Abstract
Background
The tryptophan catabolizing enzyme, indoleamine 2,3, dioxygenase (IDO) is one of two mammalian enzymes, which can catabolize the rarest essential amino acid, tryptophan. IDO is inducible by cytokines such as interferon-γ and plays a role in inflammation and maternal tolerance of fetal allografts, although its exact mode of action is unclear. Therefore, we investigated the circumstances under which IDO is expressed in vitro together with the effects of overexpression of IDO on the growth and morphology of cells.
Results
Overexpression of IDO in the murine macrophage cell line RAW 264.7 and the murine fibrosarcoma cell line MC57, resulted in the growth of macroscopic cell foci, with altered cell adhesion properties. The expression of IDO was also detected during adhesion of wild type, nontransfected cells in tissue culture to standard cell growth substrates. Inhibition of this expression, likewise resulted in alterations in cell adhesion. Overexpression of IDO or inhibition of endogenous IDO expression was accompanied by changes in metalloproteinase expression and also in the expression and activity of the cyclooxygenase enzymes. In the case of RAW cells, IDO effects on cell growth could be reversed by adding back prostaglandins.
Conclusions
These results suggest that catabolism of the rarest essential amino acid may regulate processes such as cell adhesion and prostaglandin synthesis.
Background
Two known enzymes catabolize the essential amino acid tryptophan in mammals. Tryptophan 2, 3 dioxygenase (TDO) is expressed predominantly in hepatic tissues and was the first inducible enzyme system discovered in mammals [1]. It controls serum tryptophan homeostasis and is induced following ingestion of tryptophan. A second enzyme, IDO, is distinguished from TDO by its expression pattern, substrate specificity and inducibility. IDO is expressed in a variety of non-hepatic tissues, including placenta, lung, gut and epididymis [2,3,4]. Except for the last named tissue where IDO is expressed constitutively, IDO is inducible by inflammatory mediators, including interferons. In addition, IDO catalyzes the breakdown of a variety of compounds which contain an indole ring, including D-tryptophan and serotonin, marking another difference from TDO, which is specific for L-tryptophan. Curiously, it appears as if tryptophan itself cannot induce IDO synthesis [5]. IDO is also suggested to be the evolutionary ancestor of certain novel myoglobins which occur in molluscs, marking IDO as an evolutionarily primitive enzyme [6].
IDO is known to be expressed in cells infected with intracellular pathogens such as Toxoplasma and Chlamydia species and also by viruses [7,8,9,10]. In the case of Toxoplasma and Chlamydia it has been proposed that IDO induction is a cellular defense mechanism, designed to limit the proliferation of the invading pathogen by depleting the essential amino acid tryptophan. IDO expressed in monocyte derived macrophages has also been found to inhibit the growth of extracellular bacteria such as group B streptococci [11], and is also induced in tumors taken from cancer patients [12]. In all of these systems the proximal inducer of IDO activity is interferon-γ (IFN-γ). Response elements for this cytokine have been identified in the human IDO promoter and have been shown to be essential for IFN-γ induction of reporter gene expression in vitro [13,14,15].
The unusual tissue distribution of IDO suggests that combating infection is not its only function. Our interest in IDO arose when we observed that tryptophan depletion was responsible for macrophage-induced inhibition of T cell proliferation in vitro [16]. Furthermore, we reported that a pharmacologic inhibitor of IDO, 1-methyl tryptophan, induced maternal rejection of allogeneic but not syngeneic murine fetuses [17]. As IDO is strongly expressed at the maternal-fetal interface in pregnant mice and women, we have suggested that IDO plays a role in fetal defense against the maternal immune system and could represent a novel means of immunoregulation. The apparently diverse functions and tissue distribution of IDO may have as a common theme the fact that tryptophan is the rarest essential amino acid and could be the target for cellular regulatory mechanisms. If so, tryptophan concentrations in cellular microenvironments might play a critical role in modulating various cellular processes in a way that cannot be achieved by the hepatic enzyme TDO which regulates systemic tryptophan concentrations.
The IDO promoter contains a diverse collection of motifs together with the IFN-γ response elements. These include motifs for transcription factors that bind to collagenase and elastase genes and motifs for the transcription factor MEP-1, which regulates transcription from the stromelysin-1 (MMP-3) and metallothionein genes [18,19]. Matrix metalloproteinases (MMPs) are responsible for modification of the extracellular matrix and are involved in wound healing, tumorigenesis, pregnancy and inflammation. In general, they regulate how cells interact with each other and with the extra-cellular matrix. Evidence for a tryptophan-reversible inhibition of MMP expression by IFN-γ has previously been presented [20, 21], although the exact mechanism is unclear. Therefore we decided to directly test whether IDO plays a role in controlling interactions with other cells and also the surrounding extracellular environment.
We have identified cells expressing IDO in vitro and used IDO antisense constructs to inhibit this expression. In addition, we have constitutively overexpressed IDO in adherent and non-adherent cell lines in vitro. Our results demonstrate that tryptophan catabolism has significant effects on cell adhesion and regulates the activity and expression of cyclooxygenases 1 and 2 (COX-1 and -2).
Results
Constitutive overexpression of IDO alters cell adhesion
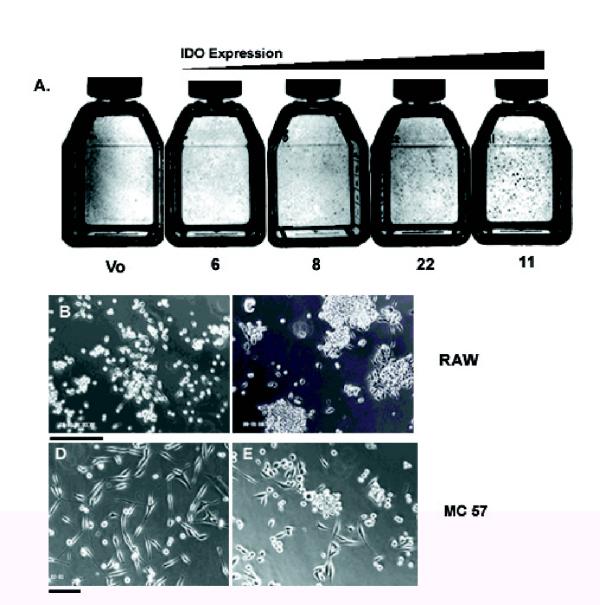
To determine whether IDO plays a role in regulating cell adhesion, we expressed a full-length IDO cDNA in cell lines in vitro. We transfected the murine macrophage cell line RAW 264.7 with a construct in which IDO was expressed under the control of the murine MHC Class II promoter (Fig 1A). IDO-transfected RAW cell clones, which expressed IDO under the control of the MHC Class II promoter, were characterized for IDO expression. We selected four clones with varying capacities for IDO expression and tryptophan depletion from culture medium (Fig 1B,C,D,E). Following 48 hours in culture, clones 22 and 11 depleted tryptophan to a greater extent than clones 6 and 8 or vector only control, consistent with the greater vector copy number of these clones. However, none of the clones depleted a substantial proportion of the tryptophan present in medium even with longer incubation times. A common feature of the tryptophan depleting clones was their tendency to form macroscopic foci, which were visible to the naked eye (Fig 2,A,B,C). At a certain point in focus growth, multicellular aggregates of RAW cells would break off from the focus and could be seen floating in suspension in the tissue culture medium. Wild type RAW cells or RAW cells transfected with vector alone and, to a lesser extent, clones 6 and 8 demonstrated a reduced ability to form macroscopic foci.
Figure 1.
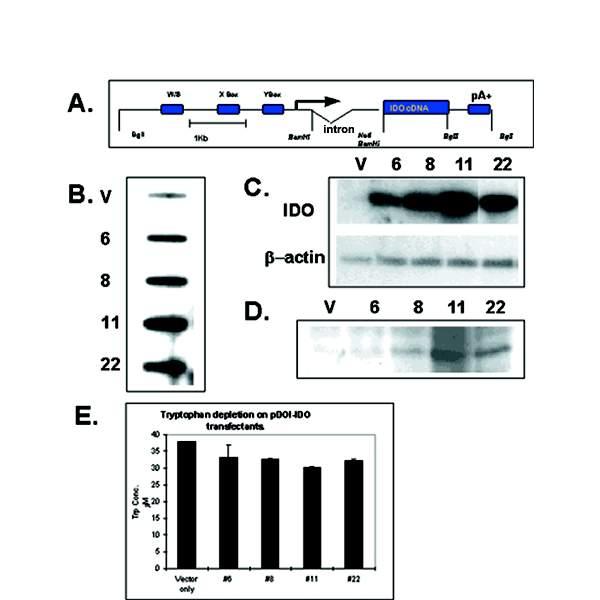
Characterization of RAW cell transfectants. (A) IDO construct used to transfect RAW cells showing X, Y and W/S boxes of MHC Class II promoter. (B) Relative copy number determination of IDO-transfected RAW cell clones. The rabbit β globin intron present in the construct was used as a probe for hybridization by slot blot. (C) RT-PCR of total RNA isolated from RAW transfectants following 15 cycles of RT-PCR, electrophoresis on 0.8% agarose, followed by Southern blotting and hybridization with an IDO specific probe. (D) Western blot of IDO-expressing RAW cell transfectants, using IDO-specific polyclonal antibody. (E) HPLC determination of tryptophan depletion from tissue culture medium by IDO-expressing RAW cells, 48 hours post-seeding into fresh medium. V; Vector-only control transfectant. Clones 6, 8, 11, 22; IDO-expressing RAW clones.
Figure 2.
Phenotype of IDO-expressing transfectants. (A) Flasks of RAW cell clones stained with trypan blue and photographed under normal light. Flasks are arranged in order of increasing IDO expression and show foci visible to the naked eye. V; Vector only control. (B) Vector-only transfected RAW cells. (C) Clone 11 IDO-expressing RAW cells. (D) Vector-only transfected MC57 cells. (E) IDO-expressing MC57 cell clone. Bar = 250 μm.
To determine if this phenomenon was unique to RAW cells, we also transfected the MC57 murine fibrosarcoma cell line [22] which grows as a monolayer. MC 57 cells are fibroblastic in appearance and disperse across the surface of a tissue culture dish in a uniform manner. Transfection of a full length, constitutively expressed IDO cDNA into MC57 cells in the pcDNA3 expression vector, resulted in MC57 cells developing a more rounded phenotype. Furthermore, cells grew as multicellular foci, in a confined area, similar to RAW cells, although the foci did not grow to as large a size before detaching from the plate (Fig 2D,E). The murine monocytic cell line P388 was also transfected and expressed IDO. It likewise exhibited a change in morphology similar to that described above and clones expressing IDO often changed from non-attached suspension cultures to adherent cultures which resembled RAW cells(not shown).
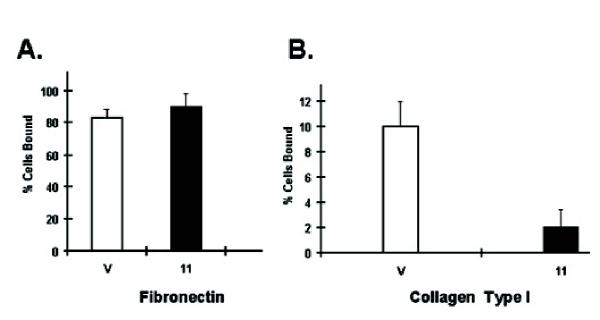
IDO-expressing clones were also slower to re-attach to tissue culture dishes following sub-culture and could be seen floating as multicellular aggregates. To quantitate the change in cell adhesion in IDO-transfected RAW cells, we performed binding studies to tissue culture plates coated with various extra-cellular matrices, including collagen, laminin, matrigel, and fibronectin. Neither vector-only controls nor IDO-expressing cells adhered significantly to laminin or matrigel coated plates in a 45 minute assay period (not shown) and both controls and IDO-expressing cells adhered strongly and to similar extents to fibronectin coated plates (Fig 3A). However, there was a substantial difference in adhesion to collagen-coated plates. Although neither sample adhered to collagen to the same extent as to fibronectin, vector only controls adhered more strongly than IDO-expressing clone 11 (Fig 3B).
Figure 3.
Adhesion properties of IDO-expressing RAW cell transfectants. (A) Adhesion of clone 11 and vector-only RAW cells to fibronectin coated tissue culture dishes. 5 × 105 RAW cells were seeded into 24 well tissue culture plates coated with fibronectin and the percentage of cells adhering to the plate 45 minutes later was determined. (B) Adhesion of clone 11 and vector-only RAW cells to type I collagen-coated tissue culture dishes. Assay system was the same as for A.
IDO expression is induced during cell attachment to growth substrates
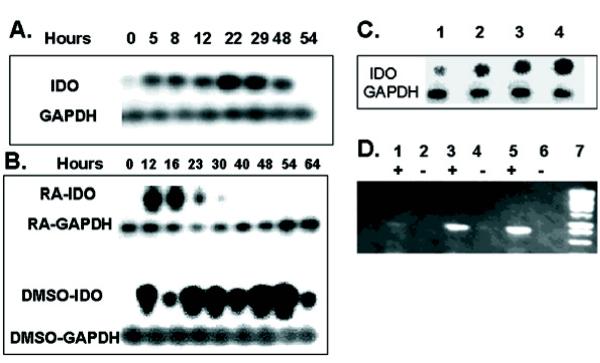
To determine if IDO normally plays a role during the course of cell adhesion, we detached log phase, wild type RAW cells from their culture flasks by scraping, reseeded them into fresh medium and assayed for IDO expression at subsequent time points. Log phase cells do not express IDO at levels detectable by standard RT-PCR methods. However, in cells detached from their normal growth substrate, IDO expression was already induced by 5 hours following reseeding into fresh tissue culture dishes and expression was detected until 48 hours (Fig 4A). Onset of expression coincided with the time when the majority of cells had begun to adhere to the plate.
Figure 4.
IDO expression in RAW and P19 cells in vitro. (A) RAW cells were harvested by scraping and 1 × 106 cells were seeded into 5 ml culture dishes. Cultures were harvested at various time points and assayed for IDO expression by RT-PCR. (B) P19 cells were aggregated in suspension cultures in the presence of either 1% DMSO or 10-6 M RA. Cultures were harvested at various time points and assayed for IDO expression by RT-PCR. (C) Effect of EGTA on expression of IDO 24 hours following seeding of P19 aggregates. P19 cells were seeded into suspension cultures in the presence of 5 mM (lane 1), 3 mM(lane 2), 1 mM (lane 3) or no EGTA (lane 4). (D) IDO antisense expression. RT-PCR products electrophoresed on 0.8% agarose. Lanes; 1&2: antisense clone C2, 3&4: antisense clone D3, 5&6: antisense clone E6, 7: molecular weight marker. Lanes marked with + or -; reverse transcriptase present or absent respectively.
To determine if IDO expression was restricted to cell interactions with standard tissue culture substrates and to further explore the possibility that IDO altered inter-cellular adhesiveness, we studied the murine embryonic carcinoma cell line P19. This cell has been characterized extensively and differentiates into skeletal and cardiac muscle or neuronal cells, depending on whether it is treated with DMSO or retinoic acid (RA) respectively [23, 24, 25]. Differentiation is dependent on an initial, 3-5 day incubation as multicellular aggregates in suspension culture, in the presence of drug, followed by a similar period growing as monolayer adherent cells in the absence of drug. Mature differentiated cells begin to appear during this subsequent growth period in the absence of drug.
IDO expression was detected when P19 cells were reseeded into bacterial petri dishes as suspension cultures and allowed to form aggregates. IDO expression was observed within 12 hours of seeding into suspension with DMSO and peaked around 48-54 hours. (Fig 4B). Thus, removing cells from their normal growth substrate and reseeding into fresh medium induced a burst of IDO expression irrespective of whether cells reassociated with tissue culture substrate or other cells. Aggregating cells in the presence of 10-6 M RA, which induces neuronal differentiation also induced a transient burst of IDO transcription but the period was shorter and the peak level observed was lower than that observed with DMSO.
To determine if IDO expression was related to the removal of cells from their normal growth substrate or the process of reattachment to new substrate, we trypsinized P19 cells and reseeded them as aggregates in suspension in the presence of various concentrations of EGTA. EGTA chelates essential Ca2+ required by cadherin molecules and inhibits cell adhesion. We observed a concentration dependent decrease in IDO expression in EGTA treated samples 24 hours after seeding, which paralleled a corresponding decrease in cell aggregation (Fig 4C). Therefore, IDO expression appeared to be induced by reattachment, rather than detachment from a previous substrate. Thus, IDO is thus expressed endogenously in various cell types and is induced during cell attachment to growth substrates.
Inhibition of endogenous IDO expression disrupts P19 cell adhesion
To examine the role of IDO expression in P19 cells, we transfected P19 cells with a DNA construct which contained a 740 bp fragment of murine IDO cDNA in the antisense orientation, under the control of the constitutively active CMV promoter in the pcDNA-3 mammalian expression vector. As control, the IDO gene fragment was cloned in a sense orientation. To confirm that transfected, G418 resistant P19 cells expressed IDO antisense transcripts, we isolated total RNA from G418 resistant clones, reverse transcribed it into cDNA using a sense primer then PCR amplified the cDNA. Three antisense-transfected clones, which expressed progressively greater amounts of IDO antisense RNA were selected for further analysis together with a sense control (Fig 4D). The ability of the sense and antisense transfectants to deplete tryptophan from culture medium was determined following 48 hours in culture. IDO sense transfected P19 cells depleted 10% of available culture tryptophan (not shown) while IDO antisense transfected P19 clones which expressed high levels of antisense (clones D3 and E6) depleted essentially no tryptophan from the medium. Clone C2, which expressed low levels of IDO antisense, depleted similar levels of tryptophan to the sense control. Therefore, the burst of IDO expression, which takes place in cells during reattachment does not result in substantial tryptophan depletion from culture medium
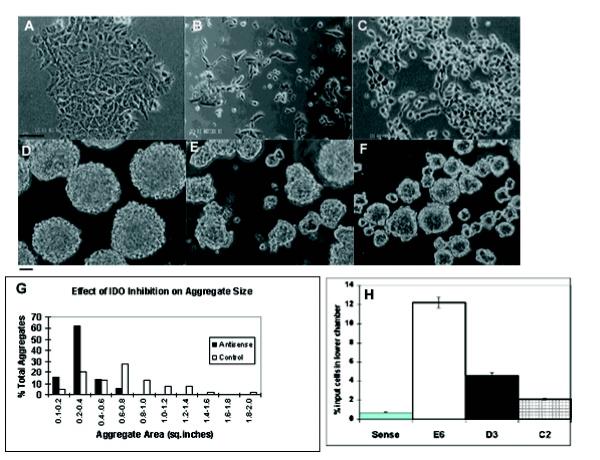
IDO antisense-transfected clones D3 and E6 exhibited a different phenotype compared to untransfected and sense transfected P19 cells (Fig 5A,B,C). IDO antisense expressing clones developed a rounded appearance with a more scattered morphology and apparent loss of cell interaction instead of the usual, adherent P19 phenotype. The degree to which this phenotype manifested, correlated with the extent of IDO antisense expression, i.e. it was prominent in clone E6 and D3 while clone C2 was indistinguishable from sense-transfected controls. However growth rates were largely unaltered. IDO-antisense and sense transfectants were aggregated in 1% DMSO and cell aggregates were visualized by phase contrast microscopy. Sense or untransfected P19 cells aggregated normally as tightly packed spheroid bodies. In contrast, antisense transfectants formed aggregates, which exhibited markedly different morphologies and differed from sense-transfected aggregates in two principal respects; shape and size (Fig 5D,E,F). At 30 hours after seeding suspension cultures, cells formed irregular shaped, non-spherical aggregates that were less tightly packed than control aggregates. Antisense clone E6 (shown in Fig. 5F) produced aggregates which were only loosely packed and a substantial number of cells which did not package into any form of aggregate while antisense clone D3 formed aggregates more diverse in shape than the uniformly spherical controls but less diverse than clone E6. Clone C2 produced aggregates similar to sense transfected controls (not shown).
Figure 5.
Phenotype of IDO-antisense transfected P19 cells. (A-C) P19 cells growing as a monolayer. (A) IDO-sense (B) IDO-antisense transfected P19 clone D3 (C) IDO antisense clone E6, growing as a monolayer. Bar:1 mm. (D-E); P19 cells growing as aggregates. (D) Aggregates from IDO sense-transfected P19 cells, (E) aggregates from IDO antisense-transfected P19 clone D3 grown in suspension for 30 hours in the presence of 1% DMSO. (F) Clone E6 treated as for D3. Bar: 50 mm (G) A representative field at 10x magnification was selected and the areas of 50 individual aggregates was calculated for both sense and antisense clone E6. Results are shown as the percentage of the total aggregates analyzed whose areas are within a given 0,2 sq. inch interval. (H) Percentage of IDO-sense and-antisense transfected P19 cells migrating to lower chamber of 24 well tissue culture plates18 hours following seeding. S: sense control, E6, D3, C2: antisense clones.
To quantitate and compare the size difference between sense and antisense-transfected aggregates we photographed sense and antisense clone 5 aggregates and calculated the area of each aggregate individually (Fig. 5G). Antisense aggregates were small, predominantly in the 0.2-0.4 sq. inch size range, whereas control aggregate sizes were spread over a much broader range. The mean size of antisense-transfected aggregates was 0.31 sq. inches, while sense transfected controls had a mean of 0.69 sq. inches (p < 0.0001). The effect of IDO inhibition on cell adhesion was demonstrated by performing cell migration assays. P19 cells were seeded into porous tissue culture inserts placed in a 24 well tissue culture plate and cell migration to the lower chamber in the absence of any stimulus was determined. Approximately 12% of clone E6 cells migrated to the lower chamber 18 hours after seeding into the upper chamber (Fig. 5H). In contrast, less than1% of control cells had migrated in the same period. Clones C2 and D3 produced intermediate levels of migration. When inserts were coated with Matrigel, no significant migration was seen in either antisense or sense transfectants, indicating that cell motility could be inhibited by supplying an extracellular matrix.
IDO axpression alters metalloproteinase expression
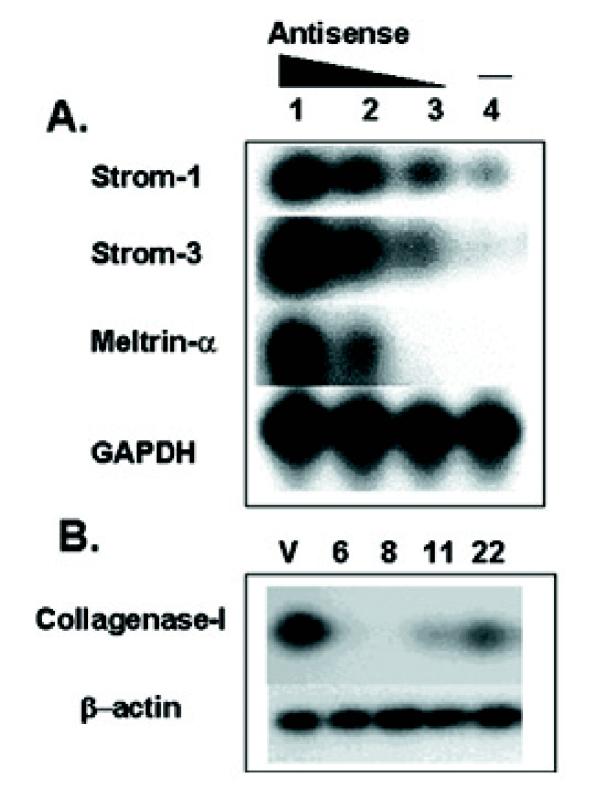
The altered adhesion of IDO-expressing RAW cells to collagen suggested that IDO might induce alterations in enzymes involved in modifying the extracellular matrix. Therefore, we investigated whether inhibition of IDO expression in the P19 in vitro aggregation system and the constitutive overexpression of IDO in RAW cells had any effect on MMP expression. We allowed the sense and three-antisense expressing clones of P19 to aggregate in 1%. DMSO for 24 hours, before harvesting total RNA and determining expression of various MMP genes, including stromelysins 1 (MMP-3), 2 (MMP-10) and 3 (MMP-11), collagenases I (MMP-1) and IV (MMP-2) and meltrins α (ADAM-12) and β (ADAM-19). Meltrin α is expressed in vivo during development in condensed mesenchymal cells that give rise to skeletal muscle while meltrin β is expressed in craniofacial and dorsal root ganglia where neuronal lineages differentiate [26]. Expression of stromelysins-1 and 3 and meltrin α, was increased in antisense-expressing aggregates, relative to the sense control (Fig 6A). Furthermore, there was a progressive increase in the expression level of these three protease genes, which correlated with the amount of IDO antisense expression. In contrast, the expression of meltrin-β and stromelysin-2 was similar in all samples and expression of collagenase I or IV was undetectable in either sense or antisense-expressing aggregates (not shown). Thus, inhibition of IDO gene expression correlated with increased expression of some but not all MMP genes in P19 cells undergoing aggregation. Furthermore, increased MMP expression coincided with decreased ability of P19 cells to aggregate in suspension culture. To determine whether IDO-expressing RAW cells also showed unusual MMP expression we tested the IDO-expressing RAW cell clones for the same group of MMPs as P19 cells. Expression of all MMPs was undetectable except for collagenase I. This showed significant expression in vector-only controls but little or no expression in IDO-expressing clones. All IDO-expressing clones demonstrated reduced expression with no correlation to the level of IDO expression (Fig 6B). Pharmacological inhibition of MMP activity in P19 cells using the broad spectrum, hydroxamic acid-based MMP inhibitor GM 6001 at concentrations ranging from 1-30 μM, resulted in partial reversal of the poor aggregation shown by IDO-AS expressing cells, with a maximal effect shown at 20 μM, indicating that changes in MMP expression were responsible, at least in part for altered cell adhesion.
Figure 6.
Effect of IDO on metalloproteinase expression. (A) IDO sense and antisense-transfected P19 cells were aggregated in 1% DMSO for 30 hours before total RNA was isolated and metalloproteinase gene expression assayed by RT-PCR. Lanes: 1-antisense clone E6, 2-antisense clone D3, 3-antisense clone C2, 4-sense. (B) Collagenase gene expression in IDO-expressing RAW transfectants. V: vector-only, 6,8, 11, 22: IDO expressing RAW clones.
IDO regulates prostaglandin synthesis
To understand the mechanism of IDO induced alterations in cell adhesion and MMP expression, we attempted to reverse IDO effects on cell adhesion. As previously mentioned, tryptophan is not significantly depleted in culture medium of RAW cells overexpressing IDO, suggesting that tryptophan deprivation is not the cause of the IDO effect. Consistent with this, adding back tryptophan to IDO-expressing RAW cells did not reverse the growth of macroscopic foci. As tryptophan is not the only substrate for IDO, we also investigated whether adding serotonin would overcome the effects of IDO expression. There was a similar lack of effect of this compound. This suggested that depletion or reduction of an IDO substrate was probably not responsible for the effects described here. An alternative possibility was that a biologically active downstream catabolite of IDO could be the cause. Therefore, we tested the tryptophan catabolites, picolinic acid and quinolinic acid to see if they could reproduce the effects of IDO overexpression. Picolinic acid (1-6 mM) produced morphological changes in both MC57 and RAW cells and also substantial reductions in growth rate but did not mimic the effects of IDO expression. In particular, at a concentration of 2 mM, picolinic acid induced a more flattened phenotype. At concentrations above 6 mM, picolinic acid-induced apoptosis was observed. Quinolinic acid was essentially without effect at concentrations up to 10 mM. Therefore, the exact mode of action of IDO therefore remains to be determined.
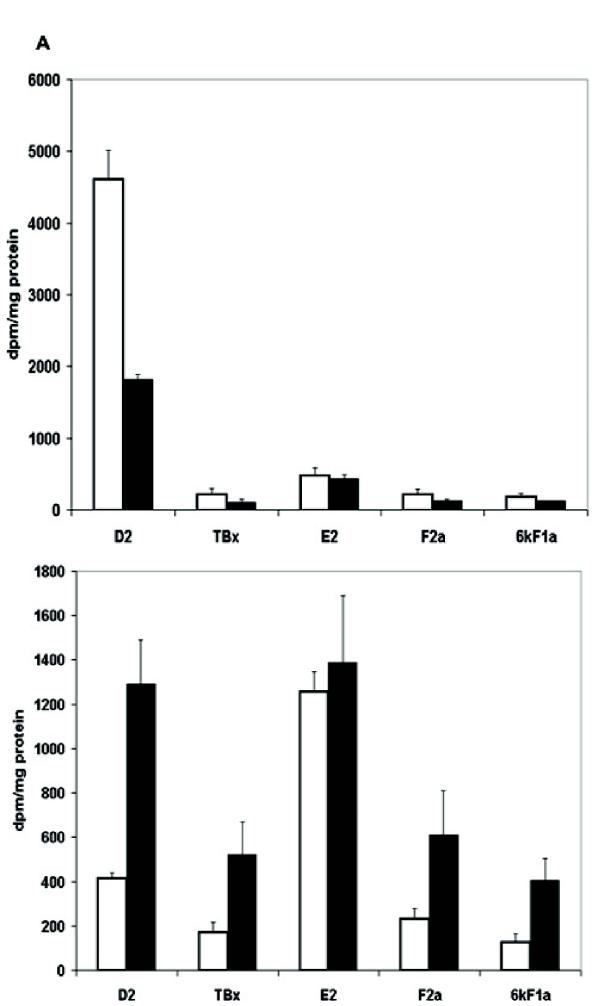
Despite the uncertainty about the proximal mediator of IDO's effect, it is known that alterations in cell adhesion and metalloproteinase activity are often associated with changes in prostaglandin synthesis [27,28,29,30]. Therefore, we analyzed the spectrum of PGs produced by IDO-expressing RAW cells using thin layer chromatography. PG D2 was the major product of both vector-only controls and IDO-expressing clone 11, consistent with reports that D2 production is typical of antigen-presenting cells [31]. There was a greater than 50% reduction in PG D2 production in clone 11 compared with the vector only control and a similar decrease in levels of PGs F2α, 6keto-F1α and thromboxane B2 in this clone (Fig 7A). However, PG E2 production was affected relatively little compared to the other PGs. Thus IDO overexpression resulted in an increase in PG E2 relative to the other PGs. However, in MC57 cells, the prostaglandin profile was quite different from that seen in RAW cells. PG E2 was the dominant prostaglandin (Fig 7B) and overexpression of IDO resulted in a relative increase in the amount of PG D2 and other PGs, relative to E2.
Figure 7.
Effect of IDO expression on prostaglandin production. (A) Analysis of prostaglandin production by vector-only transfected RAW cells () and IDO-expressing clone 11 () (B) Analysis of prostaglandin production by vector-only transfected () and clone 24 () IDO-expressing MC57 cells.
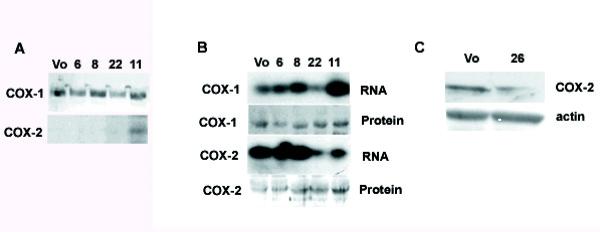
In IDO-expressing RAW cells, COX-1 protein levels were unchanged compared to the vector-only control (Fig 8A). In contrast, COX-2 was not expressed by vector only controls or RAW clones 6, 8 and 22 but COX-2 mRNA and protein was induced in the RAW clone expressing the greatest amount of IDO (clone 11) (Fig 8A). Although COX-2 is not usually expressed in RAW cells, it can be strongly induced with lipopolysaccharide (LPS) [32, 33]. Therefore, we treated IDO transfected RAW cells and controls with LPS and measured COX-2 expression 24 hours later. COX-2 mRNA was most strongly induced in vector only or low IDO-expressing clones (Fig 8B). Curiously, clones expressing higher levels of IDO (clones 22 and 11) showed lower levels of COX-2 mRNA induction. In contrast, COX-2 protein levels were higher in clones expressing lower amounts of IDO mRNA and lower in vector only controls, whereas COX-1 protein levels were unchanged by LPS treatment. MC57 cells expressed COX-2 constitutively, consistent with the domination of the PG profile by PG E2. However, IDO overexpressing clone 26 showed a reduced amount of COX-2 protein compared to the vector only control (Fig 8C).
Figure 8.
Effect of IDO on cyclooxygenase expression clones. (A) Expression of COX-1 and COX-2 protein by vector-only transfected (Vo) and IDO expressing clones. (B) Effect of LPS treatment on expression of COX-1 and COX-2 in IDO-expressing clones. Vector-only and IDO-expressing RAW cells were treated with 1 ng/ml LPS for 12 hours. (C) COX-2 expression in vector-only or IDO-expressing MC57 cells.
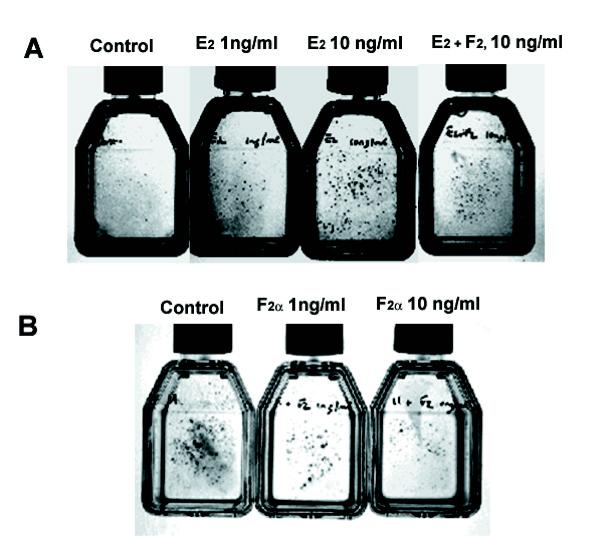
If uniformly diminished PG synthesis by IDO was responsible for growth of RAW cell macroscopic foci, inhibiting PG synthesis with a pharmacological inhibitor of COX-1 and -2 ought to reproduce the effect of IDO expression. Therefore we treated vector-only transfected RAW cells with various concentrations of indomethacin ranging from 0.1 μM to 100 mM. Although some effect of indomethacin on cell growth rate was observed, there was no sign of macroscopic foci (data not shown). Thus IDO expression does not mimic the effects of a global COX inhibitor. To test the hypothesis that alterations in the relative levels of PGs were responsible for the growth of macroscopic foci, we added PGs directly to vector-only transfected RAW cells. The phenotype produced by IDO expression could be reproduced by adding PG E2 alone to the cultures. PG E2 addition resulted in a dose-dependent increase in the appearance of macroscopic foci, with visible foci appearing at 1 ng/ml (3 nM) and becoming abundant at 10 ng/ml (30 nM) (Fig. 9A). Adding PG F2α at the same time as E2 resulted in a reduction of the number of foci. Surprisingly, addition of PG D2 also resulted in a slight increase in focus numbers (not shown).
Figure 9.
Effect of exogenously added PGs on the growth of RAW cells. (A) Vector-only transfected RAW cells were incubated with the indicated concentrations of PGs for 48 hours and stained with trypan blue. (B) Clone 11 RAW cells were incubated with the indicated concentrations of PGs for 48 hours and stained as for A.
We next attempted to reverse the phenotype seen in clone 11 cells. If increased PG E2 production relative to other PGs was responsible for the appearance of macroscopic foci, then adding back increasing amounts of other PGs such as D2 and F2α should restore the phenotype of clone 11 to that of vector only controls. As PG F2α attenuated the focus forming ability of PG E2 in the experiment shown in Fig. 9A, we added PG F2α in various concentrations to clone 11 cells. PG F2α at 10 ng/ml (30 nM) substantially reduced the focus forming ability of IDO expressing clone 11 (Fig 9B). Thus an alteration in the PG E2/F2α ratio plays an important role in mediating IDO's effects on cell growth and morphology.
Discussion
Tryptophan catabolism by cells expressing IDO is something of an enigma and has resulted in speculation as to why the body requires two enzymes with different tissue specificities to degrade the rarest essential amino acid [5, 34]. The inability of IDO to be induced by its own substrate exemplifies this puzzle. While IDO's role in controlling intracellular pathogens is well documented, there is little understanding of the reasons for IDO expression at sites in the body unlikely to be related to this function, such as the epididymis. The data we present here reveal that IDO expression is an important determinant of the way in which cells interact with their extracellular environment in vitro. In particular, cell adhesion is altered dramatically by overexpressing IDO in cells which do not otherwise express it, or inhibiting IDO expression in cells in which it is naturally induced following cell passage. Specifically, overexpression of IDO in RAW and MC57 cells resulted in the growth of macroscopic foci and other phenotypic alterations. The cell foci were multicellular aggregates, which grew vertically as well as horizontally across the plate surface and contained significant numbers of necrotic cells within their interior, as judged by trypan blue exclusion. Conversely, in P19 cell aggregates in which IDO expression was inhibited, there was a more dispersed phenotype with cells losing the ability to interact with each other. We have recently confirmed that IDO expression in RAW cells following cell passage is likewise important for correct cell adhesion (results not shown).
Our data support the hypothesis that IDO-induced alterations in PG synthesis can modify cell adhesion. We observed changes in the relative amounts of PGs in IDO-transfected RAW cells and reversal of the effects of IDO-expression by PG F2α while PG E2 stimulated focus formation. COX-2 was upregulated in IDO-expressing RAW cells. Similar effects of PG E2 on cell morphology have recently been reported in the human embryonic kidney cell line HEK 293, which overexpressed COX-2 and PG E2 synthase [35]. COX-2/PG E2 synthase-expressing cells were highly aggregated, piled up and exhibited round shape morphology similar to the RAW cells described here. MC57 cells, which demonstrated similar changes in cell adhesion to RAW cells following IDO expression, exhibited lower levels of COX-2 synthesis upon IDO expression and lower levels of PG E2 relative to other PGs such as D2. Furthermore, adding back PGE2 to IDO-expressing MC57 cells did not reproduce the wild type phenotype (not shown). Thus, similar effects on cell morphology were produced by opposite effects on COX-2 expression and PG E2 production in these two cell lines. We are presently attempting to determine if products of COX-2 activity other than E2 may be responsible for IDO effects in this cell line.
The mechanism of PG-induced changes in cell adhesion and morphology may involve MMP activity. Synthesis of MMPs such as collagenase I (MMP-1), gelatinase B (MMP-9) and matrilysin (MMP-7) has been shown to be dependent on the synthesis of PG E2, suggesting that alterations in MMP expression may be instigated by alterations in PG synthesis [27,28,29,30]. Furthermore, both COX-1 and COX-2 have recently been shown to mediate adhesion of various cell types in vivo and in vitro [36, 37]. Thus, one possibility is that alterations in MMP expression and activity could modify cellular interactions with the extracellular matrix following IDO expression. Consistent with this possibility is the observation that MMP expression in P19 cells was correlated with the degree of IDO-antisense expression. RAW transfectants over-expressing IDO showed reduced expression of collagenase I (MMP-1), and also bound less well to collagen-coated plates than controls. Collagen is one of the principal components of the extracellular matrix and RAW cells bind poorly to fibrillar type I collagen unless it is denatured or activated by collagenase [38]. Thus, the diminished expression of MMP-1 in IDO transfectants could explain their weaker binding to this substrate. The mechanism by which IDO regulates prostaglandin synthesis is yet to be determined. Tryptophan is a stimulatory co-factor for COX and degradation of tryptophan in the intracellular environment could alter COX activity. Alternatively, IDO might influence COX activity and expression through competition for or release of heme, which both enzymes require. In vitro, arachidonic acid stimulates the dissociation of heme from IDO and this correlates with IDO stimulatory effects on COX [39], providing circumstantial support for the latter possibility.
The alterations in COX-2 expression observed in IDO-expressing RAW and MC57 cells are a particularly interesting feature of our results. COX-2 is inducible by a number of inflammatory mediators including IFN-γ [40] and LPS [32]. These also induce IDO. Treatment of IDO-expressing RAW clones with a known inducer of COX-2 (LPS), revealed a lack of correlation between COX-2 RNA and protein levels. Clones 11 and 22 showed low levels of COX-2 message but high levels of protein following LPS treatment. This suggests that COX-2 RNA and/or protein turnover may be affected by IDO expression. Other workers have noted that non-steroidal anti-inflammatory drugs, which inhibit COX activity result in increased COX protein expression [41], while differences between COX protein expression and activity have been reported to be produced by some cytokines, including tumor necrosis factor-α, and also nitric oxide donors [42, 43]. Although not well understood, evidence for regulation of COX expression at the post-transcriptional level is increasing [44, 45]. The down regulation of COX-2 transcripts in LPS-treated, IDO-expressing RAW cells is reminiscent of the endotoxin tolerance effect observed in human THP-1 promonocytic cells. Cells pretreated with LPS and thus expressing COX-2 showed down regulation of COX-2 mRNA when subjected to a second LPS exposure [46]. In addition, the COX-2 inhibitor flufenamic acid induced COX-2 expression in RAW cells but inhibited TNF-α or LPS-induced COX-2 expression in the same cell type [47].
As both MMPs and COX-2 are important factors in tumor development [48, 49] IDO's role in tumorigenesis bears investigating. We have observed IDO expression routinely in murine tumors in vivo, and are presently investigating the growth properties of tumors with altered IDO expression. In addition, our recent work indicates a role for IDO during pregnancy. Pharmacological inhibition of IDO results in pronounced inflammation, complement activation and fetal loss [17, 50]. Prostaglandins may provide a common link between these important biological phenomena.
Conclusions
IDO regulates adhesion of cells to normal growth substrates. In so doing it modulates the expression and activity of COX-2 and certain MMPs. RAW cells and MC57 cells overexpressing IDO grew as multicellular foci. In the case of RAW cells, this was due to elevated PGE relative to other prostaglandins. P19 cells in which endogenousIDO expression was disrupted by antisense expression, showed lower adhesiveness. Thus, tryptophan catabolism exerts control over fundamental cellular functions.
Materials and Methods
Cells
P19 cells were obtained from the American Type Culture Collection and cultured as described [23]. Cells were differentiated into myocytes or neurones using 1% DMSO and 10-6 M RA respectively as previously reported [23, 24]. RAW 264.7 cells were a gift of Dr. D. Greaves (Oxford, England) and were cultured in Iscove's Modified Dulbecco's Medium supplemented with 10% fetal calf serum. MC57 cells were obtained from Dr. Dimitrios Moskiphidis, Medical College of Georgia and grown in Iscove's Modified Dulbecco's Medium.
IDO expression
A full length, 1.2 kb IDO cDNA was amplified from IFN-γ stimulated RAW cells and cloned into pGEM T-Easy (Promega), using primers; 5' TAG CGG CCG CGT AGA CAG CAA TGG CAC TC 3' forward, 5' TAA GAT CTT ACA CTA AGG CCA ACT CAG 3' reverse, which contain Not I and Bgl II sites respectively. The 1.2 kb IDO PCR fragment was excised with Not I and Bgl II and cloned into the Not I-Bgl II site in the pDOI vector [51], previously modified by the introduction of a Not I site in front of the Eco RI cloning site. Plasmid DNA was linearized and transfected into RAW cells by electroporation. Stably transfected lines were selected in 400 mg/ml G418 and thereafter maintained in 200 mg/ml G418. MC57 cells were also transfected by electroporation and selected in 1.2 mg/ml G418.
RT-PCR
Analysis of gene expression in P19 or RAW cells was performed using semi-quantitative RT-PCR. Total RNA was isolated from cells using RNA STAT-60 (Tel-Test Inc.) and 1 mg was amplified for 25 cycles unless otherwise stated, following reverse transcription in a one step reaction (RT-PCR "Access", Promega). 5 μl of the 50 ml reaction volume was electrophoresed on 0.8% agarose gels prior to Southern blotting and hybridization with a specific probe. Primers and amplification conditions for IDO amplification have been described elsewhere [17]. Primers for amplification of other gene specific transcripts were as follows; stromelysin-1; 5' GATGACAGGGAAGCTGGA forward, 5' ACTGCGAAGATCCACTGA reverse. Stromelysin-2; 5' GATGTACCCAGTCTACAGGT 3' forward, 5' TGTCTTGTCTCATCATTACT 3' reverse. Stromelysin-3; 5' CTGCTGCTCCTGTTGCTGCT 3' forward, 5' ACCTTGGAAGAACCAAATC 3' reverse. Meltrin-α; 5' TGCATCAGTGGTCAGCCTCA 3' forward, 5' CTTTCTCTGCGGCCATTCTG 3' reverse. Meltrin-β; 5' TTCAGTTTACACATCAGAC 3' forward, 5' AGGTCACATTGCCGAACCT 3' reverse. Collagenase I; 5' GATTGTGAACTATACTCCT 3' forward, 5' CCATAGTCTGGTTAACATCA 3' reverse. Collagenase IV; 5' GTATGGAGCGACGTCACT 3' forward, 5' CGCTCCAGAGTGCTGGCA 3' reverse. GAPDH; 5' TGCAGTGGCAAAGTGGAG 3' forward 5' CCATCCACAGTCTTCTG 3' reverse.
Antisense inhibition of IDO expression
Constructs which expressed either sense or antisense IDO RNA were produced by cloning a 740 bp RT-PCR fragment of the IDO gene, described in [17], into the T-tailed cloning vector pGEM T-Easy (Promega). This fragment was excised with Not I and subcloned into the Not I site of the mammalian expression vector pcDNA3 (Invitrogen), in either the sense or antisense orientation. Following linearization with Bgl II, the constructs were transfected into the P19 cell line using Lipofectamine (Gibco-BRL) at a concentration of 25 μl per 100 ml of serum free medium. Stable transfectants were selected in 400 mg/ml G418 over a period of 4 weeks and subsequently maintained in the absence of G418 in normal growth medium. Periodic checks of G418 resistance revealed no significant loss of the resistance phenotype. Confirmation that resisitant clones expressed IDO antisense RNA was obtained by isolating total RNA from G418 resistant clones, treating with ribonuclease free DNase RQ1 (Promega) and reverse transcribing RNA into cDNA in the presence of an IDO sense primer [17]. An antisense primer was then added and the cDNA PCR amplified for 25 cycles. Products were electrophoresed in 0.8% agarose.
Western blotting
RAW cells expressing IDO and vector only controls were harvested in cell lysis buffer (PBS, 1%NP40, 0.5% sodium deoxycholate, 0.1% SDS, 150 ng/ml PMSF, 100 ng/ml aprotinin) and 25 μg of cell protein was electrophoresed on 10% polyacrylamide gels overlayed with a 5% stacking gel. Protein was quantitated using the BCA assay (Pierce). COX-1 and COX-2 antibodies (Santa Cruz Biotechnology Inc) were used in combination with standard ECL techniques. Rabbit polyclonal IDO-specific antibody was generated against a C-terminal peptide of 42 amino acids; KPSKKKPTDGDKSEEPSNVESRGTGGTNPMTELRSVKDTTEK.
Measurement of tryptophan depletion by HPLC
Supernatants from cell cultures were extracted with HPLC grade methanol and analyzed on a Beckman Phenomenix C18(2) HPLC column and eluted with a 0-80% gradient of acetonitrile over 20 minutes. To validate retention times and for the construction of a concentration curve a standard mixture of kynurenine and tryptophan was analyzed for each assay.
Analysis of prostaglandin production
Prostaglandin synthesis was measured by pulsing IDO-expressing RAW cells and vector only controls with 14C arachidonic acid (Sigma). 5 × 106 cells were harvested and resuspended in PBS and incubated at 37°C with 1.3 mCi arachidonic acid (53 mCi/mmol) for 30 mins. Following ether extraction, samples were dissolved in ethyl acetate and spotted onto thin layer chromatography plates. Plates were developed in ethyl acetate: acetic acid, 90:1, together with unlabeled standards. Individual spots were excised from the chromatogram and radioactivity determined by scintillation counting.
Cell adhesion assay
Cell adhesion assays were performed essentially as described [36]. Briefly, cells were seeded into the wells of a 24 well plate coated with various growth substrates Following incubation at 37°C, for 45 minutes, cells unattached cells were removed by PBS washes and the remaining cells were counted.
Cell migration assay
P19 cells in log phase growth were trypsinized and 105 were seeded in quadruplicate into Falcon cell culture inserts, with or without Matrigel coating (Becton Dickinson, Franklin Lakes, NJ, 8.0 μm pore size, 1 × 105pores/sq.cm), in a volume of 0.2 ml, in a 24 well tissue culture plate. The lower chamber contained a volume of 0.8 ml growth medium, while the final volume in the upper chamber was 0.35 ml. Chambers were incubated for 18 hours after which time the number of cells in the lower chamber was determined.
Image analysis
The size of individual P19 aggregates was determined by capturing fields of 40-50 aggregates at 10x magnification and then calculating the area of each aggregate using the NIH Image (1.62) analysis program (http://rsb.info.nih.gov/nih-image/download.html).
Abbreviations
CMV: cytomegalovirus
COX: cyclooxygenase
IDO: indoleamine 2,3 dioxygenase
IFN-γ interferon gamma
LPS: lipopolysaccharide
MMP: matrix metalloproteinase
NSAID: non-steroidal anti-inflammatory drug
PG: prostaglandin
TDO: tryptophan 2, 3 dioxygenase
Acknowledgments
Acknowledgements
We wish to thank Anita Wylds, Carolyn Leithner and John Nechtman for expert technical assistance and Dr. Steve Vogel for assistance with image analysis. These studies were supported by grants AA44219 and AI 42247 from the National Institutes of Health to ALM, the Departments of Medicine, Medical College of Georgia and generous support from the Carlos and Marguerite Mason Trust.
Contributor Information
Brendan Marshall, Email: marshall@immagene.mcg.edu.
Derin Benerci Keskin, Email: mp3940@medmail.mcg.edu.
Andrew L Mellor, Email: mellor@immagene.mcg.edu.
References
- Kotake Y, Masayama T. Uber den mechanismus der kynurein-bildung aus tryptophan. Hoppe-Seyler's Z Physiol Chem. 1937;243:237–244. [Google Scholar]
- Shimizu T, Nomiyama S, Hirata F, Hayaishi O. Indoleamine 2,3 dioxygenase. Purification and some properties. J Biol Chem. 1978;253:4700–4706. [PubMed] [Google Scholar]
- Yoshida R, Nukiwa Y, Watanabe Y, Fujiwara M, Hirata F, Hayaishi O. Regulation of indoleamine 2,3 dioxygenase activity in the small intestine and epididymis of mice. Arch Biochem Biophys. 1980;203:343–351. doi: 10.1016/0003-9861(80)90185-X. [DOI] [PubMed] [Google Scholar]
- Yamazaki F, Kuroiwa T, Takikawa O, Kido R. Human indolylamine 2,3 dioxygenase. Its tissue distribution, and characterization of the placental enzyme. Biochem J. 1985;230:635–638. doi: 10.1042/bj2300635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MW, Feng G. Relationship between interferon-γ, indoleamine 2,3 dioxygenase and tryptophan catabolism. FASEB J. 1991;5:2516–2522. [PubMed] [Google Scholar]
- Suzuki T, Takagi T. A myoglobin evolved from indoleamine 2,3 dioxygenase. J Mol Biol. 1992;228:698–700. doi: 10.1016/0022-2836(92)90854-D. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Urade Y, Tokuda M, Hayaishi O. Induction of indoleamine 2,3 dioxygenase in mouse lung during virus infection. Proc Natl Acad Sci USA. 1979;76:4084–4086. doi: 10.1073/pnas.76.8.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne GI, Lehman LK, Landry GJ. Induction of tryptophan catabolism is the mechanism for gamma interferon-mediated inhibition of intracellular Chlamydia psittaci replication in T24 cells. Infect Immun. 1986;53:347–351. doi: 10.1128/iai.53.2.347-351.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer-Avni Y, Wallach D, Sariv I. Reversal of the antichlamydial effect of tumor necrosis factor by tryptophan and antibodies to beta interferon. Infect Immun. 1989;57:3484–3490. doi: 10.1128/iai.57.11.3484-3490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferkorn ER. Interferon-γ blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cell to degrade tryptophan. Proc Natl Acad Sci USA. 1994;81:908–912. doi: 10.1073/pnas.81.3.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie CR, Hadding U, Daubener W. Interferon-gamma-induced activation of indoleamine 2,3-dioxygenase in cord blood monocyte-derived macrophages inhibits the growth of group B streptococci. J Infect Dis. 1998;178:875–878. doi: 10.1086/515347. [DOI] [PubMed] [Google Scholar]
- Yasui H, Takai K, Yoshida R, Hayaishi O. Interferon enhances tryptophan metabolism by inducing pulmonary indoleamine 2, 3 dioxygenase. Proc Natl Acad Sci USA. 1986;83:6622–6626. doi: 10.1073/pnas.83.17.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanain HH, Chon SY, Gupta SL. Differential regulation of human indoleamine 2,3 dioxygenase gene expression by interferons γ and α. J Biol Chem. 1993;268:5077–5084. [PubMed] [Google Scholar]
- Chon SY, Hassanain HH, Gupta SL. Cooperative role of interferon regulatory factor 1 and p91(STAT 1) responsive elements in interferon-γ inducible expression of indoleamine 2, 3 dioxygenase. J Biol Chem. 1996;271:17247–17252. doi: 10.1074/jbc.271.29.17247. [DOI] [PubMed] [Google Scholar]
- Konan KV, Taylor MW. Importance of two interferon-stimulated response element sequences in the regulation of the human indoleamine 2, 3 dioxygenase gene. J Biol Chem. 1996;271:19140–19145. doi: 10.1074/jbc.271.32.19140. [DOI] [PubMed] [Google Scholar]
- Munn DH, Shafizadeh E, Attwood JT, Bondarev I, Pashine A, Mellor AL. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J Exp Med. 1999;189:1363–1372. doi: 10.1084/jem.189.9.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn DH, Zhou M, Attwood JT, Bondarev I, Conway SJ, Marshall B, Brown C, Mellor AL. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281:1191–1193. doi: 10.1126/science.281.5380.1191. [DOI] [PubMed] [Google Scholar]
- Labbe S, Larouche L, Mailhot D, Seguin C. Purification of mouse MEP-1 nuclear protein which binds to the metal regulatory elements of genes encoding metallothionein. Nucl Acids Res. 1993;94:1549–1554. doi: 10.1093/nar/21.7.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee J, Kuncio GS, Bhandari B, Shihab FS, Neilson EG. Identification of promoter activity and differential expression of transcripts encoding the murine stromelysin gene in renal cells. Kidney Int. 1997;52:120–129. doi: 10.1038/ki.1997.311. [DOI] [PubMed] [Google Scholar]
- Varga J, Yufit T, Brown RR. Inhibition of collagenase and stromelysin gene expression by interferon-γ in human dermal fibroblasts is mediated in part via tryptophan degradation. J Clin Invest. 1995;96:475–481. doi: 10.1172/JCI118058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J, Yufit T, Hitraya E, Brown RR. Control of extracellular matrix degradation by interferon gamma. The tryptophan connection. Adv Exp Med Biol. 1996;398:143–148. doi: 10.1007/978-1-4613-0381-7_23. [DOI] [PubMed] [Google Scholar]
- Cornain S, Klein E. Characteristics and in vitro growth influencing effects of the spleen cell population in a methylcholanthrene induced mouse sarcoma system. Z Immunitatsforsch Immunobiol. 1978;154:101–114. [PubMed] [Google Scholar]
- McBurney MW, Jones-Villeneuve EMV, Edwards MKS, Anderson J. Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature. 1982;299:165–167. doi: 10.1038/299165a0. [DOI] [PubMed] [Google Scholar]
- Jones-Villeneuve EMV, McBurney MW, Rogers KA, Kalnins VI. Retinoic Acid induces embryonal carcinoma cells to differentiate into neurons and glial cells. J Cell Biol. 1982;94:253–262. doi: 10.1083/jcb.94.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MK, Harris JF, McBurney MW. Induced muscle differentiation in an embryonal carcinoma cell line. Mol Cell Biol. 1983;3:2280–2286. doi: 10.1128/mcb.3.12.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisaki T, Masuda A, Osumi N, Nabeshima Y. Spatially and temporally restricted expression of meltrin alpha (ADAM 12) and beta (ADAM 19) in mouse embryo. Mech Dev. 1998;73:211–215. doi: 10.1016/S0925-4773(98)00043-4. [DOI] [PubMed] [Google Scholar]
- Wahl LM, Corcoran ML, Mergenhausen SE, Finbloom DS. Inhibition of phospholipase activity in human monocytes by IFN-γ blocks endogenous prostaglandin E2 dependent collagenase production. J Immunol. 1990;144:3518–3522. [PubMed] [Google Scholar]
- Corcoran ML, Stetler-Stevenson WG, Brown PD, Wahl LM. Interleukin 4 inhibition of prostaglandin E2 synthesis blocks interstitial collagenase and 92-kDa type IV collagenase/gelatinase production by human monocytes. J Biol Chem. 1992;267:515–519. [PubMed] [Google Scholar]
- Mertz PM, DeWitt DL, Stetler-Stevenson WG, Wahl LM. Interleukin 10 suppression of monocyte prostaglandin H synthase-2. Mechanism of inhibition of prostaglandin dependent matrix metalloproteinase production. J Biol Chem. 1994;269:21322–21329. [PubMed] [Google Scholar]
- Busiek DF, Baragi V, Nehring LC, Parks WC, Welgus HG. Matrilysin expression by human mononuclear phagocytes and its regulation by cytokines and hormones. J Immunol. 1995;154:6484–6491. [PubMed] [Google Scholar]
- Urade Y, Ujihara M, Horiguchi Y, Ikai K, Hayaishi O. The major source of endogenous prostaglandin D2 production is likely antigen presenting cells. J Immunol. 1989;143:2982–2989. [PubMed] [Google Scholar]
- Lee SH, Soyoola E, Chanmugam P, Hart S, Sun W, Zhong H, Liou S, Simmons D, Hwang D. Selective expression of mitogen-inducible cyclooxygenase in macrophages stimulated with lipopolysaccharide. J Biol Chem. 1992;267:25934–25938. [PubMed] [Google Scholar]
- Endo T, Ogushi F, Sone S. LPS-dependent cyclooxygenase-2 induction in human monocytes is down regulated by IL-13 but not by IFN-gamma. J Immunol. 1996;156:2240–2246. [PubMed] [Google Scholar]
- Mellor A, Munn DH. Tryptophan catabolism and T cell tolerance: immunosuppression by starvation. Immunol Today. 1999;20:469–473. doi: 10.1016/S0167-5699(99)01520-0. [DOI] [PubMed] [Google Scholar]
- Murakami M, Naraba H, Tanioka T, Semmyo N, Nakatani Y, Kojima F, Ikeda T, Fueki M, Ueno A, Oh-ishi S, Kudo I. Regulation of prostaglandin E2 biosynthesis by inducible membrane-associated prostaglandin E2 synthase in concert with cyclooxygenase-2. J Biol Chem. 2000;275:32783–32792. doi: 10.1074/jbc.M003505200. [DOI] [PubMed] [Google Scholar]
- Tsujii M, DuBois RN. Alterations in cellular adhesion and apoptosis in epithelial cells overexpressing prostaglandin endoperoxide synthase 2. Cell. 1995;83:493–501. doi: 10.1016/0092-8674(95)90127-2. [DOI] [PubMed] [Google Scholar]
- Rocca B, Spain LM, Ciabattoni G, Patrono C, FitzGerald GA. Differential expression and regulation of cyclooxygenase isozymes in thymic stromal cells. J Immunol. 1999;162:4589–4597. [PubMed] [Google Scholar]
- Gowen BB, Borg TK, Ghaffar A, Mayer EP. Selective adhesion of macrophages to denatured forms of type I collagen is mediated by scavenger receptors. Matrix Biol. 2000;19:61–71. doi: 10.1016/S0945-053X(99)00052-9. [DOI] [PubMed] [Google Scholar]
- Ueno R, Shimizu T, Kondo K, Hayaishi O. Activation mechanism of prostaglandin endoperoxide synthetase by hemoproteins. J Biol Chem. 1982;257:5584–5588. [PubMed] [Google Scholar]
- Blanco JCG, Contursi C, Salkowski CA, DeWitt DL, Ozato K, Vogel SN. Interferon regulatory factor (IRF)-1 and IRF-2 regulate interferon γ-dependent cyclooxygenase 2 expression. J Exp Med. 2000;191:2131–2144. doi: 10.1084/jem.191.12.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade EA, McIntyre TM, Zimmerman GA, Prescott SM. Peroxisome proliferators enhance cyclooxygenase-2 expression in epithelial cells. J Biol Chem. 1999;274:8328–8334. doi: 10.1074/jbc.274.12.8328. [DOI] [PubMed] [Google Scholar]
- Pang L, Knox AJ. Effect of interleukin 1β, tumor necrosis factor-α and interferon-γ on the induction of cyclooxygenase-2 in cultured human airway smooth muscle cells. Br J Pharmacol. 1997;121:579–587. doi: 10.1038/sj.bjp.0701152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitahige M, Morita I, Murota S. Existence of endogenous inhibitor(s) of prostaglandin endoperoxide H synthase activities in murine NIH 3T3 fibroblasts. Prostaglandins Leukot Essent Fatty Acids. 1999;61:97–103. doi: 10.1054/plef.1999.0077. [DOI] [PubMed] [Google Scholar]
- Dixon DA, Kaplan CD, McIntyre TM, Zimmerman GA, Prescott SM. Post-transcriptional control of cyclooxygenase expression. J Biol Chem. 2000;275:11750–11757. doi: 10.1074/jbc.275.16.11750. [DOI] [PubMed] [Google Scholar]
- Sheng H, Shao J, Dixon DA, Williams CS, Prescott SM, DuBois RN, Beauchamp RD. Transforming growth factor β1 enhances Ha-ras-induced expression of cyclooxygenase-2 in intestinal epithelial cells via stabilization of mRNA. J Biol Chem. 2000;275:6628–6635. doi: 10.1074/jbc.275.9.6628. [DOI] [PubMed] [Google Scholar]
- Fernando LP, Fernando AN, Ferlito F, Halushka PV, Cook JA. Suppression of Cox-2 and TNF-alpha mRNA in endotoxin tolerance: effect of cycloheximide, actinomycin D and oakadaic acid. Shock. 2000;14:128–133. doi: 10.1097/00024382-200014020-00009. [DOI] [PubMed] [Google Scholar]
- Paik JH, Ju JH, Boudreau MD, Hwang DH. Two opposing effects of nonsteroidal anti-inflammatory drugs on the expression of the inducible cyclooxygenase. Mediation through different signaling pathways. J Biol Chem. 2000;275:28173–28179. doi: 10.1074/jbc.M002329200. [DOI] [PubMed] [Google Scholar]
- Oshima M, Dinchuk JE, Kargman SL, Oshima H, Hancock B, Kwong E, Trzaskos JM, Evans JF, Taketo MM. Suppression of intestinal polyposis in Apc delta716 knockout mice by inhibition of cyclooxygenase 2 (COX-2). Cell. 1996;87:803–809. doi: 10.1016/S0092-8674(00)81988-1. [DOI] [PubMed] [Google Scholar]
- Williams CS, Tsujii M, Reese J, Dey SK, DuBois RN. Host cyclooxygenase modulates carcinoma growth. J Clin Invest. 2000;105:1589–1594. doi: 10.1172/JCI9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor AL, Sivakumar J, Chandler P, Smith K, Molina H, Mao D, Munn DH. Tryptophan catabolism suppresses T cell induced inflammation and B-cell independent complement activation during pregnancy. Nature Immunol. 2001;2:64–68. doi: 10.1038/83183. [DOI] [PubMed] [Google Scholar]
- Kousakoff V, Fehling HJ, Lemeur M, Benoist C, Mathis D. A vector driving the expression of foreign cDNAs in the MHC classII-positive cells of transgenic mice. J Immunol Meth. 1993;166:287–291. doi: 10.1016/0022-1759(93)90370-M. [DOI] [PubMed] [Google Scholar]