Abstract
Integrase (IN) is a retroviral enzyme that catalyzes the insertion of viral DNA (vDNA) into host chromosomal DNA, which is necessary for efficient viral replication. The crystal structure of prototype foamy virus IN bound to cognate vDNA ends, a complex referred to as the intasome, has recently been resolved. Structure analysis of the intasome revealed a tetramer structure of IN that was required for its catalytic function, and also showed the inhibitory mechanism of the IN inhibitor. Genetic analysis of IN has revealed additional non-enzymatic roles during viral replication cycles at several steps other than integration. However, the higher order structure of IN that is required for its non-enzymatic functions remains to be delineated. This is the next major challenge in the field of IN structural biology hoping to be a platform for the development of novel IN inhibitors to treat human immunodeficiency virus type 1 infectious disease.
Keywords: HIV-1, integrase, reverse transcriptase, pol, reverse transcription, intasome, Gemin2
Introduction
Reverse transcription of viral RNA into double-stranded (ds) DNA, and the subsequent insertion of the synthesized viral DNA (vDNA) into a host chromosome, are characteristic features of retroviruses including human immunodeficiency virus type 1 (HIV-1). The reverse transcription and integration of the viral genome is sequentially catalyzed by the enzymes reverse transcriptase (RT) and integrase (IN), respectively. These retroviral enzymes are originally packaged in the viral particle along with the viral genomic RNA. After synthesis of vDNA by RT, IN acts on the termini of vDNA and catalyzes insertion of the vDNA into the host chromosome through two sequential enzymatic reactions: 3′-end processing and strand-transfer (Katz and Skalka, 1994). Currently, a clinically approved IN inhibitor specifically targets strand-transfer activity but not 3′-end processing activity (Summa et al., 2008). Therefore, the IN inhibitor is referred to as a strand-transfer inhibitor (STI). The STI shows very potent antiviral activity; however, emergence of STI-resistant variants is inevitable (Metifiot et al., 2010), as seen in patients treated with a combination of inhibitors against RT and protease activities. Efforts to develop novel drugs with distinct inhibitory mechanisms must continue so that we can effectively treat HIV-1 infections. For development of novel IN inhibitors, in addition to its enzymatic action, other non-enzymatic functions of IN, as described below, might be the next target(s) (Luo and Muesing, 2010).
Structure of HIV-1 IN
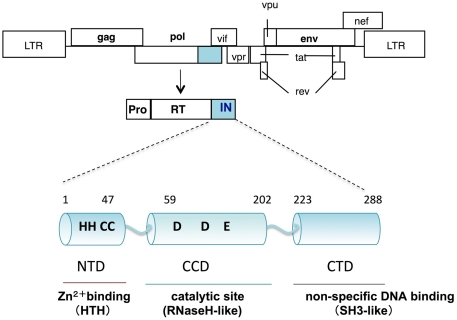
Human immunodeficiency virus type 1 IN is composed of 288 amino acids with three structurally distinct domains (Li et al., 2011): an N-terminal domain (NTD), a central catalytic core domain (CCD), and a C-terminal domain (CTD; Figure 1). The NTD contains a highly conserved (His-His-Cys-Cys, HHCC) motif, which binds to zinc ions (Zn2+) and folds a helix-turn-helix (HTH) structure. Through a tetrahedral attachment to the HHCC motif, Zn2+ enhances both multimerization and enzymatic activities of HIV-1 IN in vitro (Burke et al., 1992; Ellison et al., 1995; Cai et al., 1997). The CCD contains the highly conserved Asp, Asp, and Glu (DDE) residues directly involved in the catalytic activities of IN (Engelman and Craigie, 1992; Kulkosky et al., 1992; LaFemina et al., 1992; Bushman et al., 1993). Overall topology of the CCD is similar to those of ribonuclease H (RNaseH), the Holliday junction resolvase RuvC, and bacteriophage transposase Mu. Despite lack of sequence similarity between the CCD and RNaseH, there is remarkable similarity in the positioning of the two Asp catalytic residues (Dyda et al., 1994). The CTD, consisting of a structure that closely resembles Src homology 3 domains (SH3-like), possesses sequence- and metal ion-independent DNA binding activity (Eijkelenboom et al., 1995; Lodi et al., 1995). Each domain has been demonstrated to form a dimer and higher multimerization states (Dyda et al., 1994; Eijkelenboom et al., 1995; Cai et al., 1997), which might be required for all the enzymatic functions of IN.
Figure 1.
Schematic diagram of HIV-1 IN. The genetic organization of HIV-1 is shown at the top. HIV-1 IN is encoded by the pol region and composed of 288 amino acids with three structurally distinct domains: an N-terminal domain (NTD); a central catalytic core domain (CCD); and a C-terminal domain (CTD). The CCD contains the highly conserved DDE motif, which is directly involved in the catalytic activities of IN. Overall topology of the CCD is similar to that of ribonuclease H (RNaseH). The NTD, contains a highly conserved HHCC motif, which binds to zinc and folds a helix-turn-helix (HTH) structure. The CTD, consisting of a structure that closely resembles Src homology 3 domains (SH3), possesses sequence- and metal ion-independent DNA binding activity.
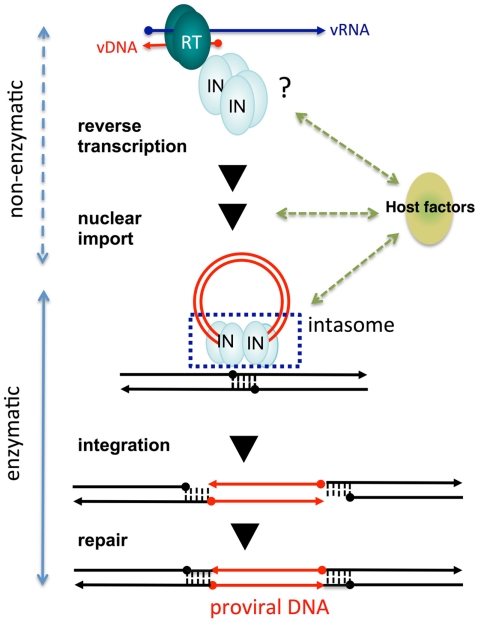
Recently, the entire prototype foamy virus (PFV) IN in a complex with its cognate vDNA ends, referred to as the intasome, has been successfully crystallized (Hare et al., 2010). The crystal structure analysis of the PFV intasome revealed an unprecedented tetramer structure for IN (see Cherepanov et al., 2011; Li et al., 2011 for recent review). The IN tetramer structure observed in the PFV intasome demonstrated that two sets of IN dimer acts on each vDNA end (Figure 2). The inner subunits of each IN dimer contact with vDNA and form a tetramer. The outer subunits of each IN dimer might be speculated to have supportive or other functions, such as engagement of target DNA or interaction with host factors. Several models for the IN tetramer have been proposed from previous structure analysis using partial IN fragments possessing the NTD–CCD or CCD–CTD (Chen et al., 2000; Wang et al., 2001; Hare et al., 2009a). However, these IN tetramer models are different from those observed in the active PFV intasome (Craigie, 2010). Stable interaction of IN with 3′-end processed vDNA in the intasome might be a plausible explanation for the difference. The stable IN tetramer formation observed in the intasome reflects the IN-DNA complex required for proper concerted integration of both vDNA ends into the proximal sites of the target host chromosomal DNA. Furthermore, analysis of the PFV intasome interacting with the STI elucidated its inhibitory mechanism. Based on the PFV intasome structure as a template, structural modeling of the HIV-1 intasome has also been reported (Krishnan et al., 2010). Structural analysis of this intasome revealed numerous details of retroviral integration and will contribute to the design of the next generation of HIV-1 IN catalytic inhibitors. The functional significance of the DNA-independent IN tetramer as observed by analysis of partial IN fragments (Chen et al., 2000; Wang et al., 2001; Hare et al., 2009a) remains unclear (Cherepanov et al., 2011).
Figure 2.
Non-enzymatic and enzymatic roles of HIV-1 IN that help establish proviral DNA. After entry into cells, retroviral genomic RNA (vRNA) is reverse transcribed into DNA (vDNA) by RT. Then, vDNA is transported into the nucleus (nuclear import) and finally integrated into host chromosomal DNA (black lines). IN forms a functional tetramer with vDNA ends (intasome) to assist with integration. Subsequent repair processes by host DNA repair machinery establishes the proviral DNA, in which retrovirus DNA is stably integrated into host chromosomal DNA. IN tetramer structures for its enzymatic (integration) and non-enzymatic functions (reverse transcription and nuclear import) and possible involvement of host factors are schematically depicted. The vRNA, vDNA, and host DNA are shown as blue, red, and black lines, respectively. An arrowhead shows the direction of each genome from the 5′- to the 3′-end.
Non-Enzymatic Functions of IN
Originally, we found that introduction of amino acid substitutions at conserved HHCC residues in the NTD of HIV-1 IN resulted in almost complete abrogation of proviral DNA formation, concomitant with a severe reduction in vDNA synthesis. This suggests the mutations in the IN affected the viral life cycle at steps prior to integration (Masuda et al., 1995). Further genetic analysis of HIV-1 IN revealed that the pleiotropic effects of IN mutations affected uncoating (Masuda et al., 1995; Leavitt et al., 1996; Nakamura et al., 1997; Briones et al., 2010), reverse transcription (Engelman et al., 1995; Masuda et al., 1995; Wu et al., 1999; Tsurutani et al., 2000; Nishitsuji et al., 2009) nuclear import of vDNA (Gallay et al., 1997; Tsurutani et al., 2000; Ikeda et al., 2004), and protein processing during viral particle assembly and maturation (Mohammed et al., 2011). Importantly, HIV-1 carrying point mutations at the catalytic sites of IN (DDE) affected the integration step, but not vDNA synthesis (Masuda et al., 1995). Thus, the pleiotropic effects of IN mutations might not be directly related to the loss of its catalytic function. These experiments suggest that IN may possess non-enzymatic roles throughout the viral replication cycle (Figure 2).
Among the possible non-enzymatic roles of IN, there has been an accumulation of evidence to suggest involvement of retroviral IN during reverse transcription (Engelman et al., 1995; Masuda et al., 1995; Tsurutani et al., 2000; Lu et al., 2005; Dobard et al., 2007). The contribution of IN during reverse transcription has also been noticed in a retrovirus-like element of Saccharomyces cerevisiae, Ty3 (Nymark-McMahon and Sandmeyer, 1999; Nymark-McMahon et al., 2002). A previous study from our laboratory showed that reverse transcription of HIV-1 was abrogated by knocking down a host factor, survival motor neuron (SMN)-interacting protein 1 (SIP1/Gemin2), which binds to HIV-1 IN (Hamamoto et al., 2006). Gemin 2 is a component of the SMN complex that mediates the assembly of spliceosomal small nuclear ribonucleoproteins and nucleolar ribonucleoproteins (Fischer et al., 1997; Liu et al., 1997; Buhler et al., 1999; Jablonka et al., 2001; Meister et al., 2001). In a subsequent study, we demonstrated that HIV-1 IN and Gemin2 synergistically stimulate RT activity by enhancing the assembly of RT on viral RNA in vitro (Nishitsuji et al., 2009). Chow and colleagues have also reported that HIV-1 IN stimulates RT activity through physical interactions with RT (Zhu et al., 2004). Thus, IN might possess a direct function to support efficient reverse transcription by RT.
Insight on Putative Structure of IN for Its Non-Enzymatic Function
Delineation of the IN mutant structure could provide clues for depicting the IN conformation required for non-enzymatic functions. Nuclear magnetic resonance (NMR) analysis of an isolated NTD has shown that the NTD exists in two conformational states, the E and D forms (Cai et al., 1997). A previous study using NMR spectroscopy indicated that the NTD mutant protein, in which the Tyr 15 residue was replaced with Ala (Y15A), folds correctly but only as the E form (Nomura et al., 2006). The IN tetramer structure was formed through interaction of NTD–CCD between the inner subunits in the intasome (Hare et al., 2010). The residues 13–26 and 40–45 in the NTD interact extensively with residues 150–196 in the CCD of the other subunit. The importance of the NTD–CCD interaction to form the IN tetramer has been proposed from previous crystal structure analysis using the NTD–CCD IN fragments (Wang et al., 2001; Hare et al., 2009b). As observed in these previous analyses, the hydrogen-bond contacts between the side chains of CCD residues Gln164 and Arg187 and the backbones of the NTD residues Lys14 and Tyr15 also persist in the recent structure-based model of the HIV-1 IN intasome (Krishnan et al., 2010). HIV-1 carrying IN mutations at the Lys186, Arg187, and Lys188 residues exhibited a reverse transcription-defective phenotype (Tsurutani et al., 2000) as found in the NTD mutants including Y15A. These experimental data suggest that the functional tetramer form, stabilized with the NTD–CCD interaction, might be critical for the non-enzymatic function of IN during reverse transcription.
Impact of Host Factors on IN Structure and Non-Enzymatic Function
Numerous host factors that interact with HIV-1 IN have been reported (Al-Mawsawi and Neamati, 2007). The best characterized factor is lens epithelium-derived growth factor/transcription co-activator p75 (LEDGF/p75; Cherepanov et al., 2003) for chromosomal targeting of HIV-1 IN (Maertens et al., 2003, 2004; Shun et al., 2007). The crystal structure analysis of the PFV intasome (Hare et al., 2010) together with results from previous studies (Li et al., 2006; Hare et al., 2009a) suggest that synapsis formation with DNA ends of the virus through tetramerization of IN might be critical for proper assembly and/or a stable and functionally active intasome. Importantly, LEDGF/p75 stabilizes the functional tetramer of IN for its enzymatic function. Meanwhile, a reduction of the tetramer form of IN was reproduced when wild-type IN was expressed in cells in which endogenous Gemin2 was knocked down by RNA interference (Nishitsuji et al., 2009). We also noticed that the intracellular stability and multimer formation of IN, especially the tetramer formation of IN, were dramatically reduced by IN mutations that led to a reverse transcription-defective phenotype. Thus, in the absence of vDNA, host factors might be involved in forming or maintaining highly ordered structures of IN, which is critical for non-enzymatic function. Therefore, inhibition of the interaction between IN and host factors could be a novel therapeutic approach for the design and development of new classes of IN inhibitors targeting non-enzymatic functions.
Conclusion
It is obvious that IN must be closely associated with the viral genome complex to form an active intasome upon the completion of reverse transcription. Physical interaction of IN with RT (Zhu et al., 2004; Wilkinson et al., 2009) could contribute to keeping IN close to the viral genome complex. How is the IN structure in the absence of vDNA before and during reverse transcription maintained? What is the contribution of host factors to IN conformation that is required for non-enzymatic functions? The highly ordered IN structure and/or its interface structure for interaction with host factors required for non-enzymatic IN function(s) should be determined.
Finally, it should be emphasized that IN mutations affecting non-enzymatic function resulted in severe deleterious affects to HIV-1 replication compared with IN mutations that specifically affected catalytic activity. Clinical efficacy of the STI that blocks IN catalytic activity guarantees that a greater impact on HIV-1 control would be achieved with a novel inhibitor that blocks the non-enzymatic function of IN.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan and a grant for HIV/AIDS research from the Ministry of Health, Labor, and Welfare of Japan.
References
- Al-Mawsawi L. Q., Neamati N. (2007). Blocking interactions between HIV-1 integrase and cellular cofactors: an emerging anti-retroviral strategy. Trends Pharmacol. Sci. 28, 526–535 10.1016/j.tips.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Briones M. S., Dobard C. W., Chow S. A. (2010). Role of human immunodeficiency virus type 1 integrase in uncoating of the viral core. J. Virol. 84, 5181–5190 10.1128/JVI.02382-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler D., Raker V., Luhrmann R., Fischer U. (1999). Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum. Mol. Genet. 8, 2351–2357 10.1093/hmg/8.13.2351 [DOI] [PubMed] [Google Scholar]
- Burke C. J., Sanyal G., Bruner M. W., Ryan J. A., Lafemina R. L., Robbins H. L., Zeft A. S., Middaugh C. R., Cordingley M. G. (1992). Structural implications of spectroscopic characterization of a putative zinc finger peptide from HIV-1 integrase. J. Biol. Chem. 267, 9639–9644 [PubMed] [Google Scholar]
- Bushman F. D., Engelman A., Palmer I., Wingfield P., Craigie R. (1993). Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc. Natl. Acad. Sci. U.S.A. 90, 3428–3432 10.1073/pnas.90.8.3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M., Zheng R., Caffrey M., Craigie R., Clore G. M., Gronenborn A. M. (1997). Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat. Struct. Biol. 4, 567–577 10.1038/nsb0797-567 [DOI] [PubMed] [Google Scholar]
- Chen J. C., Krucinski J., Miercke L. J., Finer-Moore J. S., Tang A. H., Leavitt A. D., Stroud R. M. (2000). Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl. Acad. Sci. U.S.A. 97, 8233–8238 10.1073/pnas.120156997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z. (2003). HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278, 372–381 10.1074/jbc.M209278200 [DOI] [PubMed] [Google Scholar]
- Cherepanov P., Maertens G. N., Hare S. (2011). Structural insights into the retroviral DNA integration apparatus. Curr. Opin. Struct. Biol. 21, 249–256 10.1016/j.sbi.2010.12.005 [DOI] [PubMed] [Google Scholar]
- Craigie R. (2010). Structural biology: when four become one. Nature 464, 167–168 10.1038/464167a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobard C. W., Briones M. S., Chow S. A. (2007). Molecular mechanisms by which human immunodeficiency virus type 1 integrase stimulates the early steps of reverse transcription. J. Virol. 81, 10037–10046 10.1128/JVI.00519-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyda F., Hickman A. B., Jenkins T. M., Engelman A., Craigie R., Davies D. R. (1994). Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266, 1981–1986 10.1126/science.7801124 [DOI] [PubMed] [Google Scholar]
- Eijkelenboom A. P., Lutzke R. A., Boelens R., Plasterk R. H., Kaptein R., Hard K. (1995). The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat. Struct. Biol. 2, 807–810 10.1038/nsb0995-807 [DOI] [PubMed] [Google Scholar]
- Ellison V., Gerton J., Vincent K. A., Brown P. O. (1995). An essential interaction between distinct domains of HIV-1 integrase mediates assembly of the active multimer. J. Biol. Chem. 270, 3320–3326 10.1074/jbc.270.7.3320 [DOI] [PubMed] [Google Scholar]
- Engelman A., Craigie R. (1992). Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 66, 6361–6369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A., Englund G., Orenstein J. M., Martin M. A., Craigie R. (1995). Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69, 2729–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U., Liu Q., Dreyfuss G. (1997). The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell 90, 1023–1029 10.1016/S0092-8674(00)80368-2 [DOI] [PubMed] [Google Scholar]
- Gallay P., Hope T., Chin D., Trono D. (1997). HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. U.S.A. 94, 9825–9830 10.1073/pnas.94.18.9825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto S., Nishitsuji H., Amagasa T., Kannagi M., Masuda T. (2006). Identification of a novel human immunodeficiency virus type 1 integrase interactor, Gemin2, that facilitates efficient viral cDNA synthesis in vivo. J. Virol. 80, 5670–5677 10.1128/JVI.02471-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S., Di Nunzio F., Labeja A., Wang J., Engelman A., Cherepanov P. (2009a). Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog. 5, e1000515. 10.1371/journal.ppat.1000515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S., Shun M. C., Gupta S. S., Valkov E., Engelman A., Cherepanov P. (2009b). A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 5, e1000259. 10.1371/journal.ppat.1000259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S., Gupta S. S., Valkov E., Engelman A., Cherepanov P. (2010). Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464, 232–236 10.1038/nature08784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T., Nishitsuji H., Zhou X., Nara N., Ohashi T., Kannagi M., Masuda T. (2004). Evaluation of the functional involvement of human immunodeficiency virus type 1 integrase in nuclear import of viral cDNA during acute infection. J. Virol. 78, 11563–11573 10.1128/JVI.78.21.11563-11573.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka S., Bandilla M., Wiese S., Buhler D., Wirth B., Sendtner M., Fischer U. (2001). Co-regulation of survival of motor neuron (SMN) protein and its interactor SIP1 during development and in spinal muscular atrophy. Hum. Mol. Genet. 10, 497–505 10.1093/hmg/10.5.497 [DOI] [PubMed] [Google Scholar]
- Katz R. A., Skalka A. M. (1994). The retroviral enzymes. Annu. Rev. Biochem. 63, 133–173 10.1146/annurev.bi.63.070194.001025 [DOI] [PubMed] [Google Scholar]
- Krishnan L., Li X., Naraharisetty H. L., Hare S., Cherepanov P., Engelman A. (2010). Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc. Natl. Acad. Sci. U.S.A. 107, 15910–15915 10.1073/pnas.1002346107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkosky J., Jones K. S., Katz R. A., Mack J. P., Skalka A. M. (1992). Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol. 12, 2331–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFemina R. L., Schneider C. L., Robbins H. L., Callahan P. L., Legrow K., Roth E., Schleif W. A., Emini E. A. (1992). Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 66, 7414–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt A. D., Robles G., Alesandro N., Varmus H. E. (1996). Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J. Virol. 70, 721–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Mizuuchi M., Burke T. R., Jr., Craigie R. (2006). Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 25, 1295–1304 10.1038/sj.emboj.7601289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Krishnan L., Cherepanov P., Engelman A. (2011). Structural biology of retroviral DNA integration. Virology 411, 194–205 10.1016/j.virol.2010.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Fischer U., Wang F., Dreyfuss G. (1997). The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell 90, 1013–1021 10.1016/S0092-8674(00)80367-0 [DOI] [PubMed] [Google Scholar]
- Lodi P. J., Ernst J. A., Kuszewski J., Hickman A. B., Engelman A., Craigie R., Clore G. M., Gronenborn A. M. (1995). Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry 34, 9826–9833 10.1021/bi00031a002 [DOI] [PubMed] [Google Scholar]
- Lu R., Ghory H. Z., Engelman A. (2005). Genetic analyses of conserved residues in the carboxyl-terminal domain of human immunodeficiency virus type 1 integrase. J. Virol. 79, 10356–10368 10.1128/JVI.79.19.12584-12591.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Muesing M. A. (2010). Prospective strategies for targeting HIV-1 integrase function. Future Med. Chem. 2, 1055–1060 10.4155/fmc.10.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens G., Cherepanov P., Debyser Z., Engelborghs Y., Engelman A. (2004). Identification and characterization of a functional nuclear localization signal in the HIV-1 integrase interactor LEDGF/p75. J. Biol. Chem. 279, 33421–33429 10.1074/jbc.M404700200 [DOI] [PubMed] [Google Scholar]
- Maertens G., Cherepanov P., Pluymers W., Busschots K., De Clercq E., Debyser Z., Engelborghs Y. (2003). LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278, 33528–33539 10.1074/jbc.M303594200 [DOI] [PubMed] [Google Scholar]
- Masuda T., Planelles V., Krogstad P., Chen I. S. (1995). Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J. Virol. 69, 6687–6696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Buhler D., Pillai R., Lottspeich F., Fischer U. (2001). A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 3, 945–949 10.1038/ncb1101-945 [DOI] [PubMed] [Google Scholar]
- Metifiot M., Marchand C., Maddali K., Pommier Y. (2010). Resistance to integrase inhibitors. Viruses 2, 1347–1366 10.3390/v2071347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed K. D., Topper M. B., Muesing M. A. (2011). Sequential deletion of the integrase (Gag-Pol) carboxyl terminus reveals distinct phenotypic classes of defective HIV-1. J. Virol. 85, 4654–4666 10.1128/JVI.02374-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Masuda T., Goto T., Sano K., Nakai M., Harada S. (1997). Lack of infectivity of HIV-1 integrase zinc finger-like domain mutant with morphologically normal maturation. Biochem. Biophys. Res. Commun. 239, 715–722 10.1006/bbrc.1997.7541 [DOI] [PubMed] [Google Scholar]
- Nishitsuji H., Hayashi T., Takahashi T., Miyano M., Kannagi M., Masuda T. (2009). Augmentation of reverse transcription by integrase through an interaction with host factor, SIP1/Gemin2 Is critical for HIV-1 infection. PLoS ONE 4, e7825. 10.1371/journal.pone.0007825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura Y., Masuda T., Kawai G. (2006). Structural analysis of a mutant of the HIV-1 integrase zinc finger domain that forms a single conformation. J. Biochem. 139, 753–759 10.1093/jb/mvj114 [DOI] [PubMed] [Google Scholar]
- Nymark-McMahon M. H., Beliakova-Bethell N. S., Darlix J. L., Le Grice S. F., Sandmeyer S. B. (2002). Ty3 integrase is required for initiation of reverse transcription. J. Virol. 76, 2804–2816 10.1128/JVI.76.6.2804-2816.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nymark-McMahon M. H., Sandmeyer S. B. (1999). Mutations in nonconserved domains of Ty3 integrase affect multiple stages of the Ty3 life cycle. J. Virol. 73, 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shun M. C., Raghavendra N. K., Vandegraaff N., Daigle J. E., Hughes S., Kellam P., Cherepanov P., Engelman A. (2007). LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 21, 1767–1778 10.1101/gad.1565107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summa V., Petrocchi A., Bonelli F., Crescenzi B., Donghi M., Ferrara M., Fiore F., Gardelli C., Gonzalez Paz O., Hazuda D. J., Jones P., Kinzel O., Laufer R., Monteagudo E., Muraglia E., Nizi E., Orvieto F., Pace P., Pescatore G., Scarpelli R., Stillmock K., Witmer M. V., Rowley M. (2008). Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J. Med. Chem. 51, 5843–5855 10.1021/jm800245z [DOI] [PubMed] [Google Scholar]
- Tsurutani N., Kubo M., Maeda Y., Ohashi T., Yamamoto N., Kannagi M., Masuda T. (2000). Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J. Virol. 74, 4795–4806 10.1128/JVI.74.10.4795-4806.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Y., Ling H., Yang W., Craigie R. (2001). Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 20, 7333–7343 10.1093/emboj/20.4.880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson T. A., Januszyk K., Phillips M. L., Tekeste S. S., Zhang M., Miller J. T., Le Grice S. F., Clubb R. T., Chow S. A. (2009). Identifying and characterizing a functional HIV-1 reverse transcriptase-binding site on integrase. J. Biol. Chem. 284, 7931–7939 10.1074/jbc.M806241200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Liu H., Xiao H., Conway J. A., Hehl E., Kalpana G. V., Prasad V., Kappes J. C. (1999). Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J. Virol. 73, 2126–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K., Dobard C., Chow S. A. (2004). Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J. Virol. 78, 5045–5055 10.1128/JVI.78.10.5045-5055.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]