Abstract
Human UDP-glucuronosyltransferases (EC 2.4.1.17) (UGTs) are major phase II metabolism enzymes that detoxify a multitude of endo- and xenobiotics through the covalent addition of a glucuronic acid moiety. UGTs are promiscuous enzymes that regulate the levels of numerous important endobiotics in a range of tissues, and inactivate most therapeutic compounds in concert with phase I enzymes. In spite of the importance of these enzymes, we have only a limited understanding of the molecular mechanisms governing their substrate specificity and catalytic activity. Until recently, no three-dimensional structural information was available for any mammalian UGT. The 1.8-Å resolution apo crystal structure of the UDP-glucuronic acid binding domain of human UGT2B7 (2B7CT) is the only structure of a mammalian UGT target determined to date. In this review, we summarize what has been learned about human UGT function from the analysis of this and other related glycosyltransferase (GT) crystal structures.
Keywords: Homology modeling, metal dependence, binding site, catalytic mechanism, contact residues
Introduction
UDP-glucuronosyltransferases (UGTs) are characterized by the ability of each isoform to accept a remarkable number of structurally diverse, endogenous, and exogenous substrates. A large number of compounds, such as bilirubin (BR), steroids, fatty acids (FAs), bile acids (BAs), retinoids, drugs, and environmental pollutants, are UGT substrates. These conjugation enzymes catalyze the transfer of glucuronic acid (GlcUA) from UDP-GlcUA to lipophilic substrates containing specific functional groups that serve as nucleophilic acceptors. The second-order nucleophilic substitution mechanism is commonly recognized as the mode of action for the majority of glucuronidation reactions. However, the formation of quaternary ammonium glucuronides does not require proton extraction essential for this mechanism. The major function of glucuronidation is to change hydrophobic compounds into soluble derivatives, thereby facilitating their detoxification and excretion. In addition, UGTs can also synthesize glucuronides that are biologically active, some of which demonstrate increased toxicity (Bock, 1992; Oelberg et al., 1984; Vore et al., 1983).
UGTs are membrane glycoproteins located in the endoplasmic reticulum (ER) and can be divided into two parts: the N- and C-terminal domains. UGTs are synthesized as precursors of ~530 residues containing an N-terminal signal peptide that mediates the integration of the polypeptide chain into the ER (Mackenzie and Owens, 1984; Ouzzine et al., 1999a, 1999b). The signal peptide is subsequently cleaved and the protein is N-glycosylated. The mature protein of about 505 residues is classified as a type I ER transmembrane protein (Meech and Mackenzie, 1998) with a lumenal domain consisting of about 95% of the polypeptide chain and a cytoplasmic domain of only ~20 residues (Mackenzie, 1986, 1987). A conserved region of 17 hydrophobic residues between an N-terminal Asp and a C-terminal Lys connects the two domains through the ER lipid bilayer (Iyanagi et al., 1986; Mackenzie, 1986). Experiments with chimeric and truncated proteins demonstrated that there are other regions in the lumenal domain of the protein, including the N-terminal end, that are involved in strong interactions with the lipid bilayer (Meech et al., 1996; Meech and Mackenzie, 1997). Thus, it is possible that part of the lumenal domain is buried in the lipid bilayer. Experimental evidence in support of this model has been provided by studies of microsomal UGTs using photoaffinity probes, antibodies, proteases, and detergents (Radominska-Pandya et al., 1999; Shepherd et al., 1989; Vanstapel and Blanckaert, 1988; Yokota et al., 1992) and on UGTs synthesized in vitro in the presence and absence of membranes (Mackenzie, 1987; Mackenzie et al., 1984; Mackenzie and Owens, 1984). These studies show that when the lipid bilayer is intact, most of the mature enzymes are shielded from proteolytic attack and recognition by antibodies and chemical probes. Disruption of the bilayer by detergents, however, renders the protein susceptible to these agents.
UGTs have been a subject of intense research during the past several decades. Even though these enzymes have been investigated from the perspectives of regulation, toxicology, oncology, endocrinology, and drug development, few studies have examined the structural properties of UGTs. A full length mammalian UGT crystal structure is not available, and there are few reports of computer-aided molecular modeling being applied to this system (Coffman et al., 2001, 2003; Locuson and Tracy, 2007; Xiong et al., 2008). This lack of structural knowledge has resulted in the prediction of UGT structures by using selective inhibitors, amino acid–specific chemical modification reagents, amino acid alignments, site-directed mutagenesis, and photoaffinity labeling. Fortunately, this structural knowledge gap is beginning to shrink. The high-resolution crystal structure of the UDP-GlcUA binding domain of the human UGT isoform 2B7 was determined recently (Miley et al., 2007). Furthermore, a number of related plant flavonoid glucosyltransferase and bacterial glycosyltransferase x-ray crystal structures in complex with substrates have also been determined. These new structural data are allowing us, for the first time, to start developing a molecular picture of how UGTs function.
Comparisons between GT-B and GT-A super families
Glycosyltransferases (GTs) have been divided into 91 families (GTx) on the basis of amino acid similarities and the types of donor ligand used (Campbell et al., 1997; Coutinho et al., 2003) (http://www.CAZY.org). Significant new structural information is emerging about the GT families, revealing that, in addition to the previously recognized distinct structural folds, GT-A and GT-B, there is a third GT-A-like fold family (Breton et al., 2006). GT-A proteins consist of a single α/β/α sandwich that resembles a Rossman-like fold and contains a divalent metal that is important for donor ligand binding. GT-B enzymes are composed of two Rossman-like domains that associate to form a catalytic cleft at their interface. Although there is cross talk between the domains, the C-terminal domain mainly interacts with the sugar donor, while the N-terminal domain mainly interacts with the acceptor. In contrast to GT-A fold containing GTs, the activities of GT-B enzymes are not dependent on divalent metals; however, these metals do enhance the activity of some GT-B enzymes (Hu and Walker, 2002; Miley et al., 2007; Morera et al., 2001). In spite of very low primary amino acid sequence conservation, the secondary and tertiary structures of all crystallized GT proteins show great similarity within their fold family (Hu et al., 2003; Unligil and Rini, 2000).
The catalytic mechanism among GT fold families is also highly conserved and is defined as either inverting or retaining, depending on whether the stereochemistry of the carbon atom of the sugar donor in the new glucosidic bond is inverted or retained. These distinctions define the four subgroups or “clans” of GTs (Clan I: GT-A inverting; Clan II: GT-B inverting; Clan III: GT-A retaining; Clan IV: GT-B retaining) (Coutinho et al., 2003). Human UGTs belong to the GT1 family and are members of Clan II.
At the beginning of 2009, there were 53 different GT crystal structures, representing 26 different CAZY families, in the three-dimensional glycosyltransferase database (http://www.cermav.cnrs.fr/glyco3d/). Twenty-six of the GTs with solved crystal structures adopt the GT-B fold and 10 of these belong to the GT1 family (Osmani et al., 2009). The structures of GT-B fold containing glycosyltransferases from several GT classes have been determined; these include phage T4 DNA glucosyltransferase (GT63) (Lariviere et al., 2003; Vrielink et al., 1994), MurG (GT28) (Ha et al., 2000; Hu et al., 2003), which is involved in peptidoglycan formation, and the Gtf family of enzymes (GT1), which are involved with vancomycin synthesis (Mulichak et al., 2001, 2003, 2004). Four recent structures of GT1 family plant flavonoid glucosyltransferases, MtUGT71G1 (Shao et al., 2005), VvGT1 (Offen et al., 2006), MtUGT85H2 (L. Li et al., 2007), and AtUGT72B1 have been reported (Brazier-Hicks et al., 2007). These crystal structures, which were generated in complex with different acceptor or donor substrates or analogs, have yielded important information on the composition of the donor and acceptor binding sites as well as enzyme–substrate interactions, including information about their catalytic mechanisms.
Identification of the donor binding site of human UGTs
Structural examination of UGTs has been limited by the difficulties associated with their overexpression, solubilization, and purification, making all efforts at crystallization unsuccessful until recently. Specifically, soluble, full length mammalian UGTs have yet to be generated despite our efforts and those of several other research groups (Meech and Mackenzie, 1998; Miley et al., 2007; Ouzzine et al., 1999b). Recently, however, the 1.8 Å resolution x-ray crystal structure of the C-terminal domain of human UGT2B7 was elucidated (Miley et al., 2007). The structural and functional data presented there provided novel information and clarified numerous aspects of human UGT donor–substrate binding and catalytic activity.
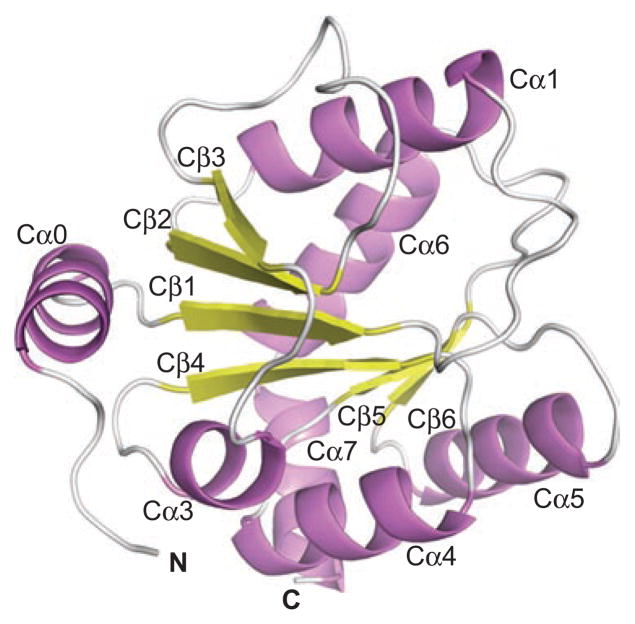
The crystallography of 2B7CT revealed it as a globular domain with a Rossman-type fold (Figure 1) meaning that at the core of the protein is a single parallel β-sheet consisting of six individual strands surrounded by seven α-helices (Miley et al., 2007). This discovery confirms that human UGTs belong to the GT-B fold family of glycosyltransferases.
Figure 1.
Ribbon cartoon of 2B7CT with labeled secondary structure elements. Reprinted from Miley et al. (2007 with permission from Elsevier.
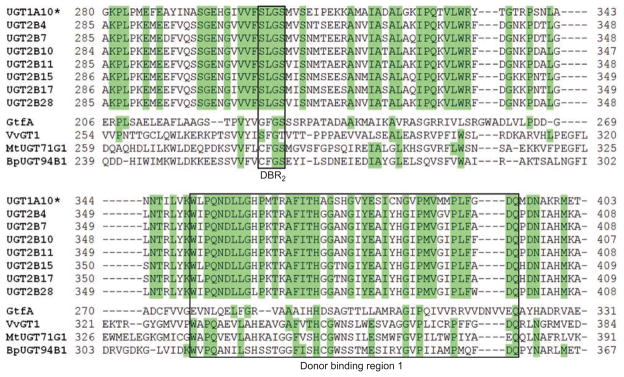
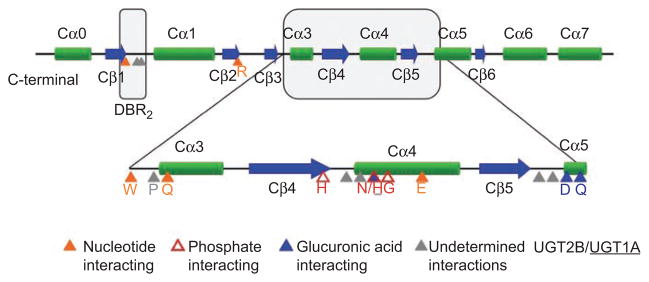
As we have anticipated for some time (Radominska-Pandya et al., 1999), the UDP-GlcUA binding site of UGT2B7 was found to be formed mainly by residues from the C-terminal domain (Miley et al., 2007). The majority of these interactions are with amino acid residues in the human Donor Binding Region 1 (DBR1) (Figures 2 and 3). Although the primary amino acid sequence of this region varies among human, plant, and bacterial GTs, the secondary and tertiary structures show great similarity. The residues predicted to interact with the sugar donor are also relatively conserved. An additional C-terminal motif, Ser-Lys-Gly-Ser (DBR2), is conserved in all human UGTs and was also found to interact with the sugar donor (Figures 2 and 3).
Figure 2.
Sequence alignment of selected GT1 family enzymes. UGT1A10 is shown as a representative member of the UGT1A family due to shared C-terminal domain sequences. The DBR1 and DBR2 are boxed. Green: Invariant in human UGTs.
Figure 3.
Predicted human UGT C-terminal domain secondary structures. Highly conserved residues of the C-terminal domain that are predicted to interact with UDP-GlcUA based on homology modeling are indicated. DBR1 and DBR2 are boxed in gray.
Specific amino acid interactions with UDP-GlcUA
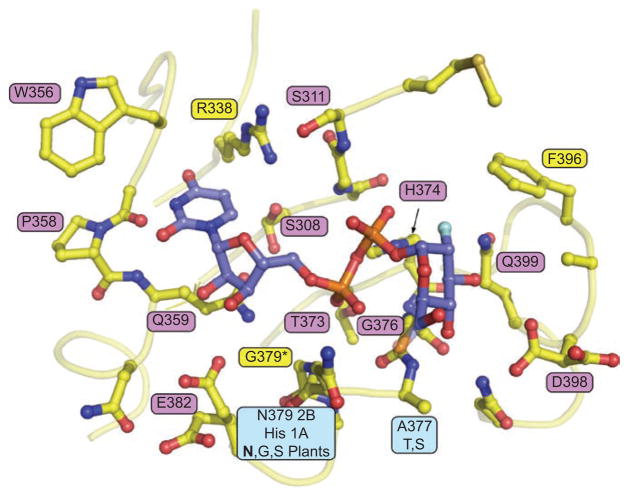
Although the crystal structure of UGT2B7CT was solved without the nucleotide-sugar donor, using homology modeling, a number of highly conserved residues of the C-terminal domain have been predicted to interact with UDP-GlcUA (Miley et al., 2007) (Figure 3 and Table 1).
Table 1.
Amino acids predicted to be important for 2B7UGT activity.
| Residue | Putative function | Mutation | Remaining activity (%)
|
|
|---|---|---|---|---|
| HDCA | TCC | |||
| Nucleotide interacting residues | ||||
| S308 | Structural scaffold (no direct interactions) | S308A | 83 | 70 |
| Q359 | Structural scaffold, “under” UDP | Q359A | 28 | 36 |
| E382 | Hydrogen bond with O2′ of ribose | E382A | 10 | 3 |
| W356 | Helps to position UDP-GlcUA | W356A | 20 | 43 |
| W356H | 27 | 60 | ||
| Phosphate interacting residues | ||||
| T373 | α-Phosphate via H2O | T373V | 1 | 5 |
| H374 | β-Phosphate | H374A | 3 | 7 |
| H374E | 10 | 6 | ||
| N378 | α-Phosphate | N378A | 25 | 10 |
| G379 | Forms a pocket under the α-phosphate | G379D | 18 | 5 |
| G379S | 90 | 134 | ||
| Glucuronic acid interacting residues | ||||
| D398 | O2′/O3′ of sugar | D398A | 10 | 5 |
| D398N | 2 | 11 | ||
| Q399 | O3′/O4′ of sugar | Q399A | 0 | 3 |
| Residues involved in catalysis | ||||
| H35 | Deprotonates acceptor facilitating nucleophilic attack at C1 of GlcUA | H35A | 0 | 4 |
| H35D | 0 | 3 | ||
| D151 | Stabilizes deprotonated His | D151A | 0 | 1 |
| D151N | 5 | 1 | ||
| Undetermined interactions | ||||
| S34 | Predicted to hydrogen bond to β-phosphate | S34A | 80 | 45 |
| R49* | Carboxyl function of GlcUA* | — | — | — |
| R259 | Carboxyl function of GlcUA | R259A | 3 | 1 |
| R338 | Ser in plant UGTs, interacts with nucleotide | R338S | 66 | 104 |
Predicted interactions and the effect of point mutations in UGT2B7 on activity toward hyodeoxycholic acid (HDCA) and tetrachlorocatechol (TCC) are shown. Activity data expressed as percent of wild-type activity.
Interactions inferred from literature not UGT2B7 homology model (Senay et al., 1997).
— No mutations done.
Using the 2B7CT-UDP-GlcUA homology model as a guide, the binding site was broken down into three groups: residues predicted to interact with the uracil base, the diphosphate group, or the glucuronic acid moiety (Table 1). The majority of these residues are located in DBR1. To probe the role that these predicted interacting residues played in 2B7 activity, a panel of point mutations at these positions was generated and their activity measured with structurally distinct substrates. HDCA is one of the most studied endogenous substrates for UGT2B7 and it has been demonstrated that the glucuronide is directed to the 6-hydroxyl group of the molecule. Tetrachlorocatechol (TCC) demonstrates one of the highest rates of glucuronidation by a variety of human UGTs and its structure is significantly different from HDCA. Differences in activity between substrates could be due to different binding orientations of acceptor molecules within the N-terminal region of the enzyme. All mutants were screened with a single concentration of each substrate. The kinetic parameters were not evaluated due to the lack of significant activity of the mutated proteins (Miley et al., 2007).
Mutation of the residues surrounding the uracil base had the least effect on enzyme activity of all the mutants analyzed, with E382A exhibiting the most pronounced alterations. Mutations at residues predicted to interact with the diphosphate moiety of UDP-GlcUA had more significant effects on enzyme function than those interacting with the nucleotide base. The conserved T373 and H374, which are positioned to interact with the α- and β-phosphates via hydrogen bonds, have been shown in plant flavonoid glucosyltransferases to be directly involved in the catalytic process and crucial for activity (Osmani et al., 2008). Mutation of these residues in human 2B7 to form T373V, H374A, and H374E virtually eliminated activity. Residues predicted to interact with the GlcUA moiety were also critical for 2B7 catalytic activity, with mutations at Q399 and D398 virtually abolishing activity. In conclusion, it appears that mutations of residues that are predicted to affect the positioning or chemistry of the bond being broken are the most important to 2B7 activity. Owing to the highly conserved nature of this domain in human UGTs, we predict that all other family members function in a similar manner.
Searching for residues that interact with the GlcUA moiety of the donor
The information from modeling studies conducted with the 2B7CT crystal structure was limited to the interactions with the sugar moiety. Although two amino acids from the DQxD motif (Xiong et al., 2008), D398 and Q399, were shown to interact with the O2′/O3′ and the O3′/O4′ of the sugar, respectively, there was no amino acid shown to directly interact with the carboxylic acid function that would account for the specificity of this enzyme for UDP-GlcUA. This clearly indicates that N-terminal domain residues are also involved in determining UDP-GlcUA specificity, since in GT-B enzymes the active site is within the cleft formed between the two domains.
There has been one successful crystal structure determination of a full length GT (1,3-glucuronyltansferase: GlcAT-I) in the presence of UDP-GlcUA (Pedersen et al., 2002). These studies were followed by homology modeling of another plant UDP-GlcUA-dependent enzyme, BpUGT94B1 (Osmani et al., 2009). Both of these studies identified the amino acid responsible for the high specificity of this isoform toward GlcUA as an Arg shown to directly interact with the carboxylic function of the sugar. The positively charged side chains of R156 and R25 for GlcAT-I and BpUGT94B1, respectively, are positioned toward the negatively charged carboxyl group of the moiety. These interactions are postulated to stabilize the binding of the negatively charged sugar residue in the active site and explain these enzymes’ specificity for UDP-GlcUA. Analyzing the information obtained from these studies and comparing it to our UGT2B7 data, we propose that an Arg located in the N-terminal end of UGT2B7 interacts with the negatively charged 6′-carboxylate of UDP-GlcUA. In UGT2B7, we have identified two potential residues (R259 and R49) that are conserved in all human UGTs that may fulfill the stabilizing function needed for GlcUA specificity in human UGTs. The residue in UGT1A6 (R52) corresponding to R49 in UGT2B7 was mutated by Senay et al. (1997) and the authors concluded that it was required for functional integrity.
Comparison between UGT1A and 2B family donor binding sites
It is recognized that the UDP-sugar binding sites in human UGTs and plant flavonoid glucosyltransferases are highly conserved (Miley et al., 2007). In fact, the majority of the amino acids involved in sugar binding are invariant in both groups (Figure 4).
Figure 4.
The UGT nucleotide-sugar binding site is highly conserved in humans and plants. 2-Deoxy-2-fluoroglucose from the plant glucosyltransferases VvGT1 co-crystal complex was modeled into the 2B7CT structure by superposition of the C-terminal domains. Pink: invariant in plants and humans; yellow: invariant in humans; cyan: conserved in humans.
There are, however, a few key differences in the UDP-GlcUA binding site among human UGT family members. Of the residues inferred to interact with UDP-GlcUA based on homology modeling, two vary among human UGTs (Figure 4). A377 (2B7 numbering) is a Thr in 2B15 and 2B17 and is a Ser in UGTs from the 1A family. In addition, N378 (2B7 numbering) is conserved in all members of the 2B family, but, in the 1A family, this residue is a His. Therefore, there might be differences between the tertiary structures and functions of the C-terminal domain of the UGT1A and UGT2B families as a whole. On the other hand, the DQxD motif is highly conserved in all human UGTs from both the families. To investigate the significance of this motif, we selected two representative isoforms from the 1A family (UGT1A6 and -1A10) in order to compare with UGT2B7.
The effects of donor site mutations on UGT1A6, -1A10, and -2B7 activity
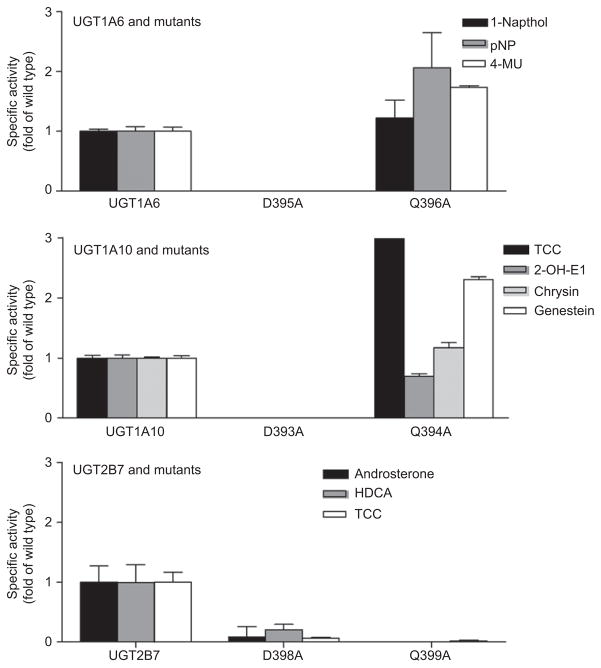
On the basis of the data obtained for UGT2B7, which showed the importance of D398 and Q399 to the activity of the enzyme, we used site-directed mutagenesis followed by activity assays to define the role of these amino acids in UGT1A6 and -1A10. The proteins were expressed as His-tag proteins in baculovirus-infected Sf9 cells and assayed for activity toward appropriate substrates.
Figure 5 compares the effect of Asp to Ala and Gln to Ala mutations in UGT1A6, -1A10, and 2B7 on activity toward isoform-specific substrates. The Asp to Ala mutations totally abolished catalytic activity in the two UGT1A isoforms and severely impaired the activity in UGT2B7, but the Gln to Ala mutation affected the two isoenzyme families differently. In UGT2B7, the Q399A mutation resulted in total loss of activity, whereas in UGT1A6 and -1A10, the mutant retained activity, although with significant variability depending on substrate.
Figure 5.
Comparison of the effects of DQ mutation on UGT1A10 and UGT2B7 activities. UGTs were expressed as His-tag proteins in baculovirus infected Sf9 insect cells and activity was measured using typical UGT1A6, -1A10, and -2B7 substrates.
In summary, these results suggest that UDP-GlcUA interacts with the first Asp of the DQxD motif and mutation of this amino acid results in the total loss of enzyme activity for the three UGTs analyzed. In addition, the prediction of this Asp having a critical role in UDP-GlcUA binding can be extrapolated to all UGTs, as evidenced by crystal structure data (Miley et al., 2007). However, the Gln, which is totally conserved in UGTs and a number of other GT enzymes, has a very different function that seems to vary between the UGT1A and –2B family isoforms. With all the substrates tested, the Q339A mutation in UGT2B7 totally inactivated the enzyme, whereas in UGT1A6 and -1A10 the activity was the same or higher than that of the wild-type enzyme, depending on the substrate used.
The information on the amino acids predicted to directly interact with the GlcUA moiety as revealed by homology modeling based on the partial crystal structure of UGT2B7 is relatively limited as compared to that for GlcAT-I (Pedersen et al., 2002). In that study, it was documented that the position of the GlcUA is determined through extensive interactions with several highly conserved residues. Each of the hydroxyl groups of the sugar (O2′, O3′, and O4′) form hydrogen bonds with a specific amino acid, and one of these also interacts with the substrate potentially stabilizing the interactions between the donor and the acceptor. Moreover, there is a specific Arg that interacts with the oxygen from the carboxylic acid group of GlcUA. If a similar pattern of interactions also applies to the binding of GlcUA by human UGTs, the contact amino acids that interact with each of the hydroxyl groups and the carboxyl function of the GlcUA moiety, which would define the sugar specificity for both UGT families, have yet to be identified.
The effects of metal ions on UGT1A6, -1A10, and -2B7 activity
GT-B superfamily glycosyltransferase activity is divalent metal ion independent; however, some GT-B enzymes achieve optimal activity in the presence of divalent metal ions (Hu and Walker, 2002; Morera et al., 2001). It is clear from the structural data thus far accumulated that a divalent metal is not involved in nucleotide-sugar binding, as it is in GT-A superfamily glycosyltransferases. To evaluate the role of metal ions in human UGT activity, UGT1A6, -1A10, and -2B7 activities were assayed in the presence of Mg and compared to identical experiments where excess EDTA was included to chelate the metal. Significant differences were observed between UGT2B7 and the 1A isoforms. UGT2B7 activity experiments with and without EDTA did not show any gain or loss of activity, suggesting divalent metals do not play a role with the acceptor substrates tested. However, UGT1A6, and -1A10 show doubled activity in the presence of Mg (Figure 6).
Figure 6.
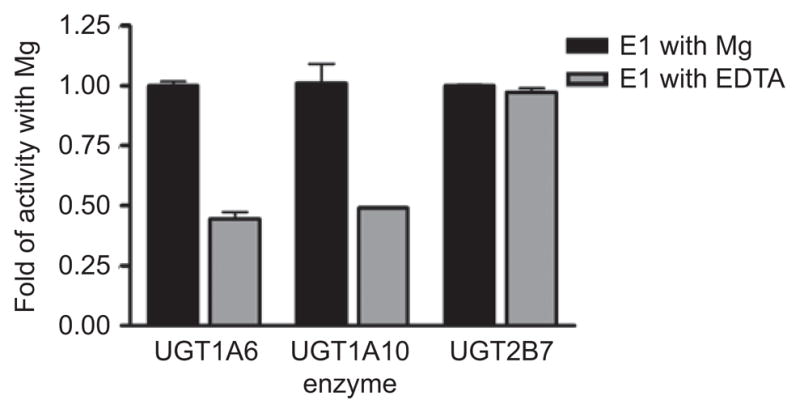
Comparison of the activity of UGT1A6, -1A10, and -2B7 toward estrone (E1) in the presence and absence of metal.
At this moment, the differences in response to divalent metals in these enzymes are difficult to explain. We can only speculate based on comparisons to information available for other GT enzymes. It has been suggested that these metals could be playing a role in product release (Lariviere et al., 2003). Alternatively, it is postulated that for some members of the GT-B family enhancement of activity by divalent metals occurs via interactions with acceptor substrates and their subsequent N-terminal domain binding sites. Both phage T4 DNA β-glucosyltransferase and MurG are activated by divalent metal ions as we see for UGT1A6 and -1A10. However, structural data describing any kind of acceptor–substrate complexes in these enzymes is, unfortunately, still lacking.
Comparative homology modeling of UDP-GlcUA binding sites in UGT2B7 and UGT1A10
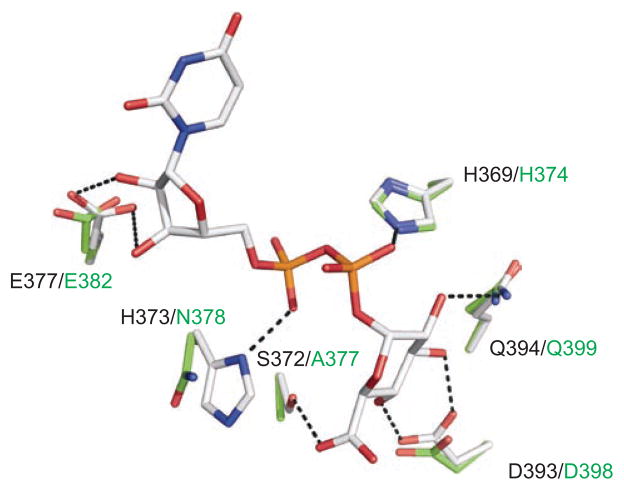
Homology modeling was also applied to further characterize the apparent differences between the UDP-GlcUA interacting residues of UGT1A10 and UGT2B7. Using the crystal structure of UGT2B7CT as a template (Miley et al., 2007), a model of the UDP-GlcUA binding site of UGT1A10 was generated (Figure 7) (Xiong et al., 2006). This model illustrates that their UDP-GlcUA binding sites are very similar. There are, however, two notable exceptions, residues S372 and H373 (UGT1A10 numbering). These two residues are variable in human UGTs (Figures 2 and 4), and are predicted by our homology model to hydrogen bond to the UDP-GlcUA. These differences may help to explain why mutations in the DQ motif in UGT1A10 and -2B7 yield different results despite the total conservation of these residues in all human UGTs. Hydrogen bonding between S372 and the sugar moiety of UDP-GlcUA in UGT1A10 is not possible in UGT2B7, potentially explaining why the Q394A mutant of UGT1A10 retains some level of activity compared to the same mutation in UGT2B7 that resulted in abolished activity.
Figure 7.
A ball-and-stick model representation of the UDP-GlcUA binding site in UGT1A10 (carbon, gray) and UGT2B7 (carbon, green). The bound co-substrate and the location of key residues within hydrogen bonding distance in either UGT1A10 or UGT2B7. In both models: nitrogen, blue; oxygen, red; phosphate, orange. Dashed black lines represent possible hydrogen bonds. The residue types and numbering are indicated for both UGT1A10 (black) and UGT2B7 (green). Model produced in PyMOL (http://www.pymol.org). Reprinted from Xiong et al. (2008) with permission from ASPET.
Catalytic mechanism of human UGTs
Using the information obtained from the analysis of the crystal structure of UGT2B7CT and that from two separate groups, which published crystal structures of plant glucosyltransferases (Offen et al., 2006; Shao et al., 2005), a catalytic mechanism for human UGTs was proposed (Miley et al., 2007). Analysis of the UGT2B7 catalytic center revealed that UGT2B7 has residues analogous to plant glucosyltransferases at key catalytic positions (H35, D151) in the N-terminal domain that are conserved in the majority in human UGTs. This model predicts that human UGTs use a serine hydrolase-like catalytic triad for catalysis.
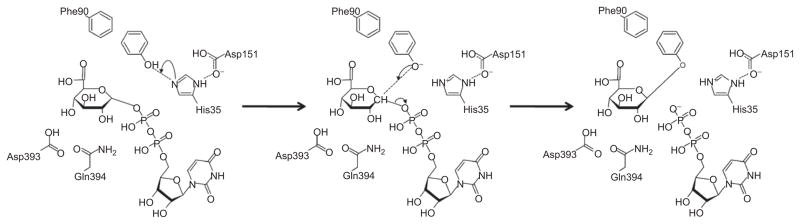
Combining the information from the crystal structure analysis of UGT2B7 and data generated for UGT1A10 using photoaffinity labeling, followed by proteolytic digestion, MALDI and LCMS/MS analysis, and site-directed mutagenesis, we were able to expand this model and include the F90 residue, which has been shown to be crucial for interaction with specific phenolic substrates in UGT1A10 (Xiong et al., 2006) (Figure 8).
Figure 8.
Schematic representation of proposed catalytic mechanism for human UGT1A10. Phenol is shown as example substrate.
In the mechanism presented here, H35 deprotonates the lipophilic substrate, phenol, which is bound in the active site though ring-stacking interactions with F90. This deprotonated substrate then serves as a nucleophilic acceptor, facilitating the nucleophilic attack at the C1 atom of GlcUA. The resulting protonated His is stabilized by neighboring D151.
To validate the significance of these two catalytic residues, H35 and D151, in UGT2B7, corresponding mutations to either Ala and Asp or Ala and Asn, respectively, were generated (Miley et al., 2007). These mutations resulted in a catalytically inactive enzyme (Table 1). Similarly mutations have been generated in UGTs from the 1A family (D. Li et al., 2007). When each of the corresponding His and Asp residues for UGT1A1 and -1A6 were mutated to Ala, all activities were abolished. This is similar to what was seen for the corresponding mutations in plant glucosyltransferases (Offen et al., 2006; Shao et al., 2005).
It is well established that a second-order nucleophilic substitution mechanism is the mode of action for the majority of glucuronidation reactions. Specifically, proton abstraction is considered an obligatory step in the glucuronidation of all aliphatic alcohols, phenols, primary and secondary amines, acidic carbon atoms, and thiols. Interestingly, the formation of quaternary ammonium glucuronides does not require the proton extraction essential for this mechanism (Kerdpin et al., 2009). Recent studies on the influence of N-terminal domain His and Pro residues on the substrate selectivity of human UGT1A1, -1A4, -1A6, -1A9, -2B7, and -2B10 identified the pivotal role of an N-terminal domain Pro in the glucuronidation of the tertiary amines by UGT1A subfamily proteins (Kerdpin et al., 2009; Kubota et al., 2007). This novel discovery sheds light on alternative catalytic mechanisms in some human UGTs, and provides a possible explanation as to why the “catalytic His” located in the N-terminal end is not present in UGT1A4 or -2B10.
Conclusions
Drug metabolizing enzymes catalyze a variety of metabolic reactions such as oxidation, reduction, and conjugation toward endogenous compounds and xenobiotics. As a member of this important group, the function of UGTs in humans is the conjugation of these compounds with UDP-GlcUA. The conversion of UGT substrates to glucuronide products and the competitive inhibition of this reaction by other compounds take place in the active site of the enzyme. It is recognized that in order to (1) develop a detailed picture of UGT substrate specificity, (2) evaluate what characteristics make a compound a suitable UGT substrate, and (3) identify the molecular mechanism of glucuronidation, a high-resolution structure of the active site will be required.
In the past, this lack of specific information has resulted in the identification of crucial binding and catalytic amino acids using selective inhibitors, amino acid–specific chemical modification reagents, amino acid alignments, and site-directed mutagenesis. More recently, homology modeling is remerging as a tool for the prediction of interactions between amino acids and substrates in the binding site. As highlighted in this review, the information gathered from the analysis of the UGT2B7CT structure provided important and novel information related to the UDP-GlcUA binding site and UGT catalysis.
Our work unambiguously confirmed that UGTs exhibit a Rossman-type folded domain that is consistent with the members of the GT-B fold family. This superfamily is characterized by a lack of dependence on a divalent metal for activity, and this also seems to be true for UGTs. The superimposition of the UGT2B7 crystal structure on to the crystal structures of plant and bacterial GTs revealed remarkable similarities in secondary and tertiary structures as well as the positioning and binding of the UDP-sugar. These models also revealed that sugar binding depends on an extended network of residues, which interact with different elements of the nucleosugars and are predominantly invariant among the compared GTs. However, two important residues were found to be unique for human UGTs. Our mutagenesis experiments showed that for some of these residues single point mutations could abolish binding/catalysis. In addition, although the majority of UDP-GlcUA binding residues are localized in the C-terminal end, it is clear that the N-terminal domain is also involved in sugar binding.
Our homology modeling also helped explain the promiscuous character of UGTs. We established that the remarkable number and variety of substrates conjugated by single UGT isoforms is apparently made possible by a serine hydrolase-like catalytic triad. This mechanism is flexible enough to accommodate compounds with different functional groups, which is the hallmark of human UGTs. With the exception of some endogenous substrates, typical Km values for the majority of human glucuronidation reactions are in the micromolar range. Rigorous identification of the serine hydrolase-like catalytic mechanism provides an explanation for the low affinity of UGT catalyzed reactions. This knowledge is already advancing structure–function relationship studies of other UGT isoforms as is evident from the number of recent papers using our discoveries as the basis for their research in the areas of homology modeling and site-directed mutagenesis to better understand UGT catalysis and substrate binding.
One of the biggest challenges facing structure–function studies of UGTs today is the unambiguous identification of contact residues for acceptor molecules in the substrate binding site. As the full length crystal structure has yet to be determined for any mammalian UGT isoform, we currently must rely on non-structural UGT data as well as structural information from other GT enzymes to help us understand its molecular details. Despite the decades of UGT study very little specific information has been gathered about this binding site. Unfortunately, gaining insight into the molecular basis of human UGT acceptor substrate binding and specificity from other GT crystal structures has also not been as informative as we would like. One of the major reasons for this is that the acceptor substrate binding domains (N-terminal) in GT-B enzymes appear to be highly specialized, so direct comparison among them is of limited utility. Human UGTs are especially unique in that they are ER localized and their N-terminal domains have been shown to physically interact with the ER membrane (Ciotti et al., 1998; Ouzzine et al., 1999b). Thus, while there are considerable structural data available for related GT-B enzymes, unfortunately we have been able to apply little to human UGT acceptor binding sites to date. It is becoming clear that only a full length crystal structure of a mammalian UGT will be able to provide information about the acceptor substrate binding site and its interactions with the UDP-GlcUA binding domain, illuminating for the first time the active site of these very important and unique enzymes.
Acknowledgments
The authors would like to acknowledge the scientific efforts of Agnieszka K. Zielinska, Yan Xiong, Jeffrey E. Keenan, Anne-Sisko Patana, and Moshe Finel in the generation of the data for this review. Moreover, they would like to thank Joanna Little for the final editing of this review.
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Bock KW. Metabolic polymorphisms affecting activation of toxic and mutagenic arylamines. Trends Pharmacol Sci. 1992;13(6):223–226. doi: 10.1016/0165-6147(92)90068-h. [DOI] [PubMed] [Google Scholar]
- Brazier-Hicks M, Offen WA, Gershater MC, Revett TJ, Lim EK, Bowles DJ, et al. Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc Natl Acad Sci U S A. 2007;104(51):20238–20243. doi: 10.1073/pnas.0706421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton C, Snajdrova L, Jeanneau C, Koca J, Imberty A. Structures and mechanisms of glycosyltransferases. Glycobiology. 2006;16(2):29R–37R. doi: 10.1093/glycob/cwj016. [DOI] [PubMed] [Google Scholar]
- Campbell JA, Davies GJ, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326(Pt 3):929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciotti M, Cho JW, George J, Owens IS. Required buried alpha-helical structure in the bilirubin UDP-glucuronosyltransferase, UGT1A1, contains a nonreplaceable phenylalanine. Biochemistry. 1998;37(31):11018–11025. doi: 10.1021/bi980747q. [DOI] [PubMed] [Google Scholar]
- Coffman BL, Kearney WR, Green MD, Lowery RG, Tephly TR. Analysis of opioid binding to UDP-glucuronosyltransferase 2B7 fusion proteins using nuclear magnetic resonance spectroscopy. Mol Pharmacol. 2001;59(6):1464–1469. doi: 10.1124/mol.59.6.1464. [DOI] [PubMed] [Google Scholar]
- Coffman BL, Kearney WR, Goldsmith S, Knosp BM, Tephly TR. Opioids bind to the amino acids 84 to 118 of UDP-glucuronosyltransferase UGT2B7. Mol Pharmacol. 2003;63(2):283–288. doi: 10.1124/mol.63.2.283. [DOI] [PubMed] [Google Scholar]
- Coutinho P, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- Ha S, Walker D, Shi Y, Walker S. The 1.9 A crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis. Protein Sci. 2000;9(6):1045–1052. doi: 10.1110/ps.9.6.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Walker S. Remarkable structural similarities between diverse glycosyltransferases. Chem Biol. 2002;9(12):1287–1296. doi: 10.1016/s1074-5521(02)00295-8. [DOI] [PubMed] [Google Scholar]
- Hu Y, Chen L, Ha S, Gross B, Falcone B, Walker D, et al. Crystal structure of the MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases. Proc Natl Acad Sci U S A. 2003;100(3):845–849. doi: 10.1073/pnas.0235749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyanagi T, Haniu M, Sogawa K, Fujii-Kuriyama Y, Watanabe S, Shively JE, et al. Cloning and characterization of cDNA encoding 3-methylcholanthrene inducible rat mRNA for UDP-glucuronosyltransferase. J Biol Chem. 1986;261:15607–15614. [PubMed] [Google Scholar]
- Kerdpin O, Mackenzie PI, Bowalgaha K, Finel M, Miners JO. Influence of N-terminal domain histidine and proline residues on the substrate selectivities of human UDP-glucuronosyltransferase (UGT) 1A1, 1A6, 1A9, 2B7 and 2B10. Drug Metab Dispos. 2009;37(9):1948–1955. doi: 10.1124/dmd.109.028225. [DOI] [PubMed] [Google Scholar]
- Kubota T, Lewis BC, Elliot DJ, Mackenzie PI, Miners JO. Critical roles of residues 36 and 40 in the phenol and tertiary amine aglycone substrate selectivities of UDP-glucuronosyltransferases 1A3 and 1A4. Mol Pharmacol. 2007;72(4):1054–1062. doi: 10.1124/mol.107.037952. [DOI] [PubMed] [Google Scholar]
- Lariviere L, Gueguen-Chaignon V, Morera S. Crystal structures of the T4 phage beta-glucosyltransferase and the D100A mutant in complex with UDP-glucose: glucose binding and identification of the catalytic base for a direct displacement mechanism. J Mol Biol. 2003;330(5):1077–1086. doi: 10.1016/s0022-2836(03)00635-1. [DOI] [PubMed] [Google Scholar]
- Li D, Fournel-Gigleux S, Barre L, Mulliert G, Netter P, Magdalou J, et al. Identification of aspartic acid and histidine residues mediating the reaction mechanism and the substrate specificity of the human UDP-glucuronosyltransferases 1A. J Biol Chem. 2007;282(50):36514–36524. doi: 10.1074/jbc.M703107200. [DOI] [PubMed] [Google Scholar]
- Li L, Modolo LV, Escamilla-Trevino LL, Achnine L, Dixon RA, Wang X. Crystal structure of Medicago truncatula UGT85H2—insights into the structural basis of a multifunctional (iso)flavonoid glycosyltransferase. J Mol Biol. 2007;370(5):951–963. doi: 10.1016/j.jmb.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Locuson CW, Tracy TS. Comparative modelling of the human UDP-glucuronosyltransferases: insights into structure and mechanism. Xenobiotica. 2007;37(2):155–168. doi: 10.1080/00498250601129109. [DOI] [PubMed] [Google Scholar]
- Mackenzie PI. Rat liver UDP-glucuronosyltransferase. Sequence and expression of a cDNA encoding a phenobarbital-inducible form. J Biol Chem. 1986;261:6119–6125. [PubMed] [Google Scholar]
- Mackenzie PI. Rat liver UDP-glucuronosyltransferase. Identification of cDNAs encoding two enzymes which glucuronidate testosterone, dihydrotestosterone, and β-estradiol. J Biol Chem. 1987;262:9744–9749. [PubMed] [Google Scholar]
- Mackenzie PI, Owens IS. Cleavage of nascent UDP glucuronosyltransferase from rat liver by dog pancreatic microsomes. Biochem Biophys Res Commun. 1984;122(3):1441–1449. doi: 10.1016/0006-291x(84)91252-x. [DOI] [PubMed] [Google Scholar]
- Mackenzie PI, Gonzalez FJ, Owens IS. Cell-free translation of mouse liver mRNA coding for two forms of UDP glucuronosyltransferase. Arch Biochem Biophys. 1984;230(2):676–680. doi: 10.1016/0003-9861(84)90449-1. [DOI] [PubMed] [Google Scholar]
- Meech R, Mackenzie PI. Structure and function of uridine diphosphate glucuronosyltransferases. Clin Exp Pharmacol Physiol. 1997;24(12):907–915. doi: 10.1111/j.1440-1681.1997.tb02718.x. [DOI] [PubMed] [Google Scholar]
- Meech R, Mackenzie PI. Determinants of UDP glucuronosyltransferase membrane association and residency in the endoplasmic reticulum. Arch Biochem Biophys. 1998;356(1):77–85. doi: 10.1006/abbi.1998.0750. [DOI] [PubMed] [Google Scholar]
- Meech R, Yogalingam G, Mackenzie P. Mutational analysis of the carboxy-terminal region of UDP-glucuronosyltransferase 2B1. DNA Cell Biol. 1996;15(6):489–494. doi: 10.1089/dna.1996.15.489. [DOI] [PubMed] [Google Scholar]
- Miley MJ, Zielinska AK, Keenan JE, Bratton SM, Radominska-Pandya A, Redinbo MR. Crystal structure of the cofactor-binding domain of the human phase II drug-metabolism enzyme UDP-glucuronosyltransferase 2B7. J Mol Biol. 2007;369(2):498–511. doi: 10.1016/j.jmb.2007.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morera S, Lariviere L, Kurzeck J, Aschke-Sonnenborn U, Freemont PS, Janin J, et al. High resolution crystal structures of T4 phage beta-glucosyltransferase: induced fit and effect of substrate and metal binding. J Mol Biol. 2001;311(3):569–577. doi: 10.1006/jmbi.2001.4905. [DOI] [PubMed] [Google Scholar]
- Mulichak AM, Losey HC, Walsh CT, Garavito RM. Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics. Structure. 2001;9(7):547–557. doi: 10.1016/s0969-2126(01)00616-5. [DOI] [PubMed] [Google Scholar]
- Mulichak AM, Losey HC, Lu W, Wawrzak Z, Walsh CT, Garavito RM. Structure of the TDP- epi-vancosaminyltransferase GtfA from the chloroeremomycin biosynthetic pathway. Proc Natl Acad Sci U S A. 2003;100(16):9238–9243. doi: 10.1073/pnas.1233577100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulichak AM, Lu W, Losey HC, Walsh CT, Garavito RM. Crystal structure of vancosaminyltransferase GtfD from the vancomycin biosynthetic pathway: interactions with acceptor and nucleotide ligands. Biochemistry. 2004;43(18):5170–5180. doi: 10.1021/bi036130c. [DOI] [PubMed] [Google Scholar]
- Oelberg DG, Chari MV, Little JM, Adcock EW, Lester R. Lithocholate glucuronide is a cholestatic agent. J Clin Invest. 1984;73(6):1507–1514. doi: 10.1172/JCI111356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, et al. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 2006;25(6):1396–1405. doi: 10.1038/sj.emboj.7600970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani SA, Bak S, Imberty A, Olsen CE, Moller BL. Catalytic key amino acids and UDP-sugar donor specificity of a plant glucuronosyltransferase, UGT94B1: molecular modeling substantiated by site-specific mutagenesis and biochemical analyses. Plant Physiol. 2008;148(3):1295–1308. doi: 10.1104/pp.108.128256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani SA, Bak S, Moller BL. Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry. 2009;70(3):325–347. doi: 10.1016/j.phytochem.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Ouzzine M, Magdalou J, Burchell B, Fournel-Gigleux S. Expression of a functionally active human hepatic UDP-glucuronosyltransferase (UGT1A6) lacking the N-terminal signal sequence in the endoplasmic reticulum. FEBS Lett. 1999a;454(3):187–191. doi: 10.1016/s0014-5793(99)00797-8. [DOI] [PubMed] [Google Scholar]
- Ouzzine M, Magdalou J, Burchell B, Fournel-Gigleux S. An internal signal sequence mediates the targeting and retention of the human UDP-glucuronosyltransferase 1A6 to the endoplasmic reticulum. J Biol Chem. 1999b;274(44):31401–31409. doi: 10.1074/jbc.274.44.31401. [DOI] [PubMed] [Google Scholar]
- Pedersen LC, Darden TA, Negishi M. Crystal structure of beta 1,3-glucuronyltransferase I in complex with active donor substrate UDP-GlcUA. J Biol Chem. 2002;277(24):21869–21873. doi: 10.1074/jbc.M112343200. [DOI] [PubMed] [Google Scholar]
- Radominska-Pandya A, Czernik PJ, Little JM, Battaglia E, Mackenzie PI. Structural and functional studies of UDP-glucuronosyltransferases. Drug Metab Rev. 1999;31(4):817–899. doi: 10.1081/dmr-100101944. [DOI] [PubMed] [Google Scholar]
- Senay C, Ouzzine M, Battaglia E, Pless D, Cano V, Burchell B, et al. Arginine 52 and histidine 54 located in a conserved amino-terminal hydrophobic region (LX2-R52-G-H54-X3-V-L) are important amino acids for the functional and structural integrity of the human liver UDP-glucuronosyltransferase UGT1*6. Mol Pharmacol. 1997;51(3):406–413. [PubMed] [Google Scholar]
- Shao H, He X, Achnine L, Blount JW, Dixon RA, Wang X. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell. 2005;17(11):3141–3154. doi: 10.1105/tpc.105.035055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd SR, Baird SJ, Hallinan T, Burchell B. An investigation of the transverse topology of bilirubin UDP-glucuronosyltransferase in rat hepatic endoplasmic reticulum. Biochem J. 1989;259(2):617–620. doi: 10.1042/bj2590617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unligil UM, Rini JM. Glycosyltransferase structure and mechanism. Curr Opinion Struct Biol. 2000;10:510–517. doi: 10.1016/s0959-440x(00)00124-x. [DOI] [PubMed] [Google Scholar]
- Vanstapel F, Blanckaert N. Topology and regulation of bilirubin UDP-glucuronosyltransferase in sealed native microsomes from rat liver. Arch Biochem Biophys. 1988;263:216–225. doi: 10.1016/0003-9861(88)90630-3. [DOI] [PubMed] [Google Scholar]
- Vore M, Montgomery C, Meyers M. Steroid D-ring glucuronides: characterization of a new class of cholestatic agents. Drug Metab Rev. 1983;14(5):1005–1019. doi: 10.3109/03602538308991419. [DOI] [PubMed] [Google Scholar]
- Vrielink A, Ruger W, Driessen HP, Freemont PS. Crystal structure of the DNA modifying enzyme beta- glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose. EMBO J. 1994;13(15):3413–3422. doi: 10.1002/j.1460-2075.1994.tb06646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Bernardi D, Bratton S, Ward MD, Battaglia E, Finel M, et al. Phenylalanine 90 and 93 are localized within the phenol binding site of human UDP-glucuronosyltransferase 1A10 as determined by photoaffinity labeling, mass spectrometry, and site-directed mutagenesis. Biochemistry. 2006;45(7):2322–2332. doi: 10.1021/bi0519001. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Patana AS, Miley MJ, Zielinska AK, Bratton SM, Miller GP, et al. The first aspartic acid of the DQxD motif for human UDP-glucuronosyltransferase 1A10 interacts with UDP-glucuronic acid during catalysis. Drug Metab Dispos. 2008;36(3):517–522. doi: 10.1124/dmd.107.016469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota H, Yuasa A, Sato R. Topological disposition of UDP-glucuronyltransferase in rat liver microsomes. J Biochem (Tokyo) 1992;112(2):192–196. doi: 10.1093/oxfordjournals.jbchem.a123876. [DOI] [PubMed] [Google Scholar]