracellular membrane compartments in discrete puncta (Figure 6A). Localization of LMP-1 D150A (Figure 6B) was indistinguishable from wild type LMP-1. These results indicate that substitution of D150 with alanine has no detectable effect on the subcellular localization of LMP-1, suggesting that LMP-1 D150A is properly folded and trafficked in cells.
Figure 6.
Mutation of D150 does not affect the subcellular localization of full-length LMP-1 but does affect activation of NFκB. HEp2 cells were transiently transfected with expression vectors encoding wild-type LMP-1 or full-length LMP-1 with a substitution of the D150 residue to alanine. After 48 h, cells were fixed and stained with antibody against the C-terminus of LMP-1, then imaged with confocal microscopy. (A) As expected, LMP-1 localizes to the periphery of the cell, as well as to intracellular puncta. (B) Like LMP-1, LMP-1 D150A is found in puncta at the cell surface and intracellularly. (C) NFκB reporter assay in HEp2 cells comparing the NFκB activation in cells expressing empty vector (EV), lyLMP-1, LMP-1 and LMP-1 D150A. (D) Western blot analysis of extracts from C. stained with antibodies to LMP-1.
Signal transduction by LMP-1 requires proper oligomerization. Therefore, to determine the impact of the D150A substitution on LMP-1 oligomerization, the ability of LMP-1 D150A to signal was compared to that of wild type LMP-1. LMP-1 exerts its profound effects on cell proliferation and survival in part through activation of the transcription factor, NFκB. One measurable consequence of NFκB activation is the up-regulation of NFκB stimulated gene expression. A population-based transcriptional reporter assay in HEp2 cells (Figure 6C) was used to determine if the D150A mutation attenuates LMP-1 activation of the NFκB pathway. Cells were co-transfected with LMP-1 expression vectors and an NFκB luciferase reporter plasmid and assayed later for luciferase accumulation and LMP-1 expression. LMP-1, lyLMP-1 (non-functional truncation variant of LMP-1 encoded by Epstein-Barr Virus) and LMP-1 D150A expression levels were analyzed by Western blot analysis (Figure 6D). Cells expressing LMP-1 accumulated nine-fold more luciferase than cells transfected with empty vector (Figure 6C). Luciferase levels in cells expressing lyLMP-1 were indistinguishable from empty vector transfected cells. LMP-1 D150A expressing cells accumulated 1.2 times more luciferase than empty vector transfected cells and approximately six-fold less than cells expressing LMP-1. Normalized luciferase levels (relative light units/LMP-1 protein) in LMP-1 D150A and lyLMP-1 expressing cells were ~15% those in LMP-1 expressing cells (Supporting Information Figure 4). Thus, LMP-1 D150A is deficient in its ability to activate NFκB in HEp2 cells.
These results are from a population assay, and thus they are an average of the NFκB activity in all cells within the population. Therefore, the decreased NFκB activity in LMP-1 D150A expressing cells could be the result of two different phenomena: cells expressing LMP-1 D150A are either impaired in their ability to activate NFκB, or fewer of these cells have the capacity to stimulate NFκB. To distinguish between these possibilities, the NFκB activity of single cells expressing LMP-1 was compared to cells expressing LMP-1 D150A. The translocation of the p65 component of the active NFκB complex to the nucleus is quantifiable at the single cell level. Therefore, the ability of wild type LMP-1 and LMP-1 D150A to promote p65 translocation to the nucleus was evaluated in single cells (Figure 7A). HEp2 cells were transfected with LMP-1 or LMP-1 D150A mutant expression vectors and the ratio of nuclear/cytoplasmic distribution of p65 was quantified later in single cells by immunofluorescence intensity. Each LMP-1 positive and LMP-1 negative cell within a given population of transfected cells was scored for p65 localization and the results are plotted as nuclear p65 fluorescence intensity versus cytoplasmic p65 fluorescence intensity (Figure 7B). The majority of wild type LMP-1 expressing cells contained predominantly nuclear p65 (78%), whereas a minority of lyLMP-1 expressing cells contained predominantly nuclear p65 (9%) (Figure 7C, Table 1). The number of LMP-1 D150A expressing cells with predominantly nuclear p65 was significantly less than that observed in wild type LMP-1 expressing cells (38% versus 78%, respectively). The majority of LMP-1 negative cells in each transfected population (those cells that contained no detectable immunoreactive LMP-1, lyLMP-1 or LMP-1 D150A) contained ≤14% nuclear p65. Of the subpopulation of cells with predominately nuclear p65 (nuclear/cytoplasmic ratio > 1), the ratio of nuclear to cytoplasmic p65 in LMP-1 expressing cells was 2.8 whereas this ratio was 1.6 in the analogous subpopulation of cells expressing LMP-1 D150A (Table 1 and Figure 7C), a significant difference suggesting that D150 plays important roles in the p65 translocation. When compared to cells expressing wild-type LMP-1, fewer LMP-1 D150A expressing cells have predominantly nuclear p65, and in those LMP-1 D150A expressing cells that do have a nuclear/cytoplasmic p65 ratio >1, less p65 has translocated to the nucleus as compared to LMP-1 expressing cells with a nuclear/cytoplasmic ratio >1. These results complement the NFκB luciferase assay results and confirm that LMP-1 D150A is compromised in its ability to signal to NFκB.
Figure 7.
Mutation of D150 to alanine reduces the ability of full-length LMP-1 to stimulate p65 translocation to the nucleus. (A) HEp2 epithelial cells expressing wild-type LMP-1 or LMP-1 D150A were fixed and stained for LMP-1 (or LMP-1 variants) and p65. Robust NFκB in LMP-1 expressing cells is represented by high fluorescence levels of the p65 transcription factor in the nucleus (closed arrows), while cells not expressing LMP-1 (arrowheads) or cells expressing lyLMP-1 (asterisks) have low NFκB activity and sequester p65 in the cytoplasm. NFκB activation in cells expressing LMP-1 D150A is limited to a smaller population of cells and less p65 translocates to the nucleus (when compared to the levels of p65 retained in the cytoplasm) (a, open arrows and b). (B) The ratio of nuclear p65 to cytoplasmic p65 from each cell can be plotted as a population. Cells that fall along the dashed line have equal amounts of p65 in the nucleus and cytoplasm. In each transfection, the p65 ratio of cells with no detectable LMP-1 immunoreactivity (negative cells) was scored: p65 was consistently high in the cytoplasm and low in the nucleus, thus these cells fall below the dashed line (grey points in b). The reduced NFκB activation in cell populations expressing the D150A mutant is demonstrated by the increased number of cells below the grey line and the greater disparity between high cytoplasmic p65 and low nuclear p65 (represented by the increased distances below the grey line) compared to LMP-1. (C) On the population level, LMP-1 stimulates p65 nuclear translocation in 78% of cells, but LMP-1 D150A is approximately half as efficient. Similarly, in cells with greater nuclear p65 than cytoplasmic p65, LMP-1 drives p65 translocation such that there is an approximate three-fold difference in the amount of nuclear p65 compared to cytoplasmic p65 (TRNuc), however LMP-1 D150A can only stimulate a 1.6-fold difference. These data are described further in Table 1. The p65 translocation for cells not expressing LMP-1 or LMP-1 variants are shown with white bars.
Table 1.
Summary of p65 nuclear translocation measurements. HEp2 cells were fixed and stained for LMP-1 (or LMP-1 variants) and p65 as described in the legend to Fig. 6. Each cell was scored for the fluorescence intensity of p65 staining in the cytoplasm and the nucleus. In each transfected population, measurements of cells with no detectable LMP-1 staining (exp −) provided an internal negative control, while cells with detectable LMP-1 immunoreactivity represent LMP-1 (or LMP-1 variant) expressing cells (exp +). The extent of p65 nuclear translocation is represented by the ratio of nuclear p65 to cytoplasmic p65 (Translocation Ratio, TR). Wild-type LMP-1 expression stimulates p65 translocation to the nucleus in 78% of LMP-1 exp + cells. This sub-population has approximately three-fold more p65 in the nucleus than in the cytoplasm (Average TR = 2.84). Expression of D150A results in fewer cells with a TR > 1 (38% of D150 exp + cells) and cells with a reduced average TR compared to wild-type (1.69). As expected, lyLMP-1 expression does not stimulate p65 translocation to the nucleus above the levels seen in cells with no detectable LMP-1 immunoreactivity
| DNA | expa | n | % of cells with nuc p65b | Avg TRNucc | % of cells with cyt p65d | Avg SRCyte |
|---|---|---|---|---|---|---|
| lyLMP-1 | − | 81 | 2 | 1.36 | 98 | −1.65 |
| lyLMP-1 | + | 99 | 9 | 1.27 | 91 | −1.60 |
|
| ||||||
| LMP-1 | − | 91 | 14 | 1.30 | 86 | −1.73 |
| LMP-1 | + | 82 | 78 | 2.84 | 22 | −1.55 |
|
| ||||||
| D150A | − | 93 | 0 | N/A | 100 | −2.99 |
| D150A | + | 108 | 38 | 1.69 | 62 | −1.96 |
Expression of transfected LMP-1 (or LMP-1 variant) DNA constructs is determined by detection of fluorescent signal from images of fixed and stained cells. For more details, see the Materials and Methods. Exp + represents cells with detectable LMP-1 (or LMP-1 variant) immunoreactivity, while exp − cells have no detectable fluorescent signal.
Cells with nuclear p65 are defined by a TR >1.
The mean TR of the sub-population of cells with a TR > 1.
Cells with cytoplasmic p65 are defined by a TR < 1.
In the case where cells have a nuclear p65 staining level that is less than cytoplasmic p65 (TR < 1), the efficiency of p65 sequestration in the cytoplasm is represented by the negative inverse of TR (-cytoplasmic p65 fluorescence intensity/nuclear p65 fluorescence intensity).
DISCUSSION
Although the constitutive signaling of LMP-1 is regulated in part through the oligomerization of its TMD,15–25 the oligomerization state of LMP-1 and the driving force behind its self-association remained poorly understood. Obtaining structural data for a highly hydrophobic, self-associating integral membrane protein such as LMP-1 is a daunting challenge. However, transmembrane helices have a high level of intrinsic stability, and therefore may be considered as independent domains and studied as such, making the structural analysis of polytopic membrane proteins experimentally approachable. The work presented here examines the relationship between the structure and function of LMP-1 from an innovative perspective, grounded in chemical principles: the contributions of individual LMP-1 transmembrane domains to its overall oligomeric status are determined through study of isolated peptide domains and, finally, tested in the context of the full-length protein.
The results presented demonstrate that TM5 forms a trimeric complex and suggest that the fifth transmembrane helix (TM5) of LMP-1 is a potential driving force for LMP-1 self-association. TM5 trimerization requires an aspartic acid at LMP-1 sequence position 150 (D150). Replacement of D150 with alanine in TM5 abolishes trimerization and the resulting peptide (TM5 D150A) is primarily dimeric. These results, together with the central position of D150 within the hydrophobic membrane and the lack of apparent intramolecular hydrogen bonding partners for D150 within LMP-1, suggest that D150 is protonated and forms a hydrogen bond network that drives TM5 trimerization and, by extension, LMP-1 oligomerization (Figure 5). Interestingly, CD40, which LMP-1 mimics, and other TNFR family members, require trimerization for proper signaling.47,48 X-ray crystallographic analysis of three short peptide segments from the CTAR1 domain of LMP-1 proteins bound to homotrimeric TRAF3 support our model of LMP-1 forming a homotrimer.49 This TNFR-based model, along with the data presented here, implicates a trimeric form of LMP-1 as the constitutive signaling complex. Substitution of D150 with alanine in full-length LMP-1 resulted in significant attenuation of NFκB activation, suggesting that LMP-1 D150A adopts a different oligomeric state than does LMP-1. Our results are consistent with a model in which the oligomeric state of LMP-1 is trimeric, and that of LMP-140 D150A is dimeric. Dimeric LMP-1 is likely to be compromised in its ability to interact with the homotrimeric TRAF complex, resulting in less efficient activation of downstream signaling pathways. Together, our findings support a novel molecular mechanism for LMP-1 trimerization in which TM5 plays a key role. While it has not been directly shown that the self-association of full-length LMP-1 is driven by the aspartic acid located in the fifth transmembrane helix, this paper provides the first evidence pointing to the participation of TM5 in the constitutive activation of LMP-1.
A number of questions remain, however. The mechanism underlying the constitutive activation of LMP-1 is complex, involving both homo-oligomerization and lipid raft association. It is not clear whether oligomerization and raft association are related. Although the available biophysical and biochemical data support a model in which the minimal functional LMP-1 oligomer is homo-trimeric, the possibility that higher order LMP-1 multimers (i.e. multimers of trimers) associate to mediate constitutive signaling has not been explored. Yashui et al. have previously demonstrated that a hot spot (FWLY) in the first TM contributes to the LMP-1 signaling.50 The isolated TM1 showed weaker oligomerization propensity than TM5, suggesting that the FWLY motif may regulate LMP-1 raft localization or other protein-lipid interactions, rather than self-association. Alternatively there might be multiple binding sites driving the oligomerization, which is consistent with previous observation of the formation of higher order multimers (“patching”).51 It is reasonable to propose that the mechanisms underlying higher order multimer formation will be distinct from those contributing to LMP-1 trimerization and are likely to involve aspects of subcellular localization and raft association. Nonetheless, our atypical approach to the study of LMP-1 with transmembrane peptides implied a new hot spot that drives LMP-1 oligomerization, which may be of biological relevance to the full-length protein in cellular membrane.
MATERIALS AND METHODS
General Peptide Synthesis
Peptides were synthesized on Rink Amide MBHA resin (100–200 mesh) (ChemPep, FL, USA) with a substitution level of 0.57 mmole/g. Peptides were synthesized using a CEM (NC, USA) Liberty automated synthesizer with Discovery microwave module at 0.1 mmol scales. The GGPG/GPGG sequence motif was included on the N-/C-termini of the peptides to increase solubility. The highly hydrophobic TM4 was modified with KK rather than GGPG at each termini to increase its solubility in polar solvents. An additional glycine was added to the N-terminus of each peptide to act as a spacer for the addition of a fluorophore. Activation of the free amino acids was achieved with HATU (0.40 M solution in N,N-dimethylformamide). The reaction solvent was N-methylpyrrolidone (HPLC grade, Fisher, PA, USA). Side chain deprotection and simultaneous cleavage from the resin was performed using a mixture of trifluoroacetic acid/water/1,2-ethanedithiol/triisopropylsilane (90:2.5:2.5:1 v/v) or trifluoroacetic acid/water/phenol/thioanisol/1,2-ethanedithiol/triisopropylsilane (81.5:5:5:5:2.5:1) at room temperature for two hours. The crude peptides were collected by precipitation with cold (−20 °C) diethyl ether (Sigma, MO, USA). The peptides were then purified on an Agilent (CA, USA) 1200 series semi-preparative reverse phase HPLC system with an Agilent Zorbax 300 SB-C8 column using a linear gradient of buffer A (10% 2-propanol, 0.1% trifluoroacetic acid in Millipore water) and buffer B (6:3:1 2-propanol/acetonitrile/water containing 0.1% trifluoroacetic acid). The identities of the purified peptides were confirmed as the monomeric species by MALDI-TOF mass spectroscopy on a Voyager DE-STR Biospectrometry Workstation (PerSeptive Biosystems, CA, USA).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of TM peptides
Bis-Tris gels: Bis-Tris was obtained from Sigma (MO, USA) and gels were cast in house. OG buffer (10 mM HEPES, 60 mM N-octyl β-D-glucopyranoside, 0.5 mM CaCl2, pH 7.5) was prepared, aliquoted and stored at −20 °C until required. Peptides samples were prepared in OG buffer, mixed 1:1 with 2 × Laemmli buffer and incubated overnight at room temperature. Each sample was boiled for 5 min at 100 °C prior to electrophoresis. Electrophoresis was carried out at room temperature with MES-SDS running buffer (50 mM MES, 50 mM Tris base, 0.1% SDS, 1 mM EDTA, pH 7.3, Boston Biosciences, MA, USA). The resulting gel was stained with Comassie Brilliant Blue Staining Solution (Bio-Rad, CA, USA). The Laemmli buffer contained reducing agent. Disulfide bond formation was observed in the form of higher order species, however, if samples were left longer than overnight. Boiling and rerunning the samples resulted in the loss of higher order species, confirming disulfide bond formation.
10% Tricine gels, tricine loading buffer and tricine running buffer were obtained from Invitrogen (CA, USA). Peptides samples were prepared as described above. Electrophoresis was carried out at room temperature with tricine running buffer. The resulting gel was stained with Comassie Brilliant Blue Staining Solution (Bio-Rad, CA, USA).
Fluorescence quenching assay
Peptides were labeled with coumarin using the following method: 7-Hydroxycoumarin-3-carboxylic acid (100 mg, 0.485 mmol) and HBTU (186 mg, 0.490 mmol) were dissolved in 1:9 dimethyl sulfoxide/N,N-dimethylformamide and added to the resin. N,N′-Diisopropylethylamine (185 μl, 1.06 mmol) was added and the reaction was stirred for 2–16 h until the ninhydrin test indicated that the reaction was complete.52 Cleavage and purification were performed as described for unlabelled peptides in General Peptide Synthesis.
Peptides were dissolved in 2,2,2-trifluoroethanol (Sigma, MO, USA) with C14-betaine (3-(N,N-dimethylmyristyl-ammonio)propanesulfonate, Sigma, MO, USA). The organic solvent was removed with nitrogen then under reduced pressure to generate a thin film of peptide/detergent mixture, which was then dissolved in 100 mM HEPES (pH= 7.4) buffer. Peptide concentrations were determined by UV absorbance at 400 nm using an extinction coefficient of 39300.53 Samples were prepared to peptide concentrations of 20 to 25 μM. Two samples were made, with final concentrations of 500 μM and 10 mM of C14-betaine. Peptide samples were combined in a 96-well plate to obtain samples with C-14 betaine concentrations ranging from 500 μM to 10 mM. Samples were mixed and allowed to sit at room temperature for 30 min to one h and then excited at 360 nm and emission was read at 460 nm using a Beckman-Coulter (CA, USA) DTX 880 Multimode Detector plate reader. Each data point was an average of three emissions readings.
CD Spectroscopy
CD measurements were performed on a J-810 spectropolarimeter (Jasco, MD, USA). Peptides were co-dissolved in 2,2,2-trifluoroethanol (Sigma, MO, USA) and C14-betaine (3-(N,N-dimethylmyristyl-ammonio)propanesulfonate, Sigma, MO, USA). The organic solvent was removed under reduced pressure to generate a thin film of peptide/detergent mixture, which was then dissolved in 20 mM HEPES (pH = 7.4) buffer. The final concentration of C14-betaine was 10 mM in the samples. Samples were prepared to peptide concentrations of 20 to 25 μM. Peptide concentrations were determined by UV absorbance at 280 nm. All spectra were measured at 25 °C and were an average of six scans. Data collected below 200 nm is not shown due to the high voltage that resulted from the C14-betaine buffer.
Analytical Ultracentrifugation
Equilibrium Sedimentation was used to determine the association state of the peptides and to provide an estimate of the homo-oligomeric association constants. The experiments were performed in a Beckman XL-I analytical ultracentrifuge (Beckman Coulter, CA, USA) using six-channel carbon-epoxy composite centerpieces at 25 °C. Deuterium was added to the buffer to increase the density of the buffer and thus counter-balance the mass of the micelle.54
Peptides were co-dissolved in 2,2,2-trifluoroethanol (Sigma, MO, USA) and C14-betaine (3-(N,N-dimethylmyristyl-ammonio)propanesulfonate, Sigma, MO, USA). The organic solvent was removed under reduced pressure to generate a thin film of peptide/detergent mixture, which was then dissolved in buffer previously determined to match the density of the detergent component (100 mM HEPES (pH= 7.4) buffer containing 21% D2O).54 The final concentration of C-14 betaine was 10 mM in the samples. Samples were prepared in peptide concentrations of 86 μM and 43 μM. Data were analyzed from four measurement speeds (30, 35, 40, and 45 kRPM) using global curve-fitting of radial concentration gradients (measured using optical absorption at 280 nm) to the sedimentation equilibrium equation for monomer-dimer-tetramer and monomer-trimer-hexamer equilibria among the peptides included in the solution. Peptide partial specific volumes were calculated using Sednterp (http://www.jphilo.mailway.com/download.htm). The solvent density (1.075 g/ml) was determined using Sednterp. Aqueous solution molar extinction coefficients at 280 nm were calculated using Sednterp. These coefficients were multiplied by the molar detergent concentration to provide mole ratio concentration units.
Plasmids
pTox755 (kindly provided by D. Langosch, Technische Universität München, Germany) was modified by insertion of a single base (t) following the BamHI restriction site. pTox7 TM5 and pTox7 TM1 were created by ligating double stranded synthetic oligonucleotides (below) into the NheI/BamHI sites of modified pTox7. pTox7 TM5 D150A was created using standard site directed mutagenesis with a commercially available Stratagene Quikchange II kit (Agilent, CA, USA). Double stranded oligonucleotides encoding TM5 and TM1 were obtained from Integrated DNA Technologies (IA, USA).
TM5(forward): 5′ctagctggcagctgctggcgttctttctggcgttctttctggatctgattctgctgattattgcgctgtatctgg3′
TM5(reverse): 5′gatcccagatacagcgcaataatcagcagaatcagatccagaaagaacgccagaaagaacgccagcagctgccag3′
TM1(forward): 5′ctagcagcagcctgggcctggcgctgctgctgctgctgctggcgctgctgttttggctgtatattgtgatgagcg3′
TM1(reverse): 5′gatccgctcatcacaatatacagccaaaacagcagcgccagcagcagcagcagcagcgccaggcccaggctgctg3′
pRSV-LMP-1 is a pRC-RSV-based vector (Invitrogen, CA, USA) and has been previously described.21 pRSV-lyLMP-1 encodes a truncated form of LMP-1 with the nucleotides encoding amino acids 1–122 removed such that translation begins at Met129. pRSV-LMP-1-D150A was generated using site-directed mutagenesis (Agilent Technologies 210519, CA, USA) to substitute D150 to alanine with the forward primer 5′-CCTAGCCTTCTTCCTAGCCCTCATCCTGCTC-3′ and its reverse complement.
ToxR Assay
ToxR plasmids (200 ng) were transformed into 200 μl FHK12 competent cells (kindly provided by D. Langosch, Technische Universität München, Germany) with heat shock at 42 °C for 90 s and incubation on ice for two min, followed by addition of 800 μl SOC media and incubation with shaking at 37 °C for one h. Details of plasmid preparation are described in Plasmids. 50 μl of the transformation mixture was used to inoculate 5 ml LB + arabinose (0.0025%) and chloramphenicol (30 μg/ml) in triplicate. Cultures were incubated with shaking at 37 °C for 20 h and β-galactosidase activity was measured using a Beckman Coulter DTX 880 plate reader (Beckman Coulter, CA, USA). 5 μl of each culture was transferred in quadruplicate to the wells of a Costar 3596 polystyrene 96-well plate (Corning, NY, USA) containing 100 μl Z buffer/chloroform (1% β-mercaptoethanol, 10% chloroform, 89% A buffer: 1M sodium phosphate, 10 mM KCl, 1 mM MgSO4 pH 7.0). Cell densities were recorded by measuring OD595. Cells were lysed by addition of 50 μl Z buffer/ SDS (1.6% w/v sodium dodecyl sulfate in Z buffer) and shaking at 28 °C for 10 min. 50 μl Z buffer/ONPG (0.4% w/v o-nitro phenyl galactoside in Z buffer) was added and β-galactosidase activity was measured by monitoring the reaction at 405 nm for a period of 20 min at intervals of 30 s at 28 °C. Miller Units were calculated using the following equation:
Western blotting was performed with antiserum recognizing the maltose binding protein moiety of the constructs (New England Biolabs, MA, USA) and the 60–70 kDa region of the blot is shown.
Controls for membrane integration were performed by transforming PD28 cells (kindly provided by D. Langosch, Technische Universität München, Germany) with the indicated plasmids. Cells were grown in minimal media with maltose as the sole carbon source. Cell density was monitored by OD595 and corresponds to the efficiency of membrane integration.
Protein Modeling and Molecular Dynamic Simulation
The initial structure was generated by threading the TM5 sequence onto the NMR structure of the coiled-coil domain of chicken cartilage matrix protein (PDB id 1AQ5)45 such that the D150 residues were within hydrogen bonding distance. The D150 residues were protonated and all side chains were allowed to sample alternate conformations using Monte Carlo optimization. Gradient-based minimization of backbone torsion angles, side chain torsion angles and rigid-body orientation of the helices was then performed in search of a lower energy structure. The Rosetta protein modeling program was used for these simulations,56 using a variation of the energy function that significantly dampens repulsion energies.57 Molecular dynamics (MD) simulations in explicit lipid and water environment were then performed to incorporate the protein-lipid interactions intrinsic to a lipid bilayer environment. During MD simulations the side chain of D150 was assumed to be uncharged which is reasonable considering the hydrophobic core of the membrane interior. MD simulations were performed with the peptide trimer inserted in the transmembrane orientation into an equilibrated and solvated 1,2-dimyristoyl-sn-glycero-3-phosphocholine membrane bilayer containing 64 lipids in each leaflet. MD simulations were performed using the NAMD software suite58 with the CHARMM22 protein force field,59 CHARMM27 lipid force field60 and TIP3P water model.61 The temperature was maintained in all the simulations at 310 K using Langevin dynamics. During the equilibration runs, the heavy atoms of the protein were constrained for 0.5 ns with a simulation time step of 1 fs. This was followed by production runs of ~2 ns with a timestep of 1 fs. The hydrogen atoms were constrained using the SHAKE62 and SETTLE63 algorithms. Periodic boundary conditions were applied in three dimensions. The Langevin piston Nosé-Hoover method was used to maintain a pressure of 1 atm, allowing isotropic cell fluctuations. Long-range electrostatics was taken into account via the particle mesh Ewald scheme.64,65
Mammalian cell culture and transfections
HEp2 is an adherent human cell line sub-cloned from the HeLa cell line. HEp2 cells were grown in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (D10F). All cell lines were grown in a humidified incubator at 37 °C and 5% CO2. HEp2 cells were transfected by incubating in DNA: Lipofectamine 2000 (Invitrogen, CA, USA) complexes for five h, then given fresh media and harvested 43 h later.
NFκB reporter assay
HEp2 cells were transfected with 100 ng p1242 (luciferase gene under the control of three kB binding sites from the Class I MHC locus upstream of a minimal fos promoter17), 15 ng RSV-lacZ, 100 ng LMP-1 expression vectors and the total quantity of DNA brought to 1 μg with pRC-RSV per manufacturer’s instructions (Mirus Bio, WI, USA). Cells were harvested 48 h post-transfection and extracts were assayed for luciferase activity with the Dual Light assay kit (Tropix, Applied Biosystems, MA, ISA). Data are shown as relative light units (RLU).
Western blot analysis for the NFκB reporter assay
Whole-cell lysates (5×104 cell equivalents/lane) were solubilized by sonication in 4X SDS sample buffer with 50 mM DTT and boiled. Samples were analyzed by SDS-PAGE and Western blotting as previously described.20 Samples were resolved on 10% SDS-PAGE gels and proteins were transferred to Immobilon (Millipore, Merck, NJ, USA). Membranes were then blocked for one h in phosphate-buffered saline-5% milk-0.1% Tween 20 (PBST) and incubated with 1° antibodies as follows: 1:4000 anti-LMP-1 antiserum, 1:2000 anti-actin and 1:2500 HRP-conjugated anti-rabbit immunoglobulin. All antibody incubations were performed at 37 °C for 45 min or at room temperature for two h except for anti-LMP-1 antiserum, which was incubated overnight at 4 °C. Blots were visualized by enhanced chemiluminescence according to the manufacturer’s instructions (GE Amersham, NJ, USA).
Antibodies
Anti-maltose binding protein antibody was from New England Biolabs (MA, USA). Anti-LMP-1 antiserum is an affinity-purified rabbit polyclonal serum raised against the C terminus of LMP-1 (residues 188 to 352) fused to glutathione S-transferase. Polyclonal rabbit antibodies recognizing actin (I-19-R) and p65 (14–6731) are from Santa Cruz Biotechnology (CA, USA) and eBioscience (CA, USA), respectively. Horseradish peroxidase (HRP)-labeled goat anti-rabbit (W401B) secondary antibody is from Promega (WI, USA) and HRP-labeled light-chain specific mouse anti-rabbit secondary antibody is from Millipore (MAB201P, Merck, NJ, USA). Alexa Fluor 488-conjugated goat anti-mouse (A11001) and Alexa Fluor 555-conjugated goat anti-rabbit (A31572) are from Invitrogen (CA, USA).
p65 translocation assay
HEp2 cells grown on acid-washed glass coverslips were transfected with 1 μg pRC-RSV-LMP-1 or pRC-lyLMP-1. Cells were fixed 48 h post-transfection with 4% paraformaldehyde (EGS) for 15 min, permeabilized with 0.1% Triton X-100 (Pierce, IL, USA), and stained with antibodies to LMP-1 and p65 (1:500), followed by Alexa Fluor 488-conjugated goat anti-mouse and Alexa Fluor 555-conjugated goat anti-rabbit (1:500). Coverslips were then mounted with DAPI-containing mounting media (H-1200, Vectrashield) and imaged on a Zeiss 510LSM confocal laser scanning microscope. Cells were then scored for LMP-1 or lyLMP-1 expression (Exp+) and p65 localization. The ratio of nuclear p65 fluorescence intensity to cytoplasmic p65 fluorescence intensity is referred to as the translocation ratio (TR). In the subpopulation of cells with a TR > 1, the average TR is referred to as TRNuc. In order to quantify the ratio of nuclear p65 to cytoplasmic p65 in cells with a TR < 1 on an equivalent scale, this subpopulation of cells was quantified using the negative inverse of the TR (-cytoplasmic p65/nuclear p65), which we refer to as the sequestration ratio (SRCyt).
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (1R21CA138373), Cancer League of Colorado for financial supports of this work. H.Y. is grateful for the 2009 Elion Award from the American Association of Cancer Research, a Kimmel Scholar Award from the Sidney Kimmel Foundation for Cancer Research (SKF-08-101) and a Stand Up to Cancer (SU2C) Innovative Research Award. We thank Drs. James Lear, Paul Billings and Ivan Korendovych for their instruction with AUC experiments.
References
- 1.Henle G, Henle W, Diehl V. Proc Natl Acad Sci U S A. 1968;59:94–101. doi: 10.1073/pnas.59.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niederman JC, McCollum RW, Henle G, Henle W. JAMA. 1968;203:205–209. doi: 10.1001/jama.203.3.205. [DOI] [PubMed] [Google Scholar]
- 3.Brady G, MacArthur GJ, Farrell PJ. J Clin Pathol. 2007;60:1397–1402. doi: 10.1136/jcp.2007.047977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flavell KJ, Murray PG. Mol Pathol. 2000;53:262–269. doi: 10.1136/mp.53.5.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thorley-Lawson DA. Nat Rev Immunol. 2001;1:75–82. doi: 10.1038/35095584. [DOI] [PubMed] [Google Scholar]
- 6.Sugden B. Cell. 1989;57:5–7. doi: 10.1016/0092-8674(89)90165-7. [DOI] [PubMed] [Google Scholar]
- 7.Sugden B. Seminars in Virology. 1994;5:197–205. [Google Scholar]
- 8.Kaye KM, Izumi KM, Kieff E. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young LS, Rickinson AB. Nature Reviews Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 10.Li HP, Chang YS. J Biomed Sci. 2003;10:490–504. doi: 10.1007/BF02256110. [DOI] [PubMed] [Google Scholar]
- 11.Lam N, Sandberg ML, Sugden B. J Virol. 2004;78:1657–1664. doi: 10.1128/JVI.78.4.1657-1664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam N, Sugden B. Cell Signal. 2003;15:9–16. doi: 10.1016/s0898-6568(02)00083-9. [DOI] [PubMed] [Google Scholar]
- 13.Bishop GA, Hostager BS. Immunol Res. 2001;24:97–109. doi: 10.1385/IR:24:2:097. [DOI] [PubMed] [Google Scholar]
- 14.Bishop GA, Hostager BS. Arch Immunol Ther Exp. 2001;49:129–137. [PubMed] [Google Scholar]
- 15.Kaykas A, Worringer K, Sugden B. EMBO J. 2001;20:2641–2654. doi: 10.1093/emboj/20.11.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moorthy RK, Thorley-Lawson DA. J Virol. 1993;67:1638–1646. doi: 10.1128/jvi.67.3.1638-1646.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell T, Sugden B. J Virol. 1995;69:2968–2976. doi: 10.1128/jvi.69.5.2968-2976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaykas A, Worringer K, Sugden B. J Virol. 2002;76:11551–11560. doi: 10.1128/JVI.76.22.11551-11560.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gires O, Zimber-Strobl U, Gonnella R, Ueffing M, Marschall G, Zeidler R, Pich D, Hammerschmidt W. Embo J. 1997;16:6131–6140. doi: 10.1093/emboj/16.20.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coffin WF, 3rd, Erickson KD, Hoedt-Miller M, Martin JM. Oncogene. 2001;20:5313–5330. doi: 10.1038/sj.onc.1204689. [DOI] [PubMed] [Google Scholar]
- 21.Coffin WF, 3rd, Geiger TR, Martin JM. J Virol. 2003;77:3749–3758. doi: 10.1128/JVI.77.6.3749-3758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaykas A, Sugden B. Oncogene. 2000;19:1400–1410. doi: 10.1038/sj.onc.1203365. [DOI] [PubMed] [Google Scholar]
- 23.Liebowitz D, Mannick J, Takada K, Kieff E. J Virol. 1992;66:4612–4616. doi: 10.1128/jvi.66.7.4612-4616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin J, Sugden B. J Virol. 1991;65:3246–3258. doi: 10.1128/jvi.65.6.3246-3258.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang D, Liebowitz D, Wang F, Gregory C, Rickinson A, Larson R, Springer T, Kieff E. J Virol. 1988;62:4173–4184. doi: 10.1128/jvi.62.11.4173-4184.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray JK, Gellman SH. Nat Protoc. 2007;2:624–631. doi: 10.1038/nprot.2007.23. [DOI] [PubMed] [Google Scholar]
- 27.Finger C, Volkmer T, Prodohl A, Otzen DE, Engelman DM, Schneider D. J Mol Biol. 2006;358:1221–1228. doi: 10.1016/j.jmb.2006.02.065. [DOI] [PubMed] [Google Scholar]
- 28.Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. Proc Natl Acad Sci U S A. 2009;106:1760–1765. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walkenhorst WF, Merzlyakov M, Hristova K, Wimley WC. Biochim Biophys Acta. 2009;1788:1321–1331. doi: 10.1016/j.bbamem.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daugherty DL, Gellman SH. J Am Chem. 1999;121:4325–4333. [Google Scholar]
- 31.Lemmon MA, Flanagan JM, Treutlein HR, Zhang J, Engelman DM. Biochemistry. 1992;31:12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 32.Unterreitmeier S, Fuchs A, Schaffler T, Heym RG, Frishman D, Langosch D. J Mol Biol. 2007;374:705–718. doi: 10.1016/j.jmb.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 33.Russ WP, Engelman DM. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 34.Johnson RM, Hecht K, Deber CM. Biochemistry. 2007;46:9208–9214. doi: 10.1021/bi7008773. [DOI] [PubMed] [Google Scholar]
- 35.Ridder A, Skupjen P, Unterreitmeier S, Langosch D. J Mol Biol. 2005;354:894–902. doi: 10.1016/j.jmb.2005.09.084. [DOI] [PubMed] [Google Scholar]
- 36.Gurezka R, Laage R, Brosig B, Langosch D. J Biol Chem. 1999;274:9265–9270. doi: 10.1074/jbc.274.14.9265. [DOI] [PubMed] [Google Scholar]
- 37.Lee J, Sugden B. J Virol. 2007;81:9121–9130. doi: 10.1128/JVI.00136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gratkowski H, Lear JD, DeGrado WF. Proc Natl Acad Sci U S A. 2001;98:880–885. doi: 10.1073/pnas.98.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choma C, Gratkowski H, Lear JD, DeGrado WF. Nat Struct Biol. 2000;7:161–166. doi: 10.1038/72440. [DOI] [PubMed] [Google Scholar]
- 40.Zhou FX, Merianos HJ, Brunger AT, Engelman DM. Proc Natl Acad Sci U S A. 2001;98:2250–2255. doi: 10.1073/pnas.041593698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevens TJ, Arkin IT. Proteins. 1999;36:135–143. doi: 10.1002/(sici)1097-0134(19990701)36:1<135::aid-prot11>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 42.Eisenberg D, Schwarz E, Komaromy M, Wall R. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 43.Langosch D, Brosig B, Kolmar H, Fritz HJ. J Mol Biol. 1996;263:525–530. doi: 10.1006/jmbi.1996.0595. [DOI] [PubMed] [Google Scholar]
- 44.Russ WP, Engelman DM. Proc Natl Acad Sci U S A. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiltscheck R, Kammerer RA, Dames SA, Schulthess T, Blommers MJ, Engel J, Alexandrescu AT. Protein Sci. 1997;6:1734–1745. doi: 10.1002/pro.5560060814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zscherp C, Schlesinger R, Tittor J, Oesterhelt D, Heberle J. Proc Natl Acad Sci U S A. 1999;96:5498–5503. doi: 10.1073/pnas.96.10.5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McWhirter SM, Pullen SS, Holton JM, Crute JJ, Kehry MR, Alber T. Proc Natl Acad Sci U S A. 1999;96:8408–8413. doi: 10.1073/pnas.96.15.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Werneburg BG, Zoog SJ, Dang TT, Kehry MR, Crute JJ. J Biol Chem. 2001;276:43334–43342. doi: 10.1074/jbc.M104994200. [DOI] [PubMed] [Google Scholar]
- 49.Wu S, Xie P, Welsh K, Li C, Ni CZ, Zhu X, Reed JC, Satterthwait AC, Bishop GA, Ely KR. J Biol Chem. 2005;280:33620–33626. doi: 10.1074/jbc.M502511200. [DOI] [PubMed] [Google Scholar]
- 50.Yasui T, Luftig M, Soni V, Kieff E. Proc Natl Acad Sci U S A. 2004;101:278–283. doi: 10.1073/pnas.2237224100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bloss T, Kaykas A, Sugden B. J Gen Virol. 1999;80(Pt 12):3227–3232. doi: 10.1099/0022-1317-80-12-3227. [DOI] [PubMed] [Google Scholar]
- 52.Kaiser E, Colescott RL, Bossinger CD, Cook PI. Anal Biochem. 1970;34:595–598. doi: 10.1016/0003-2697(70)90146-6. [DOI] [PubMed] [Google Scholar]
- 53.Sherman WR, Robins E. Anal Chem. 1968;40:803–805. doi: 10.1021/ac60260a045. [DOI] [PubMed] [Google Scholar]
- 54.Stouffer AL, DeGrado WF, Lear JD. Progress in Colloid and Polymer Science. 2006;131:108–115. [Google Scholar]
- 55.Lindner E, Langosch D. Proteins. 2006;65:803–807. doi: 10.1002/prot.21226. [DOI] [PubMed] [Google Scholar]
- 56.Rohl CA, Strauss CE, Misura KM, Baker D. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 57.Dantas G, Corrent C, Reichow SL, Havranek JJ, Eletr ZM, Isern NG, Kuhlman B, Varani G, Merritt EA, Baker D. J Mol Biol. 2007;366:1209–1221. doi: 10.1016/j.jmb.2006.11.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Journal of Computational Chemistry. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. Journal of Physical Chemistry B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 60.Feller SE, Yin DX, Pastor RW, MacKerell AD. Biophysical Journal. 1997;73:2269–2279. doi: 10.1016/S0006-3495(97)78259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Journal of Chemical Physics. 1983;79:926–935. [Google Scholar]
- 62.Ryckaert JP, Ciccotti G, Berendsen HJC. Journal of Computational Physics. 1977;23:327–341. [Google Scholar]
- 63.Miyamoto S, Kollman PA. Journal of Computational Chemistry. 1992;13:952–962. [Google Scholar]
- 64.Darden T, York D, Pedersen L. Journal of Chemical Physics. 1993;98:10089–10092. [Google Scholar]
- 65.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. Journal of Chemical Physics. 1995;103:8577–8593. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.