Abstract
Two known papuamides C (1) and D (2) together with two new depsipeptides, papuamides E (3) and F (4), were isolated from an undescribed sponge of the genus Melophlus collected in the Solomon Islands. The planar structures of the compounds were elucidated on the basis of spectroscopic studies. Papuamides C–F (1–4) showed cytotoxicity against brine shrimp with LD50 values between 92 and 106 μg/mL.
Keywords: Papuamide, Melophlus, Depsipeptide, Cytotoxic
1 Introduction
Marine sponges that are active against human immunodeficiency virus (HIV) have served as a rich source of cyclic depsipeptides incorporating rare amino acid residues. The marine sponges Theonella swinhoei and Theonella mirabilis (family Theonellidae) from Papua New Guinea have yielded papuamides A–C, cyclic depsipeptides with structurally unique features including unprecedented acid moieties.1 Callipeltin A, isolated from the New Caledonian marine sponge Callipelta sp.2, neamphamide A, obtained from Neamphius huxlei3 and mirabamides A–D from Siliquariaspongia mirabilis4 are also well recognized for their potent HIV-inhibitory activity. Mirabamide C and novel congeners mirabamides E–H with anti-HIV activity were also recently reported from Stelletta clavosa (family Ancorinidae).5
As part of our continuing search for bioactive metabolites from marine invertebrates, in the present study, we isolated two new depsipeptides (3–4) along with the previously reported papuamides C and D1 from the butanol extract of a marine sponge Melophlus sp. Marine sponges of the genus Melophlus (family Ancorinidae) have been reported to yield compounds belonging to tetramic acids and saponins.6–9 This is the first report of depsipeptides from Melophlus sp. In this paper, we describe the isolation, structure elucidation and bioactivity of these new depsipeptides.
2 Results and discussion
The sponge was extracted three times at room temperature using MeOH followed by CH2Cl2. The extracts were combined, evaporated in vacuo and partitioned between H2O and CH2Cl2. The aqueous layer was further partitioned with n-BuOH and the active n-BuOH extract was subjected to bioassay guided fractionation using C18 bonded silica. Further purification by reverse-phase C18 HPLC afforded compounds (1–4).
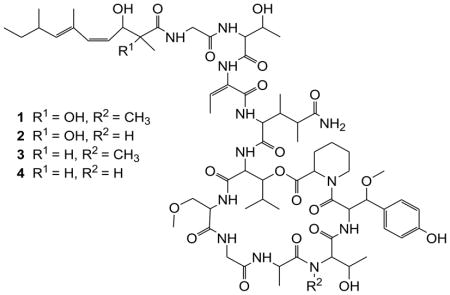
The major metabolites, compounds 1 and 2, showed protonated molecular ions [M+H]+ at m/z 1399.73678 and 1385.7203 in the HRMS (ESI), respectively. A marinlit (2010) search for the corresponding masses revealed a match to the known papuamides C and D.10 The NMR spectral data of 1 and 2 were identical to the literature values of papuamides C and D, respectively.
Compounds 3 and 4 were isolated in smaller quantities compared with 1 and 2. Comparison of the 1H NMR spectra of 3 and papuamide C (1) indicated strong similarities over most of the molecule. Detailed 2D NMR data analysis of 3 together with comparison to 1 enabled us to identify 11 amino acid residues present in 3 to be identical to the corresponding residues in 1: two glycine (Gly) residues, threonine (Thr), aminobutenoic acid (Aba), 3,4-dimethylglutamine (3,4-DiMeGln), 3-hydroxyleucine (3-OHLeu), 3-methoxyalanine (3-OMeAla), alanine (Ala), N-methylthreonine (NMeThr), β-methoxytyrosine (β-OMeTyr) and homoproline (Hpr) (Table 1).
Table 1.
NMR Spectroscopic Data (400 MHz, CD3OD), for Papuamides E (3) and F (4)
| Papuamide E (3) | Papuamide F (4) | |||
|---|---|---|---|---|
| δC | δHa | δC | δHa | |
| Homoproline (Hpr) | ||||
| 1 | n.o. | – | n.o. | – |
| 2 | 52.7 | 5.32 m | 52.7 | 5.18 d (9.6) |
| 3 | 27.3 | 2.20 m, 1.77 m | 27.5 | 2.25 m |
| 4 | 21.1 | 1.26 m | 21.7 | 1.75 s |
| 5 | 26.9 | 1.76 m | 25.8 | 1.72 ovl |
| 6 | 44.3 | 4.15 m | 44.9 | 4.06 m |
| β-Methoxytyrosine (β-OMeTyr) | ||||
| 1 | n.o. | – | n.o. | – |
| 2 | 54.0 | 5.15 d (5.2) | 54.0 | 5.22 m |
| 3 | 85.5 | 4.25 d (9.6) | 84.9 | 4.33 m |
| 3-OMe | 56.1 | 3.10 s | 56.9 | 3.15 s |
| 1′ | 129.3 | – | 129.1 | – |
| 2′, 6′ | 130.6 | 7.21 d (8.4) | 130.3 | 7.20 d (8.4) |
| 3′, 5′ | 116.0 | 6.77 d (8.4) | 116.0 | 6.76 d (8.4) |
| 4′ | 159.2 | – | 158.7 | |
| N-Methylthreonine (NMeThr/Thr-2) | ||||
| 1 | n.o. | – | n.o. | – |
| 2 | 59.3 | 3.37 m | 60.1 | 3.93 m |
| 3 | 64.1 | 3.89 m | 66.8 | 3.94 m |
| 4 | 20.0 | 0.52 d (6.4) | 19.7 | 0.76 d (6.0) |
| N-Me | 31.1 | 3.15 s | ||
| Alanine (Ala) | ||||
| 1 | n.o. | – | 175.0 | – |
| 2 | 51.1 | 4.68 m | 51.1 | 4.32 m |
| 3 | 15.8 | 1.46 d (7.2) | 17.4 | 1.47 d (7.6) |
| Glycine (Gly-1) | ||||
| 1 | n.o. | – | n.o. | – |
| 2 | 43.8 | 3.85 s | 43.9 | 4.13 m, 3.85 m |
| 3-Methoxyalanine (3-OMeAla) | ||||
| 1 | n.o. | – | n.o. | – |
| 2 | 56.0 | 4.18 m | 56.0 | 4.35 m |
| 3 | 70.9 | 3.44 m, 3.46 m | 71.9 | 3.83 m, 3.66 m |
| 3-OMe | 59.5 | 3.44 s | 59.2 | 3.38 s |
| 3-Hydroxyleucine (3-OHLeu) | ||||
| 1 | n.o. | – | n.o. | – |
| 2 | 54.0 | 4.75 m | 54.9 | 4.87 m |
| 3 | 77.4 | 5.40 m | 77.6 | 5.40 m |
| 4 | 29.6 | 2.04 m | 29.4 | 2.03 m |
| 5 | 17.6 | 0.92 m | 16.8 | 0.91 ovl |
| 5′ | 19.8 | 0.90 m | 19.6 | 0.90 ovl |
| 3,4 - Dimethylglutamine (3,4-DiMeGln) | ||||
| 1 | n.o. | – | n.o. | – |
| 2 | 58.7 | 4.21 m | 58.5 | 4.32 m |
| 3 | 39.7 | 2.15 m | 38.0 | 2.20 m |
| 4 | 42.9 | 2.64 m | 42.4 | 2.59 m |
| 5 | 180.5 | – | n.o. | – |
| 3-Me | 13.6 | 0.95 ovl | 14.0 | 0.98 d (6.8) |
| 4-Me | 15.5 | 1.19 d (7.2) | 15.5 | 1.16 d (7.2) |
| Aminobutenoic acid (Aba) | ||||
| 1 | n.o. | – | n.o. | – |
| 2 | n.o. | – | n.o. | – |
| 3 | 132.0 | 6.55 m | 131.1 | 6.57 q (7.2) |
| 4 | 13.0 | 1.77 d (4.3) | 12.9 | 1.78 d (4.0) |
| Threonine (Thr-1) | ||||
| 1 | n.o. | – | n.o. | – |
| 2 | 59.6 | 4.60 d (3.2) | 59.7 | 4.60 d (3.2) |
| 3 | 68.9 | 4.40 m | 68.9 | 4.40 dd (6.4, 3.6) |
| 4 | 19.8 | 1.24 d (6.4) | 19.8 | 1.24 d (6.4) |
| Glycine (Gly-2) | ||||
| 1 | n.o. | – | n.o. | – |
| 2 | 44.0 | 4.14 m | 45.0 | 3.39 m, 3.38 m |
| 2,3-Dihydroxy-2,6,8-trimethyl-4,6-decadienoic acid (Dhtda) | ||||
| 1 | 178.5 | – | 178.4 | – |
| 2 | 48.7 | 2.41 m | 48.7 | 2.42 m |
| 3 | 71.5 | 4.72 m | 71.3 | 4.69 t (9.6) |
| 4 | 129.8 | 5.29 ovl | 129.8 | 5.29 m |
| 5 | 137.9 | 6.09 m | 137.8 | 6.08 dd (11.6, 2.8) |
| 6 | 132.0 | – | 132.0 | – |
| 7 | 139.6 | 5.26 m | 139.7 | 5.26 m |
| 8 | 35.4 | 2.36 m | 35.4 | 2.37 m |
| 9 | 31.3 | 1.35 m, 1.30 m | 31.1 | 1.39 m, 1.29 m |
| 10 | 12.3 | 0.89 ovl | 12.3 | 0.88 ovl |
| 2-Me | 14.2 | 1.05 d (6.4) | 14.1 | 1.05 d (6.8) |
| 6-Me | 16.8 | 1.80 s | 16.8 | 1.80 s |
| 8-Me | 20.8 | 0.98 d (6.8) | 20.8 | 0.98 ovl |
Ovl: overlapped; n.o.: not observed.
Coupling constants are in parentheses and given in hertz. Although an HMBC was recorded, more assignments thus appear not possible.
A molecular formula of C66H102N12O20 for 3 was suggested by a pseudomolecular ion at 1369.7250 by HRMS (ESI) ([M+H]+; calcd. for C66H103N12O20 1369.7250; = 0.0 ppm) which differs from papuamide C (1) by loss of an oxygen atom. The quaternary oxygenated carbon signal at δC 78.8 ppm in 1 has been replaced in 3 by a methine at δH 2.41 ppm, δC 48.7 ppm as suggested by the HMBC correlation of the proton Me-2Htda (δH 1.05 ppm) to the carbon C-2Htda (δC 48.7 ppm) and the absence of any HMBC correlations in 3 to the quaternary carbon (C-2Dhtda in 1). The upfield shift of H-3Dhtda (from δH 4.88 ppm to δH 4.72 ppm) and Me-2Dhtda (from δC 22.4 ppm to δC 14.2 ppm) are further consistent with the replacement of the 2,3-dihydroxy-2,6,8-trimethyl-4,6-decadienoic acid moiety (Dhtda) of papuamide C (1) with a 3-hydroxy-2,6,8-trimethyl-4,6-decadienoic acid moiety (Htda) in 3, accounting for the difference in molecular formula between the two compounds. The chemical shifts for the Htda moiety in 3 were consistent with the proton and carbon chemical shifts assigned for the Htda moiety in mirabamide H5. However, a large (~15 Hz) coupling was not evident even if the actual coupling could not be resolved for H4 and H5 of Htda of compound 3. Furthermore, the agreement of the chemical shifts of all the NMR signals of Htda moiety in compound 3 to the Htda moiety in mirabamide H suggests a Z geometry to the C4-C5 olefin and E geometry to the C6-C7 double bond.
The HRESI(+)MS of papuamide F (4) indicated a molecular formula of C65H100N12O20 which differed from that of papuamide E (3) by loss of CH2. The N-methyl (δC 31.1 ppm, δH 3.15 ppm) signal in 3 was absent in the HSQC spectrum of 4. Compound 3 differed from compound 4 in that the threonine residue of 3 was N-methylated while 4 was not. This relationship was similar to that of papuamides C and D in that papuamide C was N-methylated while papuamide D was not. Thus, 4 and 3 are analogues of 1 and 2, respectively, in which the Dhtda moiety has been replaced with Htda. Moreover, the C4–C5 olefin of the Htda was assigned a Z geometry on the basis of coupling of approximately 11 Hz between H4 and H5 as in the papuamides1 and mirabamides5. The agreement of the chemical shifts of all NMR signals in Htda near the C6–C7 double bond suggests the same stereochemistry (E geometry) as in 1 and 2.
The optical rotation signs reported for all papuamides were same to that of the obtained for the compounds 3 and 4 indicating that all the amino acid residues in compounds 3 and 4 possessed identical configurations to papuamides A–D1. The compounds, 3 and 4, were obtained as optically active white powders that are concluded as new analogues of papuamides A–D and proposed as papuamides E and F, respectively.
Brine shrimp assay11 revealed these analogues (1–4) to be cytotoxic with LD50 values of 92, 92, 104 and 106 μg/mL, respectively. However, papuamides C–F (1–4) were found to be inactive against methicillin resistant Staphylococcus aureus (ATCC 10537), vancomycin resistant Enterococcus faecium (ATCC 12952), wild type Candida albicans (ATCC 32354) and amphotericin resistant Candida albicans (ATCC 90873).
3 Experimental section
3.1 General experimental procedures
Optical rotations were recorded on a Bellingham Stanley ADP220 polarimeter. NMR spectra were recorded on a Varian spectrometer operating at 400 MHz. Chemical shifts are referenced to residual MeOH (δC 49.0; δH 3.31) in CD3OD. High resolution ESI-MS analyses were obtained using Thermo Scientific LTQ Orbitarp Discovery LC-MS in positive electrospray ionisation mode. Reverse phase flash chromatography was performed on Bakerbond C18 40μm prep LC packing. Semi-preparative HPLC was performed using a Waters 515 HPLC system with a Alltech 10 μm C18 (250 x 10 mm) column.
3.2 Animal material (collection and taxonomy)
The marine sponge Melophlus sp. (family Ancorinidae) was collected by hand using scuba at a depth of 10 m from Karumolum Pt, Russell Island in the Solomon Islands (S8° 85.76′ and E159° 6.98′) on 21st June 2006. The marine sponge was identified by Prof. John Hooper of Queensland Museum, Australia. Voucher specimens of the sponge, SOL06-1-018, are preserved at University of Utah and The University of the South Pacific.
3.3 Extraction, isolation and purification
The frozen sponge was cut into small pieces, extracted using MeOH (3 x 1000 mL) and followed by CH2Cl2 (3 x 1000 mL). The extracts were combined and evaporated to dryness under vacuum. The crude (4.5 g) was partitioned with CH2Cl2–H2O (3:1). The aqueous layer was further partitioned with n-BuOH. The resulting biologically active n-BuOH extract was subjected to RP-silica chromatography pre-equilibrated with aqueous MeOH (20% H2O). The column was eluted with a stepwise gradient of 20–100% MeOH(aq). The active (100% MeOH) elute was rechromatographed on RP-silica using stepwise gradient 20–100% MeOH(aq) to yield 12 fractions. The eighth fraction 80% MeOH(aq) was subjected on isocratic RP-HPLC using 83% MeOH(aq) at a flow rate of 4.0 mL/min and monitoring at a wavelength of 254 nm to yield 5 fractions. The fourth fraction was further purified on RP-HPLC using isocratic elution with 42% MeCN(aq) to yield pure compounds 1 (4.2 mg, tR = 25.3 min), 2 (4.1 mg, tR = 18.5 min), 3 (1.8 mg, tR = 31.0 min) and 4 (2.0 mg, tR = 22.1 min).
3.3.1 Papuamide E (3)
white powder; [α]25D +68.5 (c 0.07, MeOH); 1H NMR (400 MHz, CD3OD), see Table 1; HRMS (ESI): m/z 1383.7407 [M + H]+ (calcd for C66H103N12O20, 1383.7406), HRMS (ESI): m/z 1405.7230 [M + Na]+ (calcd for C66H102N12O20Na, 1405.7226).
3.3.2 Papuamide F (4)
white powder; [α]25D +107.1 (c 0.03, MeOH); 1H NMR (400 MHz, CD3OD), see Table 1; HRMS (ESI): m/z 1369.7250 [M + H]+ (calcd for C65H101N12O20, 1369.7250), HRMS (ESI): m/z 1391.7067 [M + Na]+ (calcd for C65H100 N12O20Na, 1391.7069).
Supplementary Material
Acknowledgments
The authors wish to acknowledge Prof. Marcel Jaspars and Dr. Jioji Tabudravu, University of Aberdeen, Scotland, for their assistance in performing HRESIMS and NMR analyses. We are also grateful to Prof. C.M. Ireland for collection of the marine sponge. We also like to acknowledge Prof. John Hooper, Queensland Museum, Australia, for the identification of the marine sponge. This work was supported by NCDDG grant U19 CA67786 awarded to Prof. C.M. Ireland at the University of Utah by the NIH and NCI. We also thank Solomon Islands government and people of Russell Island for permission to collect.
Footnotes
1H NMR, HSQC, HMBC and HSQC-TOCSY spectra for papuamides E and F.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ford PW, Gustafson KR, McKee TC, Shigematsu N, Maurizi LK, Pannell LK, Williams DE, Silva EDD, Lassota P, Allen TM, Van-Soest R, Andersen RJ, Boyd MR. J Am Chem Soc. 1999;121:5899–5909. [Google Scholar]
- 2.Zampella A, D’Auria MV, Paloma LG, Casapullo A, Minale L, Debitus C. J Am Chem Soc. 1996;118:6202–6209. [Google Scholar]
- 3.Oku N, Gustafson KR, Cartner LK, Wilson JA, Shigematsu N, Hess S, Pannell LK, Boyd MR, McMahon JB. J Nat Prod. 2004;67:1407–1411. doi: 10.1021/np040003f. [DOI] [PubMed] [Google Scholar]
- 4.Plaza A, Gustchina E, Baker HL, Kelly M, Bewley CA. J Nat Prod. 2007;70:1753–1760. doi: 10.1021/np070306k. [DOI] [PubMed] [Google Scholar]
- 5.Lu Z, Van Wagoner RM, Harper MK, Baker HL, Hooper JNA, Bewley CA, Ireland CM. J Nat Prod. 2011;74:185–193. doi: 10.1021/np100613p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang C, Wang B, Wiryowidagdo S, Wray V, Van-Soest R, Steube KG, Guan HS, Proksch P, Ebel R. J Nat Prod. 2003;66:51–56. doi: 10.1021/np0202778. [DOI] [PubMed] [Google Scholar]
- 7.Dai HF, Edrada RA, Ebel R, Nimtz M, Wray V, Proksch P. J Nat Prod. 2005;68:1231–1237. doi: 10.1021/np050152d. [DOI] [PubMed] [Google Scholar]
- 8.Xu J, Hasegawa M, Harada K, Kobayashi H, Nagal H, Namikoshi M. Chem Pharm Bull. 2006;54:852–854. doi: 10.1248/cpb.54.852. [DOI] [PubMed] [Google Scholar]
- 9.Lee HS, Seo Y, Cho KW, Rho JR, Shin J, Paul VJ. J Nat Prod. 2000;63:915–919. doi: 10.1021/np990589j. [DOI] [PubMed] [Google Scholar]
- 10.Munro MHG, Blunt JW. Marine Lit. University of Canterbury; Feb 15, 2011. vpc 15.5. [Google Scholar]
- 11.Sam TW. Bioactive Natural Products Detection, Isolation, and Structural Determination. Chapter 18. CRC Press; Boca Raton: 1993. pp. 441–456. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


