Abstract
Glucose modulates plant metabolism, growth, and development. In Arabidopsis (Arabidopsis thaliana), Hexokinase1 (HXK1) is a glucose sensor that may trigger abscisic acid (ABA) synthesis and sensitivity to mediate glucose-induced inhibition of seedling development. Here, we show that the intensity of short-term responses to glucose can vary with ABA activity. We report that the transient (2 h/4 h) repression by 2% glucose of AtbZIP63, a gene encoding a basic-leucine zipper (bZIP) transcription factor partially involved in the Snf1-related kinase KIN10-induced responses to energy limitation, is independent of HXK1 and is not mediated by changes in ABA levels. However, high-concentration (6%) glucose-mediated repression appears to be modulated by ABA, since full repression of AtbZIP63 requires a functional ABA biosynthetic pathway. Furthermore, the combination of glucose and ABA was able to trigger a synergistic repression of AtbZIP63 and its homologue AtbZIP3, revealing a shared regulatory feature consisting of the modulation of glucose sensitivity by ABA. The synergistic regulation of AtbZIP63 was not reproduced by an AtbZIP63 promoter-5′-untranslated region::β-glucuronidase fusion, thus suggesting possible posttranscriptional control. A transcriptional inhibition assay with cordycepin provided further evidence for the regulation of mRNA decay in response to glucose plus ABA. Overall, these results indicate that AtbZIP63 is an important node of the glucose-ABA interaction network. The mechanisms by which AtbZIP63 may participate in the fine-tuning of ABA-mediated abiotic stress responses according to sugar availability (i.e., energy status) are discussed.
To optimize their growth and development as sessile organisms, plants have developed a range of efficient mechanisms to sense and respond adequately to ever-changing environmental conditions. For instance, the availability of sugar produced through photosynthesis, which relies on light access, represents an important signal that, in combination with developmental and other environmental cues such as mineral nutrition, water availability, or pathogen attacks, influence the use of energy resources to ensure survival and propagation (Corruzi and Zhou, 2001; Forde, 2002; Rolland et al., 2006; Rook et al., 2006; Gutiérrez et al., 2007; Smith and Stitt, 2007; Baena-González and Sheen, 2008).
Sugars are key metabolic signals that control gene expression (Koch, 1996; Price et al., 2004; Li et al., 2006) and modulate different developmental phases, including embryogenesis, germination, seedling development, root growth, flowering, and important processes such as photosynthesis, senescence, and stress responses (Smeekens, 2000; Rolland et al., 2002; Moore et al., 2003; Gibson, 2005). Several sugar sensing and signaling mechanisms have been described. Suc, for example, which is the main transported form of sugar, specifically regulates the mRNA translation of five members of S-group basic-leucine zipper (bZIP) transcriptional regulators in Arabidopsis (Arabidopsis thaliana; Rook et al., 1998; Wiese et al., 2004).
Glc, one of the hydrolytic hexose products of Suc, is a major sugar signaling metabolite. The characterization of glucose insensitive2 (gin2) mutants has provided compelling evidence for a Hexokinase1 (HXK1)-dependent Glc sensing and signaling pathway that is uncoupled from HXK phosphorylation activity and mediates the repression of photosynthetic gene expression and growth control in Arabidopsis (Moore et al., 2003). The molecular mechanisms responsible for Glc-dependent transcriptional repression of the chlorophyll a/b CAB2 involve a nuclear HXK1 complex that binds the CAB2 promoter (Cho et al., 2006). Complementation of the gin2 Arabidopsis mutant with rice (Oryza sativa) HXK5 and HXK6 indicates that HXK-mediated Glc sensing is conserved in angiosperms (Cho et al., 2009).
Other Glc sensing pathways have been described (for review, see Rolland et al., 2002, 2006). One of them is a glycolysis-dependent pathway that requires HXK catalytic activity and regulates the expression of the pathogenesis-related genes PR1 and PR2 (Xiao et al., 2000). A third pathway is involved in the regulation of a restricted number of genes such as those coding for chalcone synthase and cell wall invertase and is independent of increased HXK activity and HXK1 (Roitsch, 1999; Xiao et al., 2000). Genetic evidence also indicates that a HXK-independent Glc sensing and signaling mechanism involving a G protein-coupled receptor system exists in plants (Ullah et al., 2002; Chen et al., 2003a, 2006; Chen and Jones, 2004; Huang et al., 2006). Interestingly, a genetic interaction between G protein signaling and a Golgi-localized hexose transporter has been reported (Wang et al., 2006).
The use of Glc signaling mutants has revealed a relationship between Glc and the signaling pathways of hormones such as abscisic acid (ABA; Zhou et al., 1998; Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000; Rook et al., 2001; Brocard et al., 2002; Cheng et al., 2002; Brocard-Gifford et al., 2004), ethylene (Zhou et al., 1998; Gibson et al., 2001; Cheng et al., 2002; Cho et al., 2010), and auxin and cytokinin (Moore et al., 2003). HXK1-dependent Glc-induced responses require ABA synthesis and subsequent ABA signal transduction (Zhou et al., 1998; Arenas-Huertero et al., 2000; Cheng et al., 2002). Within this regulatory cascade, the ABA biosynthetic genes ABA2/GIN1 encoding a short-chain dehydrogenase/reductase (Cheng et al., 2002), ABA3/GIN5 encoding a molybdenum cofactor sulfurase (Arenas-Huertero et al., 2000), and the ABA signaling gene ABI4/GIN6 gene coding for a transcriptional regulator of the APETALA2 domain family (Finkelstein et al., 1998; Arenas-Huertero et al., 2000) have pivotal roles (for review, see León and Sheen, 2003; Rolland et al., 2006). ABI4 also represses the promoter of an rbcS gene in response to Glc or ABA (Acevedo-Hernández et al., 2005), which may partly reflect the central role of ABI4 in retrograde signaling from the chloroplast to the nucleus (Koussewitzky et al., 2007). Moreover, ABI4 is a regulator of Man-induced inhibition of seedling germination (Pego et al., 1999; Huijser et al., 2000), suggesting a general role for ABI4 in hexose signaling. ABI4 is also involved in the synergistic activation of APL3 (a large subunit of ADP-glucose pyrophosphorylase) by Glc and ABA and is an example of a Glc-induced response that is modulated by ABA (Rook et al., 2001; Li et al., 2006). Several other ABA signaling components were shown to be involved in defining Glc sensitivity (Ramon et al., 2008).
Our knowledge of the interactions between Glc and ABA signaling pathways and their integration with developmental programs is still incomplete. For instance, we still do not know whether ABA synthesis and/or signaling are involved in HXK-independent Glc-induced responses. It is also unclear to what extent short-term responses to Glc fluctuations are interrelated with ABA (Li et al., 2006). The latter point is becoming increasingly relevant to our understanding of how carbon resources are used to minimize growth inhibition under conditions of energy limitation and ABA-mediated abiotic stress responses (Baena-González and Sheen, 2008; Usadel et al., 2008). Transcription factor genes coregulated by short-term treatment with Glc or ABA have been identified and represent potential candidates for integrating transient changes in Glc and ABA (Li et al., 2006). Recently, some members of the Arabidopsis S- and C-groups of bZIP transcription factor genes (Jakoby et al., 2002; Corrêa et al., 2008) have emerged as important mediators of the adaptive responses induced by the Snf1-related kinases (SnRK1) KIN10 and KIN11 during energy or sugar limitation (Baena-González et al., 2007; Usadel et al., 2008). In addition, a restricted set of S-group proteins associates with C-group members by forming heterodimers, thus creating another level of interaction in the regulation of gene expression (Weltmeier et al., 2006; Alonso et al., 2009).
During work on the functional diversification of C-group bZIP genes, we observed that the expression of AtbZIP63 (At5g28770), one of the four Arabidopsis group C members that is likely to represent an ancestral angiosperm function (Vincentz et al., 2003), is regulated by Glc, ABA, and Man, suggesting the involvement of this gene in the cross talk between carbohydrate and ABA-mediated responses (e.g., in abiotic stress). To assess this possibility, we investigated the dependence of Glc-induced repression of AtbZIP63 on ABA synthesis and signaling. We show here that the Glc-induced repression of AtbZIP63 is independent of HXK1 and that the short-term response of AtbZIP63 to 2% Glc does not rely on ABA accumulation. Furthermore, the requirement of ABA synthesis to obtain full repression of AtbZIP63 by 6% Glc together with the synergistic repression of AtbZIP63 by Glc+ABA, which partly relies on posttranscriptional regulation, suggest that ABA modulates Glc-mediated responses. Since AbZIP63 is involved in the control of energy homeostasis (Baena-González and Sheen, 2008), mediating partially KIN10-induced responses, we also discuss the importance of the modular regulation of AtbZIP63 expression by Glc and ABA.
RESULTS
AtbZIP63 Is Repressed by Glc and ABA, and Glc-Induced Repression Is Independent of HXK1 Glc Sensing Activity
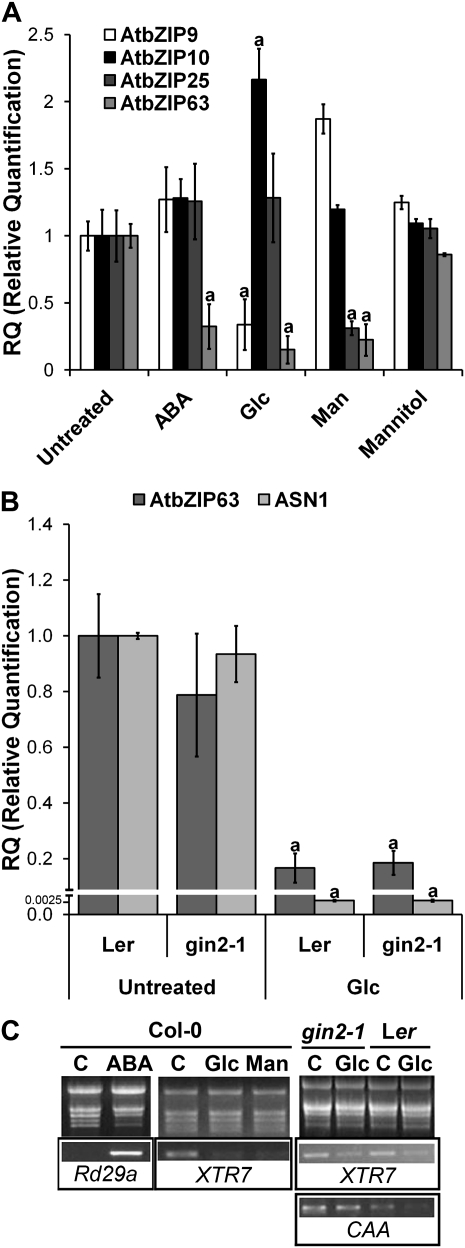
To evaluate the short-term responses of C-group bZIP genes AtbZIP9 (At5g24800), AtbZIP10 (At4g02640), AtbZIP25 (At3g54620), and AtbZIP63 (At5g28770) to Glc and ABA signals, wild-type Columbia (Col-0) seedlings grown for 6 d (growth stage 1.0, corresponding to fully opened cotyledons; Boyes et al., 2001) under constant dim light (20 μmol m−2 s−1) and in half-strength Murashige and Skoog (MS/2) liquid medium were treated for 4 h with 2% Glc or 100 μm ABA. Treatment efficiency was verified by the induction of Rd29a (At5g52310) by ABA (Arroyo et al., 2003) and repression of XTR7 (At4g14130) by Glc (Price et al., 2004). While AtbZIP63 was repressed by both Glc (6.7-fold; Fig. 1A) and ABA (3.3-fold with 100 μm and 2-fold with 10 μm; Fig. 1A; Supplemental Fig. S1), AtbZIP9 and AtbZIP10 only responded to Glc. AtbZIP9 was repressed by Glc and AtbZIP10, being induced (3.3- and 2.2-fold, respectively) by this signal (Fig. 1A). These alterations were not caused by osmotic stress, since 2% mannitol only marginally changed the mRNA levels of these genes (Fig. 1A). AtbZIP25 was not responsive to Glc or ABA (Fig. 1A).
Figure 1.
The expression of the Arabidopsis C-group bZIP genes AtbZIP9, AtbZIP10, AtbZIP25, and AtbZIP63 is regulated by ABA, Glc, or Man, and Glc-mediated regulation of AtbZIP63 is independent of HXK1 sensing activity. Wild-type Col-0 and Ler or mutant gin2-1 seedlings were grown for 6 d in MS/2 or MS/10 liquid medium for Ler and gin2-1 and were subsequently treated with 100 μm ABA, 2% Glc, 2% Man, or 2% mannitol for 4 h. Total RNA was analyzed by qRT-PCR assays. A, Relative transcript abundance of C-group bZIP genes. The data are means ± sd (error bars) of at least three independent experiments. B, Expression of AtbZIP63 and AtASN1 in response to Glc treatment in gin2-1, a HXK1 null mutant, and the corresponding Ler wild type. The data are means of at least three independent experiments. C, RNA integrity. C stands for the untreated control. The effectiveness of the treatments was verified by evaluating the induction of Rd29a by ABA and the repression of XTR7 by Glc and Man by semiquantitative RT-PCR. Carbonic anhydrase (CAA; At5g14740) was used as a control for HXK1-mediated Glc-dependent gene repression. The differences from the untreated control were considered significant at P < 0.05 (Student’s t test) and are indicated by the letter a. Expression data are normalized to the Actin2 mRNA levels, and the relative quantification refers to the respective untreated wild-type Col-0 or Ler genotype.
To determine whether the Glc-induced repression of AtbZIP9 and AtbZIP63 is mediated by HXK1 (At4g29130) Glc sensing activity (Moore et al., 2003) or requires HXK catalytic activity to produce downstream regulatory metabolites (glycolysis dependent; Xiao et al., 2000), the Glc analogue Man, which is readily phosphorylated by HXK and poorly metabolized, was used. The promotion of Glc-like responses by Man would be indicative of HXK sensing activity, whereas the absence of any response to Man would be indicative of a glycolysis-dependent or HXK-independent pathway. Man was almost as efficient as Glc in repressing AtbZIP63 mRNA (Fig. 1A), suggesting a role for HXK sensing and signaling in this regulation. In contrast, AtbZIP9 mRNA was unresponsive to Man (Fig. 1A), indicating that Glc-induced repression of AtbZIP9 was dependent on Glc metabolism or independent of HXK activity (Xiao et al., 2000; Villadsen and Smith, 2004). Interestingly, AtbZIP25 was specifically repressed by Man (Fig. 1A), revealing the possible existence of a Man-restricted signaling pathway.
To evaluate more precisely the involvement of HXK1 sensing activity in the control of AtbZIP63 expression by Glc, we analyzed the regulation of this gene in an HXK1 null mutant, gin2-1 (ecotype Landsberg erecta [Ler] background), which is deficient in Glc-dependent photosynthetic gene repression (Moore et al., 2003; Cho et al., 2006). Expression analysis was done under low-nitrogen conditions (MS/10; Moore et al., 2003). Under these conditions, there was no HXK1-mediated Glc-induced inhibition of carbonic anhydrase (At5g14740) photosynthetic gene expression in gin2-1 (Fig. 1C; Moore et al., 2003), while the HXK1-independent Glc-induced repression of the Gln-dependent Asparagine synthetase1 (ASN1; At3g47340) gene was, as expected, still effective in the gin2-1 mutant (Fig. 1B; Baena-González et al., 2007). The levels of AtbZIP63 mRNA in response to Glc were also not significantly different between gin2-1 and the wild type (Fig. 1B), indicating that a pathway independent of HXK1 Glc sensing activity was involved in the Glc-induced regulation of this gene. Similarly, the Glc-induced repression of AtbZIP9 expression was as efficient in Ler and gin2-1 and, therefore, is also independent of HXK1 Glc sensing activity (data not shown). Several lines of genetic evidence support the role of ABA synthesis and sensing in Glc-mediated responses (see the introduction; for review, see León and Sheen, 2003). Since AtbZIP63 is also regulated by ABA (Fig. 1A), we next examined the extent to which Glc-dependent control of AtbZIP63 expression is connected to ABA signaling.
The Repression of AtbZIP63 by Glc Is Modulated by ABA
The interaction between Glc and ABA to regulate gene expression may involve Glc-triggered modulation of ABA levels, as suggested previously (Arenas-Huertero et al., 2000). To assess the possibility that Glc-induced repression of AtbZIP63 could be the result of an increase in ABA content, we monitored the changes in ABA levels in response to short-term Glc treatment. In addition, the changes in ABI5 (At2g36270) mRNA levels were used as a positive control to verify the efficiency of the treatments (Fig. 2B), since this gene is induced by ABA (Lopez-Molina et al., 2003; Price et al., 2003).
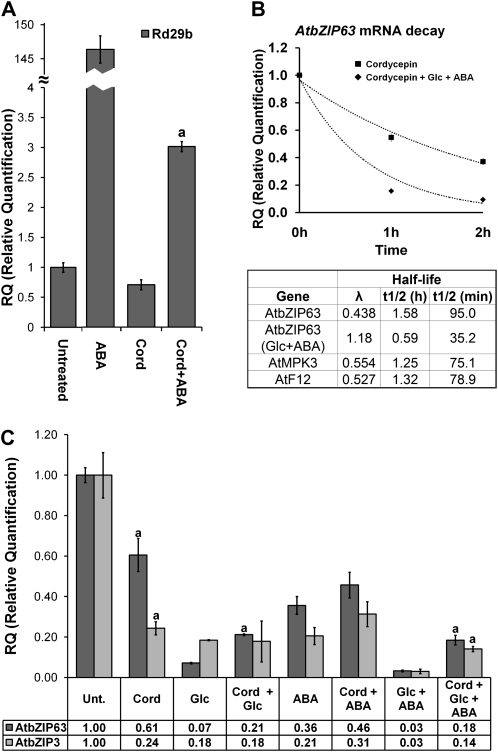
Figure 2.
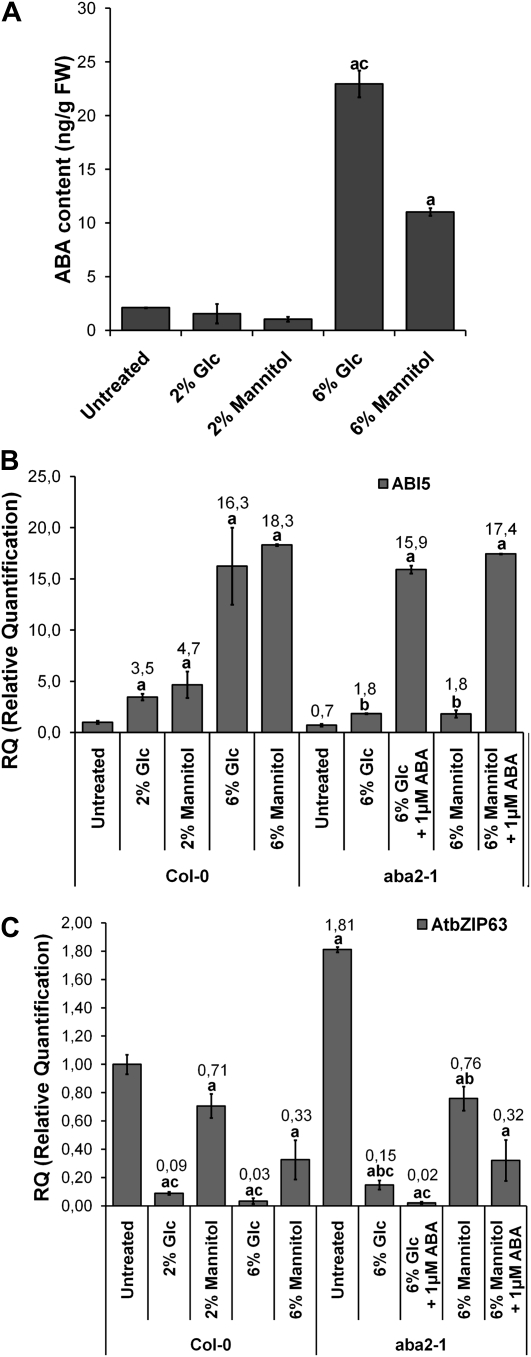
The repression of AtbZIP63 by Glc is modulated by ABA. A, Quantification of ABA in untreated whole seedlings or seedlings treated with 2% or 6% Glc or 2% or 6% mannitol. Significant differences between treated and untreated seedlings and between Glc and mannitol treatments are indicated by letters a and c, respectively (n = 6; P < 0.05). FW, Fresh weight. B and C, Relative ABI5 (B) and AtbZIP63 (C) mRNA quantification in response to 2% and 6% Glc or mannitol in aba2-1 (with or without 1 μm ABA application) and the Col-0 wild type. All experiments were performed with 6-d-old seedlings. Numbers above the bars represent mean values of the relative quantification (n = 3). Significant differences between treated and untreated seedlings of the same genotype (letter a), between equally treated aba2-1 and Col-0 (letter b), and between Glc and mannitol treatments (letter c; n = 5; P < 0.05) are indicated. Growth (MS/2), treatments, and qRT-PC analysis were performed as described in Figure 1 except that transcript levels were normalized to PDF2 mRNA.
Two percent Glc or mannitol had no substantial effect on ABA accumulation in whole seedlings after 4 h of treatment, while 6% Glc or mannitol was able to induce a 10- or 5-fold increase in ABA content, respectively (Fig. 2A). These findings indicate that the repression of AtbZIP63 by 2% Glc (Fig. 2C) was not mediated by ABA accumulation. Treatment with 2% mannitol had only a marginal effect on AtbZIP63 expression, probably because of osmotic activity (Fig. 2C), which could also be the reason for ABI5 mRNA induction by 2% Glc or mannitol (3.5- and 4.7-fold, respectively; Fig. 2B). At higher sugar concentrations (e.g., 6% Glc or mannitol), AtbZIP63 down-regulation was around three times greater than the repression caused by 2% of the corresponding hexose (Fig. 2C). As expected, 6% Glc or mannitol strongly induced ABI5, most likely as the result of an increase in ABA content (Fig. 2, A and B; Price et al., 2003). The stronger repression of AtbZIP63 by 6% Glc could be triggered by the combination of the Glc signal and an increase in ABA content that has been induced by high concentrations of sugars (Fig. 2A). This result suggests that Glc and ABA may interact to repress AtbZIP63. In the case of 6% mannitol, the AtbZIP63 repression (Fig. 2C) could be attributed to an increase in ABA content (Fig. 2A), which is in agreement with the repression observed in response to ABA application (Fig. 1A). To further assess the contribution of ABA to the repression of AtbZIP63 by 6% Glc or mannitol, we analyzed the expression of AtbZIP63 in the ABA-deficient mutant aba2-1 (Col-0 ecotype; accumulates approximately 10% of the wild-type ABA level; Léon-Kloosterziel et al., 1996). The repression of AtbZIP63 by 6% Glc was significantly less effective in aba2-1 than in the wild-type (12-fold repression versus 33-fold, respectively; Fig. 2C), and essentially the same value was obtained when 2% Glc was used (Fig. 2C). The same trend was observed in the case of 6% mannitol, although the difference between aba2-1 and the wild type was more tenuous (2.4-fold repression versus 3-fold, respectively; Fig. 2C), possibly because mannitol exerts an ABA-independent osmotic activity and/or aba2-1 is slightly leaky. Interestingly, the supply of 1 μm ABA to aba2-1 seedlings treated with 6% Glc or mannitol restored the wild-type AtbZIP63 mRNA repression level (Fig. 2C). As a control, the induction of ABI5 by 6% Glc or mannitol was reduced in aba2-1, and full induction could also be restored by exogenously applied 1 μm ABA (Fig. 2B). These results support the hypothesis of a combinatorial effect of Glc and ABA to modulate AtbZIP63 repression. We next were interested in evaluating the mechanism involved in this regulatory pattern.
The Synergistic Repression of AtbZIP63 by a Combination of Glc and ABA Partly Involves the Modulation of mRNA Decay
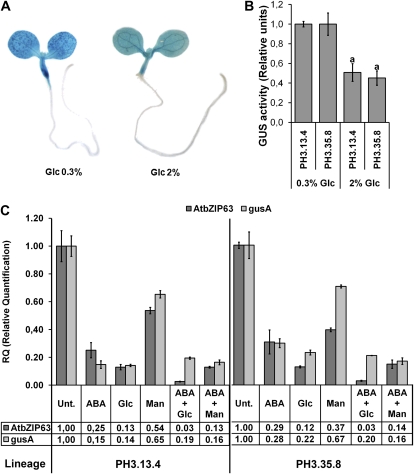
Initially, we wished to determine whether Glc- and ABA-induced repression of AtbZIP63 could be the result of transcriptional control. To this end, the expression of the GUS reporter gene under the control of the AtbZIP63 promoter and the 5′ untranslated region (UTR) sequence in response to ABA, Glc, and Man was evaluated in transgenic seedlings containing the AtbZIP63 promoter-5′UTR::GUS fusion. Two transgenic lines homozygous for a single insertion of the AtbZIP63 promoter-5′UTR::GUS transgene and that were representative of the overall expression pattern found among six independent lines were analyzed. The detection of GUS activity in situ and in plant extracts indicated that transgenic seedlings grown with 0.3% Glc accumulated more GUS activity than those grown in 2% Glc (Fig. 3, A and B), suggesting that AtbZIP63 promoter and 5′UTR sequences were apparently able to mediate Glc-induced regulation. We next measured the transient effect of Glc and ABA on GUS mRNA. The Glc- and ABA-induced repression of AtbZIP63 was partly mediated by the AtbZIP63 promoter and 5′UTR sequences, since changes in GUS mRNA accompanied those of AtbZIP63 mRNA in response to these signals (Fig. 3C). Collectively, these data provide evidence for a role of transcription and/or 5′UTR-mediated control of mRNA stability in the regulation of AtbZIP63 expression by Glc and ABA. This finding prompted us to investigate whether ABI4, which encodes for an AP2-type transcriptional regulator involved in ABA and Glc response pathways (Finkelstein et al., 1998; Acevedo-Hernández et al., 2005), would be part of both Glc- and ABA-mediated regulation of AtbZIP63 expression. To this end, we analyzed the AtbZIP63 responses to Glc and ABA in the abi4-1 mutant (Col-0 ecotype), which lacks ABI4 activity (Söderman et al., 2000). The Glc- and ABA-induced repression of AtbZIP63 was stronger in the abi4-1 mutant compared with the wild type (23.3- versus 8.3-fold for Glc and 13.6- versus 4-fold for ABA; Supplemental Fig. S2), suggesting that ABI4 antagonizes the Glc- and ABA-induced repression of AtbZIP63. This possibility is reinforced by the presence of two ABI4-binding sites, CCAC, in the AtbZIP63 promoter (Supplemental Table S1).
Figure 3.
The synergistic repression of AtbZIP63 by ABA+Glc cannot be reproduced by AtbZIP63 promoter sequences. Transgenic seedlings of the PH3.13.4 and PH3.35.8 lineages (both homozygote for one transgenic locus) containing the reporter gene GUS under the control of the AtbZIP63 promoter, 5′UTR, and 36 bp of coding sequences were grown for 5 d in MS/2 solid medium supplemented with 0.3% Glc or 2% Glc (w/v) under constant light at 22ºC for GUS activity assays (A and B) and for 6 d in MS/2 liquid medium and were treated for 4 h with 100 μm ABA, 2% Glc, 2% Man, or different combinations of these molecules (used at the same final concentration as when applied individually) for qRT-PCR assays (C). A, Histochemical analysis of in situ GUS activity in the PH3.35.8 lineage. B, Relative GUS activity in PH3.13.4 and PH3.35.8 lineages. The raw fluorescence units were normalized by total protein content. Relative GUS activity refers to the respective genotype treated with 0.3% Glc. Relative GUS activity was defined in the linear portion of the enzymatic kinetic. Significant differences are represented by the letter a (n = 3; P < 0.05). C, Relative transcript abundance of AtbZIP63 and GUS in transgenic seedlings. The data are means ± sd (error bars) of three independent experiments. Total RNA was treated with DNase and analyzed by qRT-PCR as described in Figure 1 except that transcript levels were normalized to PDF2 mRNA. The numbers at the bottom represent mean decreases of transcript amounts in response to each treatment relative to the corresponding untreated (Unt.) sample.
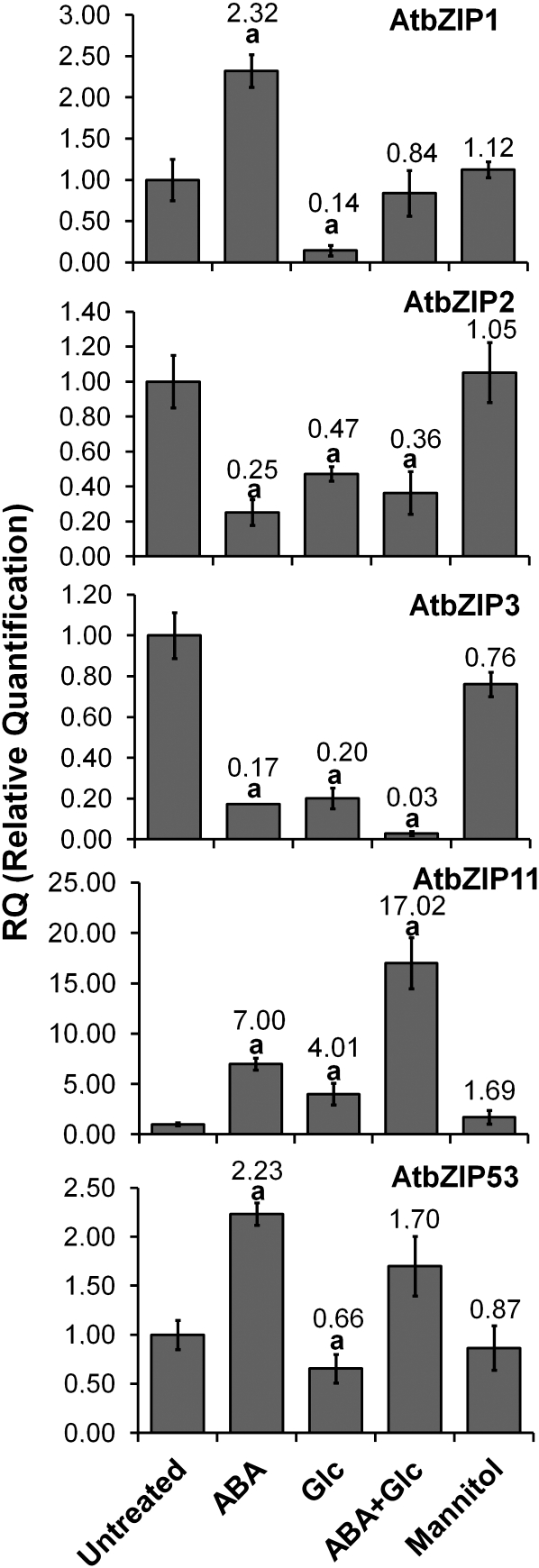
A comparison of how signals alone and in combination can regulate the expression of target genes could provide clues about how these signals interact. Therefore, we analyzed the regulation of AtbZIP63 mRNA abundance by combinations of ABA, Glc, and Man. The most important finding of this analysis was that the combination of Glc and ABA synergistically repressed AtbZIP63 (Fig. 3C), a response also observed in the Wassilewskija (Ws) ecotype (data not shown). The combination Glc+ABA repressed AtbZIP63 mRNA by approximately 33-fold, which was almost 3-fold more than the sum of the repression levels observed for each stimulus separately (approximately 3.8-fold for ABA and approximately 8.3-fold for Glc; Fig. 3C). This response was specific for the combination Glc+ABA, since no synergy was observed when the Man+ABA combination was used (Fig. 3C). Moreover, among the four members of the group-C bZIP genes, the synergistic response was restricted to AtbZIP63 (Supplemental Fig. S3). The evolutionary relatedness between C-group and S-group bZIP genes (Corrêa et al., 2008) prompted us to evaluate the Glc- and ABA-mediated regulations of the S-group genes AtbZIP1 (At5g49450), AtbZIP2 (At2g18170), AtbZIP11 (At4g34590), and AtbZIP53 (At3g62420), which are functionally related to AtbZIP63 (Baena-González et al., 2007; Dietrich et al., 2011). None of them were synergistically down-regulated by Glc+ABA, but interestingly, AtbZIP11 was synergistically induced by these signals (Fig. 4), revealing the diversification of ABA- and Glc-related regulatory output. However, AtbZIP3 (At5g15830), another S-group gene that is down-regulated by Glc and ABA (Li et al., 2006), shared synergetic down-regulation with AtbZIP63 (Fig. 4), raising the possibility that these two genes are under the control of the same regulatory mechanisms. Indeed, AtbZIP63 and AtbZIP3 promoters share common motifs related to Glc and ABA regulation (Supplemental Table S1).
Figure 4.
The expression of the Arabidopsis S-group bZIP genes AtbZIP1, AtbZIP2, AtbZIP3, AtbZIP11, and AtbZIP53 is regulated by Glc and ABA. Wild-type Col-0 seedlings were grown for 6 d in MS/2 liquid medium under constant dim light and were subsequently treated with 2% mannitol (osmotic control), 100 μm ABA, and/or 2% Glc for 4 h. Total RNA was extracted and analyzed. cDNA synthesized from total RNA treated with DNase was used in qRT-PCR assays. S-group bZIP transcript levels were normalized to the PDF2 mRNA levels, and the relative quantification refers to untreated Col-0. The data are means ± sd (error bars) of three independent experiments. Differences from the untreated control were considered significant at P < 0.05 (Student’s t test) and are indicated by the letter a. The number at the top of each column corresponds to the relative expression level.
Surprisingly, the AtbZIP63 promoter and 5′UTR sequences were not sufficient to synergistically repress the GUS reporter gene (Fig. 3). Thus, the repression of AtbZIP63 mRNA accumulation by the combination Glc+ABA may either require a transcriptional regulatory element not included in our construct or involve a control of AtbZIP63 mRNA decay. To assess this latter possibility, we determined the half-life of AtbZIP63 mRNA by using cordycepin (3-deoxyadenosine) to inhibit transcription (Holtorf et al., 1999; Gutiérrez et al., 2002; Souret et al., 2004). We designed a protocol that produced almost complete inhibition (97%) of ABA-mediated Rd29b mRNA transcription (Fig. 5A; Uno et al., 2000), indicating that conditions for the efficient inhibition of transcription were achieved. The half-lives of AtMPK3 (At3g45640) and AtF12 (At5g03430) mRNA were in good agreement with previous results (Fig. 5B; Gutiérrez et al., 2002), further indicating that our protocol was efficient. Under our conditions, the half-life of AtbZIP63 mRNA was approximately 95 min (Fig. 5B). This estimate implies that only around 17% of the synergistic repression fold of AtbZIP63 mRNA after a 4-h treatment with Glc+ABA (expected 5.6-fold based on half-life estimation versus the observed 33-fold reduction; Fig. 3C) can theoretically be accounted for by transcriptional control. The remaining 83%, therefore, must be related to mRNA degradation. The increase of AtbZIP63 mRNA decay rate by simultaneously combining cordycepin and Glc+ABA (half-life of AtbZIP63 mRNA was reduced to approximately 33 min; Fig. 5B) agrees with a control of AtbZIP63 mRNA stability by these two signals. The increased AtbZIP63 mRNA decay triggered by Glc+ABA seems to be specific, since the Actin2 reference gene used to normalize quantitative reverse transcription (qRT)-PCR data was stable among all treatments. To further confirm and refine this conclusion, we also examined the impact of Glc and ABA signals individually on the posttranscriptional control of AtbZIP63 mRNA. The addition of Glc or Glc+ABA to seedlings treated with cordycepin reduced AtbZIP63 mRNA accumulation by almost 3-fold compared with the control treatment with cordycepin alone (Fig. 5C). These results indicate that Glc alone may also control the stability of AtbZIP63 mRNA and that the expected Glc+ABA-mediated synergistic repression was hindered in the presence of cordycepin (Fig. 5C), raising the possibility that successful synergism requires transcription. This latter conclusion also applies to Glc-induced down-regulation of AtbZIP63, since this regulation was significantly weaker in the presence of cordycepin compared with the samples treated only with Glc (Fig. 5C). In contrast, the ABA-mediated reduction of AtbZIP63 mRNA in the presence of cordycepin was not significantly different from the control treatments with cordycepin or ABA alone (Fig. 5C), indicating that ABA acts mainly by limiting transcription.
Figure 5.
The control of AtbZIP63 and AtbZIP3 mRNA decay is involved in the response to Glc+ABA. Inhibition of transcription was performed with 100 μg mL−1 cordycepin (Cord). Wild-type Col-0 6-d-old seedlings were grown in MS/2 liquid medium, treated with cordycepin, and harvested after 0, 1, and 2 h for half-life measurements or pretreated with cordycepin for 1 h and then treated with 100 μm ABA and/or 2% Glc (w/v) for an additional 2 h. A, ABA-mediated induction of Rd29b was used to monitor the efficiency of transcription inhibition. B, Kinetics of mRNA decay and half-life of AtbZIP63 in the absence and presence of Glc+ABA and of two genes with known half-lives (AtMPK3 [At3g45640] and AtF12 [At5g03430]; Gutiérrez et al., 2002); λ = decay constant; t1/2 (h) = half-life in hours; t1/2 (min) = half-life in minutes. C, Repression of AtbZIP63 and AtbZIP3 by 2% Glc, 100 μm ABA, and 2% Glc + 100 μm ABA with or without cordycepin. Transcript levels were normalized to the Actin2 mRNA. Significant differences between treated samples with or without cordycepin are indicated by the letter a (n = 3; P < 0.05). The numbers at the bottom represent mean decreases of transcript amounts in response to each treatment relative to the untreated (Unt.) sample.
Since AtbZIP63 and AtbZIP3 are coregulated in response to ABA and/or Glc, we wondered whether the regulation of AtbZIP3 expression also involves the control of mRNA stability. As with AbZIP63, efficient synergistic repression of AtbZIP3 did not tolerate transcriptional inhibition by cordycepin (Fig. 5C), indicating that both genes are subjected to similar regulatory aspects. However, in contrast to AtbZIP63, the Glc- and ABA-mediated reduction of AtbZIP3 mRNA levels was attributable mainly to transcriptional repression (Fig. 5C).
Together, these data support the idea that Glc- and Glc+ABA-induced repression of AtbZIP63 mRNA levels and Glc+ABA-induced reduction of AtbZIP3 transcripts are partly attributable to accelerated mRNA decay, which requires ongoing transcription for full efficiency. For instance, continuous transcription may be necessary to maintain an active pool of a factor mediating mRNA decay in response to Glc+ABA. Alternatively, a specific transcriptional control step in response to Glc+ABA may be required to trigger posttranscriptional control of AtbZIP63 and AtbZIP3 mRNAs. Based on this reasoning, it is conceivable that the Glc+ABA-mediated transcriptional activation of an AtbZIP63-related microRNA (miRNA) gene may be involved. However, this possibility was discarded, because the synergistic repression of AtbZIP63 remained functional in miRNA pathway mutants (Supplemental Fig. S4). Moreover, this result was further supported by the lack of any AtbZIP63-related miRNA in miRBase (http://www.mirbase.org/; Griffiths-Jones et al., 2008).
To obtain some additional clues about the physiological role of AtbZIP63, we searched for putative AtbZIP63 target genes.
ASN1, SEN1, and DIN10 Are Putative Target Genes of AtbZIP63
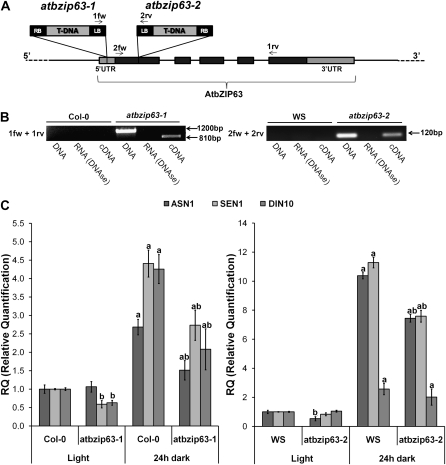
AtbZIP63 has been shown to interact with the SnRK1.1 KIN10 to promote the expression of Gln-dependent ASN1 in a transient protoplast assay system (Baena-González et al., 2007). KIN10 is a key regulator of energetic stress adaptation, which involves a large extent of transcriptional changes (Baena-González et al., 2007). To further assess the involvement of AtbZIP63 in KIN10-mediated responses, we analyzed the expression of selected genes that are known to be induced by KIN10 and highly coexpressed with AtbZIP63 (http://cressexpress.org; Srinivasasainagendra et al., 2008) in two AtbZIP63 mutants corresponding to T-DNA insertion lines (atbzip63-1 in Col-0 and atbzip63-2 in Ws; Fig. 6, A and B). We found that ASN1 and SEN1 (for senescence-associated protein 1; At4g35770) are misregulated in both atbzip63-1 and atbzip63-2 after 24 h of darkness when compared with their respective wild-type genotypes. Additionally, DIN10 (for dark inducible 10; At5g20250) expression was found to present significant differences from the wild type only in atbzip63-1 (Fig. 6C). DIN10 is a putative raffinose synthase (EC 2.4.1.82) from the GH36 family (http://www.cazy.org/GH36.html) that shares approximately 38% identity with a functionally characterized raffinose synthase from pea (Pisum sativum; European Bioinformatics Institute accession no. AJ426475; Peterbauer et al., 2002) that catalyzes the conversion of Suc and galactinol into myoinositol and raffinose. The differential expression profile between atbzip63-1 and atbzip63-2 could be due to the different genetic backgrounds, Col-0 and Ws, respectively. In conclusion, these results support the notion that AtbZIP63 participates in the KIN10-mediated transcriptional changes.
Figure 6.
Putative AtbZIP63 target genes. The expression of ASN1 (At3g47340) and SEN1 (At4g35770) is misregulated in two AtbZIP63 T-DNA insertion mutants after dark-induced energy starvation. A, Schematic representation of T-DNA insertion sites in atbzip63-1 and atbzip63-2 mutants. LB, T-DNA left border; RB, T-DNA right border. Primers used to locate T-DNA insertion and detect chimeric transcripts are indicated by the arrows (for primer sequences, see Supplemental Table S2). B, PCR amplification from genomic DNA of the T-DNA insertion region and RT-PCR after DNase treatment showing chimeric transcripts between AtbZIP63 and T-DNA in both atbzip63-1 and atbzip63-2 mutants. Size differences between amplification products from genomic DNA (1,200 bp) and cDNA (810 bp) in atbzip63-1 are due to introns that are absent in spliced AtbZIP63 mRNA. C, ASN1, SEN1, and DIN10 transcript accumulation in atbzip63-1 (Col-0 ecotype) and atbzip63-2 (Ws ecotype) 6-d-old seedlings after 24 h of dark treatment. Significant differences related to seedlings of the same genotype without dark treatment (light) are represented by the letter a (n = 3; P < 0.05) and those between equally treated mutants and their respective wild-type genotype are represented by the letter b (n = 3; P < 0.05). Growth (MS/2), treatments, and qRT-PCR analysis were performed as described in Figure 1 except that transcript levels were normalized to the PDF2 mRNA.
DISCUSSION
Higher plants have evolved strategies to use their energy resources in such a way as to optimize growth and development and ensure survival (Polge and Thomas, 2006; Smith and Stitt, 2007; Baena-González and Sheen, 2008; Baena-González, 2010). Recent evidence indicates that the SnRK1 kinases KIN10 and KIN11 are key players in the ability of plants to adjust to energy deprivation. The interaction of KIN10 with members of S-group bZIP transcription factors (AtbZIP1, AtbZIP2/GBF5, AtbZIP11, and AtbZIP53) and AtbZIP63, a C-group bZIP factor, can partially trigger the transcriptional responses involved in energy reposition (Baena-González et al., 2007; Usadel et al., 2008).
As shown here, AtbZIP63 is also an early Glc-responsive gene that is a good candidate for a role in Glc transduction pathways (Fig. 1A). The Glc-sensing activity of HXK1 is not involved in this regulation (Fig. 1B), and the participation of Glc phosphorylation and/or further metabolism requires more analysis. Moreover, the 2% Glc-induced repression of AtbZIP63 does not involve changes in ABA levels (Fig. 2). However, ABA can interact with Glc to modulate AtbZIP63 repression (Figs. 2 and 3), indicating that ABA-related processes are not limited to long-term adaptive responses to Glc but are also involved in early Glc-triggered regulation, further emphasizing the importance of the link between ABA and Glc signaling (Gibson, 2005). Recently, a HXK1-dependent early seedling developmental arrest was found to be promoted by low 2% Glc. Interestingly, this long-term response appeared to be also uncoupled from ABA synthesis but requires low-nitrogen conditions (Cho et al., 2010). Whether short-term Glc-mediated repression of AtbZIP63 can also be modulated by nitrogen supply is an interesting possibility related to nitrogen/carbon regulatory features that remains to be tested.
The strong repression of AtbZIP63 by Glc and ABA and the striking synergistic negative effect conferred by the combination Glc+ABA on the mRNA level of AtbZIP63 constitute good support for the idea that AtbZIP63 is a cross talk node between Glc and ABA signaling cascades. The repression of AtbZIP63 by Glc is consistent with the reported interaction between AtbZIP63 and KIN10, which is possibly involved in a broader regulatory scheme dedicated to optimizing energy supply under adverse conditions (Baena-González et al., 2007). The identification of the three KIN10-activated genes ASN1, SEN1, and DIN10 as putative AtbZIP63 target genes (Fig. 6C) is in agreement with the participation of AtbZIP63 in the KIN10-related signaling pathway.
We hypothesize that the Glc-induced repression of AtbZIP63 and the attenuation of KIN10-mediated processes by Glc or Suc (Baena-González et al., 2007; Usadel et al., 2008) help to tune and ultimately reset to minimum levels the KIN10/AtbZIP63-mediated energy starvation response. A further level of complexity is added to this regulatory scheme by the ABA-induced repression of AtbZIP63. This control suggests that AtbZIP63 activity is incompatible with ABA-mediated responses and, more particularly, with abiotic stress (Seki et al., 2007), an energy-consuming process (Shinozaki and Yamaguchi-Shinozaki, 2006) that requires adjustment according to the available energy level. For instance, the ABA-induced accumulation of osmolytes such as sugars and Pro (Seki et al., 2007; Kempa et al., 2008) improves tolerance to drought and salt stress but also provides an alternative source of energy, carbon, and even nitrogen that may be essential when energy is scarce.
We propose that AtbZIP63 integrates a network that coordinates the use of available energy to sustain growth with the need to correct the adverse effect of abiotic stress and that an important mechanistic aspect of AtbZIP63 participation involves the fine-tuning of AtbZIP63 mRNA levels by two mechanisms. First, ABA or low Glc concentration (2%) directly down-regulates AtbZIP63 expression partly at the transcriptional level (Figs. 1A and 3C). Direct ABA measurement revealed that short-term treatment with 2% Glc did not alter ABA levels (Fig. 2A), suggesting that Glc-mediated regulation of AtbZIP63 does not rely on a linear pathway in which the stimulation of ABA accumulation by Glc would subsequently trigger this response. However, as suggested by the observation that the stronger repression of AtbZIP63 by 6% Glc compared with 2% Glc (33- versus 11-fold) is ABA2 dependent (Fig. 2C), it is likely that ABA can modulate the sensitivity to Glc. This possibility is further supported by the synergistic repression conferred by Glc+ABA (Fig. 3C). The latter regulatory feature constitutes the second mechanism by which AtbZIP63 integrates Glc and ABA signals. The synergistic response most likely reflects a situation of unlimited energy availability that favors an optimal response to abiotic stress. Interestingly, only part of the response (15% of the total repression fold) was associated with the AtbZIP63 promoter and 5′UTR sequences (Fig. 3C), suggesting that a posttranscriptional control step acting on AtbZIP63 mRNA was also involved. Three additional pieces of evidence support this conclusion. First, the estimated half-life of AtbZIP63 mRNA (Fig. 5B) cannot explain the large decrease in AtbZIP63 mRNA in response to Glc+ABA treatment solely by invoking transcriptional inhibition. Second, the AtbZIP63 mRNA decay rate was accelerated by ABA+Glc treatment (approximately 3-fold compared with the untreated control; Fig. 5B). Third, even under preestablished transcriptional inhibition with cordycepin, Glc and Glc+ABA still significantly reduced AtbZIP63 mRNA levels by 3-fold (Fig. 5C). However, Glc+ABA-induced synergistic repression was hampered by cordycepin-induced transcriptional inhibition (Fig. 5C), indicating that an unstable or Glc+ABA transiently induced regulatory factor may be involved. Together, these results provide evidence for a pathway regulating AtbZIP63 mRNA decay that is efficiently activated by the convergence of Glc and ABA signals. Interestingly, this regulatory model for AtbZIP63 also applies to the evolutionarily related S-group AtbZIP3 gene. Since S-group genes apparently derived from C-group genes (including AtbZIP63) in the ancestral lineage of angiosperms (Corrêa et al., 2008), it remains to be defined whether this regulatory feature is an ancestrally shared derived character or reflects an event of convergence. Conserved regions were identified in the 3′UTR sequences of AtbZIP3 and AtbZIP63 (Supplemental Fig. S5) and represent putative posttranscriptional cis-regulatory sequences.
The control of mRNA metabolism during ABA-mediated stress responses has been described and may involve various mechanisms (Lu and Fedoroff, 2000; Xiong et al., 2001; Nishimura et al., 2005; Kant et al., 2007), including miRNA- or small interfering RNA-mediated regulation (Borsani et al., 2005; Reyes and Chua, 2007). However, the direct participation of miRNA in the repression of AtbZIP63 mRNA accumulation is incompatible with our observation that AtbZIP63 synergistic repression was almost the same in ago1-25 and dcl1-9 when compared with their respective wild-type genotypes (Supplemental Fig. S4). Interestingly, the synergistic response was stronger in the hyl1-2 miRNA biogenesis mutant compared with the wild type (150- versus 40-fold in the Col-0 ecotype; Supplemental Fig. S4), and the hypersensitivity of this mutant to ABA (Lu and Fedoroff, 2000) may partly explain this result. Several molecular pathways are involved in the control of mRNA decay (Narsai et al., 2007; Houseley and Tollervey, 2009), and further analysis should reveal the mechanistic aspects of the synergistic down-regulation of AtbZIP3 and AtbZIP63 mRNA levels by Glc and ABA. Suc-mediated translational control has been shown to be a relevant posttranscriptional regulatory step of a subset of S-group bZIP genes (Wiese et al., 2004), and our data highlight the importance of mRNA stability control as an additional means to regulate the expression of members of C- and S-group bZIP genes.
The previous description of an ABI4-dependent synergistic transcriptional induction of APL3 (At4g39210) by ABA+Glc (Li et al., 2006), together with the partly posttranscriptional synergistic repression of AtbZIP63 and AtbZIP3 shown here (Figs. 3C and 5), underlies the mechanistic versatility involved in the interaction between ABA and Glc.
It remains now to define to what extent the core signaling components PYR/PYL/RCAR-PP2C-Snrk2 and downstream ABF bZIP regulators (Hubbard et al., 2010; Umezawa et al., 2010) mediate the ABA-induced regulation of AtbZIP63 expression and whether ABA-, Glc-, and KIN10-related signaling converge toward specific regulatory hubs or act through distinct pathways (Halford and Hey, 2009).
AtbZIP63 may function as part of a heterodimerization network involving C- and S-group bZIPs (Ehlert et al., 2006; Kang et al., 2010). The modulation of DNA binding specificity by heterodimerization together with divergent expression patterns may contribute to establish specific gene expression profiles (Fig. 4; Weltmeier et al., 2006). For instance, DNA binding specificity of the AtbZIP1 homodimer is drastically different from that of the AtbZIP1/63 heterodimer (Kang et al., 2010), which, in addition to the opposite ABA-mediated regulation of AtbZIP1 and AtbZIP63 expression (Figs. 2 and 4), is expected to promote a shift in target gene selection. Similarly, the repression of AtbZIP63 expression by Glc and/or ABA may affect the activity of potential AtbZIP63 heterodimerization partners, such as AtbZIP10, which is a positive regulator of the hypersensitive defense response (Kaminaka et al., 2006), or AtbZIP1, AtbZIP53, and AtbZIP11, which stimulate Pro degradation and Asn synthesis (Weltmeier et al., 2006; Baena-González et al., 2007; Hanson et al., 2008; Kang et al., 2010; Dietrich et al., 2011). In any case, the finding that synergistic down-regulation by Glc+ABA among the above-mentioned bZIP genes is restricted to AtbZIP63 suggests a prominent role for this bZIP regulator in shaping the heterodimerization network of C- and S-group proteins in response to Glc and ABA.
Glc is one of the oldest signaling molecules in life’s evolutionary history, and a comprehensive knowledge of the diversity of its associated regulatory networks remains an important issue in understanding the regulation of plant growth and development. Our results highlight the interplay of transcriptional and posttranscriptional regulatory processes in integrating Glc and ABA signals.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Treatments
Arabidopsis (Arabidopsis thaliana) Col-0, Ws, and Ler wild-type ecotypes, as well as the mutants aba2-1 (Léon-Kloosterziel et al., 1996; Cheng et al., 2002), abi4-1 (Finkelstein, 1998), gin2-1 (Jang et al., 1997), atbzip63-1 (SALK_006531), and atbzip63-2 (FLAG_610A08), were obtained from the Arabidopsis Biological Resource Center. Segregation analysis of kanamycin resistance was used to isolate genotypes homozygous for one T-DNA locus. Seeds (10 mg) were surfaced sterilized and incubated in MS/2 for 72 h at 4ºC in the dark to break dormancy. Seedlings were subsequently grown for 5 d in 10 mL of liquid MS/2 or MS/10 salt medium (Sigma) adjusted to 0.3% Glc (w/v) at 22ºC, constant light (20 μmol m−2 s−1), and constant agitation (70 rpm). The medium was then replaced by 10 mL of Glc-free medium, and seedlings were further grown for 24 h followed by 2- or 4-h treatments with 2% or 6% Glc or mannitol, 2% Man, or 100 μm ±cis,trans-ABA (Sigma). Transcription inhibition assays consisted of challenging 6-d-old seedlings (MS/2) with 100 μg mL−1 (final concentration) cordycepin (3-deoxyadenosine; Sigma). Measurements of mRNA half-life were performed as described by Lam et al. (2001) and Gutiérrez et al. (2002), with samples treated with cordycepin for 1 and 2 h. Since the degradation of a mRNA obeys first-order kinetics, the difference in mRNA levels between 0 and 1 h was used to estimate the decay constant (λ), which was used in the equation t1/2 = ln(2)/λ to estimate the transcript half-life. Glc- and ABA-mediated posttranscriptional responses were evaluated by incubating seedlings with 100 μg mL−1 cordycepin for 1 h followed by 2-h treatments with 2% Glc and/or 100 μm ABA.
DNA Constructs, Plant Transformation, and GUS Detection
Transgenic lines expressing a translational fusion between the AtbZIP63 promoter and the reporter gene GUS were obtained following the strategy described by Silveira et al. (2007). The AtbZIP63 (At5g28770) promoter, 5′UTR sequence (75 bp), and 36 bp coding for the first 12 amino acids (approximately 2.8 kb) were amplified from genomic DNA with two specific primers, 5′-GGTGATTGCCCAATCTGCAGCTTTAATCG-3′ and 5′-GTGATGGGATCCGGAGATTTCTTCG-3′, in which a PstI and a BamHI restriction site, respectively, was introduced to facilitate a translational fusion with GUS in PBI121 plant transformation binary vector (Chen et al., 2003b) from which the cauliflower mosaic virus 35S promoter was removed. Poly(A) signals are from the nopaline synthase gene present in pBI121. Two homozygous lines (PH3.13.4 and PH3.35.8) for a single transgenic locus were selected based on kanamycin resistance segregation, and the presence of the chimeric gene was verified by PCR amplification. In situ detection of GUS activity was realized as described by Silveira et al. (2007). Fluorometric GUS assays were performed as described previously (Jefferson et al., 1987). GUS activity was standardized with respect to protein content in the extract determined by the bicinchoninic acid method (BCA Protein Assay Kit; Thermo Scientific) following the manufacturer’s instructions.
RNA Isolation, DNase Treatment, RT, and PCR
Total RNA was isolated with a buffer containing 8 m guanidine-HCl (Invitrogen), 50 mm Tris-HCl, pH 8.0 (Invitrogen), 20 mm EDTA, pH 8.0 (Invitrogen), and 50 mm β-mercaptoetanol (Gibco) following the methodology described by Logemann et al. (1987). When necessary, RNA was treated with DNase (Turbo DNA Free; Ambion) following the manufacturer’s instructions. cDNA synthesis from 1.5 μg of total RNA (final volume of 12.5 μL) was performed using ImProm II Reverse Transcriptase (Promega) and oligo(dT)18 essentially according to the manufacturer’s instructions. Semiquantitative PCR conditions were basically realized as described by Silveira et al. (2007). Control genes for ABA, Glc, and Man treatments were Rd29a for ABA (At5g52310; Arroyo et al., 2003) and XTR7 for Glc and Man (At4g14130; Price et al., 2004), respectively. The carbonic anhydrase gene (At5g14740) was used as a Glc-repressible HXK1-dependent positive control (Moore et al., 2003). Primers and annealing temperatures are given in Supplemental Table S2.
Real-Time PCR Analysis
qRT-PCR was performed using an ABI PRISM 7500 HT (Applied Biosystems). Gene expression was calculated with the Delta-Delta cycle threshold method (Livak and Schmittgen, 2001). Actin2 (At3g18780) or PDF2 (At1g13320) was used as the reference gene (Czechowski et al., 2005). ABI5 (At2g36270), Rd29a (At5g52310), and Rd29b (At5g52300) were used as controls for ABA treatment. Primers of all genes are given in Supplemental Table S2. For most of the genes, one primer spanning an exon-exon junction was designed. In the case of intronless genes, DNase-treated RNA was used, and a control without reverse transcriptase was included.
ABA Quantification by HPLC-Tandem Mass Spectrometry
For ABA extraction, 200 mg of powdered Col-0 seedlings was mixed with 1 mL of extraction solvent (acetone:water:acetic acid, 80:19:1 [v/v/v]) and 60 ng of (−)-5,8′,8′,8′-d4-ABA internal standard (NRC Plant Biotechnology Institute; http://www.nrc-cnrc.gc.ca/eng/ibp/pbi.html). The supernatant was lyophilized at room temperature and dissolved in 100 μL of methanol:acetic acid (99:1, v/v), combined with 900 μL of water:acetic acid (99:1, v/v), and centrifuged for 1 min. The supernatant was passed through a 1-mL solid-phase extraction cartridge (Oasis HLB 1; Waters) previously equilibrated with 1 mL of methanol and 1 mL of water:methanol:acetic acid (90:10:1, v/v/v). ABA was eluted with 1 mL of methanol:water:acetic acid (80:19:1, v/v/v) and lyophilized. Samples were resuspended in 120 μL of 0.07% acetic acid:acetonitrile (85:15, v/v) and analyzed with a HPLC system (Shimadzu; Phenomenex Mercury MS C-18 column, 20 × 4 mm, 5-μm particle size) coupled to a triple quadrupole mass spectrometer (Quattro II; Micromass) using a FCV-12AH valve (Shimadzu) to direct the flow rate, ranging from 17.4 to 21 min, to the mass spectrometer. The mobile phase consisted of 0.1% formic acid as solvent A and acetonitrile as solvent B, with a flow rate of 0.2 mL min−1 from 0 to 22.1 min, 0.6 mL min−1 from 22.3 to 34.5 min, and 0.2 mL min−1 until 35.0 min. Negative multiple reaction monitoring analyses were done monitoring the mass transition of ABA (mass-to-charge ratio 263 → 153) and ABA-d4 (mass-to-charge ratio 267 → 156) with the following parameters: source temperature at 100°C, desolvation temperature at 200°C, capillary voltage at 4.0 kV, collision energy at 15 eV, and sample cone voltage and extractor cone voltage at 20 and 5 V, respectively. For ABA determination, a standard curve containing ABA (0–0.04 ng μL−1) and the internal standard ABA-d4 (0.1 ng μL−1) was used. Data were processed by Mass Lynx NT version 3.2 software (Micromass).
Statistical Analysis
All statistical comparisons were done using Student’s t test (P < 0.05).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The expression of AtbZIP63 is repressed by 10 μm ABA.
Supplemental Figure S2. ABI4 modulates ABA- and Glc-induced AtbZIP63 repression.
Supplemental Figure S3. Regulation of the three C-group bZIP genes, AtbZIP9, AtbZIP10, and AtbZIP25, by different combinations of ABA, Glc, and Man.
Supplemental Figure S4. The synergistic repression of AtbZIP63 by Glc+ABA does not rely directly on miRNA activity.
Supplemental Figure S5. Sequence analyses of AtbZIP3 and AtbZIP63 3′UTRs.
Supplemental Table S1. Common promoter motifs between AtbZIP63 and AtbZIP3.
Supplemental Table S2. Primers used in PCR, semiquantitative RT-PCR, and qRT-PCR assays.
Supplemental Table S3. Actin2 and PDF2 qRT-PCR cycle threshold (Ct) in different treatments and mutants.
Acknowledgments
We thank S.A. Martins for technical support, the Arabidopsis Biological Resource Center stock center for the different mutant genotypes, and Hervé Vaucheret for kindly providing miRNA pathway mutants.
References
- Acevedo-Hernández GJ, León P, Herrera-Estrella LR. (2005) Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. Plant J 43: 506–519 [DOI] [PubMed] [Google Scholar]
- Alonso R, Oñate-Sanchez L, Weltmeier F, Ehlert A, Diaz I, Dietrich K, Vicente-Carbajosa J, Dröge-Laser W. (2009) A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21: 1747–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, León P. (2000) Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev 14: 2085–2096 [PMC free article] [PubMed] [Google Scholar]
- Arroyo A, Bossi F, Finkelstein RR, León P. (2003) Three genes that affect sugar sensing (Abscisic Acid Insensitive 4, Abscisic Acid Insensitive 5, and Constitutive Triple Response 1) are differentially regulated by glucose in Arabidopsis. Plant Physiol 133: 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E. (2010) Energy signaling in the regulation of gene expression during stress. Mol Plant 3: 300–313 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelin JM, Sheen J. (2007) A central integrator of transcription networks in plant stress and energy signaling. Nature 448: 938–943 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Sheen J. (2008) Convergent energy and stress signaling. Trends Plant Sci 13: 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard IM, Lynch TJ, Finkelstein RR. (2002) Regulation and role of the Arabidopsis Abscisic Acid-Insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol 129: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard-Gifford I, Lynch TJ, Garcia ME, Malhotra B, Finkelstein RR. (2004) The Arabidopsis thaliana abscisic acid-insensitive8 encodes a novel protein mediating abscisic acid and sugar responses essential for growth. Plant Cell 6: 406–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Jones AM. (2004) AtRGS1 function in Arabidopsis thaliana. Methods Enzymol 389: 338–350 [DOI] [PubMed] [Google Scholar]
- Chen JG, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP. (2003a) A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301: 1728–1731 [DOI] [PubMed] [Google Scholar]
- Chen P-Y, Wang C-K, Soong S-C, To K-Y. (2003b) Complete sequence of the binary vector pBI121 and its application in cloning T-DNA insertion from transgenic plants. Mol Breed 11: 287–293 [Google Scholar]
- Chen Y, Ji F, Xie H, Liang J, Zhang J. (2006) The regulator of G-protein signaling proteins involved in sugar and abscisic acid signaling in Arabidopsis seed germination. Plant Physiol 140: 302–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W-H, Endo A, Zhou L, Penney J, Chen H-C, Arroyo A, Leon P, Nambara E, Asami T, Seo M, et al. (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14: 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Ryoo N, Eom J, Lee D, Kim H, Jeong S, Lee Y, Kwon Y, Cho M, Bhoo SH, et al. (2009) Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol 149: 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Sheen J, Yoo S. (2010) Low glucose uncouples hexokinase1-dependent sugar signaling from stress and defense hormone abscisic acid and C2H4 responses in Arabidopsis. Plant Physiol 152: 1180–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y-H, Yoo S-D, Sheen J. (2006) Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127: 579–589 [DOI] [PubMed] [Google Scholar]
- Corrêa LGG, Riaño-Pachón DM, Guerra Schrago C, Vicentini dos Santos R, Mueller-Roeber B, Vincentz M. (2008) The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS ONE 3: e2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corruzi GM, Zhou L. (2001) Carbon and nitrogen sensing and signaling in plants: emerging “matrix effect.” Curr Opin Plant Biol 4: 247–253 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich K, Weltmeier F, Ehlert A, Weiste C, Stahl M, Harter K, Dröge-Laser W. (2011) Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell 23: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlert A, Weltmeier F, Wang X, Mayer CS, Smeekens S, Vicente-Carbajosa J, Dröge-Laser W. (2006) Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J 46: 890–900 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG. (2002) Local and long-range signaling pathways regulating plant responses to nitrate. Annu Rev Plant Biol 53: 203–224 [DOI] [PubMed] [Google Scholar]
- Gibson SI. (2005) Control of plant development and gene expression by sugar signaling. Curr Opin Plant Biol 8: 93–102 [DOI] [PubMed] [Google Scholar]
- Gibson SI, Laby RJ, Kim D. (2001) The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1. Biochem Biophys Res Commun 280: 196–203 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res (Database Issue) 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez RA, Ewing RM, Cherry JM, Green PJ. (2002) Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA 99: 11513–11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez RA, Lejay LV, Dean D, Chiaromonte F, Shasha DE, Coruzzi GM. (2007) Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol 8: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford NG, Hey SJ. (2009) Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem J 419: 247–259 [DOI] [PubMed] [Google Scholar]
- Hanson J, Hanssen M, Wiese A, Hendriks MMWB, Smeekens S. (2008) The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J 53: 935–949 [DOI] [PubMed] [Google Scholar]
- Holtorf H, Schöb H, Kunz C, Waldvogel R, Meins F., Jr (1999) Stochastic and nonstochastic post-transcriptional silencing of chitinase and β-1,3-glucanase genes involves increased RNA turnover: possible role for ribosome-independent RNA degradation. Plant Cell 11: 471–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. (2009) The many pathways of RNA degradation. Cell 136: 763–776 [DOI] [PubMed] [Google Scholar]
- Huang J, Taylor JP, Chen JG, Uhrig JF, Schnell DJ, Nakagawa T, Korth L, Jones AM. (2006) The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell 18: 1226–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. (2010) Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev 24: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser C, Kortstee A, Pego J, Weisbeek P, Wisman E, Smeekens S. (2000) The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: involvement of abscisic acid in sugar responses. Plant J 23: 577–585 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Döge-Laser W, Vicent-Carbajosa J, Tiedemann J, Kroj T, Parcy F. (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Jang JC, Leon P, Zhou L, Sheen J. (1997) Hexokinase as a sugar sensor in higher plants. Plant Cell 9: 5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminaka H, Näke C, Epple P, Dittgen J, Schütze K, Chaban C, Holt BF III, Merkle T, Schäfer E, Harter K, et al. (2006) bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J 25: 4400–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SG, Price J, Lin P-C, Hong JC, Jang J-C. (2010) The Arabidopsis bZIP1 transcription factor is involved in sugar signaling, protein networking, and DNA binding. Mol Plant. 3: 361–373 [DOI] [PubMed] [Google Scholar]
- Kant P, Kant S, Gordon M, Shaked R, Barak S. (2007) Stress response suppressor1 and stress response suppressor2, two dead-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses. Plant Physiol 145: 814–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempa S, Krasensky J, Dal Santo S, Kopka J, Jonak C. (2008) A central role of abscisic acid in stress-regulated carbohydrate metabolism. PLoS ONE 3: e3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KE. (1996) Carbohydrates modulate gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 509–540 [DOI] [PubMed] [Google Scholar]
- Koussewitzky S, Not A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. (2007) Signal from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Laby RJ, Kincaid MS, Kim D, Gibson SI. (2000) The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J 23: 587–596 [DOI] [PubMed] [Google Scholar]
- Lam LT, Pickeral OK, Peng AC, Rosenwald A, Hurt EM, Giltnane JM, Averett LM, Zhao H, Davis RE, Sathyamoorthy M, et al. (2001) Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol 2: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León P, Sheen J. (2003) Sugar and hormone connections. Trends Plant Sci 8: 110–116 [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M. (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10: 655–661 [DOI] [PubMed] [Google Scholar]
- Li Y, Lee KK, Walsh S, Smith C, Hadingham S, Sorefan K, Cawley G, Bevan MW. (2006) Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a relevance vector machine. Genome Res 16: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. (1987) Improved method for the isolation of RNA from plant tissues. Anal Biochem 163: 16–20 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH. (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Fedoroff N. (2000) A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12: 2351–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng W-H, Liu Y-X, Hwang I, Jones T, Sheen J. (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light and hormonal signaling. Science 3000: 332–336 [DOI] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Millar AH, O’Toole N, Small I, Whelan J. (2007) Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19: 3418–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Kitahata N, Seki M, Narusaka Y, Narusaka M, Kuromori T, Asami T, Shinozaki K, Hirayama T. (2005) Analysis of ABA Hypersensitive Germination2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant J 44: 972–984 [DOI] [PubMed] [Google Scholar]
- Pego JV, Weisbeek PJ, Smeekens SCM. (1999) Mannose inhibits Arabidopsis germination via a hexokinase-mediated step. Plant Physiol 119: 1017–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterbauer T, Mach L, Mucha J, Richter A. (2002) Functional expression of a cDNA encoding pea (Pisum sativum L.) raffinose synthase, partial purification of the enzyme from maturing seeds, and steady-state kinetic analysis of raffinose synthesis. Planta 215: 839–846 [DOI] [PubMed] [Google Scholar]
- Polge C, Thomas M. (2006) SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci 12: 20–27 [DOI] [PubMed] [Google Scholar]
- Price J, Laxmi A, Saint Martin S, Jang J-C. (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16: 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Li T-C, Kang SG, Na JK, Jang J-C. (2003) Mechanisms of glucose signaling during germination of Arabidopsis. Plant Physiol 132: 1424–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon M, Rolland F, Sheen J. (2008) Sugar sensing and signaling. The Arabidopsis Book 6: e0117, doi/10.1199/tab.0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Chua NH. (2007) ABA induction of miRNA159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J 49: 592–606 [DOI] [PubMed] [Google Scholar]
- Roitsch T. (1999) Source-sink regulation by sugar and stress. Curr Opin Plant Biol 2: 198–206 [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-González E, Sheen J. (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J. (2002) Sugar sensing and signaling in plants. Plant Cell (Suppl) 14: S185–S205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW. (2001) Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J 26: 421–433 [DOI] [PubMed] [Google Scholar]
- Rook F, Gerrits N, Kortstee A, Van Kampen M, Borrias M, Weisbeek P, Smeekens S. (1998) Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J 15: 253–263 [DOI] [PubMed] [Google Scholar]
- Rook F, Hadingham SA, Li Y, Bevan MW. (2006) Sugar and ABA response pathways and the control of gene expression. Plant Cell Environ 29: 426–434 [DOI] [PubMed] [Google Scholar]
- Seki M, Umezawa T, Urano K, Shinozaki K. (2007) Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol 10: 296–302 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. (2006) Gene networks involved in drought stress response and tolerance. J Exp Bot 58: 221–227 [DOI] [PubMed] [Google Scholar]
- Silveira AB, Gauer L, Tomaz JP, Cardoso PR, Carmello-Guerreiro S, Vincentz M. (2007) The Arabidopsis AtbZIP9 protein fused to VP16 transcriptional activation domain alters leaf and vascular development. Plant Sci 172: 1148–1156 [Google Scholar]
- Smeekens S. (2000) Sugar-induced signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol 51: 49–81 [DOI] [PubMed] [Google Scholar]
- Smith AM, Stitt M. (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Söderman EM, Brocard IM, Lynch TJ, Finkelstein RR. (2000) Regulation of the Arabidopsis-insensitive 4 gene in seed and abscisic acid response signaling network. Plant Physiol 124: 1752–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souret FF, Kastenmayer JP, Green PJ. (2004) AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell 15: 173–183 [DOI] [PubMed] [Google Scholar]
- Srinivasasainagendra V, Page GP, Mehta T, Coulibaly I, Loraine AE. (2008) CressExpress: a tool for large-scale mining of expression data from Arabidopsis. Plant Physiol 147: 1004–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Wang S, Jones AM. (2002) Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol 129: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K. (2010) Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol 51: 1821–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Blasing OE, Gibon Y, Retzlaff K, Hohne M, Gunther M, Stitt M. (2008) Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol 146: 1834–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villadsen D, Smith S. (2004) Identification of more than 200 glucose-responsive Arabidopsis genes none of which responds to 3-O-methylglucose or 6-deoxyglucose. Plant Mol Biol 55: 467–477 [DOI] [PubMed] [Google Scholar]
- Vincentz MGA, Bandeira-Kobarg C, Gauer L, Schlögl P, Leite A. (2003) Evolutionary pattern of angiosperm bZIP factors homologous to the maize Opaque-2 regulatory protein. J Mol Evol 55: 1–12 [DOI] [PubMed] [Google Scholar]
- Wang HX, Weerasinghe RR, Perdue TD, Cakmakci NG, Taylor JP, Marzluff WF, Jones AM. (2006) A Golgi-localized hexose transporter is involved in heterotrimeric G protein-mediated early development in Arabidopsis. Mol Biol Cell 17: 4257–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltmeier F, Ehlert A, Mayer CS, Dietrich K, Wang X, Schütze K, Alonso R, Harter K, Vicente-Carbajosa J, Dröge-Laser W. (2006) Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J 25: 3133–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese A, Elzinga N, Wobbes B, Smeekens S. (2004) A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16: 1717–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Sheen J, Jang JC. (2000) The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol Biol 44: 451–461 [DOI] [PubMed] [Google Scholar]
- Xiong L, Gong Z, Rock CD, Subramanian S, Guo Y, Xu W, Galbraith D, Zhu J-K. (2001) Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell 1: 771–781 [DOI] [PubMed] [Google Scholar]
- Zhou L, Jang J-C, Jones TL, Sheen J. (1998) Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci USA 95: 10294–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]