Abstract
Purine salvage enzymes have been implicated, but not proven, to be involved in the interconversion of cytokinin (CK) bases, ribosides, and nucleotides. Here, we use Arabidopsis (Arabidopsis thaliana) lines silenced in adenosine kinase (ADK) expression to understand the contributions of this enzyme activity to in vivo CK metabolism. Both small interfering RNA- and artificial microRNA-mediated silencing of ADK led to impaired root growth, small, crinkled rosette leaves, and reduced apical dominance. Further examination of ADK-deficient roots and leaves revealed their irregular cell division. Root tips had uneven arrangements of root cap cells, reduced meristem sizes, and enlarged cells in the elongation zone; rosette leaves exhibited decreased cell size but increased cell abundance. Expression patterns of the cyclinB1;1::β-glucuronidase and Arabidopsis Response Regulator5::β-glucuronidase reporters in the ADK-deficient background were consistent with altered cell division and an increase in CK activity, respectively. In vivo feeding of ADK-deficient leaves with radiolabeled CK ribosides of isopentenyladenosine and zeatin showed a decreased flux into the corresponding CK nucleotides. Comprehensive high-performance liquid chromatography-tandem mass spectrometry analysis detected significantly higher levels of active CK ribosides in both sense ADK and artificial microADK. Taken together, these metabolic and phenotypic analyses of ADK-deficient lines indicate that ADK contributes to CK homeostasis in vivo.
Energy-dependent shifts in metabolism are vital to numerous cellular processes such as growth, production of storage compounds, and stress responses, making the maintenance of available ATP crucial for survival. In plants, adenylate nucleotides are either synthesized de novo or recycled from nucleosides and nucleobases via salvage pathways; the resulting nucleotide monophosphates are used in the synthesis of ATP, nucleotide cofactors, as well as cytokinins (CKs; Zrenner et al., 2006). Adenosine kinase (ADK; EC 2.7.1.20) catalyzes the phosphorylation of adenosine (Ado) to AMP and is an essential component for maintaining purine nucleotide pools in Arabidopsis (Arabidopsis thaliana; Moffatt et al., 2000; Zrenner et al., 2006).
The Arabidopsis genome encodes two ADK isoforms designated as ADK1 (At3g09820) and ADK2 (At5g03300). Silencing both ADK genes by overexpressing the ADK1 cDNA results in dwarf stature, wavy leaves, and short internodes leading to clustered siliques that exhibit impaired dehiscence (Moffatt et al., 2002). The phenotype of the ADK-deficient lines has been linked to an accumulation of Ado, causing subsequent feedback inhibition of S-adenosyl-Met-dependent methylation reactions, including those affecting pectin and DNA (Moffatt et al., 2002). However, since ADK also accepts CK ribosides as substrates (Moffatt et al., 2000), it is possible that the phenotype of ADK-deficient plants may also reflect changes in CK homeostasis.
Numerous plant processes, including cellular division (Hoth et al., 2003), meristem development (Huang et al., 2003), chloroplast biogenesis (Hoth et al., 2003), and programmed senescence (Carimi et al., 2003), are CK dependent and thus are sensitive to changes in endogenous CK profiles. In addition to biosynthesis, degradation, and conjugation, interconversion is also a means by which plants can regulate CK activity. Given that free bases and ribosides are believed to be the biologically active forms of adenine (Ade)-derived CKs (Astot et al., 2000; Romanov et al., 2006; Kurakawa et al., 2007), the conversion of CK ribosides to nucleotides would lead to their inactivation (Kwade et al., 2005). Thus, the phosphorylation of CK ribosides via ADK could be one way by which plants modulate intracellular CKs.
A role for ADK in CK metabolism was first proposed on the basis of in vitro assays. The first of such assays found isopentenyladenosine (iPR) to be a substrate for recombinant ADK isolated from mouse Sarcoma 180 cells (Divekar and Hakala, 1971). A comparison of its apparent Km values suggests that ADK has a preference for Ado (11.4 μm iPR versus 0.5 μm Ado). Subsequently, other in vitro assays using plant extracts from wheat germ (Triticum aestivum; Chen and Eckert, 1977), Physcomitrella patens (von Schwartzenberg et al., 1998), Arabidopsis (Moffatt et al., 2000), and tobacco (Nicotiana tabacum; Kwade et al., 2005) have also identified CK ribosides as substrates of ADK. In addition to the in vitro experiments, in vivo assays have also investigated this issue. Feeding studies using chloronemal tissues of Physcomitrella revealed that exogenous iPR is converted into iPR monophosphate (iPRMP) via an ADK-dependent pathway (von Schwartzenberg et al., 1998, 2003). Despite these studies, however, it is unclear how essential ADK is to CK regulation in vivo. Based on kinetic analyses of recombinant Arabidopsis ADKs, they have a 10-fold higher affinity for Ado than for iPR (Moffatt et al., 2000), suggesting a secondary or minor role for ADK in CK interconversion. However, the ADK isoforms of tobacco BY-2 cells actually have a greater affinity for iPR and zeatin riboside (ZR) than Ado (Kwade et al., 2005). In fact, iPR-induced apoptosis of BY-2 cells is dependent on the intracellular phosphorylation of iPR to iPMP by ADK (Mlejnek and Procházka, 2002).
Here, we investigate the functional significance of ADK activity in Arabidopsis development and CK metabolism using ADK-deficient lines created by transgene silencing (artificial microADK [amiADK] and sense ADK [sADK]). Based on phenotypic analyses as well as CK in vivo labeling and profiling experiments, we show that ADK deficiency results in elevated CK riboside levels. Thus, despite its high Km for CK ribosides, ADK plays a significant role in CK metabolism in vivo in Arabidopsis.
RESULTS
In an effort to disrupt ADK expression in Arabidopsis, homozygous T-DNA insertion lines adk1-1 (SAIL_597_D09; insertion in the fourth exon) and adk2-1 (SALK_000565; insertion in the 10th exon) were examined. Eliminating either ADK1 or ADK2 expression caused no discernible phenotype despite substantially reducing ADK activity (Supplemental Table S1). To create a complete knockout of ADK, these two lines were crossed; however, recovery of the double mutant was unsuccessful. Closer analysis of the F2 population showed that 5.5% (33 out of 600) of embryos failed to fully develop into seeds. Given that a homozygous T-DNA insertion in both ADK genes will occur in 6.25% of all F2 seeds and that no double mutants were identified by PCR analysis of F2 individuals, we reasoned that the aborted seeds were the double mutants (χ2 test; P < 0.05, χ2 = 3.69). Based on these data, we concluded that complete removal of ADK activity in Arabidopsis caused embryo lethality. As a result, gene silencing was used to generate lines with a partial reduction in ADK activity.
ADK-deficient lines created by overexpression of the ADK1 cDNA in the sense orientation (sADK) were previously reported; the construct silenced the expression of both ADK genes (Moffatt et al., 2002). However, due to the possibility of nontarget silencing, alternate ADK-deficient lines were pursued. Artificial microRNAs (amiRNAs) targeted to reduce ADK transcript accumulation were selected using the online tool at Weigelworld (wmd2.weigelworld.org). To design specific 21-mer amiRNA sequences targeting ADK1 and ADK2 transcripts, both genes were submitted to the MicroRNA Designer; two different amiRNA sequences directed against different conserved regions of ADK were selected. Transgenic plants expressing the amiRNAs from the 35S promoter were generated and analyzed for ADK expression, with a minimum of 15 T1 individuals being studied for each line. Plants expressing amiRNA2 displayed no distinct phenotype, whereas those expressing amiRNA1 displayed similar patterns of abnormal morphology to that of sADK lines, including small, wrinkled leaves and decreased apical dominance. Of the 15 amiRNA1 T1 lines recovered, several were advanced to be homozygous and one representative line (amiRNA 7-7) was selected for further study; this is referred to as amiADK hereafter. sADK 4-2 was used as the representative sADK line due it having the most affected phenotype and lowest levels of ADK.
Homozygous ADK-Silencing Lines Exhibit Generational Phenotypic Variation
Despite being homozygous for their respective constructs, both sADK and amiADK populations segregated with respect to the severity of their phenotypes. We have called this “generational phenotypic variation.” Among the progeny of each generation, a portion of the plants exhibited wild-type-like morphology, while others had an intermediate or severely affected phenotype. Mature 5-week-old sADK individuals exhibiting crinkled leaves, a primary shoot less than 10 cm, and clustered inflorescences were classified as having a severe phenotype. Plants displaying a primary shoot between 10 and 20 cm in length and a clustered inflorescence were classified as intermediate. In a representative population (n = 50) of sADK (Table I), 29.8% had a severe phenotype, 40.4% were intermediate, and 29.8% were wild type like in appearance. With the exception of clustered inflorescences, the above-mentioned criteria were also used when analyzing amiADK populations (Table I): 38% were severe, 32% were intermediate, and 30% were wild type like. The proportions of phenotypes varied slightly in different generations, although phenotypic variation was always observed. Interestingly, in both sADK and amiADK, individuals exhibiting a normal phenotype still gave rise to plants with a severe phenotype in the next generation. As both modes of silencing resulted in generational phenotypic variation, this event appears to be associated with ADK activity. As documented previously for sADK lines (Moffatt et al., 2002), analysis of ADK activity in 4-week-old plants revealed that those with the most severe phenotypes had the lowest residual activity (8.7% sADK and 6.3% amiADK; Table I). Furthermore, retained ADK activity in intermediate phenotypes ranged from 10% to 40%, and wild-type-like plants exhibited activity closer to that of the wild type (Supplemental Table S1).
Table I. ADK activity correlated to phenotype in ADK-silencing lines.
Not all of the homozygous sADK or amiADK exhibited a phenotype. In a representative population, 29.8% of sADK and 38% of amiADK had severe phenotypes consisting of small, crinkled leaves and decreased apical dominance. Further examination of ADK activity demonstrated that the individuals exhibiting a severe ADK-deficient phenotype had the lowest levels of retained activity.
| Parameter | Wild Type | sADK | amiADK |
| Proportion of severe phenotypes in a segregating homozygous population | – | 29.8% | 38% |
| ADK activity (nmol AMP mg−1 protein min−1) | 4.6 ± 0.34 | 0.4 ± 0.08 | 0.29 ± 0.06 |
| Retained ADK activity | 100% | 8.7% | 6.3% |
Discernible morphological phenotypes only become apparent in either the sADK or amiADK lines around 17 d after germination (DAG). Closer examination of protein activity and expression showed that silencing was not detectable in the leaves of 2-week-old plants, suggesting that reduction of ADK activity only began after this point (data not shown). Thus, studies were conducted on plants older than 2.5 weeks to ensure that only plants effectively silencing ADK were used. The following experiments were performed with ADK-deficient plants exhibiting severe phenotypes.
Abnormal Cellular Division Is Evident in ADK-Deficient Tissues
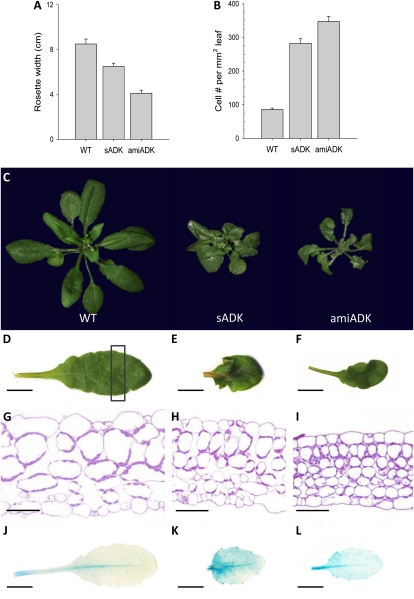
At 2.5 weeks, leaf size was noticeably reduced in both sADK and amiADK plants. As the leaves developed, crinkling began to occur in the leaf margins, and by 24 DAG, the differences between wild-type and ADK-deficient plants was readily apparent (Fig. 1, C–F). At maturity, the average sADK rosette width was 6.5 ± 0.29 cm and amiADK rosettes were 4.11 ± 0.27 cm, compared with wild-type rosettes of 8.5 ± 0.44 cm (Fig. 1A). To determine the cause of this decreased leaf size, the seventh rosette leaves of 3.5-week-old plants were cleared and mesophyll cells were counted (Fig. 1B). In comparison with the 85.3 cells mm−2 present in the wild type, sADK had over a 3-fold increase (282.3 cells mm−2) and amiADK had four times the cell number (347 cells mm−2). This trend was also observed in the epidermis; however, the difference was less pronounced: both ADK-deficient lines had double the epidermal cells of the wild type.
Figure 1.
Rosette width decreased (A), while cell number increased (B), in ADK-deficient rosette leaves. Phenotypic analysis showed that ADK-deficient plants have smaller, curled leaves (C). Upon sectioning and staining ADK-deficient rosette leaves with toluidine blue O and observing smaller and more abundant developing mesophyll cells (sADK [H] and amiADK [I]), cycB1;1::GUS expression was studied in leaves of 3.5-week-old sADK (K) and amiADK (L). Bars = 1 cm (D–F and J–L) and 60 μm (G–I). WT, Wild type.
For a more detailed analysis of cell morphology, the distal portions of leaves from 3.5-week-old wild-type and ADK-deficient plants were fixed and sectioned. As was observed in the cleared leaves, sADK (Fig. 1H) and amiADK (Fig. 1I) cells were noticeably smaller and with greatly reduced air space between them. Of particular interest in the leaf sections was the higher occurrence of undifferentiated mesophyll cells in ADK-deficient tissue. This increase in small, undifferentiated cells suggested a change in cell division rather than impaired cell expansion.
To monitor cell division in an ADK-deficient background, both the sADK and amiADK lines were crossed to the cell division marker line cyclinB1;1 (cycB1;1)::GUS (Colón-Carmona et al., 1999). Histochemical detection of GUS activity in the seventh rosette leaf of 3.5-week-old plants showed stronger cycB1;1 expression in sADK and amiADK (Fig. 1, J–L). Further comparison revealed cycB1;1::GUS to be distributed throughout the entire leaf and not just limited to the vasculature in ADK-deficient backgrounds. Thus, in accordance with the increased number of cells, higher cycB1;1::GUS expression in rosette leaves supported increased cell division in ADK-deficient leaves.
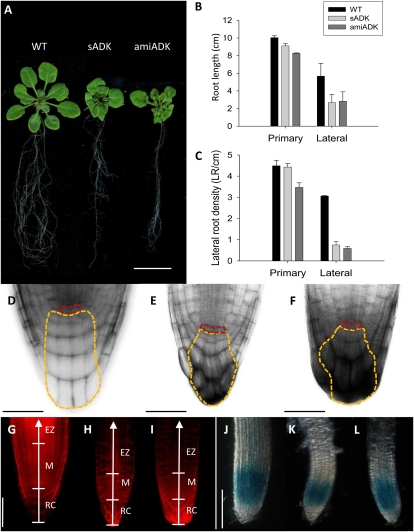
Irregular cell division was also observed in roots of 3-week-old sADK and amiADK plants. Abnormal root development was reported previously for sADK lines, with mature plants exhibiting decreased root length and an increased number of lateral roots of the same length (Moffatt et al., 2002). Preliminary analysis of amiADK roots also indicated impaired root development (Fig. 2A). Both sADK and amiADK had shorter primary roots, but the more significant differences were in the lateral roots, which were half the length of wild-type roots (Fig. 2B). The difference between lateral and primary root length in ADK-deficient seedlings may be due to the fact that much of the primary root being measured was established before the onset of silencing: mature plants of sADK have noticeably shorter primary roots (Moffatt et al., 2002). In addition to root length, the lateral root density (the number of lateral roots per centimeter) on secondary roots was 3-fold lower in sADK and amiADK (Fig. 2C).
Figure 2.
At 3 weeks, ADK-deficient roots (A) exhibited decreased secondary growth in both length (B) and lateral root formation (C). n = 4. Further propidium staining revealed uneven cell arrangement in both the sADK (E) and amiADK (F) root cap compared with the wild type (WT; D; yellow outlines, columella root cap; red outlines, quiescent center). Propidium iodide-stained roots also showed that the size of the root cap (RC) and meristem (M) was reduced in sADK (H) and amiADK (I) in comparison with the wild type (G); additionally, cells in the elongation zone (EZ) of ADK-deficient roots were noticeably larger than the wild type. To compare cellular division, cycB1;1::GUS expression was also observed in sADK (K) and amiADK (L); both ADK-deficient lines exhibited smaller areas of GUS activity in comparison with cycB1;1::GUS alone (G). Bars = 2 cm (A), 50 μm (D–F), and 100 μm (G–L).
To ascertain why root development was impaired in ADK-deficient backgrounds, cellular organization was visualized with propidium iodide. When compared with the uniform cellular arrangement of the wild-type columella root cap (Fig. 2D), both sADK (Fig. 2E) and amiADK (Fig. 2F) root tips were abnormally organized; additionally, root caps were much smaller when measured relative to the location of the quiescent center. Further examination of propidium iodide-stained roots (Fig. 2, H–J) revealed that ADK-deficient roots had a smaller meristem and root cap, with enlarged cells in the elongation zone (layers including epidermis and endodermis). As suggested by the decreased size of the root meristem, the localization of cycB1;1::GUS expression was changed, with GUS detected in a smaller area of the sADK and amiADK roots (Fig. 2, L and M). Therefore, based on the anatomical structure and cycB1;1::GUS expression, cellular division appeared to be lowered in ADK-deficient roots; contrary to what was observed in rosette leaves.
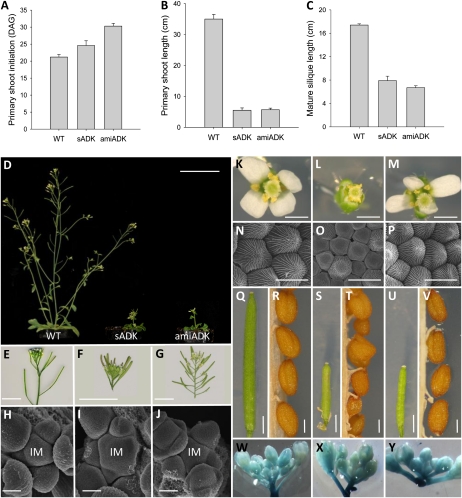
Aerial organ development was also affected in sADK and amiADK. In both ADK-silencing lines, shoot initiation (DAG to achieve greater than 1 cm) was noticeably delayed, with amiADK being the most adversely affected: sADK was 25 ± 1.4 DAG and amiADK was 30 ± 0.9 DAG versus 21 ± 0.8 DAG for the wild type (Fig. 3A). Primary shoot length 2 weeks after bolting was also greatly reduced, with the wild type being seven times the height of sADK and amiADK (Fig. 3B). Upon reaching maturity, reduced apical dominance (Fig. 3D) was consistently observed in the ADK-silencing lines. Closer examination of aerial organs revealed differences between sADK and amiADK lines. Although amiADK plants had shortened internodes (Fig. 3G), they were not clustered to the same extent as sADK (Fig. 3F). Scanning electron microscopy (SEM) showed that the inflorescence meristem of sADK (Fig. 3I) was enlarged and irregularly shaped compared with the wild type (Fig. 3H) and amiADK (Fig. 3J). Abnormal development was also observed in later bud inflorescence stages, with sADK flowers exhibiting no visible petals (Fig. 3L) and amiADK flowers having an irregular petal arrangement (Fig. 3M); further dissection of sADK flowers revealed small, underdeveloped petals that did not extend past the sepals (Supplemental Fig. S1). SEM analysis of the adaxial portion of the sADK petal (Fig. 3O) was consistent with the impaired cell division observed in leaf tissue, with the majority of cells being smaller and lacking the distinctive conical shape of the wild type (Fig. 3N). Adaxial petals of amiADK (Fig. 3P) also contained smaller, irregularly shaped cells, but not as severe as sADK.
Figure 3.
Primary shoot initiation (A), primary shoot length 2 weeks after shoot initiation (B), and mature stage 17b silique length (C) were measured. An overview of 4.5-week-old plants (D) showed that ADK-deficient lines have a dwarf phenotype. Wild-type (WT; E, H, K, N, Q, R, and W), sADK (F, I, L, O, S, T, and X), and amiADK (G, J, M, P, U, V, and Y) inflorescences (E–G), SEM inflorescence meristems (IM; H–J), stage 14 flowers (K–M), adaxial surface of petals by SEM (N–P), mature stage 17b siliques (Q, S, and U), seeds (R, T, and V), and cycB1;1::GUS expression (W–Y) were also compared. Bars = 5 cm (D), 1 cm (E–G), 30 μm (H–J), 0.4 cm (K–M), 10 μm (N–P), 1.5 cm (Q, S, and U), and 0.5 mm (R, T, and V).
Other aerial organs also displayed abnormal development, including mature stage 17b siliques that were half the length of the wild type in sADK and amiADK plants (Fig. 3, C, Q, S, and U). In addition to silique length, mature siliques of sADK did not exhibit silique shattering: even after 2 weeks of dehydration, sADK siliques showed no evidence of opening. Impaired silique shattering was unique to sADK and not observed in amiADK. Further variation between sADK and amiADK was noted in their seeds: sADK (Fig. 3T) seeds were smaller and wrinkled in appearance compared with both wild-type (Fig. 3S) and amiADK (Fig. 3U) seeds. Overall, sADK seeds were on average 0.55 ± 0.089 mm in length, with 35.6% exhibiting a wrinkled phenotype, versus the 0.67 ± 0.057 mm observed in the wild type. While some sADK siliques had an increased number of seeds relative to their size, on average there was no significant difference from the wild type: sADK exhibited 3.6 ± 0.5 seeds per mm of silique, and the wild type had 3.2 ± 0.2 (n = 20). As with the rosette leaves and roots, cycB1;1::GUS activity was detected in the ADK-deficient inflorescence. Unlike the other organs tested, however, cycB1;1::GUS expression remained unchanged in both ADK-deficient inflorescences (Fig. 3, W–Y) and mature siliques (Supplemental Fig. S1).
ADK-Deficient Tissues Exhibit Altered ARR5 and ARR7 Expression
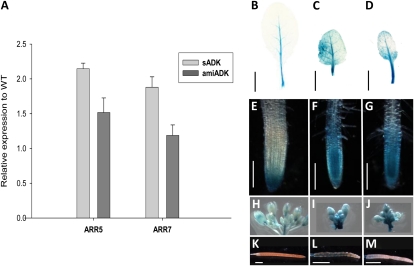
Altered cell division, as implied by cycB1;1::GUS expression in leaves and roots, suggested that ADK deficiency may be contributing to a CK imbalance. To provide a quantitative assessment of the CK response pathway, transcript levels of Arabidopsis Response Regulator5 (ARR5) and ARR7 (another A-type CK regulator) were measured in young rosette leaves by quantitative PCR (Fig. 4A). sADK leaves displayed a 2-fold increase in ARR5 and ARR7 transcript levels, while amiADK showed less change (ARR7 increased by 1.2-fold and ARR5 by 1.5-fold).
Figure 4.
ARR5 and ARR7 expression was tested in young leaves using reverse transcription-PCR (A). WT, Wild type. GUS staining was performed on ARR5::GUS (B, E, and H), ARR5::GUS in the sADK background (C, F, and I), and ARR5::GUS in the amiADK background (D, G, and J) in seventh developed rosette leaves (B–D), root tips (E–G), inflorescence buds (H–J), and stage 17b siliques (K–M). Bars = 1 cm (B–D), 100 μm (E–G), and 1.5 cm (K–M).
Further examination of the ARR5 response in ADK-deficient tissue was performed by introducing the ARR5::GUS transgene into both sADK and amiADK backgrounds. As was observed for cycB1;1::GUS, the pattern and intensity of ARR5::GUS expression was altered in the leaves (Fig. 4, B–D) and roots (Fig. 4, E–G). Rather than being limited to the vasculature, GUS staining of the seventh rosette leaf exhibited patchy GUS staining throughout the tissue in sADK and amiADK. ARR5::GUS expression was also considerably stronger in ADK-deficient roots (Fig. 4, F and G); histochemical staining was not limited to the root cap, as was observed in wild-type plants (Fig. 4E), but in the meristems and elongation zone as well. Unlike cycB1;1::GUS in ADK-deficient lines, ARR5::GUS expression was elevated in sADK (Fig. 4I) and amiADK (Fig. 4J) inflorescences and mature siliques (Fig. 4, L and M). ARR5::GUS staining in sADK was particularly strong, with GUS detected in both the replum and carpel (Fig. 4L) compared with the pedicel in amiADK (Fig. 4M) and undetectable in the wild-type background (Fig. 4H). Taken together, these data suggest that sADK and amiADK plants have elevated CK levels in numerous organs.
In Vivo Labeling with Radiolabeled CK Ribosides
To test whether ADK affects CK metabolism under in vivo conditions, we carried out labeling experiments with detached leaves of sADK plants using tritiated iPR and ZR. After labeling over a period of 4 h, CKs were extracted and separated by HPLC; the effluent was analyzed for radiolabeled metabolites using online liquid scintillation counting. The relative proportion of radiolabeled degradation products (metabolites without the N6 side chain) was higher when labeling with [3H]iPR compared with [3H]ZR. However, for each substrate, a comparable proportion of degradation product was measured in sADK and wild-type leaves; this would suggest that CK oxidase/dehydrogenase-mediated inactivation was not significantly affected by ADK silencing (Fig. 5).
Figure 5.
In vivo labeling with CK ribosides [3H]ZR (A) and [3H]iPR (B). Detached leaves of 4-week-old plants were incubated at a riboside concentration of 60 nm for 4 h. Extractable radiolabeled compounds were separated by reverse-phase HPLC coupled to liquid scintillation counting. ZX1-P, Unknown [3H]ZR-derived compound, presumably phosphate; ZX2-(R), unknown [3H]ZR-derived compound, presumably nucleoside; WT, wild type.
After a 4-h incubation with [3H]ZR, it was evident that the substrate had been metabolized to the expected major compounds, [3H]ZRMP and [3H]Z, in addition to the labeled degradation products. Unknown radiolabeled metabolites were also obtained, one of which we called ZX1-P, as treatment with alkaline phosphatase converted it to another unknown compound, ZX1-(R).
The relative amount of ZR-bound radioactivity was found to be only 4% in the wild type but 16% in sADK, indicating a slower metabolism of [3H]ZR. Additionally, the labeled ZR nucleotide ZRMP was reduced in the sADK leaves: 17% versus 27% in the wild type. Both the reduced metabolism of the substrate [3H]ZR and the lowered formation of the nucleotide showed that ADK phosphorylates CK ribosides in vivo (Fig. 5A).
An unknown compound, ZX2-(R), was present in higher abundance in sADK compared with the wild type (4.6% compared to 2.7%). The proportion of the unknown phosphorylated compound ZX1-(P) was reduced in the sADK leaves, which led us to the conclusion that ZX1-(P) is a nucleotide and ZX2-(R) is a riboside closely related to ZRMP and ZR, respectively (Fig. 5A). A similar reduction of nucleotide formation was found when [3H]iPR was used as the radiolabeled substrate, although a lower overall number of labeled compounds was detectable with this substrate.
In experiments using radiolabeled [3H]iPR, sADK leaves once again had a higher proportion of [3H]iPR, indicating a slower metabolism of this substrate compared with the wild type (Fig. 5B). In this representative experiment, 5.4% of the radioactivity was present in the iPR fraction of sADK leaves versus 2.6% in the wild type. Overall, sADK had a decreased amount of [3H]iPR phosphorylated to [3H]iPRMP when compared with the wild type: 17% versus 30% (Fig. 5B). The reduced recovery of radiolabeled nucleotides ZRMP and iPRMP in sADK indicated that CK ribosides are indeed substrates for ADK in planta and that ADK has an impact in CK riboside phosphorylation.
CK Profiles Are Altered in the ADK-Deficient Background
To assess purine and CK profiles in an ADK-deficient background, rosette leaves of 4.5-week-old plants were evaluated using HPLC-tandem mass spectrometry (MS/MS) analysis. As expected, reduced ADK activity resulted in elevated Ado levels (Fig. 6A), with a 3-fold increase in sADK and a 7-fold increase in amiADK. Unexpectedly, there was no significant reduction in the product of ADK activity, AMP, nor was there a significant change in Ade abundance (Fig. 6A).
Figure 6.
Contents of adenylates and CK compounds as determined by HPLC-MS/MS in leaves of 4-week-old plants. se was used to indicate variation; n = 4. Values with no or the same letters were not significantly different (Fisher’s lsd; P < 0.05). Nucleotides shown in D are composed of monophosphates, diphosphates, and triphosphates. FW, Fresh weight; WT, wild type.
As in the in vivo labeling experiments, CK riboside levels were most affected in the ADK-deficient leaf samples. Free CK bases were minimally altered in the ADK-deficient leaves, with only cis-zeatin (cZ) showing a modest but statistically supported increase in sADK (Fig. 6B); there were no significant changes in Z or iP levels for either ADK-deficient line. All of the CK ribosides analyzed (iPR, ZR, cZR, and dihydrozeatin riboside [DZR]) showed significant accumulation in both sADK and amiADK plants (Fig. 5C). The largest increase was observed in ZR levels, with a greater than 50-fold increase in sADK leaves and a 23-fold increase in amiADK. Consistent with ARR5 transcript levels, CK base and riboside levels were higher in the sADK background compared with amiADK.
No significant change was observed in CK nucleotides (Fig. 5D), but multiple O-glucoside forms of ribosylated zeatin, ZROG, cZROG, and DZROG (Fig. 5E), were greatly increased in both sADK and amiADK. The ring glucoside CKs, trans-zeatin 9-glucoside and iP7G, were higher only in sADK lines.
DISCUSSION
Constitutive overexpression of the ADK1 cDNA in the sense orientation (sADK) or amiRNA transcripts targeted to both ADK genes (amiADK) substantially reduced ADK activity (6%–8% of the wild type). In both types of ADK-deficient lines, decreased ADK activity was first detected 2.5 weeks after germination, limiting the stages of development that could be analyzed. This observed delay in silencing is not uncommon in transgene silencing. Previous reports have documented that the onset and severity of small interfering RNA (siRNA) silencing is influenced by intrinsic factors such as transgene dosage and insert loci as well as environmental influences (Vaucheret et al., 2001). Furthermore, plants homozygous for a silencing transgene, as in the case of sADK and amiADK lines, do not necessarily silence upon germination. For example, in Arabidopsis, homozygous Rps10-silencing lines exhibit silencing at 3 weeks (Majewski et al., 2009), while in the homozygous tobacco silencing line gn1, GN1 mRNA levels do not decrease until 6 to 7 weeks after germination (Niebel et al., 1995).
While in the majority of plants, ADK was silenced at 2.5 weeks, approximately 30% of homozygous sADK and amiADK displayed wild-type-like morphology and ADK activity, in a phenomenon we termed generational phenotypic variation. It has been previously reported that posttranscriptional gene-silencing lines can undergo spontaneous and developmentally regulated reactivation in a process known as resetting (Meins and Kunz, 1994; Kunz et al., 1996; Kanazawa et al., 2007; Majewski et al., 2009). In homozygous Rps10-silencing lines, it was found that 17% of the plants were able to switch off silencing in the late vegetative growth stage (Majewski et al., 2009). In petunia (Petunia hybrida), transgenic white-flowering plants gave rise to branches with wild-type purple pigmentation as a result of epigenetic modification (Kanazawa et al., 2007). In both cases, resetting occurred in newly formed tissues of plants that had already successfully induced silencing. This is in contrast with our observations of the homozygous ADK-deficient lines, in which a portion of progeny appear to never experience silencing. Moreover, in the studies performed by Majewski et al. (2009) and Kanazawa et al. (2007), progeny of reverted transformants do not display posttranscriptional gene silencing, but in ADK-deficient lines, wild-type-appearing plants give rise to progeny with ADK silencing. The molecular basis for the ADK generational phenotypic variation remains unclear, but several explanations are possible. Perhaps the most likely is that RNA silencing itself relies on ADK or CK activity. For example, it has been previously reported that ADK is necessary for the RNA silencing of viral infection in tobacco (Wang et al., 2005). It is also possible that this variation is due to epigenetic silencing triggered to maintain viability, since complete loss of ADK activity is lethal. Additionally, this phenomenon may be an example of allele-specific DNA restoration (Lolle et al., 2005). Given the substantial metabolic stress associated with low ADK activity, any one of the above-mentioned mechanisms could be involved; however, none of these scenarios provide insight as to why gene silencing “returns” in subsequent generations.
For those sADK and amiADK that exhibited severe silencing, common traits were observed: impaired secondary root growth, irregular cell size and arrangement in root tips and leaves, crinkled leaves, reduced apical dominance with decreased internode length, and shortened siliques. Some of the phenotypes, particularly those affecting the inflorescence organs, were specific to sADK. For example, the amiADK line did not have enlarged inflorescence meristems, impaired petal development and dehiscence, delayed silique shattering, or small, wrinkled seeds. Since amiRNA-induced silencing is believed to be more specific than siRNA silencing (Schwab et al., 2006), it is possible that the floral phenotype observed in sADK is the result of off-target silencing transcripts unrelated to ADK. Alternatively, siRNAs are more effective than amiRNAs in silencing ADK in the floral organs, thereby leading to a more severe phenotype.
Closer study of the small, crinkled leaves observed in both sADK and amiADK lines revealed their mesophyll and epidermal cells to be greatly reduced in size. Mesophyll cell counts showed increased cell density when compared with the wild type (3-fold more mesophyll cells in sADK and 4-fold more in amiADK). As a result of these increased cell numbers, intercellular spaces were greatly reduced, to the point that callus-like patches of cells were occasionally observed within the leaves. The basis for this increased cell density was evaluated by examining the expression of the cell division reporter cycB1;1::GUS. In both sADK and amiADK, cycB1;1::GUS expression was elevated and dispersed throughout a larger area of the leaf in comparison with the wild-type background. Irregular cell division was also implicated in ADK-deficient roots, but in this case, decreased division appeared to be occurring: root cap and meristem had fewer cells, while the elongation zone contained enlarged cells. cycB1;1::GUS staining of the ADK-deficient roots occurred in a smaller, more concentrated area, consistent with their smaller meristems and zone of cell division. No changes were observed for cycB1;1::GUS expression in the floral organs.
Since increased CK activity is known to enhance cell division and impair cellular expansion in leaves (Werner et al., 2003) and inhibit root development by decreasing the size of the root meristem (Dello Ioio et al., 2007), markers for CK expression were examined. A-type Arabidopsis response regulator genes (e.g. ARR5 and ARR7) are highly induced in the presence of active CKs and are negative regulators of CK activity (D’Agostino et al., 2000). The bases Z, benzyladenine, iP, and kinetin in addition to the riboside iPR have all been shown to equally induce ARR5::GUS transcription (Romanov et al., 2002). Conversely, structurally related compounds, namely Ade and Ado, do not induce ARR5::GUS expression (Romanov et al., 2002). ADK-deficient leaves, roots, inflorescences, and mature siliques all showed an increase in ARR5::GUS expression. Interestingly, mature sADK siliques had considerably higher ARR5::GUS activity than either amiADK or the wild type. This pattern of ARR5::GUS expression suggested that the unique floral phenotype of sADK (enlarged meristem, undeveloped petals, and failed silique shattering) results from elevated CK levels and not off-target silencing. Whether the differences in ARR5::GUS activity between sADK and amiADK is due to the efficiency of ADK silencing in these organs will be the focus of future study. When measured quantitatively, ARR5 and ARR7 transcript levels of sADK and amiADK were only modestly higher than wild-type levels: between 1.2- and 2-fold increase. In comparison, previous quantitative PCR analysis of single ADK1 and ADK2 knockouts with no discernible phenotype showed a 1.5-fold increase in ARR5 and ARR16 transcript levels (Baliji et al., 2010). Thus, only a small reduction in ADK activity results in the down-regulation of CKs, but further lowering of ADK did not correlate to substantially increased A-type ARR signaling. This observation may suggest that modest decreases of ADK activity that disturb CK homeostasis can be compensated by down-regulation of CK signaling, but more substantial reduction of ADK leads to additional deactivation of CKs (by O-glucosylation and potentially by CKX). Nevertheless, increased ARR5 and ARR7 transcript abundance and altered ARR5::GUS expression suggest elevated CK levels in sADK and amiADK lines.
The phenotypes of sADK and amiADK are consistent with those reported for other CK mutants. Overexpression of an aldose-like enzyme (ALL) believed to be involved in Arabidopsis CK biosynthesis (all-1D) results in elevated CK signaling (increased ARR4 and ARR5 expression), dwarfism, reduced apical dominance, and dark green, rolled leaves (Jung et al., 2005). Conversely, reduced CK levels in Arabidopsis transgenic lines has been linked to decreased cellular division and impaired expansion throughout the plant (Werner et al., 2003; Argyros et al., 2008; Heyl et al., 2008). Overexpression of CK oxidase (AtCKX) in Arabidopsis increases CK degradation (55%–70% of CK activity retained), causing CK bases, ribosides, and glucosides showing significant decreases. As a result of their reduced CK activity, AtCKX-overexpressing plants have smaller leaf primordia and apical meristems as well as an increased root system (Werner et al., 2003). Insensitivity to active CKs arising from loss of the B-type Arabidopsis response regulator 1 via gene silencing (35S::ARR1-SDRX; Heyl et al., 2008), triple knockout of ARR1, ARR10, and ARR12 (arr1-3 arr10-5 arr12-1; Argyros et al., 2008), and double knockout of sensor His kinases AHK2 and AHK3 (ahk2, ahk3; Riefler et al., 2006) are correlated with smaller rosette leaves (decreased cell division), increased root mass (with the exception of 35S::ARR1-SDRX), and enlarged seeds in Arabidopsis. Thus, phenotypic analysis in both sADK and amiADK lines were consistent with increased CK activity.
However, as well as contributing to AMP synthesis, ADK activity is crucial in maintaining plant transmethylation reactions that rely on S-adenosyl-Met as the methyl donor (Moffatt et al., 2000). In the absence of ADK, methyltransferases would be subject to product inhibition by S-adenosyl-l-homo-Cys. Accordingly, sADK plants have decreased methylation in the seed mucilage (Moffatt et al., 2002) and lower pectin methylation in the leaf cell wall (Pereira et al., 2006). As transmethylation has far-reaching effects, there is little reason to doubt that other processes are not also affected in ADK-deficient plants. The distinction between methylation and CK is also further blurred by the fact that CKs can be used as signals for transmethylation. Previous research has shown that feeding CKs to sahh1-1 Arabidopsis mutants, which exhibit reduced DNA methylation, increases the global DNA methylation status (Li et al., 2008). This can be investigated by examining the effect of introducing mutations in the CK receptors to the ADK-deficient background (Riefler et al., 2006). This should serve to make the plant CK insensitive and perhaps reveal which characteristics of sADK are the direct result of elevated CK activity.
Nonetheless, in vivo labeling experiments showed that ADK is in fact phosphorylating CK ribosides. Providing [3H]ZR or [3H]iPR to detached sADK leaves led to a reduction in nucleotide formation, relative to their metabolism, in wild-type leaves. The advantage of feeding radiolabeled ribosides in low concentrations over a short period of time was seen in the fact that biosynthesis could be ruled out as a major source of labeled CK nucleotides. To directly examine steady-state CK profiles resulting from all relevant CK metabolic and biosynthetic processes, LC-MS/MS analysis was performed. Both ADK-silenced lines accumulated significantly more Ado compared with the wild type, confirming that the loss of ADK is detrimental to nucleoside recycling. However, AMP levels were not significantly affected in either sADK or amiADK, suggesting that ADK-deficient plants can still derive AMP from other routes. One possible source of AMP is the degradation of excess CK ribosides that were detected in ADK-deficient leaves. Statistically significant increases in the ribosides ZR and iPR were measured in both ADK-silencing lines, with sADK being the most affected: ZR accumulated more than 50-fold in sADK and 23-fold in amiADK. This result was in full agreement with the [3H]ZR- and [3H]iPR-feeding experiments (Fig. 4), where the same substrates also accumulated in the sADK leaves due to their reduced phosphorylation.
Furthermore, an increase in the cZ base was detected in sADK, but no significant changes in any CK base were documented for amiADK. Thus, we concluded that the phenotypes associated with ADK deficiency were the result of increased CK riboside levels, not CK bases. While bases are considered the most active CKs in plants (Kurakawa et al., 2007), ribosides are also thought to contribute CK activity (Romanov et al., 2006); therefore, the very large increases in ZR alone could explain much of the phenotypic alteration seen in the ADK-silenced lines.
Attempts by the ADK-deficient plants to maintain normal CK homeostasis were evidenced by increased levels of inactive CKs. While no significant change was observed in CK nucleotides, ADK-deficient plants appeared to be shuttling excess active CKs to glucosides. Specifically, increases in ZR and cZR forms were echoed in the corresponding side chain Glc conjugates ZROG, cZROG, and DZROG. Glc conjugate formation via zeatin-O-glucosyltransferases (Mok et al., 2005) was most likely increased as a way to compensate for the loss of phosphorylation-mediated inactivation normally provided by ADK. Unlike the in vivo feeding studies, however, no significant change was observed in CK nucleotides in either ADK-silencing line. The absence of lowered CK nucleotides indicates that other processes like CK biosynthesis overlaid the effect of ADK loss, and this was more clearly visible in the short-term in vivo labeling experiments.
Technically, CK profiles from wild-type plants were consistent with those reported in several other Arabidopsis studies (Catterou et al., 2002; Corbesier et al., 2003; Heyl et al., 2008; Novák et al., 2008; Zhang et al., 2009). Accordingly, concentrations of all CKs were typically in the range of 1 to 20 pmol g−1 fresh weight of shoot tissue. In a few studies, there were occasionally forms that were as high as 25 to 50 pmol g−1 fresh weight, and these were either nucleotides or glucoside conjugates (Corbesier et al., 2003; Zhang et al., 2009). The concentration ratios of the different groups of CKs found in this study were also consistent with other reports. CK nucleotides were more highly concentrated than the free bases or ribosides, the latter forms being equally abundant. We found N-glucoside CKs to be the most concentrated forms, as documented previously by others (Catterou et al., 2002; Novák et al., 2008; Zhang et al., 2009). Furthermore, the approximately equal amounts of cis- and trans-isomers and the either low or undetectable levels of dihydro-type CKs are also consistent with publications on profiles of Arabidopsis leaf CKs (Corbesier et al., 2003; Novák et al., 2008; Izumi et al., 2009).
In conclusion, based on phenotypic and metabolic analysis of ADK-deficient lines, our findings here prove that ADK activity metabolizes CK ribosides in vivo. Considering that CKs contribute to numerous fundamental plant growth and development processes and that the conversion of CK ribosides to CK nucleotides is thought to deactivate CKs, these findings highlight the significance of CK interconversion in the regulation of plant growth and development processes.
MATERIALS AND METHODS
Growth Conditions
All experiments were performed with Arabidopsis (Arabidopsis thaliana) ecotype Columbia. Soil-grown plants were potted on a 50:50 mixture of Sunshine LC1 Mix and Sunshine LG3 Germination Mix (JVK). Seedlings for root studies were grown on half-strength Murashige and Skoog basal medium salts with Gamborg’s vitamins (Sigma). Upon being planted, seeds were imbibed in the dark at 4°C for 48 h before being put under fluorescent light (150 ± 20 μmol m−2 s−1 photosynthetically active radiation) in an 18-h-light/6-h-dark photoperiod set at 21°C.
Generation of ADK-Deficient Lines
The creation of sADK lines is described by Moffatt et al. (2002). They express a transgene composed of the complete ADK1 open reading frame downstream of an enhanced 35S promoter. The amiADK sequence used for microRNA silencing was generated using WMD2 Web MicroRNA Designer (wmd2.weigelworld.org). Currently, WMD version 3 is released (wmd3.weigelworld.org). Two amiRNA sequences (1, TATGGGCAAATGTATGTCCTC; 2, TTAACTGGTTGCCCCAGGGAC) located in different regions of the ADK transcripts were chosen. The amiRNA sequence 1 targets 5′-GAGGACAAACATTTGCCCATG-3′ (ADK1) and 5′-GAAGACAAACATTTGCCCATG-3′ (ADK2), and sequence 2 targets 5′-GTTCCTGGGGCAACCAGTTAC-3′ (ADK1) and 5′-ATTCCTGGGGCAACCAGTTAC-3′ (ADK2). Both amiRNA sequences were then engineered into an endogenous microRNA precursor for expression. The approximately 400 bp of amiADK precursor was introduced into the Columbia wild-type background under the control of the 35S promoter. Homozygous lines were obtained by screening subsequent generations for glufosinate resistance. Single T-DNA insertion lines adk1-1 (SALK_597_D09) and adk2-1 (SALK_000565) were obtained from the SALK Institute and screened by PCR using either adk1 (ADK1L, 5′-CAAGTGGGTTGGTGGGATAG-3′; ADK1R, 5′-CCAAGCCAAAACACACACTC-3′) or adk2 (ADK2L, 5′-CAAGACGCTTCACTCAGCAG-3′; ADK2R, 5′-CAAAACTGAGGGTAACCCAA-3′) with the conditions provided by the SALK Institute.
Enzyme Assay
To perform the assay for ADK activity, protein was first extracted from rosette leaves of 4-week-old plants: leaves seven to nine were sampled. Collected tissue was homogenized in 100 mm HEPES (pH 7.4) at a ratio of 4:1 (400 mg of tissue with 100 μL of buffer) and desalted, and protein concentrations were determined using a Bradford assay (Bradford, 1976) with bovine serum albumin as the standard. During all steps of extraction, protein was stored on ice or kept at 4°C. ADK activity was then measured in 50-μL reactions using tritiated Ado (Sigma; 49.9 Ci mmol−1, 1 mCi mL−1) as a substrate (Moffatt et al., 2000). Each sample was measured in triplicate with three different protein concentrations per set.
Microscopic Analysis
To compare cell size, rosette leaves of the seventh position from 3.5-week-old plants were cleared overnight at room temperature in 1:3 ethanol:acetic acid and subsequently softened for 4 h in 8 m NaOH. To ensure that the same area of leaf was being sampled each time, tissue was collected based on a constant ratio of leaf length to distance from the base of 0.7 to 0.8. Cell counting was then performed in three different areas on both the right and left sides of the primary rib in four leaf replicates of each plant line using a Zeiss Axio-Imager.D1. For sectioning, leaf samples of 3.5-week-old wild-type, sADK, and amiADK plants were immediately transferred to fixative solution (0.05 m sodium cacodylate, 2% paraformaldehyde [w/v], pH 7, and 0.1% glutaraldehyde). After overnight incubation at 4°C, the samples were dehydrated with ethanol and slowly infiltrated into LR White plastic resin (hard grade; Canemco-Marivac). The samples were then cured in LR White using a 60°C to 65°C oven for 2 to 24 h. Tissue was sectioned at 4 μm thickness using a Reichert Ultracut E microtome and stained with toluidine blue O for visualization on a Zeiss Axiophot microscope.
Root tips were studied in 3-week-old roots by staining fresh tissue in 100 μg mL−1 (w/v) propidium iodide and visualized using a Zeiss Axio-Imager.D1 (536-nm excitation filter, 617-nm emission filter).
To prepare samples for SEM analysis, tissue was fixed in 4% (w/v) paraformaldehyde (pH 7) and 2% (w/v) glutaraldehyde dissolved in 1× phosphate-buffered saline overnight at 4°C. Once the samples were fixed, the tissue was fully dehydrated in ethanol and placed in HPLC-grade acetone for 24 h at 4°C. The samples were then dried using critical point freezing, sputter coated with gold particles, and visualized using a Hitachi S570 SEM system.
To study GUS expression in ARR5::GUS and cycB1;1::GUS, tissue was prepared according to Jefferson et al. (1987). Prestaining, tissue was cleared in 90% acetone for 1 h and washed with water twice. The samples were incubated with 5-bromo-4-chloro-3-indolyl-β-glucuronic acid for 18 h in the dark at 37°C and then fully cleared with 90% ethanol. Leaves were visualized with a Canon440 scanner.
Quantitative Reverse Transcription-PCR
Younger rosette leaves (11–13 developed) were collected from individual 4.5-week-old plants in three biological replicates and flash frozen in liquid nitrogen prior to RNA extraction. Total RNA was isolated using TriPure isolation reagent according to the manufacturer’s instructions (Roche). The RNA was then quantified at 260 nm so 2.5 μg could be DNase (Ambion) treated; the success of DNase treatment was confirmed via PCR. Upon making cDNA with SuperScript III reverse transcriptase (Invitrogen), quantitative reverse transcription-PCR was performed according Argyros et al. (2008) with three technical replicates per sample. Primers used are as follows: ARR5-F, 5′-GCCTCGTATCGATAGATGTCTTGAAGAAGG-3′; ARR5-R, 5′-TCTGATAAACTCAGATCTTTGCGCGT-3′; ARR7-F, 5′-AGAGTGGAACTAGGGCTTTGCAGT-3′; ARR7-R, 5′-CTCCTTCTTTGAGACATTCTTGTATACGAGG-3′; Tubulin-F, 5′-TTCCGTACCCTCAAGCTCGCTAAT-3′; Tubulin-R, 5′-ATCCTCTCGATGTCAATGGTGCGA-3′.
Sample Collection and Preparation for HPLC-MS/MS
Each sample composed of 100 to 300 mg of leaf tissue was collected from 4.5-week-old plants and immediately flash frozen in liquid nitrogen. Phytohormones and adenylates were extracted concomitantly using a modified protocol as described (Emery et al., 1998). Liquid nitrogen-frozen tissue was ground to a powder in a 2-mL microcentrifuge tube with a zirconium oxide grinding ball (diameter, 5 mm) with the aid of a tissuelyser (MM 301 Mixer Mill; Retsch). After pulverization, the following stable isotope-labeled compounds were added to each sample: 250 ng of [13C5]Ado (Omicron Biochemicals), [U-15N5]AMP (Cambridge Isotopes), and [13C6]Ade (OlChemIm); and 10 ng of [2H6]iP, [2H6][9R]iP, [2H6][9R-MP]iP, [2H5]Z, [2H3][9R]Z, [2H6][9R-MP]Z, [2H3]DZ, [2H3][9R]DZ, [2H6][9R-MP]DZ, [2H7][9R-OG]DZ, and [2H5][9R-OG]Z (OlChemIm).
Because labeled standards were not commercially available, the quantification of cZR and cZRMP was estimated based on the recovery of the corresponding deuterium isotopically labeled transisomers.
Pulverized tissue was soaked in 1 mL of precooled (−80°C) modified Bieleskie’s reagent (methanol:water:HCOOH, 15:4:1 [v/v/v]) at 5 mL g−1 fresh weight for a minimum of 16 h to allow for passive extraction. After passive extraction, solids were separated by centrifugation (4°C, 20,800g, 15 min; Eppendorf 5417C), the resulting pellet was reextracted with 500 μL of extraction buffer for 30 min at −80°C, and solids were then removed by centrifugation (see above). The resulting supernatant was pooled with that of the first extraction. Pooled supernatants were dried in a speed vacuum concentrator at 40°C and kept at −80°C.
To remove interfering compounds, extraction residues were reconstituted in 1 m formic acid and purified according to Dobrev and Kamínek (2002) on a mixed-mode, reverse-phase, cation-exchange cartridge (Oasis MCX 60 μm 6cc, 150 mg). The pH of the extract was adjusted to be between 1.4 and 2.8 with 1 m HCl, which ensured complete protonation of all CKs and adenylates (Dobrev and Kamínek, 2002). After a wash with analytical-grade methanol, the CK nucleotides Ade and AMP were eluted in 0.35 m NH4OH. Finally, Ado, CK free bases, CK ribosides (CKR), and CK glucosides (CK-O-G) eluted with 0.35 m NH4OH in 60% methanol. After purification, each fraction was completely evaporated at 40°C and kept at −80°C until HPLC-MS/MS analysis.
Prior to being measured, CK nucleotides were dephosphorylated and detected as their corresponding CK forms. Dephosphorylation was accomplished using 3.4 units of bacterial alkaline phosphatase (Sigma) in 1 mL of 0.1 m ethanolamine-HCl (pH 10.4) for 12 h at 37°C (Emery et al., 2000). The resulting CKRs were brought to dryness in a speed vacuum concentrator at 40°C. Samples were reconstituted in 1 mL of ultrapure water and further purified using water-wettable, reverse-phase cartridges (60 mg; Oasis HLB). Columns were activated with 1 mL of HPLC-grade methanol and equilibrated with 1 mL of ultrapure water. After column equilibration, the samples were loaded onto the HLB cartridge and allowed to pass through the cartridge by gravity. The sorbent was washed with 1 mL of ultrapure water, and analytes were eluted using 1 mL of methanol:water (80:20, v/v). All sample eluents were dried in a speed vacuum concentrator at 40°C and stored at −80°C until analysis by HPLC-MS/MS.
HPLC-MS/MS
All metabolites were separated and analyzed using a Dionex Ultimate 3000 HPLC system coupled to a Qtrap 5500 triple quadrupole hybrid ion-trap mass spectrometer (MDS Sciex) equipped with a turbo V-spray source. Positive ion mode was used for all analytes. The Ade and AMP MCX fraction was injected on a Synergi Fusion reverse-phase column (4 μm, 2.1 × 150 mm; Phenomenex). The purines were eluted with an increasing gradient of acetonitrile:20 mm CH3COONH4 (A; pH 4.5, 95:5 [v/v]) and 20 mm CH3COONH4 (B; pH 4.5) at a flow rate of 500 μL min−1. The initial conditions were 5% A and 95% B, changing linearly to 40% A and 60% B in 4 min, and then immediately to 95% A at 4.1 min.
The Ado, CKR, CK free bases, and CK-O-G MCX fraction was injected on a Kinetex C18 column (2.6 μm, 2.1 × 50 mm; Phenomenex), and CKs were eluted with an increasing gradient of methanol (A) and 0.08% acetic acid (B) at a flow rate of 500 μL min−1. The initial conditions were 5% A and 95% B, changing linearly to 45% A and 55% B in 4 min, and then changing to 75% A and 25% B in 5 min. Ado was analyzed from a diluted second aliquot using the same conditions as for the CKs. HPLC effluents were introduced into the turbo V-spray source using conditions specific for each analyte, where quantification was obtained by multiple reaction monitoring of the protonated intact precursor molecule [M+H]+ and a specific product ion (Supplemental Table S2). All data were analyzed and processed using Analyst version 1.5 software. Concentrations were calculated on the basis of the peak areas for the endogenous compounds related to those determined for the internal standards.
In Vivo Labeling of Detached Leaves with Tritiated CKs
Leaves of 4-week-old plants showing the typical sADK phenotype were detached and submerged in half-strength Murashige and Skoog medium. Leaves were submitted to moderate vacuum of approximately 300 mbar by a sink aspirator in order to effectively infiltrate the leaf material with medium. Tritiated CKs were administered to the submerged leaves in a final concentration of 60 nm [2-3H]iPR or [2-3H]ZR (specific radioactivity of 40 Ci mmol−1 each; OlChemIm). The labeling solution was constantly aerated with an air flux of approximately 0.1 L min−1. After 4 h, the labeling was stopped by separating the leaf from the solution using a mesh. Leaves were rinsed with half-strength Murashige and Skoog medium and immediately transferred into –20°C cold Bieleskie’s reagent (for composition, see above). Samples were stored at –20°C until extraction.
Extraction and HPLC separation of labeled metabolites were carried out according to the protocol previously described by von Schwartzenberg et al. (2003, 2007). Briefly, 100 mg of leaf tissue was homogenized in 1 mL of Bieleskie’s reagent using a plastic pestle. The homogenate was centrifuged to separate solids from the CK-containing supernatant at 15,000g. Solids were reextracted twice in fresh Bieleskie’s reagent, and the supernatants were combined. Approximately 600 μL of distilled water was added to cause a phase separation. The mixture was vortexed vigorously, and the CK-containing upper aqueous phase was withdrawn from the lower organic phase. The aqueous phase was evaporated to almost dryness by rotary evaporation and subsequently redissolved in solvent A (10% methanol containing 10 mm triethylamine, pH 5.8). The sample was then divided into equal aliquots. The first was subjected directly to reverse-phase HPLC to separate and quantify radioactivity in the fractions of degradation products, CKR monophosphate, riboside, and base. The second aliquot was adjusted to pH 7.5 and then subjected to treatment with calf intestine alkaline phosphatase (Fermentas) to convert CK nucleotides into ribosides prior to HPLC separation.
The HPLC apparatus was equipped with a diode array detector (DAD 540+; Bio-TEK) in combination with an online liquid scintillation counter (LSC Radiomatic 500 TR Series; Canberra-Packard). A scintillation cocktail Ultima-Flo M (Perkin-Elmer) was used. Separation of CK metabolites was performed on a LiChrospher 60, RP-Select B, 5-μm column (Merck) at a flow rate of 0.8 mL min−1 using a gradient of solvent A and methanol. A gradient of solvent A (10 mm tetraethylammonium and 10% methanol [v/v]) and 100% methanol was applied over a run time of 35 min. The quantity of radioactive degradation products, CKR monophosphate, riboside, and base, was measured by HPLC of the first aliquot. The quantity of radioactive diphosphates and triphosphates of iPR or ZR was deduced from the difference of radioactivity between in the riboside fraction in the two HPLC runs. Three independent labeling experiments were carried out for iPR and ZR. One representative experiment is presented for each substrate.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Morphology and cycB1;1::GUS expression in ADK-deficient floral tissue.
Supplemental Table S1. Residual ADK activity in sADK-deficient Arabidopsis plants of select ages.
Supplemental Table S2. MS conditions for CK and adenylate compounds in Arabidopsis.
Acknowledgments
We thank Steven Chatfield for providing cycB1;1::GUS, Joseph Kieber for ARR5::GUS, and the Arabidopsis Stock Center for seed stocks. We also thank Susanne Bringe for skillful technical help.
References
- Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE. (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astot C, Dolezal K, Nordström A, Wang Q, Kunkel T, Moritz T, Chua NH, Sandberg G. (2000) An alternative cytokinin biosynthesis pathway. Proc Natl Acad Sci USA 97: 14778–14783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliji S, Lacatus G, Sunter G. (2010) The interaction between geminivirus pathogenicity proteins and adenosine kinase leads to increased expression of primary cytokinin-responsive genes. Virology 402: 238–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Carimi F, Zottini M, Formentin E, Terzi M, Lo Schiavo F. (2003) Cytokinins: new apoptotic inducers in plants. Planta 216: 413–421 [DOI] [PubMed] [Google Scholar]
- Catterou M, Dubois F, Smets R, Vaniet S, Kichey T, Van Onckelen H, Sangwan-Norreel BS, Sangwan RS. (2002) hoc: an Arabidopsis mutant overproducing cytokinins and expressing high in vitro organogenic capacity. Plant J 30: 273–287 [DOI] [PubMed] [Google Scholar]
- Chen CM, Eckert RL. (1977) Phosphorylation of cytokinin by adenosine kinase from wheat germ. Plant Physiol 59: 443–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P. (1999) Spatio-temporal analysis of mitotic activity with labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Prinsen E, Jacqmard A, Lejeune P, Van Onckelen H, Périlleux C, Bernier G. (2003) Cytokinin levels in leaves, leaf exudate and shoot apical meristem of Arabidopsis thaliana during floral transition. J Exp Bot 54: 2511–2517 [DOI] [PubMed] [Google Scholar]
- D'Agostino IB, Deruere J, Kieber JJ. (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17: 678–682 [DOI] [PubMed] [Google Scholar]
- Divekar AY, Hakala MT. (1971) Adenosine kinase of sarcoma 180 cells N6-substituted adenosines as substrates and inhibitors. Mol Pharmacol 7: 663–673 [PubMed] [Google Scholar]
- Dobrev PI, Kamínek M. (2002) Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J Chromatogr A 950: 21–29 [DOI] [PubMed] [Google Scholar]
- Emery RJ, Leport L, Barton JE, Turner NC, Atkins CA. (1998) cis-Isomers of cytokinins predominate in chickpea seeds throughout their development. Plant Physiol 117: 1515–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery RJ, Ma Q, Atkins CA. (2000) The forms and sources of cytokinins in developing white lupin seeds and fruits. Plant Physiol 123: 1593–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A, Ramireddy E, Brenner WG, Riefler M, Allemeersch J, Schmülling T. (2008) The transcriptional repressor ARR1-SRDX suppresses pleiotropic cytokinin activities in Arabidopsis. Plant Physiol 147: 1380–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth S, Ikeda Y, Morgante M, Wang X, Zuo J, Hanafey MK, Gaasterland T, Tingey SV, Chua NH. (2003) Monitoring genome-wide changes in gene expression in response to endogenous cytokinin reveals targets in Arabidopsis thaliana. FEBS Lett 554: 373–380 [DOI] [PubMed] [Google Scholar]
- Huang S, Cerny RE, Qi Y, Bhat D, Aydt CM, Hanson DD, Malloy KP, Ness LA. (2003) Transgenic studies on the involvement of cytokinin and gibberellin in male development. Plant Physiol 131: 1270–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y, Okazawa A, Bamba T, Kobayashi A, Fukusaki E. (2009) Development of a method for comprehensive and quantitative analysis of plant hormones by highly sensitive nanoflow liquid chromatography-electrospray ionization-ion trap mass spectrometry. Anal Chim Acta 648: 215–225 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Yun J, Seo YH, Park CM. (2005) Characterization of an Arabidopsis gene that mediates cytokinin signaling in shoot apical meristem development. Mol Cells 19: 342–349 [PubMed] [Google Scholar]
- Kanazawa A, O’Dell M, Hellens RP. (2007) Epigenetic inactivation of chalcone synthase-A transgene transcription in petunia leads to a reversion of the post-transcriptional gene silencing phenotype. Plant Cell Physiol 48: 638–647 [DOI] [PubMed] [Google Scholar]
- Kunz C, Hanspeter S, Maike S, Kooter J, Frederick M. (1996) Developmentally regulated silencing and reactivation of tobacco chitinase transgene expression. Plant J 10: 437–450 [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Kwade Z, Swiatek A, Azmi A, Goossens A, Inzé D, Van Onckelen H, Roef L. (2005) Identification of four adenosine kinase isoforms in tobacco BY-2 cells and their putative role in the cell cycle-regulated cytokinin metabolism. J Biol Chem 280: 17512–17519 [DOI] [PubMed] [Google Scholar]
- Li CH, Yu N, Jiang SM, Shangguan XX, Wang LJ, Chen XY. (2008) Down-regulation of S-adenosyl-L-homocysteine hydrolase reveals a role of cytokinin in promoting transmethylation reactions. Planta 228: 125–136 [DOI] [PubMed] [Google Scholar]
- Lolle SJ, Victor JL, Young JM, Pruitt RE. (2005) Genome-wide non-mendelian inheritance of extra-genomic information in Arabidopsis. Nature 434: 505–509 [DOI] [PubMed] [Google Scholar]
- Majewski P, Wołoszyńska M, Jańska H. (2009) Developmentally early and late onset of Rps10 silencing in Arabidopsis thaliana: genetic and environmental regulation. J Exp Bot 60: 1163–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meins F, Kunz C. (1994) Silencing of chitinase expression in transgenic plants: an autoregulatory model. Paszkowski J, , Homologous Recombination and Gene Silencing in Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 335–348 [Google Scholar]
- Mlejnek P, Procházka S. (2002) Activation of caspase-like proteases and induction of apoptosis by isopentenyladenosine in tobacco BY-2 cells. Planta 215: 158–166 [DOI] [PubMed] [Google Scholar]
- Moffatt BA, Stevens YY, Allen MS, Snider JD, Pereira LA, Todorova MI, Summers PS, Weretilnyk EA, Martin-McCaffrey L, Wagner C. (2002) Adenosine kinase deficiency is associated with developmental abnormalities and reduced transmethylation. Plant Physiol 128: 812–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt BA, Wang L, Allen MS, Stevens YY, Qin W, Snider J, von Schwartzenberg K. (2000) Adenosine kinase of Arabidopsis: kinetic properties and gene expression. Plant Physiol 124: 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok MC, Martin RC, Dobrev PI, Vanková R, Ho PS, Yonekura-Sakakibara K, Sakakibara H, Mok DW. (2005) Topolins and hydroxylated thidiazuron derivatives are substrates of cytokinin O-glucosyltransferase with position specificity related to receptor recognition. Plant Physiol 137: 1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebel FdC, Frendo P, Van Montagu M, Cornelissen M. (1995) Post-transcriptional cosuppression of β-1,3-glucanase genes does not affect accumulation of transgene nuclear mRNA. Plant Cell 7: 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O, Hauserová E, Amakorová P, Dolezal K, Strnad M. (2008) Cytokinin profiling in plant tissues using ultra-performance liquid chromatography-electrospray tandem mass spectrometry. Phytochemistry 69: 2214–2224 [DOI] [PubMed] [Google Scholar]
- Pereira LA, Schoor S, Goubet F, Dupree P, Moffatt BA. (2006) Deficiency of adenosine kinase activity affects the degree of pectin methyl-esterification in cell walls of Arabidopsis thaliana. Planta 224: 1401–1414 [DOI] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T. (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov GA, Kieber JJ, Schmülling T. (2002) A rapid cytokinin response assay in Arabidopsis indicates a role for phospholipase D in cytokinin signalling. FEBS Lett 515: 39–43 [DOI] [PubMed] [Google Scholar]
- Romanov GA, Lomin SN, Schmülling T. (2006) Biochemical characteristics and ligand-binding properties of Arabidopsis cytokinin receptor AHK3 compared to CRE1/AHK4 as revealed by a direct binding assay. J Exp Bot 57: 4051–4058 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Béclin C, Fagard M. (2001) Post-transcriptional gene silencing in plants. J Cell Sci 114: 3083–3091 [DOI] [PubMed] [Google Scholar]
- von Schwartzenberg K, Kruse S, Reski R, Moffatt B, Laloue M. (1998) Cloning and characterization of an adenosine kinase from Physcomitrella involved in cytokinin metabolism. Plant J 13: 249–257 [DOI] [PubMed] [Google Scholar]
- von Schwartzenberg K, Nunez MF, Blaschke H, Dobrev PI, Novak O, Motyka V, Strnad M. (2007) Cytokinins in the bryophyte Physcomitrella patens: analyses of activity, distribution, and cytokinin oxidase/dehydrogenase overexpression reveal the role of extracellular cytokinins. Plant Physiol 145: 786–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwartzenberg K, Pethe C, Laloue M. (2003) Cytokinin metabolism in Physcomitrella patens: differences and similarities to higher plants. Plant Growth Regul 39: 99–106 [Google Scholar]
- Wang H, Buckley KJ, Yang X, Buchmann RC, Bisaro DM. (2005) Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J Virol 79: 7410–7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T. (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Vankova R, Malbeck J, Dobrev PI, Xu Y, Chong K, Neff MM. (2009) AtSOFL1 and AtSOFL2 act redundantly as positive modulators of the endogenous content of specific cytokinins in Arabidopsis. PLoS ONE 4: e8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrenner R, Stitt M, Sonnewald U, Boldt R. (2006) Pyrimidine and purine biosynthesis and degradation in plants. Annu Rev Plant Biol 57: 805–836 [DOI] [PubMed] [Google Scholar]