Abstract
Numerous auxiliary nuclear factors have been identified to be involved in the dynamics of the photosystem II (PSII) complex. In this study, we characterized the high chlorophyll fluorescence243 (hcf243) mutant of Arabidopsis (Arabidopsis thaliana), which shows higher chlorophyll fluorescence and is severely deficient in the accumulation of PSII supercomplexes compared with the wild type. The amount of core subunits was greatly decreased, while the outer antenna subunits and other subunits were hardly affected in hcf243. In vivo protein-labeling experiments indicated that the synthesis rate of both D1 and D2 proteins decreased severely in hcf243, whereas no change was found in the rate of other plastid-encoded proteins. Furthermore, the degradation rate of the PSII core subunit D1 protein is higher in hcf243 than in the wild type, and the assembly of PSII is retarded significantly in the hcf243 mutant. HCF243, a nuclear gene, encodes a chloroplast protein that interacts with the D1 protein. HCF243 homologs were identified in angiosperms with one or two copies but were not found in lower plants and prokaryotes. These results suggest that HCF243, which arose after the origin of the higher plants, may act as a cofactor to maintain the stability of D1 protein and to promote the subsequent assembly of the PSII complex.
PSII is a multisubunit protein-pigment complex embedded in the thylakoid membrane that harnesses light energy to split water into oxygen, protons, and electrons. It comprises more than 20 different subunits, most of which are integral membrane proteins and bind numerous cofactors (Wollman et al., 1999; Iwata and Barber, 2004; Nelson and Yocum, 2006). The PSII reaction center complex is composed of the D1 and D2 proteins, the α- and β-subunits of cytochrome b559, and the PsbI protein. The D1 and D2 heterodimers bind all the essential redox components of PSII required for primary charge separation and subsequent electron transfer (Nanba and Satoh, 1987). In addition, PSII core complexes also contain the intrinsic chlorophyll a-binding proteins (CP43 and CP47), the extrinsic oxygen-evolving complex (33-, 23-, and 17-kD proteins), and the other low-molecular-mass proteins (Bricker and Ghanotakis, 1996). The functional form of PSII in the thylakoid membrane consists of the PSII core and the associated light-harvesting complex (Nelson and Yocum, 2006).
Despite considerable advances in the elucidation of the structure and function of PSII, knowledge of the assembly of this multiprotein complex is only in its infancy. A number of studies have proved that this assembly is likely to involve multistep processes (Rokka et al., 2005). The first step is formation of the PSII reaction center, in which the D1 protein is incorporated into a precomplex, probably consisting of the D2, cytochrome b559, and PsbI proteins (Adir et al., 1990; van Wijk et al., 1997; Müller and Eichacker, 1999; Zhang et al., 1999). Subsequently, CP47 and CP43, core antenna subunits that bind chlorophyll a, are recruited to form the PSII core complex, which allows further binding of the oxygen-evolving enhancer proteins, including PsbO, PsbP, and PsbQ. D1, D2, and CP47 appear to accumulate in a coordinated manner (Jensen et al., 1986; de Vitry et al., 1989; Yu and Vermaas, 1990). CP43 is synthesized independently and is a dynamic component of PSII, with dissociation and reassociation constantly cycling (de Vitry et al., 1989; Zhang et al., 2000). Integration of the low-molecular-mass proteins into PSII has been found to occur at different stages of the PSII assembly process (Hager et al., 2002; Suorsa et al., 2004; Rokka et al., 2005). The final establishment of a functional PSII involves the dimerization of PSII monomers and the association of LHCII trimers (Rokka et al., 2005).
Because of the structural complexity of PSII, the functional assembly of this highly complex oligomeric protein should require precisely controlled mechanisms. Specific mechanisms should operate to allow the stoichiometric accumulation of the various subunits encoded by the two genetic compartments required for PSII assembly (Choquet and Vallon, 2000). So far, a number of nucleus-encoded auxiliary and regulatory proteins have been identified that should be involved in this dynamic process (Goldschmidt-Clermont, 1998; Barkan and Goldschmidt-Clermont, 2000; Leister, 2003). For example, although plastid-encoded proteins are normally synthesized in the high chlorophyll fluorescence136 (hcf136) mutant of Arabidopsis (Arabidopsis thaliana), the assembly of PSII reaction centers is blocked and no stable PSII complexes appear to accumulate because of the lack of the HCF136 protein (Meurer et al., 1998; Plücken et al., 2002). Similarly, accumulation of PSII supercomplexes is defective in the low psii accumulation2 (lpa2) mutant of Arabidopsis, and the LPA2 protein directly interacts with ALB3 and may form a complex that is specifically involved in the efficient assembly of CP43 within PSII (Ma et al., 2007).
Furthermore, photosynthetic water splitting is inevitably coupled with the formation of reactive oxygen species followed by the photooxidative damage and repair cycle of PSII (Andersson and Aro, 1997; Keren et al., 2005). During this progress, D1 protein is the main target for light-induced damage among PSII proteins (for review, see Prasil et al., 1992; Aro et al., 1993), and it undergoes a very high light-dependent turnover and shows very high rates of both synthesis and degradation (Sundby et al., 1993). The damaged D1 protein depletion should be considered as the first phase in the repair cycle of PSII, and much effort has been made to identify the proteases responsible for the degradation of the D1 protein (Adam and Clarke, 2002; Yoshioka and Yamamoto, 2011). The photodamaged D1 protein is replaced constantly with newly synthesized D1 protein, which has been described as a cotranslational event to maintain PSII in a functional state (Aro et al., 1993). Therefore, many studies were focused on D1 turnover, and numerous regulation factors have been reported. For example, PAM68, a conserved integral membrane protein found in cyanobacterial and eukaryotic thylakoids, has also been suggested to play a role in the stability and maturation of D1 (Armbruster et al., 2010). LPA1 appears to be an integral membrane chaperone that assists efficient PSII assembly through direct interaction with D1 (Peng et al., 2006), and REP27 is essential for D1 protein turnover, permitting completion of the translation process, maturation, and activation of D1 into a functional PSII reaction center complex (Park et al., 2007). Ossenbühl et al. (2006) further found that Srl1471p, a homolog of ALB3, is essential for integrating the D1 precursor into the thylakoid membrane, leading to accumulation of the D1 precursor in the membrane phase.
Despite these advances, the assembly and stabilization of the D1 protein in PSII and its regulation mechanism remain poorly understood (Aro et al., 1993; Yokthongwattana and Melis, 2006). Screening mutants with altered chlorophyll fluorescence has proved to be a specific and efficient way to dissect the molecular mechanisms underlying the biogenesis of photosystems (Peng et al., 2006), since alterations in chlorophyll fluorescence indicate defects in the photosynthetic electron transport chain, which may result from changes in the structure or function of the thylakoid membrane (Miles, 1994). In this study, we characterized the hcf243 mutant of Arabidopsis. RNA gel-blot and immunoblot analyses revealed that plastid-encoded mRNAs for PSII core subunits were present in the mutant, but the corresponding subunits were dramatically reduced. Protein-labeling studies revealed that the accumulation of D1 protein was significantly reduced in the hcf243 mutant. These results indicate that the HCF243 gene encodes a cofactor that is involved in D1 dynamics and subsequently the stability and assembly of the PSII complex.
RESULTS
Phenotype of the hcf243 Mutant
To investigate genes involved in the biogenesis of the PSII complex, we screened the T-DNA mutant collection from the Arabidopsis Biological Resource Center (Weigel et al., 2000) for the high chlorophyll fluorescence phenotype, which was reported previously (Meurer et al., 1996; Peng et al., 2006), and isolated hcf243, a previously unidentified mutant. Genetic analysis showed that the hcf243 mutation is recessive. The phosphinothricin resistance marker carried by the T-DNA and the hcf243 mutant phenotype cosegregated, indicating that the mutation was due to the T-DNA insertion (data not shown).
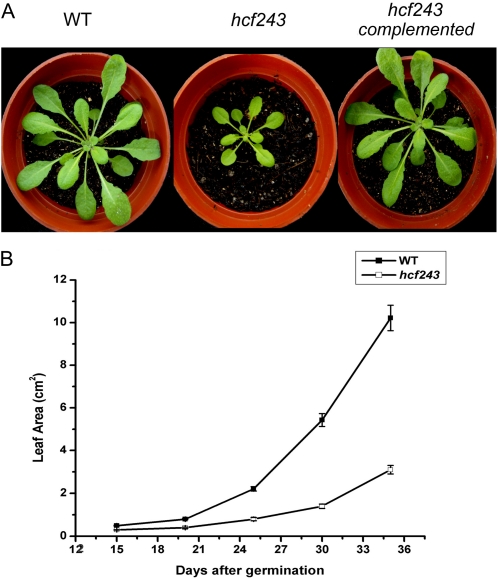
In addition to the high chlorophyll fluorescence phenotype, we found that plant growth was also affected in the hcf243 mutant (Fig. 1A). The inflorescence stems of the hcf243 mutant were shorter in height, and its rosette leaves were paler and smaller in size (Fig. 1A). The leaf areas of 6-week-old hcf243 mutant plants were approximately 70% smaller than in the wild type (Fig. 1B). The high chlorophyll fluorescence phenotype in hcf243 indicates impaired photosynthesis, which in turn results in the phenotypes of pale leaf and reduced plant growth.
Figure 1.
Phenotypes of wild-type (WT), hcf243, and hcf243 complemented plants. A, Six-week-old wild-type (left), hcf243 (middle), and hcf243 complemented (right) plants. B, Growth kinetics of hcf243 mutants compared with wild-type plants. Values shown are means ± se of three biological replicates; each replicate represents six transgenic plants.
PSII Activity Is Dramatically Reduced in the hcf243 Mutant
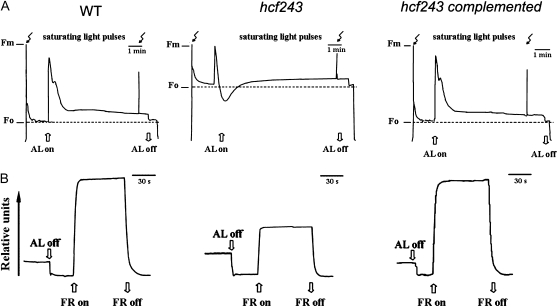
Noninvasive fluorometric analyses were performed to investigate the photosynthetic characteristics of the hcf243 mutant. Chlorophyll fluorescence induction experiments revealed that the ratio of variable fluorescence to maximum fluorescence (Fv/Fm; indicating the maximum potential of photochemical reactions of PSII) was dramatically reduced in the hcf243 mutant (0.42 ± 0.02) compared with that of wild-type plants (0.82 ± 0.03; Fig. 2A), indicating that the hcf243 mutant has defects in energy transfer within PSII. Furthermore, it is noteworthy that P700 content was lower in the hcf243 mutant than in wild-type plants (Fig. 2B), suggesting that P700 might be partially oxidized, but PSI is functional in the hcf243 mutant, as observed in both lpa1 and lpa2 mutants (Peng et al., 2006; Ma et al., 2007). Clearly, these findings demonstrate that the hcf243 mutation causes a dramatic decrease in PSII activity.
Figure 2.
Spectroscopic analysis of wild-type (WT), hcf243, and hcf243 complemented plants. A, Chlorophyll fluorescence induction. The minimum fluorescence yield (F0) of dark-adapted plants was measured using a pulsed measuring beam of red light. The maximum fluorescence yield (Fm) was determined after a saturating pulse of white light in dark-adapted leaves. B, P700 redox kinetics. The redox kinetics were investigated by measuring absorbance changes of P700 at 820 nm induced by far-red light (FR; 720 nm). AL, Actinic light (120 μmol m−2 s−1).
HCF243 Is Involved in the Induction Kinetics of Chlorophyll Fluorescence
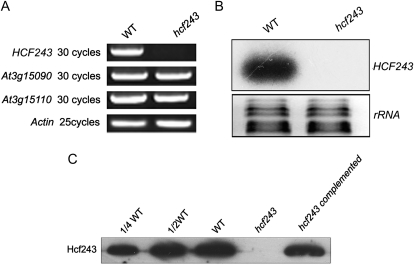
To determine the genetic basis of the hcf243 phenotype, the genomic regions flanking the left border of the T-DNA were isolated by thermal asymmetric interlaced-PCR. Sequence analysis showed that the T-DNA was inserted in the 5′ untranslated region of At3g15095. There are three different gene models in The Arabidopsis Information Resource database for At3g15095. The cloned sequence of the HCF243 gene is consistent with At3g15095.1 and has been submitted to GenBank (accession no. HM748832). To evaluate the effect of the T-DNA insertion on gene expression, reverse transcription (RT)-PCR and northern-blot analysis revealed that expression of the HCF243 gene in the isolated mutant was barely detectable compared with that in wild-type plants (Fig. 3, A and B). Further immunoblot analyses with the HCF243 polyclonal antibody, which was raised against recombinant HCF243 protein (amino acids 221–408), also showed that no signal was detected in the total protein preparations (Fig. 3C).
Figure 3.
Characterization of the hcf243 mutant. A, RT-PCR analysis of HCF243 gene expression. RT-PCR was performed with actin-specific primers and the specific primers for At3g15095, At3g15090, and At3g15110. B, Northern-blot analysis of HCF243 gene expression in wild-type (WT) and hcf243 plants. Thirty micrograms of total RNA from wild-type and hcf243 plants was size fractionated by agarose gel electrophoresis, transferred to a nylon membrane, and probed with 32P-labeled HCF243 cDNA probe. rRNAs were detected by ethidium bromide staining. C, Immunodetection of HCF243 with the specific HCF243 antibody.
We recovered homologous genes or protein sequences from all angiosperm species with complete genome sequences. However, no corresponding homologous genes were identified in lower plants and prokaryotes, for example, Selaginella moellendorffii, Physcomitrella patens, and cyanobacteria. The predicted protein sequences of all homologous genes are highly variable, and no distinct domain was identified (Supplemental Fig. S1). These aligned proteins are 36% to 86% (averaged 48%) identical to one another. Two copies were found for four species, Zea mays, Populus trichocarpa, Glycine max, and Manihot esculenta, while only one was recovered for the others. It remains unknown whether the homologous genes exist in the gymnosperms, because the complete genome sequence of this group is not reported. The comparison suggests that HCF243 is a novel gene and may be found only in higher plants. Phylogenetic analysis further suggested that two copies in a few angiosperm species due to duplication occurred independently after the origin of each separate lineage (Supplemental Fig. S2). In addition, in some such lineages (e.g. the Sorghum-Zea lineage), one duplicated copy seems to have been lost again (Supplemental Fig. S2).
To prove that the disruption of At3g15095.1 was responsible for the phenotypes observed in the hcf243 mutant, the isolated full-length cDNA of HCF243 was fused to the cauliflower mosaic virus 35S promoter in the plant transformation vector pSN1301 and introduced into a homozygous hcf243 mutant using the floral dip method by means of Agrobacterium tumefaciens. Twenty-five independent transgenic plants were regenerated. The protein levels of HCF243 in the complemented plants were comparable with those in wild-type plants (Fig. 3C), and their chlorophyll fluorescence induction kinetics were also indistinguishable from those of wild-type plants (Fig. 2A). These results unequivocally demonstrate that the disruption of the At3g15095.1 gene is responsible for the phenotypes of the hcf243 mutant.
Thylakoid Membrane Protein Composition in the hcf243 Mutant
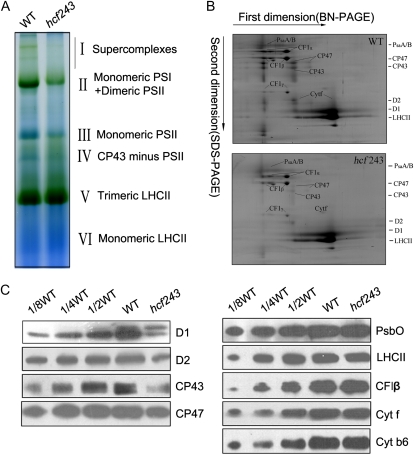
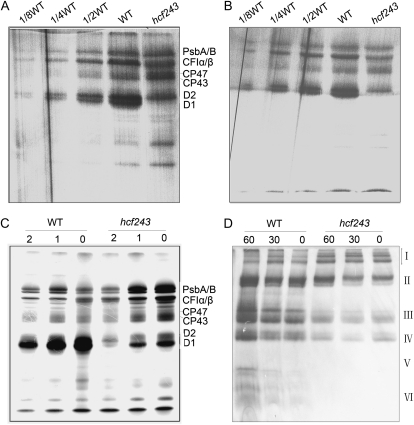
A block in energy transfer within PSII found in the hcf243 mutant could be the result of a defect in the PSII complex. To address this possibility, we assayed putative structural alterations of photosynthetic protein complexes in the mutant, and the chlorophyll-protein complexes were subjected to blue native (BN)/SDS-PAGE analysis. After the first-dimensional separation in the presence of Coomassie blue G-250 dye, six major chlorophyll-protein complexes, marked I to VI, were revealed (Fig. 4A). The positions of numerous thylakoid membrane complexes were identified from similar gels by immunoblot analysis with distinct antibodies and apparently represented PSII supercomplexes (band I), monomeric PSI and dimeric PSII (band II), monomeric PSII (band III), CP43 minus PSII (band IV), trimeric LHCII (band V), and monomeric LHCII (band VI; Li et al., 2003; Guo et al., 2005). As clearly shown in Figure 4A, PSII supercomplexes were almost absent, and both band II (monomeric PSI and dimeric PSII) and band III (monomeric PSII) were dramatically reduced in the mutant compared with wild-type plants. Analyses of the two-dimensional SDS-urea-PAGE gels after Coomassie blue staining also confirmed that the relative amounts of subunits of PSII protein complexes were greatly reduced in the mutant (Fig. 4B). In addition, to verify the steady-state levels of the thylakoid protein change, immunoblot analysis with specific antibodies against the subunits of photosynthetic protein complexes was performed. Our results showed that levels of the plastid-encoded PSII core subunits D1, D2, CP47, and CP43 were dramatically reduced in the hcf243 mutant compared with those in wild-type plants; interestingly, altered accumulation of the D1 precursor was absolutely detected in the mutant (Fig. 4C), whereas no significant changes were found in either plastid-encoded proteins (PsaA/B, Cytf, Cytb6) or nucleus-encoded proteins (LHCII, PsbO; Fig. 4C), which is consistent with the results of two-dimensional SDS-urea-PAGE (Fig. 4B). Altogether, the accumulation of PSII and its core subunits is significantly affected due to the interruption of HCF243, which should confirm the partially inefficient PSII activity proved above by spectroscopic analyses (Fig. 2).
Figure 4.
Analysis of thylakoid proteins from wild-type (WT) and hcf243 plants. A, Two-dimensional separation of protein complexes in the thylakoid membranes. BN-PAGE-separated thylakoid proteins in a single lane from a BN gel were separated in a second dimension by 15% SDS-urea-PAGE and stained with Coomassie blue. B, BN gel analysis of thylakoid membrane (10 μg of chlorophyll) from wild-type and hcf243 plants was solubilized and separated by BN gel electrophoresis. The positions of protein complexes were identified with appropriate antibodies (Guo et al., 2005). C, Immunoblot analysis of thylakoid proteins from the wild type and hcf243. The thylakoid membrane proteins were separated by SDS-urea-PAGE, and the blots were probed with specific anti-D1, anti-D2, anti-CP47, anti-CP43, anti-CF1β, anti-LHCII, anti-PsbO, anti-Cytf, and anti-Cytb6 antibodies.
Steady-State mRNA Levels and Polysome Association of the hcf243 PSII Core Subunits
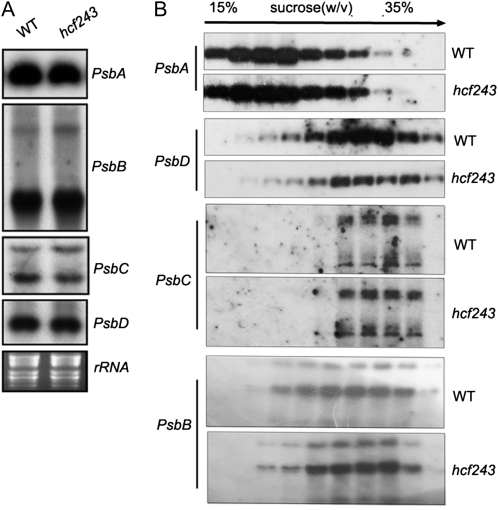
To test whether the significant reductions observed in the mutant’s PSII complexes resulted from one or more impaired transcriptions of PSII core subunits, the effect of the hcf243 mutation on the transcription of PSII core subunits was investigated by northern-blot analysis. The results clearly showed that the amounts of psbA, psbB, psbC, and psbD (encoding the D1, CP47, CP43, and D2 subunits of PSII, respectively) were almost identical in the mutant and wild-type plants (Fig. 5A). The protein synthesis capacity of chloroplasts was further studied by analyzing changes in the polysome association of psbA, psbB, psbC, and psbD transcripts following Suc gradient fractionation. Interestingly, no obvious differences were observed in the association of these transcripts with polysomes between the mutant and wild-type plants (Fig. 5B).
Figure 5.
mRNA expression in chloroplasts and polysome association of chloroplast mRNA. A, Northern-blot analysis of transcripts in wild-type (WT) and hcf243 plants. Transcripts of psbA, psbB, psbC, and psbD genes were detected by probing the filter with the appropriate gene-specific probes. B, Polysome association in wild-type and hcf243 plants of mRNAs encoding the chloroplast proteins (psbA, psbB, psbC, and psbD). Total extracts were prepared and separated on Suc gradients. Ten fractions of equal volume were collected from the top to the bottom of the Suc gradients, and northern-blot analysis was performed on equal proportions of the RNA purified from each fraction. rRNAs were detected by ethidium bromide staining.
In Vivo Synthesis and Stabilization of the PSII Core Subunits in the hcf243 Mutant
To determine whether the impaired accumulation of the subunits of the PSII complex is caused by decreased translation or accelerated degradation, the rates of synthesis of chloroplast-encoded thylakoid membrane proteins were studied by pulse labeling of mutant leaves with [35S]Met in the presence of cycloheximide as an inhibitor of cytoplasmic translation. As shown in Figure 6, A and B, the rates of synthesis of subunits CP43 and CP47 of PSII, proteins A/B of the PSI reaction center, and the subunits of the chloroplast ATP synthase (CF1α/β) in the 12-d-old and 4-week-old mutant were comparable to those in their wild-type counterparts. In contrast, the incorporation of [35S]Met label into the D1 and D2 polypeptides was reduced significantly, especially D1 protein. When the time of pulse labeling was extended to 30 min, the amount of radioactivity incorporated into D1 in the hcf243 mutant was obviously increased compared with that occurring after 15 min of labeling (Fig. 6C). Pulse labeling for 30 min was followed by a chase with unlabeled Met to monitor the turnover rates of plastid-encoded proteins in young seedlings. The results showed that the turnover rate of D1 protein is much more strongly affected than that of other subunits in the hcf243 mutant (Fig. 6C).
Figure 6.
In vivo [35S]Met labeling of thylakoid proteins from wild-type (WT) and hcf243 plants. A and B, Pulse labeling of thylakoid membrane proteins in 14-d-old young seedlings (A) and 4-week-old plants (B). After pulse labeling young Arabidopsis seedlings in the presence of cycloheximide for 20 min, thylakoid membranes were isolated, and the proteins were separated by SDS-urea-PAGE and visualized autoradiographically. C, Pulse and chase labeling of thylakoid membrane proteins. After 30 min of pulse labeling in 14-d-old young seedlings in the presence of cycloheximide followed by 1 or 2 h of chase with cold Met, thylakoid proteins were separated by SDS-urea-PAGE and visualized autoradiographically. D, BN -PAGE analysis of the incorporation of [35S]Met into thylakoid membrane protein complexes. A 15-min pulse in Arabidopsis young seedlings in the presence of cycloheximide was followed by a chase of cold unlabeled Met for 30 and 60 min. The thylakoid membranes were isolated and solubilized with DM, and the protein complexes were separated by BN-PAGE and visualized by autoradiography. Bands corresponding to various PSII assembly complexes of PSII supercomplexes (band I), monomeric PSI superimposed on the PSII dimer (band II), monomeric PSII (band III), CP43-free PSII monomer (band IV), reaction center (band V), and free proteins (band VI) are indicated at right.
To study the assembly of photosynthetic protein complexes, the [35S]Met-labeled thylakoid membrane proteins were separated by BN gel electrophoresis. After a 15-min pulse, most of the radiolabeling was found in protein complexes, as shown in Figure 6D. Compared with wild-type plants, the incorporation of radioactivity into monomeric PSI superimposed on the PSII dimer (band II), monomeric PSII (band III), and CP43-free PSII monomer (band IV) was dramatically reduced in the hcf243 mutant. During the chase period, those protein complexes significantly increased in the autoradiogram obtained after a 60-min chase in the wild-type plants. However, as the chase time increased, the accumulation of radioactivity in those complexes was barely increased in the hcf243 mutant. These results indicated that the assembly of PSII is partially inefficient in the hcf243 mutant because of the lack of HCF243 protein.
The HCF243 Protein Is Targeted to Chloroplast and Interacts with the D1 Protein in Vivo
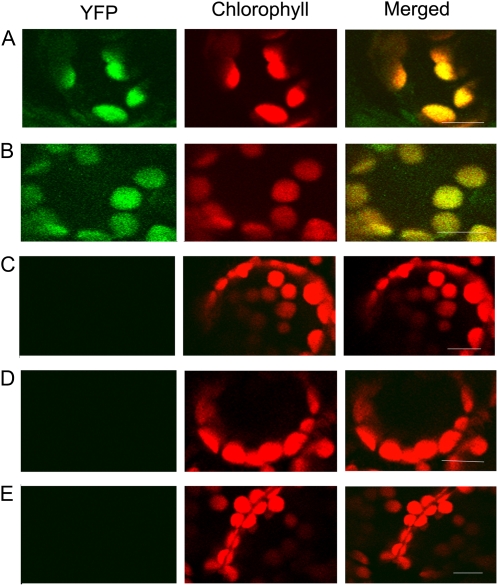
Sequence analyses predicted that the N-terminal sequence of the HCF243 protein is rich in positive and hydroxylated amino acid residues, which is characteristic of chloroplast transit peptides. Further sequence analysis predicted HCF243 to be a chloroplast-localized protein using the ChloroP 1.1 server (http://www.cbs.dtu.dk/services/ChloroP/; data not shown). To determine the actual subcellular location of HCF243, a yellow fluorescent protein (YFP)-tagged HCF243 construct was transformed into Nicotiana benthamiana leaf epidermal cells. It was demonstrated that HCF243-YFP signals were colocalized with the chloroplast autofluorescence signals in the same cell (Fig. 7A). Therefore, our results demonstrate that HCF243 is a chloroplast-localized protein.
Figure 7.
BiFC detection of HCF243 and D1 protein interactions and their subcellular localization. GFP-tagged HCF243, nYFP-tagged HCF243, and cYFP-tagged D1 constructs were transiently expressed in leaf epidermal cells of N. benthamiana plants, and their interaction and subcellular locations were examined with a laser scanning confocal microscope. Green fluorescence indicates GFP, red fluorescence shows chloroplast autofluorescence, and orange/yellow fluorescence shows images with the two types of fluorescence merged. A, N. benthamiana leaf epidermal cells expressing YFP-tagged HCF243 constructs. B, Interaction between nYFP-tagged HCF243 and cYFP-tagged D1 in chloroplast of N. benthamiana leaf epidermal cells. C, nYFP and cYFP constructs were coexpressed in N. benthamiana leaf epidermal cells as a negative control, showing no signal. D and E, nYFP-tagged HCF243 (D) and cYFP-tagged D1 (E) constructs were expressed alone in N. benthamiana leaf epidermal cells as a negative control, showing no signal. Bars = 10 μm.
To test whether HCF243 interacts with the D1 protein in plant cells in vivo, bimolecular fluorescence complementation (BiFC) analysis was performed by using N. benthamiana leaf epidermal cells as a transient expression system. In this approach, the YFP molecule is split into N-terminal (nYFP) and C-terminal (cYFP) nonfluorescent portions. Restoration of the YFP fluorescence signal is observed when nYFP and cYFP are brought together as fusions with interacting proteins (Tzfira et al., 2004). Coexpression of both nYFP-tagged HCF243 and cYFP-tagged D1 resulted in significant fluorescence in N. benthamiana leaf epidermal cells (Fig. 7B). This result suggests that the HCF243 protein interacts with the D1 protein. Moreover, coexpression of nYFP-HCF243 with cYFP-D1 further verified the chloroplast localization of the HCF243 protein. No fluorescence was found in N. benthamiana leaf epidermal cells with transformation of nYFP-HCF243, cYFP-D1, or both nYFP and cYFP (Fig. 7, C–E). Collectively, these results demonstrate that HCF243 is a chloroplast-localized protein and interacts with the D1 protein.
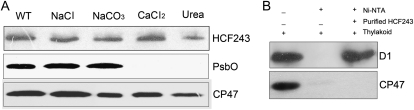
To further investigate whether HCF243 is a transmembrane protein, the wild-type membrane fractions were isolated and then the protein was subjected to immunoblot analysis. HCF243 protein was still retained in the membranes when membrane preparations were sonicated in the presence of various salts, as described in “Materials and Methods” (Fig. 8A). During these treatments, PsbO (the 33-kD luminal protein of PSII) and CP47 (the PSII core protein) were used as controls. The above results showed that HCF243 is an intrinsic membrane protein.
Figure 8.
Immunolocalization and pull-down assay of HCF243. A, Immunolocalization of HCF243. The wild-type (WT) thylakoid membranes were sonicated in the presence of 250 mm NaCl, 200 mm Na2CO3, 1 m CaCl2, and 6 m urea for 30 min at 4°C. PsbO (the 33-kD luminal protein of PSII) and CP47 (the PSII core protein) were used as markers. Membranes that had not been subjected to any salt treatment were used as controls. B, Pull-down assay of HCF243 and D1 protein interaction. About 10 μg of HCF243 His tag fusion protein coupled to Ni-NTA resin was incubated with 100 mg of DM-solubilized thylakoid membranes. Bound proteins were eluted, separated by SDS-PAGE, and subjected to immunoblot analysis with D1 and CP47 antibodies. Similar results were obtained in two additional independent experiments.
To provide further evidence for such interaction, pull-down experiments were performed with recombinant HCF243 protein fused with N-terminal His tags. The purified His-HCF243 fusion proteins were incubated with dodecyl-β-d-maltopyranoside (DM)-solubilized thylakoid membranes, and then nickel-nitrilotriacetic acid agarose (Ni-NTA) resin-bound proteins, after washing with the buffer described in “Materials and Methods,” were separated by SDS-PAGE and examined by immunoblot analysis. As shown in Figure 8B, D1 protein was detected with D1 antibody when His-HCF243 fusion proteins were used in the assay, but no signal was detected when only the solubilized thylakoid membrane and the resin were used. However, the PSII core protein CP47 was not detected when His-HCF243 fusion proteins were used. These results indicated a direct interaction between HCF243 and D1 proteins.
DISCUSSION
Considerable progress has been made in recent years in identifying and defining nuclear genes that regulate the biogenesis and assembly of both the chloroplast- and nucleus-encoded proteins into PSII of Arabidopsis. These nuclear genes act as additional auxiliary and regulatory factors to assist multistep processing in the biogenesis and assembly of the PSII complex (Goldschmidt-Clermont, 1998; Barkan and Goldschmidt-Clermont, 2000; Rochaix, 2001; Leister, 2003). The identification and molecular characterization of these factors are essential for our understanding of the ways in which photosynthetically active protein complexes are assembled and functionally maintained. Here, we present evidence that HCF243, a novel nucleus-encoded gene, is also critical for the biogenesis and assembly of Arabidopsis PSII, which marks an important step toward further understanding of this process.
Sequence analyses predict that HCF243 is localized to the chloroplast, which then was confirmed by the actual subcellular location (Figs. 7A and 8A). This location further suggested that the chloroplast-localized HCF243 protein might have a biological function involved in the biogenesis of the PSII complex. This implication is in fact substantiated by the T-DNA mutation. Mutation of the HCF243 gene resulted in a severe defect in the leaf size and growth of plants (Fig. 1A). In addition, this mutation caused a dramatic reduction in PSII activity, implying a deficiency in PSII function (Fig. 2A). Thorough analyses of the thylakoid membrane complexes isolated from leaves of the hcf243 mutant indicate a dramatic reduction in the content of both the PSII complex and its reaction core subunits (D1, D2, CP43, and CP47) compared with wild-type plants (Fig. 4). Interestingly, a dramatic increase in the accumulation of the D1 precursor was detected in the mutant (Fig. 4C), which has also been observed in pam68 (Armbruster et al., 2010) and hcf136 (Meurer et al., 1998) plants. In contrast, no significant difference in the contents of either plastid-encoded proteins (PsaA, Cytf, and Cytb6) or nucleus-encoded proteins (LHCII and PsbO) was observed between the mutant and wild-type plants (Fig. 4C). Taken together, these results indicate that HCF243 should have been involved in the biogenesis of the PSII complex. This is consistent with previous observations that LPA1 and LPA2 are specifically localized in the chloroplast and are required for the biogenesis of the PSII complex (Peng et al., 2006; Ma et al., 2007).
To gain insights into the molecular function of HCF243, it was critical to pinpoint the malfunctioning in the accumulation of the PSII core subunits in the hcf243 mutant. There are several possible explanations for the reduced content of the PSII core subunits. Similar abundance and patterns of PSII core subunit gene transcripts in both the hcf243 mutant and wild-type plants were detected, indicating that the reduced PSII core subunits are not due to the absence of transcripts encoding one or more of these PSII proteins. Further polysome analyses indicated that initiation of the translation of PsbA, PsbB, PsbC, and PsbD mRNA was not altered in the hcf243 mutant (Fig. 5B). Therefore, the reduced accumulation of the PSII core subunits may result from impaired translation or, alternatively, accelerated degradation of the PSII subunits once they have been synthesized.
As shown in Figure 6, the accumulation rate of both D1 and D2 proteins decreased severely in the hcf243 mutant, especially the significantly reduced content of the D1 protein. However, the synthesis rate of the other plastid-encoded proteins was barely affected (Fig. 6, A and B). A possible explanation for the decreased accumulation of the D1 and D2 proteins in the hcf243 mutant may be that their degradation rates were higher than those in wild-type plants. A similar phenomenon was found in cyanobacteria, whose inactivation of the PSII genes has no severe effects on the translation of their respective proteins but instead appears to accelerate the degradation of the close assembly partners (Yu and Vermaas, 1990, 1993). To monitor the degradation rates of D1 and D2 proteins in hcf243 mutants, a chase experiment with unlabeled Met was performed after pulse labeling for 30 min (Fig. 6C). The results showed that a significantly increased degradation rate of D1 protein in the hcf243 mutant was detected compared with that of wild-type plants. Interestingly, previous studies showed that a deficiency of some special proteins results in the increased degradation of D1, suggesting that these proteins may be required for D1 stability (Choquet and Vallon, 2000; Choquet and Wollman, 2002). Similar results have also been observed in the Arabidopsis lpa1 mutant, in which the synthesis of D1 is indicated to be considerably reduced owing to an accumulation of the unassembled D1 protein in the membrane (Peng et al., 2006). Furthermore, the increased degradation of D1 protein should impact the assembly of PSII. Indeed, as shown in Figure 6D, the assembly of PSII in the hcf243 mutant is retarded significantly. Taken together, it can be speculated that the rapid degradation of the D1 protein may result from the impaired stability of the protein in the hcf243 mutant, which is further supported by HCF243 localization and its interaction with the D1 protein (Figs. 7 and 8). Our bioinformatics study of the known homologs of HCF243 in plants suggested that homologous genes were found only in angiosperms, with one or two copies existing in each species, but no such homologous genes in lower plants or prokaryotes (Supplemental Fig. S2). Our results suggest that HCF243, which arose after the origin of higher plants, may act as an important cofactor to maintain the stability of the D1 protein and to promote the subsequent assembly of the PSII complex.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana ecotype Columbia) was grown under short-day conditions (10-h-light/14-h-dark cycle) with a photon flux density of 120 μmol m−2 s−1 in soil at a constant temperature of 20°C. To ensure synchronized germination, the seeds were sown in darkness for 48 h at 4°C. The hcf243 mutant was isolated as a recessive high chlorophyll fluorescence plant in a collection of pSKI015 T-DNA-mutagenized Arabidopsis lines from the Arabidopsis Biological Resource Center (Weigel et al., 2000). Mutants exhibiting the high chlorophyll fluorescence phenotype were initially selected in the dark under strong long-wavelength UV light as described elsewhere (Miles, 1994; Meurer et al., 1996). Measurement of leaf area was performed with LI-3000 A (Li-Cor) using standard protocols.
Measurements of Chlorophyll Fluorescence
Fluorescence measurements were performed as described by Peng et al. (2006) using a portable fluorometer (PAM-2000; Walz). Before measurement, leaves were dark adapted for 30 min. The minimum fluorescence yield (F0) was measured under measuring light (650 nm) with very low intensity (0.8 μmol m−2 s−1). To estimate the maximum fluorescence yield (Fm), a saturating pulse of white light (3,000 μmol m−2 s−1 for 1 s) was applied. The maximum photochemical efficiency of PSII was determined from the ratio of variable (Fv) to maximum (Fm) fluorescence [Fv/Fm = (Fm − F0)/Fm]. All the above measurements were performed in a dark room with stable ambient conditions. For recording P700 absorbance changes, the PAM fluorometer was equipped with an ED 800 T emitter detector unit (Walz) and the measurements were performed according to Meurer et al. (1996). The relative amount of photooxidizable P700 was reflected from the absorbance changes induced by saturating far-red light.
BN-PAGE, SDS-PAGE, and Immunoblot Analysis
The thylakoid membrane was prepared in accordance with standard methods (Zhang et al., 1999). BN-PAGE was performed as described previously (Schägger et al., 1994) with some modifications (Cline and Mori, 2001; Li et al., 2003). The thylakoid membrane was solubilized with 1% (w/v) dodecyl-β-d-maltoside in 20% glycerol and 25 mm BisTris-HCl, pH 7.0, at 1.0 mg chlorophyll mL−1. After incubation at 4°C for 5 min, insoluble material was removed by centrifugation at 13,000g for 10 min. The supernatant was combined with one-tenth volume of 5% Serva blue G in 100 mm BisTris-HCl, pH 7.0, 0.5 m 6-amino-n-caproic acid, and 30% (w/v) glycerol and applied to 0.75-mm-thick 6% to 12% acrylamide gradient gels in a Hoefer Mighty Small vertical electrophoresis unit connected to a cooling circulator. For two-dimensional analysis, excised BN-PAGE lanes were soaked in SDS sample buffer with 5% β-mercaptoethanol for 30 min and layered onto 1-mm-thick 15% SDS polyacrylamide gels containing 6 m urea (Laemmli, 1970).
Thylakoid proteins were solubilized and separated by SDS-PAGE (Laemmli, 1970) on 15% (w/v) acrylamide gels with 6 m urea. After electrophoresis, the proteins were transferred to nitrocellulose membranes and probed with specific antibodies.
Nucleic Acid and Polysome Analysis
Genomic DNA was extracted from Arabidopsis leaves as described by Liu et al. (1995). Total RNA was extracted from 0.5 g of leaves of wild-type and hcf243 mutant plants using an RNA Isolation Kit (U-gene) according to the manufacturer’s protocol. After separation of 15 μg of total RNA on a formaldehyde denaturing 1.5% agarose gel, RNA was transferred onto Hybond-N+ nylon membranes (Sambrook and Russell, 2001). The membranes were probed with 32P-labeled cDNA probes specific for HCF243, PsbA, PsbB, PsbC, and PsbD. Following high-stringency hybridization and washing, all the blots were exposed to x-ray film for 1 to 3 d.
RT-PCR of HCF243 and two genes located around the insertion site was performed using the following primers: At3g15095, 5′-TTTGGTATTGTTCATGCTTCGC-3′ and 5′-AATTACCCAGGTAGAGTTCGAG-3′; At3g15090, 5′-GACTGCTTGGCGTGCTTT-3′ and 5′-CCCGGAATCTGTTTCTTCTC-3′; At3g15110, 5′-ACTCCGATAGAAGGTGGT-3′ and 5′-TGATGAGTCTCCAGGTTGT-3′. To ensure equal amounts of RNA in each sample, RT-PCR analysis of actin cDNA was carried out using the following primers: sense (5′-AACTGGGATGATATGGAGAA-3′) and antisense (5′-CCTCCAATCCAGACACTGTA-3′).
Polysomes were isolated from leaf tissues of wild-type and hcf243 mutant plants under conditions that maintain polysome integrity according to Barkan (1988). RNA was isolated, fractionated, and transferred onto nylon membranes. The filters were hybridized with the same 32P-labeled cDNA probes described above.
In Vivo Labeling of Chloroplast Proteins
In vivo protein labeling was performed essentially according to Meurer et al. (1998). Primary leaves of 12-d-old and 4-week-old mutant and wild-type seedlings were preincubated for 30 min in 50 μL of double distilled water containing 100 μg mL−1 cycloheximide and radiolabeled with 1 μCi μL−1 [35S]Met (specific activity > 1,000 Ci mmol−1; Amersham Pharmacia Biotech) in the presence of 50 μg mL−1 cycloheximide, and radioactive Met was allowed to incorporate for varying periods at 25°C in ambient light. Pulse labeling of the leaves was followed by a chase in the presence of 10 mm unlabeled Met. After labeling, the thylakoid membranes were isolated and subjected to BN-PAGE and SDS-PAGE analyses by the method described by Ma et al. (2007). For autoradiography, gels were stained, dried, and exposed to x-ray films. The relative amounts of [35S]Met in proteins were quantified by scanning the x-ray films and analyzing the acquired data using the AlphaImager 2200 system.
Antiserum Production
The nucleotide sequence encoding amino acids 221 to 408 of the HCF243 protein (nucleotide positions 661–1,224 of the HCF243 gene) was amplified by RT-PCR using the primers 5′-GGATCCAGTAATTCGTGTGGTGCG-3′ and 5′-CTCGAGTCTTCCACAACTTTGAC-3′. The resulting DNA fragment was cleaved with BamHI and XhoI and fused in frame to the N-terminal His affinity tag of pET28a vector. The construct was transformed into Escherichia coli strain BL21 and harvested after the application of 0.4 mm isopropylthio-β-d-galactoside for 4 h and resuspended in 500 mm NaCl and 20 mm NaH2PO4, pH 8.0. After incubation for 30 min at 4°C in the presence of lysozyme at a final concentration of 1 mg mL−1, the bacterial lysate was sonicated for about 30 min (10-s sonication with an interval of 10 s) at 4°C. The overexpressed proteins in inclusion bodies were centrifuged at 3,000g for 30 min, and the pellet was solubilized in 500 mm NaCl, 8 m urea, and 20 mm NaH2PO4, pH 8.0. The fusion protein was purified on a Ni-NTA resin matrix, and a polyclonal antibody was raised in rabbit with purified antigen.
Cloning of HCF243
To clone the T-DNA-tagged gene in the hcf243 mutant, T-DNA left border sequences were isolated using a thermal asymmetric interlaced-PCR strategy essentially according to Liu et al. (1995). The amplified products were cloned into a pMD18-T vector (Takara) in accordance with the manufacturer’s instructions and sequenced with universal M13 primers. Briefly, preamplification was performed using the SK1 primer together with the AP1 primer, and then the amplification products were diluted 600-fold and 5-μL aliquots were used for the second step amplifications using the SK2 primer together with the AP2 primer. The specific primers SK1 and SK2 were designed according to the T-DNA left border sequences. The two-step PCR conditions were as described by Liu et al. (1995). The PCR products were cloned into a pMD18-T vector (Takara) in accordance with the manufacturer’s instructions and propagated in the Top10 strain of E. coli. Positive clones were harvested and cultured. Plasmid DNA was extracted using the Plasmid Moni Kit (U-gene) and sequenced using universal M13 primers. The primers used were as follows: AP1 (5′-GTAATACGACTCACTATAGGGC-3′), AP2 (5′-ACTATAGGGCACGCGTGGT-3′), SK1 (5′-GACTCTAGCTAGAGTCAAGCAGATCGT-3′), and SK2 (5′-GATCGACCGGCATGCAAG-3′). The full length of this gene was deposited in GenBank under accession number HM748832.
Complementation of the hcf243 Mutant
For complementation of the hcf243 mutation, the cDNA containing the coding region was amplified by PCR with the following specific primers, which include SalI and XhoI restriction sites at their 5′ end to facilitate cloning: sense (5′-CTAGTCGACTCTCAATATCTCTCCAATGGCT-3′) and antisense (5′-GATCTCGAGTCCTTTATTCCCACTCACGTCT-3′). The PCR product was subcloned into the plant expression vector of pSN1301 under the control of the cauliflower mosaic virus 35S promoter. The construct pSN1301-HCF243 was transformed into Agrobacterium tumefaciens strain C58 and introduced into hcf243 mutant plants by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected with 0.1% hygromycin. The successful complementation was confirmed by immunoblot analysis with the specific antibody against the HCF243 protein.
BiFC Assay and Subcellular Localization of HCF243
Full-length coding sequences of HCF243 and D1 were amplified by PCR with specific primers (HCF243 sense [5′-TGGCGGAAACTGAGAGACC-3′] and HCF243 antisense [5′-CTAGAACCCTACTCCGGCG-3′]; D1 sense [5′-ATGACTGCAATTTTAGAGAGACGC-3′] and D1 antisense [5′-TTATCCATTTGTAGATGGAGCCTC-3′]) and were fused in frame with YFP at the N terminus or the N terminus of either the 5′ half (nYFP) or the 3′ half (cYFP) of the YFP open reading frame dissected between codons 154 and 155 (Hu et al., 2002). YFP-tagged HCF243, nYFP-tagged HCF243, and cYFP-tagged D1 were constructed using the Gateway recombination system. All vectors were transformed into A. tumefaciens strain GV3101. Different combinations of A. tumefaciens containing each construct were prepared and mixed to an optical density at 600 nm of 0.5:0.5. Bacterial infiltration of Nicotiana benthamiana leaf epidermal cells was performed as described by Lavy et al. (2002). YFP fluorescence and chloroplast autofluorescence were visualized with an Olympus Fluoview Fv1000 spectral confocal scanning microscope after 48 h of incubation.
Pull-Down Assays
Fragments encoding full-length HCF243 were amplified by PCR with the following primers: sense (5′-GCTAGCATGGCGGAAACTGAGAGACCCC-3′) and antisense (5′-GGATCCGAACCCTACTCCGGCGCCGCCG-3′). The PCR products were cloned into NdeI and BamHI of the pET28a vector and transformed into E. coli BL21. The expression of fusion proteins was induced with 0.4 mm isopropylthio-β-d-galactoside for 6 h. The overexpressed recombinant proteins were purified on Ni-NTA resin matrix. About 10 μg of recombinant fusion protein coupled to 100 mL of a 50% (v/v) suspension of Ni-NTA beads in equilibration buffer for 60 min at 4°C. The wild-type thylakoid membranes (100 μg of chlorophyll) solubilized with 1% (w/v) DM in 20% (w/v) glycerol, 25 mm BisTris-HCl, pH 7.0, and 1 mm phenylmethylsulfonyl fluoride for 15 min at 4°C were centrifuged at 12,000g for 10 min at 4°C, and the supernatant was incubated with His-HCF243 coupled to the Ni-NAT resin for 4 h at 4°C. After the resin was washed five times with the buffer containing 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, and 1 mm EDTA, the bound proteins were eluted with SDS-PAGE sample buffer and resolved by SDS-PAGE followed by immunoblot analysis.
Immunolocalization Studies
The subcellular localization of HCF243 was determined essentially according to Lennartz et al. (2001). The wild-type thylakoid membranes were suspended to a final concentration of 0.1 mg chlorophyll mL−1 and incubated for 30 min at 4°C in ice-cold buffer containing 10 mm HEPES-KOH, pH 8.0, 10 mm MgCl2, 330 mm sorbitol, and 1 mm phenylmethylsulfonyl fluoride supplemented with 250 mm NaCl, 200 mm Na2CO3, 1 m CaCl2, or 6 m urea, respectively. The membrane fractions without supplements were used as a control. After treatment, the membranes were centrifuged at 100,000g for 2 h at 4°C, washed twice with suspension buffer, and then used for immunoblot analysis.
Phylogenetic Analysis of HCF243
The Arabidopsis HCF243 (At3g15095) protein sequence was used as a query to search for homologs against the protein and genome databases of the National Center for Biotechnology Information using BLASTP and TBLASTN. All significant hits with an e-value less than e−5, coverage ratio greater than 35%, and match ratio greater than 35% were considered as potential homologs. To study the evolutionary history of the HCF243 genes, we also searched plant species with complete genome sequences, including Chlamydomonas reinhardtii, Selaginella moellendorffii, Physcomitrella patens, Populus trichocarpa, Cucumis sativus, Arabidopsis, Arabidopsis lyrata, Sorghum bicolor, and Oryza sativa. All homologous genes and their predicted protein sequences were used for further analyses. We reexamined the annotation of these genes based on the original predication and the available transcriptional data (but lack of EST information for this gene in a few species, e.g. A. lyrata). Multiple nucleotide and protein sequences were aligned using the ClustalX 1.81 program followed by manual adjustment. Domain structures of these potential protein sequences were analyzed by searching the Pfam (Finn et al., 2010) and SMART (Letunic et al., 2006) protein domain databases, with the E-value thresholds set to 0.1. However, we did not identify any conservative domain in these protein sequences, so phylogenetic analyses were directly conducted using full-length protein sequences. Neighbor-joining trees were constructed using MEGA 4.0 (Tamura et al., 2007; Kumar et al., 2008), and the reliability of internal branches was assessed with 1,000 bootstrap replicates.
Sequence data used for the alignment from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: NP_188128, Arabidopsis At3g15095; XP_002882914, Arabidopsis lyrata e_gw1.3.6495.1; XP_002523264, Ricinus communis 29705 m000578; XP_002316801, Populus trichocarpa2 POPTR_0011s09990.1; NP_001057589, Oryza sativa Os06t0352900-01; XP_002467967, Sorghum bicolor Sb01g037260.1; cassava19184.valid.m1, Manihot esculenta1; cassava46811.m1, Manihot esculenta2; POPTR_0001s38210.1, Populus trichocarpa1; Glyma12g35610.1, Glycine max1; Glyma13g34820.1, Glycine max2; Cucsa.032440.1, Cucumis sativus; GRMZM2G358238_P01, Zea mays1; GRMZM2G008490_P01, Zea mays2; mgf022716m, Mimulus guttatus (from http://www.phytozome.net/). The accession number of the HCF243 gene is HM748832.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of the deduced amino acid sequences of the HCF243 gene and its homolog products in P. trichocarpa, C. sativus, M. esculenta, G. max, Z. mays, R. communis, A. lyrata, S. bicolor, O. sativa, and Arabidopsis.
Supplemental Figure S2. Phylogenetic tree of HCF243 and genic structures of all homologous genes.
Acknowledgments
We are indebted to Prof. Lixin Zhang (Institute of Botany, Chinese Academy of Sciences) and Prof. Jianquan Liu (Lanzhou University) for supervising this work.
References
- Adam Z, Clarke AK. (2002) Cutting edge of chloroplast proteolysis. Trends Plant Sci 7: 451–456 [DOI] [PubMed] [Google Scholar]
- Adir N, Shochat S, Ohad I. (1990) Light-dependent D1 protein synthesis and translocation is regulated by reaction center II: reaction center II serves as an acceptor for the D1 precursor. J Biol Chem 265: 12563–12568 [PubMed] [Google Scholar]
- Armbruster U, Zühlke J, Rengstl B, Kreller R, Makarenko E, Rühle T, Schünemann D, Jahns P, Weisshaar B, Nickelsen J, et al. (2010) The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22: 3439–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson B, Aro EM. (1997) Proteolytic activities and proteases of plant chloroplasts. Physiol Plant 100: 780–793 [Google Scholar]
- Aro EM, Virgin I, Andersson B. (1993) Photoinhibition of Photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta 1143: 113–134 [DOI] [PubMed] [Google Scholar]
- Barkan A. (1988) Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J 7: 2637–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Goldschmidt-Clermont M. (2000) Participation of nuclear genes in chloroplast gene expression. Biochimie 82: 559–572 [DOI] [PubMed] [Google Scholar]
- Bricker TM, Ghanotakis DF. (1996) Introduction to oxygen evolution and the oxygen-evolving complex. Ort DR, Barber J, , Oxygenic Photosynthesis: The Light Reactions, Vol 4. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 113–136 [Google Scholar]
- Choquet Y, Vallon O. (2000) Synthesis, assembly and degradation of thylakoid membrane proteins. Biochimie 82: 615–634 [DOI] [PubMed] [Google Scholar]
- Choquet Y, Wollman FA. (2002) Translational regulations as specific traits of chloroplast gene expression. FEBS Lett 529: 39–42 [DOI] [PubMed] [Google Scholar]
- Cline K, Mori H. (2001) Thylakoid DeltapH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J Cell Biol 154: 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Vitry C, Olive J, Drapier D, Recouvreur M, Wollman FA. (1989) Posttranslational events leading to the assembly of photosystem II protein complex: a study using photosynthesis mutants from Chlamydomonas reinhardtii. J Cell Biol 109: 991–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunesekaran P, Ceric G, Forslund K, et al. (2010) Database issue. Nucleic Acids Res 38: 211–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. (1998) Coordination of nuclear and chloroplast gene expression in plant cells. Int Rev Cytol 177: 115–180 [DOI] [PubMed] [Google Scholar]
- Guo JK, Zhang ZZ, Bi YR, Yang W, Xu YN, Zhang LX. (2005) Decreased stability of photosystem I in dgd1 mutant of Arabidopsis thaliana. FEBS Lett 579: 3619–3624 [DOI] [PubMed] [Google Scholar]
- Hager M, Hermann M, Biehler K, Krieger-Liszkay A, Bock R. (2002) Lack of the small plastid-encoded PsbJ polypeptide results in a defective water-splitting apparatus of photosystem II, reduced photosystem I levels, and hypersensitivity to light. J Biol Chem 277: 14031–14039 [DOI] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Iwata S, Barber J. (2004) Structure of photosystem II and molecular architecture of the oxygen-evolving centre. Curr Opin Struct Biol 14: 447–453 [DOI] [PubMed] [Google Scholar]
- Jensen KH, Herrin DL, Plumley FG, Schmidt GW. (1986) Biogenesis of photosystem II complexes: transcriptional, translational, and posttranslational regulation. J Cell Biol 103: 1315–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren N, Ohkawa H, Welsh EA, Liberton M, Pakrasi HB. (2005) Psb29, a conserved 22-kD protein, functions in the biogenesis of photosystem II complexes in Synechocystis and Arabidopsis. Plant Cell 17: 2768–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9: 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lavy M, Bracha-Drori K, Sternberg H, Yalovsky S. (2002) A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14: 2431–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. (2003) Chloroplast research in the genomic age. Trends Genet 19: 47–56 [DOI] [PubMed] [Google Scholar]
- Lennartz K, Plücken H, Seidler A, Westhoff P, Bechtold N, Meierhoff K. (2001) HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b6f complex in Arabidopsis. Plant Cell 13: 2539–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. (2006) SMART 5: domains in the context of genomes and networks. Nucleic Acids Res 34: D257–D260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BB, Guo JK, Zhou Y, Zhang ZZ, Zhang LX. (2003) Blue native gel electrophoresis analysis of chloroplast pigment protein complexes. Prog Biochem Biophys 30: 639–643 [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Ma JF, Peng LW, Guo JK, Lu QT, Lu CM, Zhang LX. (2007) LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 19: 1980–1993 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Meurer J, Meierhoff K, Westhoff P. (1996) Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198: 385–396 [DOI] [PubMed] [Google Scholar]
- Meurer J, Plücken H, Kowallik KV, Westhoff P. (1998) A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J 17: 5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles D. (1994) The role of high chlorophyll fluorescence photosynthesis mutants in the analysis of chloroplast thylakoid membrane assembly and function. Maydica 39: 35–45 [Google Scholar]
- Müller B, Eichacker LA. (1999) Assembly of the D1 precursor in monomeric photosystem II reaction center precomplexes precedes chlorophyll a-triggered accumulation of reaction center II in barley etioplasts. Plant Cell 11: 2365–2377 [PMC free article] [PubMed] [Google Scholar]
- Nanba O, Satoh K. (1987) Isolation of a photosystem II reaction center consisting of D-1 and D-2 polypeptides and cytochrome b-559. Proc Natl Acad Sci USA 84: 109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N, Yocum CF. (2006) Structure and function of photosystems I and II. Annu Rev Plant Biol 57: 521–565 [DOI] [PubMed] [Google Scholar]
- Ossenbühl F, Inaba-Sulpice M, Meurer J, Soll J, Eichacker LA. (2006) The Synechocystis sp PCC 6803 oxa1 homolog is essential for membrane integration of reaction center precursor protein pD1. Plant Cell 18: 2236–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Khamai P, Garcia-Cerdan JG, Melis A. (2007) REP27, a tetratricopeptide repeat nuclear-encoded and chloroplast-localized protein, functions in D1/32-kD reaction center protein turnover and photosystem II repair from photodamage. Plant Physiol 143: 1547–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng LW, Ma JF, Chi W, Guo JK, Zhu SY, Lu QT, Lu CM, Zhang LX. (2006) LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plücken H, Müller B, Grohmann D, Westhoff P, Eichacker LA. (2002) The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett 532: 85–90 [DOI] [PubMed] [Google Scholar]
- Prasil O, Adir N, Ohad I. (1992) Dynamics of photosystem II: mechanisms of photoinhibition and recovery process. Barber J, , The Photosystems: Structure, Function and Molecular Biology, Vol 11. Elsevier Science Publishers, Amsterdam, pp 295–348 [Google Scholar]
- Rochaix JD. (2001) Assembly, function, and dynamics of the photosynthetic machinery in Chlamydomonas reinhardtii. Plant Physiol 127: 1394–1398 [PMC free article] [PubMed] [Google Scholar]
- Rokka A, Suorsa M, Saleem A, Battchikova N, Aro EM. (2005) Synthesis and assembly of thylakoid protein complexes: multiple assembly steps of photosystem II. Biochem J 388: 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schägger H, Cramer WA, von Jagow G. (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 217: 220–230 [DOI] [PubMed] [Google Scholar]
- Sundby C, McCaffery S, Anderson JM. (1993) Turnover of the photosystem II D1 protein in higher plants under photoinhibitory and nonphotoinhibitory irradiance. J Biol Chem 268: 25476–25482 [PubMed] [Google Scholar]
- Suorsa M, Regel RE, Paakkarinen V, Battchikova N, Herrmann RG, Aro EM. (2004) Protein assembly of photosystem II and accumulation of subcomplexes in the absence of low molecular mass subunits PsbL and PsbJ. Eur J Biochem 271: 96–107 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V. (2004) Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature 431: 87–92 [DOI] [PubMed] [Google Scholar]
- van Wijk KJ, Roobol-Boza M, Kettunen R, Andersson B, Aro EM. (1997) Synthesis and assembly of the D1 protein into photosystem II: processing of the C-terminus and identification of the initial assembly partners and complexes during photosystem II repair. Biochemistry 36: 6178–6186 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blázquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrándiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al. (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman FA, Minai L, Nechushtai R. (1999) The biogenesis and assembly of photosynthetic proteins in thylakoid membranes. Biochim Biophys Acta 1141: 21–85 [DOI] [PubMed] [Google Scholar]
- Yokthongwattana K, Melis A. (2006) Photoinhibition and recovery in oxygenic photosynthesis: mechanism of a photosystem II damage and repair cycle. Demmig-Adams B, Adams WW, III, Mattoo AK, , Photoprotection, Photoinhibition, Gene Regulation and Environment. Springer, Dordrecht, The Netherlands, pp 175–191 [Google Scholar]
- Yoshioka M, Yamamoto Y. (2011) Quality control of photosystem II: where and how does the degradation of the D1 protein by FtsH proteases start under light stress? Facts and hypotheses. J Photochem Photobiol B Biol 104: 229–235 [DOI] [PubMed] [Google Scholar]
- Yu J, Vermaas W. (1990) Transcript levels and synthesis of photosystem II components in cyanobacterial mutants with inactivated photosystem II genes. Plant Cell 2: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vermaas WF. (1993) Synthesis and turnover of photosystem II reaction center polypeptides in cyanobacterial D2 mutants. J Biol Chem 268: 7407–7413 [PubMed] [Google Scholar]
- Zhang LX, Paakkarinen V, van Wijk KJ, Aro EM. (1999) Co-translational assembly of the D1 protein into photosystem II. J Biol Chem 274: 16062–16067 [DOI] [PubMed] [Google Scholar]
- Zhang LX, Paakkarinen V, van Wijk KJ, Aro EM. (2000) Biogenesis of the chloroplast-encoded D1 protein: regulation of translation elongation, insertion, and assembly into photosystem II. Plant Cell 12: 1769–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]