Abstract
CENP-A is a histone H3-like protein specific to centromeres that is essential for kinetochore formation and accurate chromosome segregation in eukaryotes. Recent studies (Dunleavy et al., 2009; Foltz et al., 2009; Perpelescu et al., 2009; Pidoux et al., 2009; Williams et al., 2009) analyze CENP-A binding proteins required for the recruitment of CENP-A to centromeres in humans and in fission yeast, bringing us closer to understanding how centromere identity is faithfully propagated.
Genome stability in cells and organisms relies on the accurate partitioning of chromosomes into daughter cells during division. Centromeres are the chromatin regions associated with kinetochores, which mediate chromosome segregation and the mitotic checkpoint. Surprisingly, centromeres are formed and propagated through cell division by epigenetic mechanisms in most eukaryotes (Allshire and Karpen, 2008). Centromeric chromatin contains the centromere-specific histone H3-like protein CENP-A (also known as CenH3), which replaces histone H3 in centromeric nucleosomes. CENP-A is required for the recruitment of nearly all known components of the centromere and kinetochore, placing it at or near the top of the hierarchy for centromere assembly. When CENP-A is absent, kinetochore assembly and chromosome segregation fail entirely. Thus, determining how the “ring” CENP-A is exclusively and faithfully delivered to centromeric chromatin by a fellowship of proteins is key to understanding centromere identity and propagation. The findings reported in this issue of Cell (Dunleavy et al., 2009; Foltz et al., 2009) and recently in Molecular Cell (Pidoux et al., 2009; Williams et al., 2009) and the Journal of Cell Biology (Perpelescu et al., 2009) identify new CENP-A binding partners that act as “ring bearers” that bring CENP-A to centromeres for assembly.
The CENP-A Fellowship
The segregation of existing nucleosomes to sister chromatids during DNA replication creates gaps that must be filled by assembly of new nucleosomes. In contrast to the rapid addition of canonical histone H3.1/H4 dimers (followed by histone H2A/H2B dimers) to newly replicated DNA during the S phase of the cell cycle, histone variants such as histone H3.3 are assembled into chromatin by replication-independent mechanisms (Groth et al., 2007). Although centromeric DNA is replicated during S phase, CENP-A replenishment occurs independently of replication and displays unusual cell cycle dynamics (Allshire and Karpen, 2008). CENP-A is recruited to centromeric DNA in anaphase in early fly embryos, during late telophase through G1 phase in human cells, and shortly after mitosis in both S and G2 phases in the fission yeast Schizosaccharomyces pombe. Thus, CENP-A replenishment coincides with the timing of kinetochore formation and chromosome segregation and must be coordinated with mitotic progression.
Although a connection between cell cycle regulators and centromere formation has been discovered in Drosophila (Erhardt et al., 2008), it has yet to be established in human and S. pombe cells. Interestingly, components of the human and S. pombe Mis18 complex, which are required for CENP-A assembly, are transiently depleted from centromeres during mitosis. In human cells, the Mis18 complex reassociates with centromeres during late telophase, coinciding with the timing of new CENP-A recruitment (Fujita et al., 2007; Jansen et al., 2007). S. pombe Mis18 reassociates with centromeres after mitosis, and persists throughout S and G2 when CENP-A (Cnp1 or CENP-ACnp1) is recruited. It has been suggested that the Mis18 complex may “prime” the centromere for the loading of new CENP-A, a process that may involve acetylation of centromeric chromatin (Fujita et al., 2007). However, centromeric chromatin is hypoacetylated in S. pombe (Hayashi et al., 2004). Thus, the molecular functions of the Mis18 complex need further investigation.
Central to understanding centromere deposition is knowing which “cis” elements within CENP-A and which “trans” factors are required for new CENP-A assembly after replication. A key feature is the CENP-A targeting domain (CATD) that lies at the interface between CENP-A and histone H4 (Black et al., 2007). In human cells, replacing the equivalent region in H3 with the CATD is sufficient to target chimeric H3CATD to the centromere, and to sustain kinetochore assembly and chromosome segregation (Black et al., 2007). Several trans-acting proteins that are required for targeting CENP-A nucleosomes to centromeres have been identified in organisms that have “regional” epigenetically regulated centromeres (Allshire and Karpen, 2008). These factors include the Mis16, Mis18, Mis6, and Sim3 proteins in fission yeast (Dunleavy et al., 2007; Hayashi et al., 2004) and the Mis18 complex, RbAp46/48, and CENP-H/I complex in humans (Foltz et al., 2006; Fujita et al., 2007). However, little is known about the molecular roles of these proteins in CENP-A deposition and whether they directly bind to CENP-A. For example, although Mis16 and Mis18 interact with each other and are reciprocally required for centromere localization (Hayashi et al., 2004), a direct interaction between Mis16, Mis18, and CENP-A has not been established. Furthermore, it is unclear whether these proteins directly regulate centromere assembly, as opposed to other steps in the pathway (Allshire and Karpen, 2008). Thus, the factors directly responsible for CENP-A assembly at “regional” centromeres has remained elusive.
SpScm3, the Fellow Linking CENP-A and Mis18?
In Saccharomyces cerevisiae, Scm3 (ScScm3) is required for centromeric localization of CENP-A (Cse4 or CENP-ACse4). ScScm3 mediates the release of histone H2A/H2B from preformed CENP-ACse4 nucleosomes and forms a stable 1:1:1 hexameric complex with CENP-A/H4 in vitro, and H2A/H2B are depleted at centromeres in vivo (Zhang et al., 2007). These results suggested that ScScm3 participates in the formation of a unique type of CENP-ACse4 nucleosome that might exist in other organisms with Scm3 homologs.
Two recent studies in Molecular Cell by Pidoux et al. (2009) and Williams et al. (2009) suggest that the S. pombe homolog of ScScm3 (SpScm3) may be the “missing link” between Mis16/Mis18 and CENP-ACnp1. SpScm3 is essential for the localization of endogenous and newly synthesized CENP-ACnp1. Most importantly, SpScm3 coimmunoprecipitates with Mis16, Mis18, and CENP-ACnp1 (Pidoux et al., 2009; Williams et al., 2009) and interacts directly with CENP-ACnp1, Mis18, and itself in vitro (Pidoux et al., 2009). However, SpScm3 is unlikely to be an integral part of CENP-ACnp1 nucleosomes. First, SpScm3 displays identical localization dynamics as Mis16 and Mis18 and leaves the centromere during mitosis, whereas CENP-ACnp1 is always present at the centromere (Pidoux et al., 2009; Williams et al., 2009). Second, SpScm3 is more tightly bound to chromatin than CENP-ACnp1 (Pidoux et al., 2009). Chromatin immunoprecipitation (ChIP) experiments do show that histone H2B levels are diminished across the S. pombe centromere central core, but this could result from inefficient crosslinking during ChIP or dynamic association of H2B.
SpScm3 localization to centromeric chromatin does not require CENP-ACnp1, but it does require Sim4, Mis6, Mis16, and Mis18, placing it downstream of these factors in the CENP-ACnp1 loading pathway (Pidoux et al., 2009; Williams et al., 2009). These studies suggest that SpScm3 is recruited to centromeric chromatin by Sim4, Mis6, Mis16, and Mis18, and that it in turn recruits CENP-ACnp1 to centromeres (Figure 1A). These exciting results raise new questions about the molecular function of SpScm3 and the mechanism for propagating centromere identity in S. pombe. SpScm3 may act as a “receptor” that captures or stabilizes CENP-ACnp1 molecules delivered to centromeres by the general histone chaperone Sim3/NASP (Dunleavy et al., 2007; Pidoux et al., 2009) (Figure 1A). SpScm3 may also be directly involved in CENP-ACnp1 nucleosome assembly or may recruit another specific or common chromatin assembly factor. It is also unclear which proteins recruit the Mis18 complex to centromeres and whether Mis18 targeting is sufficient to drive CENP-ACnp1 assembly. Future studies should determine whether SpScm3 is sufficient to recruit CENP-ACnp1 to centromeric chromatin and whether the interactions between SpScm3, Sim3/NASP, and CENP-ACnp1 are mediated by the CATD.
Figure 1. CENP-A Recruitment to S. pombe and Human Centromeres.
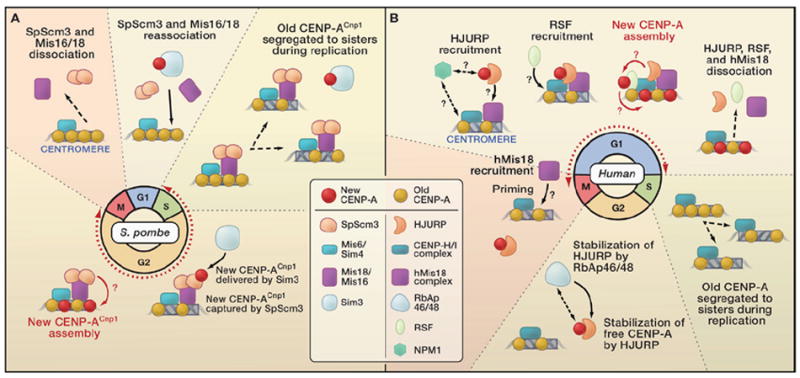
(A) In Schizosaccharomyces pombe, centromeric DNA is replicated and existing CENP-ACnp1 is diluted by nucleosome segregation to sister chromatids during S phase. Recruitment of new CENP-ACnp1 occurs during both S and G2 phases (red dotted line/double arrowheads). The Sim3/NASP histone chaperone interacts with free CENP-ACnp1 and delivers it to the centromere, where it is received by SpScm3 and assembled into nucleosomes by unknown factors and mechanisms. Nucleosome gaps could be filled or H3 nucleosomes could be replaced. SpScm3 is shown as a dimer interacting directly with Mis18. SpScm3 recruitment at centromeres requires the Sim4/Mis6 and Mis16/Mis18 complexes. Mis16/Mis18 and SpScm3 are removed from centromeres during mitosis and reassociate starting in late anaphase. (B) In humans, centromere replication during S phase creates CENP-A nucleosome “gaps” (with or without H3 nucleosome deposition) that are replenished only when new CENP-A assembly occurs in the following late mitosis through G1 phases (red dotted line/double arrowheads). Interactions between chromatin-free CENP-A and HJURP, which ensures the stability of free CENP-A, occur throughout the cell cycle but levels of both proteins increase closer to mitosis. Stabilization of HJURP is mediated by RbAp46/48 through an unknown mechanism. In telophase, the human Mis18 complex is recruited to centromeres, followed by HJURP. The ATP-dependent remodeling and spacing factor (RSF) complex is recruited to the centromere in mid-G1 phase and interacts with CENP-A nucleosomes. It could mediate the assembly of CENP-A into chromatin, or stabilize already assembled CENP-A nucleosomes by promoting incorporation of H2A/H2B or H2A variants or regulating nucleosome phasing. The proteins responsible for HJURP, RSF, and Mis18 recruitment to centromeres are not known, nor is the function of Npm1 in centromere assembly.
Is HJURP CENP-A’s Frodo?
In this issue of Cell, Dunleavy et al. (2009) and Foltz et al. (2009) provide insights into a new CENP-A binding partner in human cells called HJURP (Holliday junction-recognition protein) (Kato et al., 2007). These studies sought to identify the elusive CENP-A loading factor by isolating proteins that specifically associate with prenucleosomal CENP-A and not histone H3.1. The common CENP-A binding partners found by both studies were HJURP and nucleophosmin (Npm1), a nucleolar protein also required for chromosome segregation (Amin et al., 2008). HJURP homologs are present in several mammals, and the human HJURP protein contains several tryptophans that weakly resemble the WD40 domains of the histone chaperones CAF1-p60 and HIRA (Foltz et al., 2009). Additional CENP-A interactors identified include RbAp48 (Dunleavy et al., 2009), RuvB like-1 (RuvBL1), and replication protein A1 (RPA1) (Foltz et al., 2009).
Both groups focused on examining HJURP’s role in CENP-A localization to centromeres because HJURP depletion results in the loss of centromeric CENP-A, which did not occur with partial depletion of Npm1 (Dunleavy et al., 2009; Foltz et al., 2009). HJURP directly associates with CENP-A/H4 heterotetramers in vitro, independent of the presence of H2A/H2B dimers (Foltz et al., 2009). It is not known whether HJURP and CENP-A can interact independently of H4, as was observed for SpScm3 and CENP-ACnp1 (Pidoux et al., 2009). Interestingly, HJURP interacts with chimeric H3CATD in coimmunoprecipitation and in vitro binding assays (Foltz et al., 2009). Thus, HJURP specificity depends on the CENP-A/H4 structure conferred by the CATD domain required for endogenous CENP-A recruitment to centromeres.
The hypothesis that HJURP is a factor directly required for CENP-A localization is further strengthened by its distribution pattern during the cell cycle. HJURP is found throughout the nucleus, but detergent extraction revealed punctate centromere localization during cytokinesis and G1 (Dunleavy et al., 2009; Foltz et al., 2009). This localization pattern is intriguing, as it roughly coincides with the deposition of new CENP-A (Jansen et al., 2007). HJURP becomes centromeric slightly later than Mis18α, supporting the possibility that the Mis18 complex primes the centromere to receive CENP-A and that HJURP acts as the likely CENP-A delivery vehicle (Foltz et al., 2009; Fujita et al., 2007; Hayashi et al., 2004) (Figure 1B). It will be interesting to determine if HJURP localization is dependent on the Mis complex and whether they physically interact.
In agreement with a central role in CENP-A localization, depletion of HJURP causes a dramatic loss of CENP-A from centromeres and results in chromosome segregation defects in mitosis (Dunleavy et al., 2009; Foltz et al., 2009). However, total cellular CENP-A protein levels dramatically decrease when HJURP is absent. Previous studies showed that over-expressed, mislocalized CENP-A is subjected to proteasome-mediated degradation in budding yeast and Drosophila (Allshire and Karpen, 2008). These observations raise the possibility that HJURP’s role in centromere assembly is to act as a protein chaperone that ensures CENP-A stability. Indeed, Dunleavy et al. notice that overexpressed green fluorescent protein (GFP)-CENP-A is stable after depletion of HJURP, in contrast to the rapid degradation of endogenous CENP-A. Despite its stability, GFP-CENP-A (and yellow fluorescent protein-CENP-A in Foltz et al., 2009) still failed to localize to centromeres after HJURP depletion, though it was readily incorporated into noncentromeric regions. These observations suggest that HJURP promotes CENP-A stability and regulates the specificity of CENP-A incorporation at centromeres, rather than directly mediating chromatin assembly. We speculate that HJURP delivers a high concentration of CENP-A to centromeres, and that another chromatin assembly factor mediates its incorporation into chromatin (Figure 1B). Misincorporation of overexpressed CENP-A could result from the broad activity of such assembly factors when targeting specificity is lost after HJURP depletion.
A candidate CENP-A chromatin assembly factor is the ATP-dependent remodeling and spacing factor (RSF) complex. Perpelescu et al. (2009) recently showed that RSF components physically interact with CENP-A nucleosomes and are required for stable incorporation of CENP-A. However, enrichment of the RSF complex component Rsf1 at centromeres is not visible until mid-G1 phase, after CENP-A assembly initiates in late mitosis. Furthermore, the RSF complex regulates the replacement of the H2Av variant in Drosophila (Hanai et al., 2008), in addition to nucleosome remodeling and spacing functions. Thus, it is not yet known whether RSF directly impacts CENP-A assembly or ensures the stability of incorporated CENP-A nucleosomes through H2A/B/variant assembly or altered nucleosome positioning (Figure 1B).
Dunleavy et al. (2009) also provide some insights into the role of human RbAp46/48 proteins found in several chromatin remodeling complexes, such as CAF1. Human RbAp46/48 proteins, like S. pombe Mis16, have not been shown to physically bind CENP-A but are required for CENP-A localization in vivo (Fujita et al., 2007). RbAp46/48 are not detected in HJURP immunoprecipitates, but Dunleavy et al. observed that HJURP protein levels are dramatically reduced after depletion of the two proteins. This raises the possibility that RbAp46/48 promote CENP-A localization indirectly through stabilization of HJURP (Figure 1B). Finally, although some components previously identified in complexes with CENP-A nucleosomes (e.g., the CENP-H/I complex) are required for targeting of new CENP-A (Mellone et al., 2006), they are absent from prenucleosomal CENP-A complexes. Thus, it is possible that they contribute to CENP-A delivery independently of HJURP, or by transiently regulating HJURP. Elucidating the interplay between HJURP, other CENP proteins, RbAp46/48, and RSF will be crucial to establishing which component is the elusive CENP-A loading factor.
Approaching Mordor: Linking SpScm3 and HJURP
The identification and characterization of proteins that interact directly with CENP-A and are required for CENP-A localization represent important breakthroughs. In S. pombe, Sim3/NASP associates with soluble CENP-ACnp1 to deliver it to centromeres (Dunleavy et al., 2007), whereas SpScm3 localizes to centromeres and directly interacts with CENP-ACnp1 to promote its proper localization (Pidoux et al., 2009; Williams et al., 2009) (Figure 1A). In human cells, however, it is intriguing that both the delivery and localization of CENP-A may be mediated by HJURP, although the RSF complex or other factors could mediate actual CENP-A assembly (Dunleavy et al., 2009; Foltz et al., 2009; Perpelescu et al., 2009) (Figure 1B).
The Mis18 complex provides an interesting connection between human HJURP, S. pombe Scm3, and centromere assembly in these two widely diverged eukaryotes. The cell cycle localization dynamics of these proteins remain a fascinating phenomenon that deserves more in depth examination. Both SpScm3 and HJURP are absent from centromeres during mitosis and only reassociate with the region after chromosome segregation is accomplished (Figure 1). Even though the timing of new CENP-A incorporation differs greatly between human and S. pombe cells, the commonality is that these CENP-A partners are absent from centromeres during much of mitosis. It will be important to identify the factors that regulate their removal and the significance of this process to centromere assembly.
The unusual behaviors of HJURP and SpScm3 during mitosis, the initiation of CENP-A loading during mitosis in humans and Drosophila, and the links between centromere assembly and mitotic cell cycle regulators are intriguing, especially given that the primary function of centromeres is to form the kinetochore and promote chromosome segregation during mitosis. Although new CENP-A can be targeted to centromeres after massive disruption of spindle microtubules (Jansen et al., 2007), it is possible that satisfying the mitotic checkpoint and mitotic exit serve as signals for epigenetic propagation of centromere identity. Determining what cell cycle factors and events in mitosis are necessary for centromere “priming” by Mis18 and for CENP-A recruitment will be crucial to elucidating this essential process. It would be interesting, for example, to determine whether regulators such as HJURP, SpScm3, RSF, and the Mis18 complex have differential associations with centromeres in properly versus improperly segregated chromosomes.
In conclusion, the advancements provided by the studies discussed here have moved us significantly closer to understanding how CENP-A is specifically recruited to centromeres. Now that the ring’s “fellows” are revealed, it is crucial that we determine what proteins and mechanisms are directly responsible for the assembly of CENP-A nucleosomes into centromeric chromatin. Further studies directed toward the isolation of HJURP and SpScm3 complexes at different stages of the cell cycle and dissection of the relationship between Mis18 complexes, HJURP, RSF, and CENP-A are essential to elucidate the molecular mechanisms responsible for the propagation of centromere identity. It will also be important to determine whether SpScm3 and HJURP functions or domains are conserved and represent activities or mechanisms used at centromeres in other eukaryotes.
Acknowledgments
We are grateful to The Journal of Cell Biology for providing the Perpelescu et al. manuscript prior to publication and for insightful comments from the reviewers. W.Z. is supported by Susan Komen Foundation. Centromere studies in G.K.’s lab are supported by NIH R01 GM066272.
References
- Allshire RC, Karpen GH. Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin MA, Matsunaga S, Uchiyama S, Fukui K. FEBS Lett. 2008;582:3839–3844. doi: 10.1016/j.febslet.2008.10.023. [DOI] [PubMed] [Google Scholar]
- Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Pidoux AL, Monet M, Bonilla C, Richardson W, Hamilton GL, Ekwall K, McLaughlin PJ, Allshire RC. Mol Cell. 2007;28:1029–1044. doi: 10.1016/j.molcel.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. Cell. 2009 doi: 10.1016/j.cell.2009.02.040. this issue. [DOI] [PubMed] [Google Scholar]
- Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF. J Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Bailey AO, Yatee JR, III, Bassett EA, Wood S, Black BE, Cleveland DW. Cell. 2009 doi: 10.1016/j.cell.2009.02.039. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Hanai K, Furuhashi H, Yamamoto T, Akasaka K, Hirose S. PLoS Genet. 2008;4:e1000011. doi: 10.1371/journal.pgen.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Sato N, Hayama S, Yamabuki T, Ito T, Miyamoto M, Kondo S, Nakamura Y, Daigo Y. Cancer Res. 2007;67:8544–8553. doi: 10.1158/0008-5472.CAN-07-1307. [DOI] [PubMed] [Google Scholar]
- Mellone B, Erhardt S, Karpen GH. Nat Cell Biol. 2006;8:427–429. doi: 10.1038/ncb0506-427. [DOI] [PubMed] [Google Scholar]
- Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K. J Cell Biol. 2009 doi: 10.1083/jcb.200903088. in press 10.1083/jcb.200903088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X. Mol Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Mellone BG, Karpen GH. Cell. 2007;129:1047–1049. doi: 10.1016/j.cell.2007.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]


