Abstract
We report here the low-resolution structure of the complex formed by the endo-polygalacturonase from Fusarium phyllophilum and one of the polygalacturonase-inhibiting protein from Phaseolus vulgaris after chemical cross-linking as determined by small-angle x-ray scattering analysis. The inhibitor engages its concave surface of the leucine-rich repeat domain with the enzyme. Both sides of the enzyme active site cleft interact with the inhibitor, accounting for the competitive mechanism of inhibition observed. The structure is in agreement with previous site-directed mutagenesis data and has been further validated with structure-guided mutations and subsequent assay of the inhibitory activity. The structure of the complex may help the design of inhibitors with improved or new recognition capabilities to be used for crop protection.
Plant innate immunity relies on a wide array of defense strategies that are based on plant structural features as, for example, the degradability of the cell wall, and on the plant capability of perceiving the presence of pathogens and activating defense responses (Bent and Mackey, 2007). To gain access to the plant tissue, pathogens secrete cell wall-degrading enzymes (CWDEs), among which endo-polygalacturonase (PG) and other enzymes capable of hydrolyzing homogalacturonan (main component of pectin) are secreted prior to other CWDEs attacking their substrates (Lionetti et al., 2010). PGs are well-known virulence factors of phytopathogenic fungi and some isoforms have been shown to be necessary for pathogenicity of Botrytis cinerea, Sclerotinia sclerotiorum, and other fungal species (Isshiki et al., 2001; Kars et al., 2005; Zuppini et al., 2005).
To counteract PG activity, plants have evolved small gene families encoding PG-inhibiting proteins (PGIPs; Frediani et al., 1993; Federici et al., 2006). PGIPs inhibit the activity of PGs and favor the accumulation of homogalacturonan fragments, called oligogalacturonides (OGs) that act as damage-associated molecular patterns (Boller and Felix, 2009; De Lorenzo et al., 2011). OGs are signaling molecules specifically recognized by receptor proteins belonging to the wall-associated kinase family and trigger a wide transcriptional defense program (Brutus et al., 2010). In Arabidopsis (Arabidopsis thaliana) the genes activated by OGs largely overlap with those activated by the necrotroph B. cinerea (Ferrari et al., 2007). The importance of PGIPs in defense is well documented by studies showing that transgenic plants overexpressing the inhibitor are more resistant to fungi (Powell et al., 2000; Ferrari et al., 2003; Manfredini et al., 2005; Joubert et al., 2006; Janni et al., 2008) and, conversely, plants where the expression of PGIPs is partly silenced are more susceptible to fungal infection (Ferrari et al., 2006).
The PG-PGIP interaction is considered a paradigm that describes some of the recognition events determining plant immunity (Misas-Villamil and van der Hoorn, 2008). PGIP belongs to the Leu-rich repeat (LRR) family of proteins, like the majority of the proteins encoded by the so-called resistance genes (Komjanc et al., 1999; De Lorenzo et al., 2001), and is the only plant extracellular LRR protein with a solved crystal structure (Di Matteo et al., 2003). Distinct PGIP isoforms that display different specificity of recognition for PGs are produced by plants (D’Ovidio et al., 2004). In some cases a single PGIP isoform inhibits different PGs with a different mechanism, i.e. competitive, noncompetitive, and/or mixed (King et al., 2002; Sicilia et al., 2005; Bonivento et al., 2008). Genes encoding PGIPs are under selective pressure for diversification and a number of hot spots for the interaction with PGs have been identified on the inhibitor concave surface and validated by site-directed mutagenesis (Casasoli et al., 2009). Unlike for other CWDE inhibitors that engage many contacts in the interaction with their partners (Payan et al., 2004; Raiola et al., 2004; Di Matteo et al., 2005), only a few residues, sometimes only one, are critical for maintaining the stability of the PG-PGIP interaction (Leckie et al., 1999; Casasoli et al., 2009). This is probably why, despite the efforts made during the past years, the crystal structure of the complex is not available yet. Some amino acid positions may be engaged in the interaction, depending on the PG partner and on the different structural domains recognized in fungal PGs (Federici et al., 2006). However a detailed analysis of the many aspects of the PG-PGIP interaction may be performed only if the structure of the complex becomes available. This is particularly important in view of possible genetic modifications or selection of PGIP genes with new recognition capabilities toward fungal PGs that are unable to be inhibited by any PGIP isolated so far.
To obtain useful structural information on the PG-PGIP complex, we have used small-angle x-ray scattering (SAXS), a diffractometric technique that allows analysis of proteins directly in solution (Svergun et al., 2001; Koch et al., 2003; Lipfert and Doniach, 2007). A limitation of SAXS is the lower resolution that characterizes this technique as compared to x-ray crystallography. On the other hand, a main advantage of SAXS lies in the possibility of studying a protein complex formed in solution at physiological pH and ionic strength conditions, with a minimal perturbation of the protein structures with respect to the native environment (Jacobsen et al., 2006).
We present here a low-resolution three-dimensional structure of the complex formed by PG from Fusarium phyllophilum (FpPG; formerly known as Fusarium moniliforme PG; Mariotti et al., 2008) and the PGIP2 isoform from Phaseolus vulgaris (PvPGIP2). The crystal structures of both FpPG and PvPGIP2 have been reported in previous articles (Federici et al., 2001; Di Matteo et al., 2003). The native and a chemically cross-linked complex were analyzed as well as the complex formed by FpPG with PvPGIP2.Q224K, i.e. a variant inhibitor carrying a point mutation that causes a strong increase of its dissociation constant with FpPG (Leckie et al., 1999). The structure obtained by SAXS analysis is in agreement with and complements previous site-directed mutagenesis studies. This study considerably increases our understanding of the recognition versatility of PGIP and paves the way to the design of new and improved PGIP inhibitors for biotechnological purposes.
RESULTS
SAXS Analysis of the FpPG-PvPGIP2 Complex
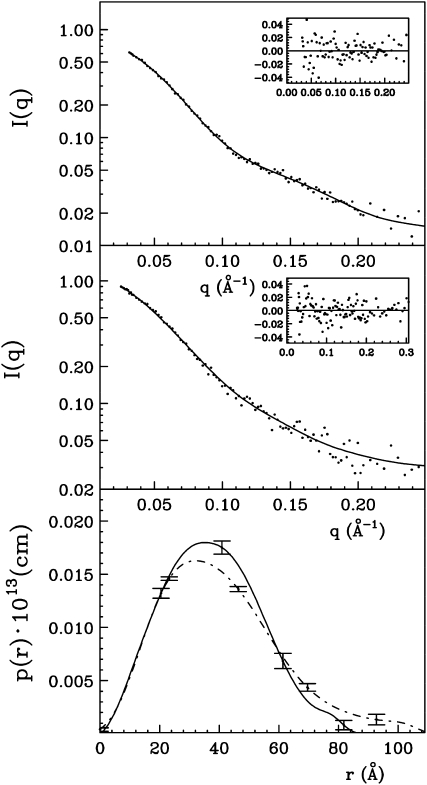
FpPG and PvPGIP2 were used because their crystal structures are known (Federici et al., 2001; Di Matteo et al., 2003) and their affinity is high enough (KD = 47.7 nm; Leckie et al., 1999) to allow the formation of the complex at the concentrations normally used for SAXS analysis. The analysis was performed in a buffer mimicking the physiological pH and ionic strength conditions found in the plant cell wall at the beginning of the infection, i.e. before pathological pH changes occur in the infected tissue. For comparison, the combination of FpPG and PvPGIP2.Q224K, a variant inhibitor that does not inhibit FpPG (Leckie et al., 1999), was used. The scattering intensities and the corresponding p(r) functions, reported in Figure 1, show differences between the two combinations of proteins. In both cases, Guinier plots allow the presence of aggregates to be excluded (Glatter and Kratky, 1982; Svergun et al., 2001; Koch et al., 2003; Jacques and Trewhella, 2010). The complex formed by FpPG and the wild-type PvPGIP2 is characterized by a Dmax = 80 ± 5 Å and a Rg = 29.1 ± 0.4 Å, whereas the complex of the enzyme with the variant PvPGIP2Q224K has a Dmax = 100 ± 5 Å and a Rg = 32.5 ± 0.5 Å.
Figure 1.
Experimental scattered intensities (dots) and the calculated ones with the ITP program relative to the complex between FpPG and PvPGIP2 (top section) and between FpPG and PvPGIP2.Q224K (middle section). The residuals are reported in the insets and the corresponding p(r) functions in the bottom section (continuous line for the complex between FpPG and PvPGIP2 and dot dash line for the complex between FpPG and PvPGIP2.Q224K). For clarity only a few error bars are shown.
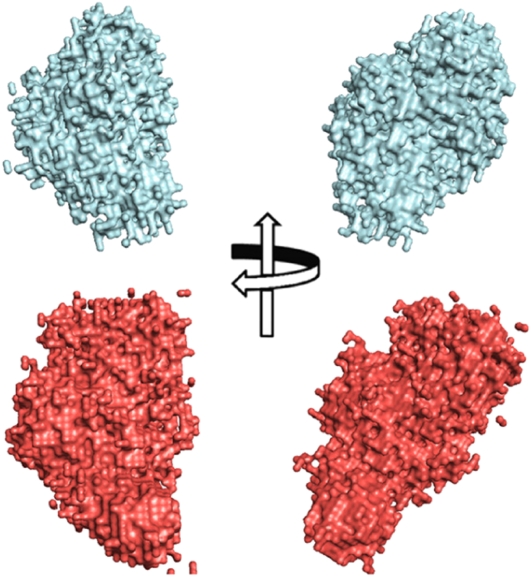
Scattering patterns were analyzed by the GA_STRUCT and SASREF programs for shape reconstruction, using ab initio and rigid body docking protocols, respectively. Both approaches did not give unambiguous solutions for both samples. The most represented envelopes, derived from the analysis performed with the GA_STRUCT program, are reported in Figure 2. The envelope relative to the combination FpPG-PvPGIP2.Q224K is more expanded than that of the combination FpPG-wild-type inhibitor. The difficulties of obtaining a reliable recovered structure for both combinations did not allow a deeper analysis of the complex. This might be explained with a fluctuation in the shape of the complex, due to a continuous dissociation and association of the two proteins during the time course of data acquisition.
Figure 2.
Consensus envelopes for the complex between FpPG and PvPGIP2 (cyan) and between FpPG and PvPGIP2.Q224K (red) as calculated with the GA_STRUCT program. Two orthogonal viewpoints are represented.
We tested, therefore, the possibility of stabilizing the protein complex through chemical cross-linking. Formaldehyde was used as a chemical cross-linker because it is a short-arm linker that should avoid nonnative interactions. Chemical cross-linking was performed at pH 4.6 and low ionic strength, i.e. an environment resembling that of the plant cell wall, where the interaction takes place. The reaction produced the heterodimeric complex (approximately 67 kD) with a yield greater than 95% while only a low amount of free proteins were observed in solution (Fig. 3A). The cross-linking reaction was also performed by using FpPG or PvPGIP2 alone to assess that homopolymerization of the single proteins does not take place under the experimental condition used (Fig. 3B).
Figure 3.
A, SDS-PAGE of the complex between FpPG and PvPGIP2 upon cross-linking with formaldehyde. A band of approximately 67 kD corresponding to the molecular mass of the complex accounted for more than 95% of the total proteins detectable on the gel. The cross-linking reaction was also performed in the presence of PvPGIP2 alone (approximately 31 kD; B) or FpPG alone (approximately 35 kD; C), to exclude the possibility that cross-linked homodimers or multimers are formed. In both cases, no high molecular weight bands were detected.
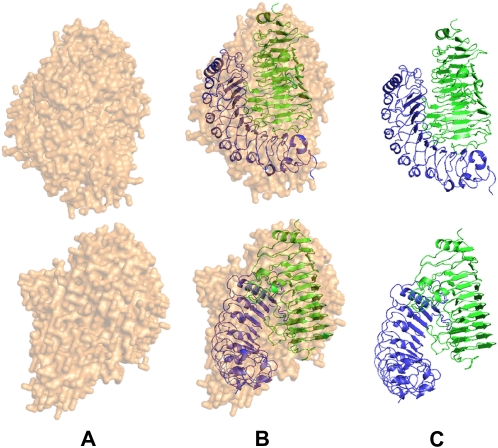
The heterocomplex obtained by chemical cross-linking was subjected to SAXS analysis. The intensity and the corresponding p(r) functions for the cross-linked complex (Dmax = 80 ± 5 Å and a Rg = 28.6 ± 0.4 Å; see Fig. 4) were very close to the values obtained with the same sample in the absence of cross-linker (described above), indicating that the cross-linking reaction favors the formation of a stable complex. The analysis of the scattered intensity, performed by the GA_STRUCT and SASREF algorithms, gave unambiguous results and a reliable structure of the complex. Figure 5 shows the electron density map obtained by using the GA_STRUCT program superimposed to the recovered reference structure obtained by using the SASREF program. A Protein Data Bank file with the coordinates of this reference reconstruction is included as Supplemental File S1. The obtained normalized spatial discrepancy (NSD) value of 1.89 is indicative of a good superposition (Kozin and Svergun, 2001) of the two structures obtained via two independent computational procedures. Electron density is compatible with a model in which the two proteins contact each other in a head-to-head orientation and the concave surface of PvPGIP2 interacts with the loops of FpPG surrounding the active site cleft.
Figure 4.
Experimental scattered intensities (dots) and the calculated ones with the ITP program relative to the complex between FpPG and PvPGIP2 in the presence of the cross-linker (top section). The residuals are reported in the insets and the corresponding p(r) function in the bottom section (dot dash line) in comparison with the p(r) function relative to the complex between FpPG and PvPGIP2 in the absence of the cross-linker (line). For clarity only a few error bars are shown.
Figure 5.
Three-dimensional structure of the chemically cross-linked complex between FpPG (in green) and PvPGIP2 (in blue). A, The GA_STRUCT consensus envelope. B, The superimposition between the consensus envelope and the three-dimensional structure obtained with the SASREF code. C, The corresponding three-dimensional structures are shown. Two orthogonal viewpoints are reported.
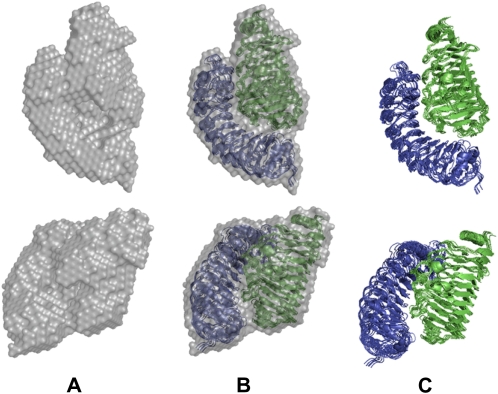
Since a potential limitation of SAXS analysis is the possibility of obtaining multiple and divergent solutions, the SASREF protocol was repeated seven times. The mean agreement index value of the superimpositions of six structures with the seventh taken as reference (Petoukhov and Svergun, 2005) is 0.34 ± 0.04. A check with the experimental data gives a mean agreement index value between observed and calculated intensities of 0.58 ± 0.01; this means a sd of less than 2%. The results were combined and analyzed using the DAMAVER program. The seven superimposed structures and the corresponding calculated consensus envelope is represented in Figure 6. A remarkable degree of superimposition (NSD = 1.09) is observed.
Figure 6.
A, Seven superimposed envelopes obtained for the chemically cross-linked complex between FpPG (green) and PvPGIP2 (blue) with the DAMAVER program. B, The superimposition between the consensus envelopes and the three-dimensional structures calculated with the SASREF code. C, The corresponding three-dimensional structures are shown. Two orthogonal viewpoints are reported.
Structure-Assisted Design of PvPGIP2 Site-Directed Mutants
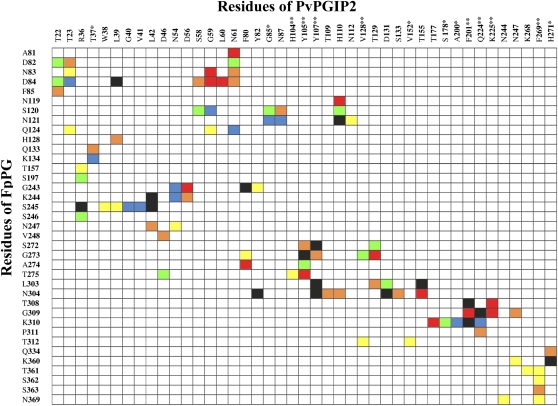
The interfaces of the FpPG-PvPGIP2 model structures obtained by the SASREF protocol were analyzed using the contact map tool within the SPACE suite of programs (Sobolev et al., 2005; http://ligin.weizmann.ac.il/cma/). A contact map that reports the interactions observed in the seven model complexes and their relative frequency is shown in Figure 7.
Figure 7.
Contact map of the residues involved in the FpPG-PvPGIP2 interaction. Contacts between residues of the two proteins are shown as squares in the map. The map was calculated using the SPACE server (http://ligin.weizmann.ac.il/cma/) and represents the average between the single contact maps obtained from the seven measurements and structures of Figure 6. The colors of the squares refer to the frequency with which each contact appears: yellow represents the presence of the contact in two out of seven structures; orange, red, green, blue, and black represent the presence of the contact in three, four, five, six, and seven out of seven structures, respectively. PvPGIP residues identified by Casasoli et al. (2009) as positively selected sites in Fabaceae and hotspots for the interaction with FpPG are indicated by a single and a double asterisk, respectively.
Remarkably, many residues that were previously selected and validated as hot spots for this interaction (Casasoli et al., 2009) were found at the protein-protein interface of the complex (see “Discussion”). The active site of FpPG is located in a deep cleft that is open on both sides to allow the accommodation of the substrate (homogalacturonan) and the release of the hydrolysis products. The low-resolution structure obtained by SAXS analysis indicates that an area near the N terminus of the inhibitor binds the enzyme loops surrounding the active site cleft (Figs. 6 and 7). Interestingly, previous analyses also suggested that the first and the second LRRs of PvPGIP2 are involved in the interaction (Casasoli et al., 2009).
Two Gly residues, G59 and G85 located in the first and second LRRs, respectively, were mutated into Phe to assess their effect on the interaction in a loss-of-function approach. Inhibition assays using the single variant inhibitors showed that the mutations PvPGIP2.G59F and PvPGIP2.G85F affect the inhibition by 5-fold and 15-fold, respectively, confirming their involvement in the interaction with FpPG (Table I). To exclude that these effects were due to protein instability, the mutated proteins were also tested against PGII of Aspergillus niger. Both mutations did not cause any loss of inhibitory activity against this enzyme. We also mutated T177, a residue of the sixth LRR predicted to be involved in recognition and located in proximity of the functionally relevant F201 (Casasoli et al., 2009). This residue was mutated into Asp and a dramatic loss of function was observed (Table I).
Table I. Inhibitory activities of native and site-directed mutants of PvPGIP2.
Inhibitory activity is expressed as the amount of PGIP (in ng) causing 50% inhibition of one agarose diffusion unit of FpPG (this enzyme is the same as that indicated as F. moniliforme PG in a previous article; D’Ovidio et al., 2004). The symbol ∞ indicates >700 ng. The asterisk indicates cases where inhibitory activities of PvPGIP2 and the mutants are significantly different (P < 0.003). WT, Wild type.
| PvPGIP2 Mutants | Inhibitory Activity |
| ng | |
| WT | 7 |
| G59F | 47* |
| G85F | 140* |
| H110E | 130* |
| T177D | ∞* |
The structure obtained by SAXS analysis indicates that N121 of FpPG interacts with H110 of PvPGIP2, through a hydrogen bond involving the main chain of H110 and the side chain of N121. The amino acid N121 belongs to a four-residue loop (SNSN) of FpPG that is absent in the PGII from A. niger (Federici et al., 2001) and is subject to a considerable variation as well as to a positive selection for diversification in other PGs (Mariotti et al., 2008). For example, PG from Fusarium verticillioides, which is not inhibited by PvPGIP2, has a Lys in this topological position (Mariotti et al., 2008). A marked loss of inhibition against FpPG activity was observed when H110 was mutated into a Glu, indicating that loop 120-123 in FpPG is an important target of PvPGIP2 recognition.
DISCUSSION
In this work, we have analyzed the interaction between a fungal PG and a plant PGIP using SAXS. SAXS is a method for the analysis of biological macromolecules in solution that, due to the progress in computational methods of the last decade, has become a powerful tool to decipher three-dimensional low-resolution structures (Svergun et al., 2001; Koch et al., 2003; Lipfert and Doniach, 2007). By using ab initio and rigid body modeling (Svergun, 1999; Chacón et al., 2000; Heller et al., 2003), the shape of monomeric forms, the multimeric state of proteins, and protein complexes can be assessed (Putnam et al., 2007; Mertens and Svergun, 2010; Nogales et al., 2010; Pons et al., 2010). In addition, SAXS allows the quantitative analysis of flexible systems including multidomain and intrinsically unfolded proteins (Bernadó et al., 2005), mechanisms of denaturation and unfolding (Segel et al., 1998; Galantini et al., 2008; Leggio et al., 2009), the stabilizing role of the ligands in denaturating conditions (Galantini et al., 2010), and molecular mechanisms of ligand release (Tabarani et al., 2009).
The PG-PGIP interaction has been previously investigated through site-directed mutagenesis, surface plasmon resonance, inhibition assays, and a variety of bioinformatics tools including protein-protein docking (Federici et al., 2006). The structure obtained by SAXS analysis, albeit at low resolution, is, to our knowledge, the first three-dimensional structure of a plant extracellular LRR protein in complex with its target. PvPGIP2 interacts with the enzyme by means of the concave surface of its LRR scaffold. This feature, anticipated by functional as well as evolutionary analysis, finds now a direct structural confirmation. An extended surface of PvPGIP2 accounting for 810 Å2 as calculated by the PROTORP server (http://www.bioinformatics.sussex.ac.uk/protorp/) is buried during the formation of the complex with FpPG. This area comprises residues belonging to the 10 LRRs of the inhibitor and therefore spans over the entire LRR domain. The enzyme and inhibitor interact in a head-to-head orientation with the two N termini located on the same side. Within this orientation, the inhibitor mainly uses residues located at the end of the loops connecting the convex-side 310 helices with the β-strands located on the concave side as well as the initial residues of the β-strands themselves. It is worth noting that some of the inhibitor residues appearing as contact points in the SAXS structure, have been previously predicted to be subjected to positive selection by codon variation analysis and to possess favorable desolvation energy for protein-protein interactions (Casasoli et al., 2009). Some of these, i.e. residues H104, Y105, Y107, D131, F201, Q224, K225, and F269, have been already shown to play a role in the interaction with FpPG in previous mutagenesis studies (Leckie et al., 1999; Casasoli et al., 2009; Spinelli et al., 2009). In particular, the involvement of Q224 shown to be critical for the interaction (Leckie et al., 1999), confirms the validity of our SAXS analysis. The analysis of the FpPG-PvPGIP2.Q224K complex having a dissociation constant approximately 75-fold higher than that of the wild-type complex (Leckie et al., 1999), shows, in fact, a tendency of the two proteins to interact in a less tight manner, possibly due to the charge repulsion effects of the substitution of Q224 with a Lys. An increase of both Dmax and Rg is reflected in a more extended envelope and suggests a nonoptimal interaction of the two proteins in the complex.
To further confirm the value of the SAXS structural data, we performed site-directed mutagenesis on a few other untested residues that appear to be buried in the complex. First we mutated G59 and G85, located in the first and second LRR, respectively, since the involvement of these two repeats has been previously suggested without an experimental demonstration (Casasoli et al., 2009). Then we mutated residue T177, located in the middle of the concave surface. Mutation of G59 and G85 resulted in a markedly reduced functionality of the protein while mutation of T177 completely abolished the interaction.
On the other hand, FpPG interacts with PvPGIP2 through residues located on the loops surrounding both sides of the active site cleft. This is in agreement both with previous desolvation energy calculations and with the competitive mechanism of inhibition played by PvPGIP2 against FpPG (Federici et al., 2001). By covering both sides of the enzyme active site cleft, PvPGIP2 prevents the entry of the large polymeric substrate. However different PGs, like those from A. niger and Colletotrichum lupini, are inhibited noncompetitively by PvPGIP2. This suggests that the inhibitor recognizes different structural domains on different PGs and that single residues of the PvPGIP2 concave surface interact with different enzyme residues depending on the PG partner (Casasoli et al., 2009). The structure obtained by SAXS analysis supports the idea that the interaction between FpPG and PvPGIP2 is the result of a specific and unique set of contact points. The structure, for example, indicates that residues S120 and N121 of FpPG are contact points in the complex. These two residues belong to the loop SNSN, which is under selective pressure for diversification and is located in proximity of the active site cleft; this loop is typical of FpPG while it is generally absent or mutated in other PGs. For instance, the loop is absent in A. niger PGII and this may consequently cause the noncompetitive mechanism of inhibition of this enzyme by PvPGIP2. On the other hand, residue N121 is replaced by a Lys in a PG from F. verticillioides and this may consequently cause the lack of inhibition of this enzyme by PvPGIP2 (Mariotti et al., 2008). Mutation of H110, the inhibitor residue that faces the enzyme N121 in the SAXS structure, resulted in a marked loss of inhibition, suggesting the importance of the SNSN loop in FpPG recognition.
In conclusion, to our knowledge, we have obtained the first low-resolution structure of a PG-PGIP combination. This structure is in very good agreement with site-directed mutagenesis data and may be used for planning a mutational strategy aimed at improving the recognition properties of PvPGIP2. The SAXS approach employed in this work has a general validity and may be used to characterize different PG-PGIP combinations and their recognition versatility as well as other combinations between LRR proteins and their partners.
MATERIALS AND METHODS
FpPG Expression and Purification
The cDNA encoding FpPG was cloned in pGAPZαA (Invitrogen) using the EcoRI and XbaI restriction sites introduced by using the primers EcoFw (5′-acctgagaattcgatccctgctccgtgac-3′) and XbaRv (5′-gcctatctagactagctggggcaagtgtt-3′). The construct, generated in frame with the signal sequence for secretion of the yeast (Saccharomyces cerevisiae) α factor, was amplified by transforming Escherichia coli DH5α [genotype: F- φ80lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk−, mk+) phoA supE44 thi-1 gyrA96 relA1 λ-, provided by Invitrogen] competent cells. Transformants were selected on low-salt Luria-Bertani plates containing 25 μg/mL zeocin (Duchefa Biochemie) and analyzed by direct PCR amplification using specific primers of the α-factor signal peptide and the 3′-Alcohol Oxidase terminator sequence according to the manufacturer’s instruction. One PCR-positive colony was picked with a sterile tip and used to inoculate 5 mL of low-salt Luria-Bertani medium (1% tryptone [Duchefa Biochemie], 0.5% yeast extract [Duchefa Biochemie], 0.5% NaCl [Carlo Erba Ragenti], pH 7.4) containing 25 μg/mL zeocin. The culture was grown overnight at 37°C at 250 rpm. Plasmid DNA was extracted from the cells using a plasmid mini prep kit (Macherey & Nagel) and analyzed by digestion with EcoRI and XbaI restriction enzymes, followed by 1% agarose gel analysis. The plasmid DNA was linearized with AvrII restriction enzyme and used to transform Pichia pastoris X33 (wild-type strain provided by Invitrogen) by electroporation. The selection of the zeocin-resistant P. pastoris transformants was carried out according to the manufacturer’s instruction.
The medium used for growth of P. pastoris contained yeast extract (1%), tryptone (1%), and Glc (2%). The filtrate obtained from 3-d-old cultures was concentrated using a Vivaflow 200 (Sartorius Stedim) and dialyzed against 20 mm sodium (Na) acetate pH 4.0. The dialyzed proteins were mixed with a suspension of diethylaminoethyl cellulose (DE52, Whatman) equilibrated with 20 mm Na acetate pH 4.0. The not-absorbed proteins were loaded on a HiTrap S-sepharose column (GE Healthcare), preequilibrated with 20 mm Na acetate pH 4.0. Eluition was achieved using a linear gradient of NaCl (0 to 1 m) in 20 mm Na acetate pH 4.0. The fractions that showed the highest PG activity were pooled and dialyzed against 20 mm Na acetate pH 4.0. Ammonium sulfate was added to the dialyzed proteins to reach 2 m final concentration. The sample was loaded on a HiTrap phenyl-sepharose column (GE Healthcare) preequilibrated with 20 mm Na acetate pH 4.0 and ammonium sulfate 2 m. Elution was achieved by progressively diluting the ammonium sulfate amount (2 m to 0 in 10 min) in 20 mm Na acetate pH 4.6. The fractions that showed the highest PG activity were pooled and concentrated to a final concentration of 25 mg/mL.
PvPGIP2 Expression and Purification
Wild-type PvPGIP2 was recovered from Potato virus X-infected Nicotiana benthamiana plants and purified as previously described (Di Matteo et al., 2003). In terms of activity toward different PGs, PvPGIP2 expressed in N. benthamiana is comparable to that expressed in P. pastoris as reported below.
The PvPGIP2 gene was cloned in pGAPZαA using the EcoRI and XbaI restriction enzymes introduced by using primers EcoFw (5′-atcgatgaattcgagctatgcaacccacaa-3′) and XbaRv (5′-ggatgtctagattaagtgcaggcaggaag-3′) to generate a construct with the signal sequence of PGIP2 replaced by the yeast α-factor signal sequence for secretion. Mutagenesis of the gene was performed using the quick change site-directed mutagenesis kit (Stratagene). PCR was carried out directly on pGAPZαA/PGIP2 plasmid using internal overlap primers that hybridize at the site of the desired mutation and containing the appropriate mismatched bases. All mutations were checked by sequencing analysis.
Expression of mutated PGIPs was achieved by transforming P. pastoris with the appropriate plasmids and growing the transformants in yeast extract (1%), tryptone (1%), and Glc (2%). The filtrate obtained from 3-d-old cultures was concentrated using a Vivaflow 200 and dialyzed against 20 mm Na acetate pH 4.6. The dialyzed proteins were mixed with a suspension of diethylaminoethyl cellulose equilibrated with 20 mm Na acetate pH 4.6. The not-absorbed proteins were passed on a column HiTrap S-sepharose preequilibrated with 20 mm Na acetate pH 4.6. Elution was obtained using a linear gradient of NaCl (0–1 m in 10 min) in 20 mm Na acetate pH 4.6. The fractions that showed the highest inhibitory activity were subjected to SDS-PAGE and quantified by Blue Coomassie staining.
The capability of wild-type and mutant PvPGIPs to inhibit FpPG activity was measured by means of the agar diffusion assay as previously described (Ferrari et al., 2003). PG activity was expressed as agarose diffusion units and one unit was defined as the amount of enzyme that produced a halo of 0.5-cm radius (external to the inoculation well) after 16 h at 30°C. Each experiment was performed at least three times. In the enzymatic assays, data are reported as means. Statistical significance was calculated using the Student’s t test. P < 0.003 were considered significant.
PvPGIP2-FpPG Chemical Cross-Linking
Seventy micrograms of PvPGIP2 purified from N. benthamiana were cross-linked to 78 μg of FpPG (molar ratio 1:1) in 200 μL of a solution containing 50 mm Na-acetate pH 4.6 supplied with fresh 1% methanol-free formaldehyde (Thermo-Fisher Scientific). The reaction was incubated at T = 28°C for 16 h and finally concentrated to 2 μg of total proteins/μL. The single proteins used as negative control were cross-linked using the same reaction conditions. Cross-linked proteins and negative controls were analyzed by SDS-PAGE.
SAXS Measurements
SAXS measurements were carried out in thermostatted (25.0°C ± 0.1°C) quartz capillary of 1-mm diameter by using a Kratky Compact camera, containing a slit collimation system, equipped with a NaI scintillation counter. Nickel-filtered copper Kα radiation (λ = 1.5418 Å) was used. Scattering curves were recorded within the range of 0.01 ≤ q ≤ 0.5 Å−1 (q = 4πsinθ/λ, where 2θ is the scattering angle). The moving slit method was employed to measure the intensity of the primary beam. The collimated scattering intensities were put on an absolute scale, subtracted for the solvent and the capillary contributions, and then expressed in electron units, eu (electrons2 Å−3) per centimeter primary-beam length (Stabinger and Kratky, 1978; Glatter and Kratky, 1982). In terms of total scattering cross section of an ensemble of particles, 1 eu corresponds to 7.94056·10−2 cm−1 (Orthaber et al., 2000).
Each SAXS measurement was obtained averaging three consecutive runs. The superimposition of the patterns allowed us to exclude a successive formation of oligomer or a damage of the sample due to the x-ray radiation.
The Indirect Fourier transform method developed in the ITP program was used to interpret the spectra (Glatter, 1977). For very dilute samples (no particle interactions) the scattered intensity, I(q), can be related to the pair distribution function p(r) of the single scattering particle according to the equation
 |
On the basis of this equation, the ITP program allows the extraction of the p(r) function from the desmeared scattering pattern. The p(r) function is strongly dependent on the shape and size of the scattering particles and vanishes at the maximum particle size Dmax. Furthermore, it permits the determination of the electronic radius of gyration Rg (Glatter, 1977). The obtained values are more accurate than those derived from the Guinier approximation (Guinier and Fournet, 1955).
Analysis of SAXS Data
The methods employed in the shape reconstruction can be divided into ab initio methods and rigid body modeling. Three-dimensional envelopes that define the molecular shape of the macromolecule in solution to better than 15 Å resolution can be obtained.
The ab initio method as employed in the GA_STRUCT program was used to analyze our data (Heller et al., 2003). Starting from an aggregate of spheres, related to the expected volume and the Dmax of the scattering particle, the p(r) is calculated by means of a Monte Carlo method. A fitting parameter is determined from the calculated (Fourier transform) I(q) and the experimental one. A linear minimization is performed using a genetic algorithm that improves 50 models by means of mating, mutation, and extinction operations. At the end, all models are docked and on the basis of a docking score, from the 70% of models with the highest total docking score a consensus envelope is constructed.
Analysis of the SAXS intensity profiles was also based on rigid body modeling, taking advantage of the high-resolution structures of the single proteins (Federici et al., 2001; Di Matteo et al., 2003). A simulated annealing protocol within the program SASREF (Petoukhov and Svergun, 2005) was employed to construct an interconnected ensemble of subunits without steric clashes, while minimizing the discrepancy between the experimental scattering data and the curves calculated from the appropriate subunits assemblies. In this procedure, the scattering patterns I(q) of each protein were calculated with the program CRYSOL (Svergun et al., 1995), starting from the high-resolution crystal structures.
The models constructed by a rigid body refinement, although built from high-resolution protein structure, are still low-resolution models. A potential limitation of the technique is the possibility of obtaining multiple solutions compatible with the experimental data. In some cases, multiple runs of the programs and ranking of the models according to their biological relevance are indispensable for the cross validation of the results.
For this reason seven different SASREF reconstructions were analyzed by the DAMAVER program (Volkov and Svergun, 2003) and the various calculated complex models were superimposed by the SUPCOMB program (Kozin and Svergun, 2001). The program gives an NSD value that certifies the goodness overlap and is used as a parameter to determine the difference between two three-dimensional objects.
For each reconstruction, the average value NSDk with respect to the other models in the set was computed and the reconstruction with lowest NSDk was selected as reference. From the cross-correlation NSD table a mean value over all pairs < NSD > and dispersion Δ(NSD) were calculated.
Possible outliers with NSDk exceeding < NSD > + 2Δ(NSD) must be discarded. All seven models obtained by the SASREF analysis and subsequently subjected to the DAMAVER program, passed the test. This represents a further confirmation of the strict similarity among the SASREF structures, which were extracted without any bias.
The models were superimposed onto the reference one using SUPCOMB and the entire assembly of structures was remapped onto a densely packed grid of beads where each grid point was characterized by its occupancy factor (the number of beads in the entire assembly that are in the vicinity of the grid point). The grid points with higher occupancies were selected to yield the volume equal to the average excluded volume of all reconstructions.
Protein Data Bank files from this article can be found in the GenBank/EMBL data libraries under accession numbers 1HG8_A (FpPG) and 1OGQ (PvPGIP2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental File S1. A Protein Data Bank file with the coordinates of the reference reconstruction.
References
- Bent AF, Mackey D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Bernadó P, Blanchard L, Timmins P, Marion D, Ruigrok RWH, Blackledge M. (2005) A structural model for unfolded proteins from residual dipolar couplings and small-angle x-ray scattering. Proc Natl Acad Sci USA 102: 17002–17007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bonivento D, Pontiggia D, Di Matteo A, Fernandez-Recio J, Salvi G, Tsernoglou D, Cervone F, Lorenzo GD, Federici L. (2008) Crystal structure of the endopolygalacturonase from the phytopathogenic fungus Colletotrichum lupini and its interaction with polygalacturonase-inhibiting proteins. Proteins 70: 294–299 [DOI] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G. (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasoli M, Federici L, Spinelli F, Di Matteo A, Vella N, Scaloni F, Fernandez-Recio J, Cervone F, De Lorenzo G. (2009) Integration of evolutionary and desolvation energy analysis identifies functional sites in a plant immunity protein. Proc Natl Acad Sci USA 106: 7666–7671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacón P, Díaz JF, Morán F, Andreu JM. (2000) Reconstruction of protein form with x-ray solution scattering and a genetic algorithm. J Mol Biol 299: 1289–1302 [DOI] [PubMed] [Google Scholar]
- D’Ovidio R, Raiola A, Capodicasa C, Devoto A, Pontiggia D, Roberti S, Galletti R, Conti E, O’Sullivan D, De Lorenzo G. (2004) Characterization of the complex locus of bean encoding polygalacturonase-inhibiting proteins reveals subfunctionalization for defense against fungi and insects. Plant Physiol 135: 2424–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzo G, Brutus A, Savatin DV, Sicilia F, Cervone F. (2011) Engineering plant resistance by constructing chimeric receptors that recognize damage-associated molecular patterns (DAMPs). FEBS Lett 585: 1521–1528 [DOI] [PubMed] [Google Scholar]
- De Lorenzo G, D’Ovidio R, Cervone F. (2001) The role of polygalacturonase-inhibiting proteins (PGIPs) in defense against pathogenic fungi. Annu Rev Phytopathol 39: 313–335 [DOI] [PubMed] [Google Scholar]
- Di Matteo A, Federici L, Mattei B, Salvi G, Johnson KA, Savino C, De Lorenzo G, Tsernoglou D, Cervone F. (2003) The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein involved in plant defense. Proc Natl Acad Sci USA 100: 10124–10128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Matteo A, Giovane A, Raiola A, Camardella L, Bonivento D, De Lorenzo G, Cervone F, Bellincampi D, Tsernoglou D. (2005) Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell 17: 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federici L, Caprari C, Mattei B, Savino C, Di Matteo A, De Lorenzo G, Cervone F, Tsernoglou D. (2001) Structural requirements of endopolygalacturonase for the interaction with PGIP (polygalacturonase-inhibiting protein). Proc Natl Acad Sci USA 98: 13425–13430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federici L, Di Matteo A, Fernandez-Recio J, Tsernoglou D, Cervone F. (2006) Polygalacturonase inhibiting proteins: players in plant innate immunity? Trends Plant Sci 11: 65–70 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Denoux C, De Lorenzo G, Ausubel FM, Dewdney J. (2007) Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol 144: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Vairo D, Cervone F, De Lorenzo G. (2006) Antisense expression of the Arabidopsis thaliana AtPGIP1 gene reduces polygalacturonase-inhibiting protein accumulation and enhances susceptibility to Botrytis cinerea. Mol Plant Microbe Interact 19: 931–936 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Vairo D, Ausubel FM, Cervone F, De Lorenzo G. (2003) Tandemly duplicated Arabidopsis genes that encode polygalacturonase-inhibiting proteins are regulated coordinately by different signal transduction pathways in response to fungal infection. Plant Cell 15: 93–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frediani M, Cremonini R, Salvi G, Caprari C, Desiderio A, D’Ovidio R, Cervone F, De Lorenzo G. (1993) Cytological localization of the PGIP genes in the embryo suspensor cells of Phaseolus vulgaris L. Theor Appl Genet 87: 369–373 [DOI] [PubMed] [Google Scholar]
- Galantini L, Leggio C, Konarev PV, Pavel NV. (2010) Human serum albumin binding ibuprofen: a 3D description of the unfolding pathway in urea. Biophys Chem 147: 111–122 [DOI] [PubMed] [Google Scholar]
- Galantini L, Leggio C, Pavel NV. (2008) Human serum albumin unfolding: a small-angle X-ray scattering and light scattering study. J Phys Chem B 112: 15460–15469 [DOI] [PubMed] [Google Scholar]
- Glatter O. (1977) A new method for the evaluation of small-angle scattering data. J Appl Cryst 10: 415–421 [Google Scholar]
- Glatter O, Kratky O. (1982) Small Angle X-Ray Scattering. Academic Press, London: [DOI] [PubMed] [Google Scholar]
- Guinier A, Fournet G. (1955) Small Angle X-Rays. Wiley, New York [Google Scholar]
- Heller WT, Krueger JK, Trewhella J. (2003) Further insights into calmodulin-myosin light chain kinase interaction from solution scattering and shape restoration. Biochemistry 42: 10579–10588 [DOI] [PubMed] [Google Scholar]
- Isshiki A, Akimitsu K, Yamamoto M, Yamamoto H. (2001) Endopolygalacturonase is essential for citrus black rot caused by Alternaria citri but not brown spot caused by Alternaria alternata. Mol Plant Microbe Interact 14: 749–757 [DOI] [PubMed] [Google Scholar]
- Jacobsen RB, Sale KL, Ayson MJ, Novak P, Hong JH, Lane P, Wood NL, Kruppa GH, Young MM, Schoeniger JS. (2006) Structure and dynamics of dark-state bovine rhodopsin revealed by chemical cross-linking and high-resolution mass spectrometry. Protein Sci 15: 1303–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques DA, Trewhella J. (2010) Small-angle scattering for structural biology—expanding the frontier while avoiding the pitfalls. Protein Sci 19: 642–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janni M, Sella L, Favaron F, Blechl AE, De Lorenzo G, D’Ovidio R. (2008) The expression of a bean PGIP in transgenic wheat confers increased resistance to the fungal pathogen Bipolaris sorokiniana. Mol Plant Microbe Interact 21: 171–177 [DOI] [PubMed] [Google Scholar]
- Joubert DA, Slaughter AR, Kemp G, Becker JV, Krooshof GH, Bergmann C, Benen J, Pretorius IS, Vivier MA. (2006) The grapevine polygalacturonase-inhibiting protein (VvPGIP1) reduces Botrytis cinerea susceptibility in transgenic tobacco and differentially inhibits fungal polygalacturonases. Transgenic Res 15: 687–702 [DOI] [PubMed] [Google Scholar]
- Kars I, Krooshof GH, Wagemakers L, Joosten R, Benen JAE, van Kan JAL. (2005) Necrotizing activity of five Botrytis cinerea endopolygalacturonases produced in Pichia pastoris. Plant J 43: 213–225 [DOI] [PubMed] [Google Scholar]
- King D, Bergmann C, Orlando R, Benen JA, Kester HC, Visser J. (2002) Use of amide exchange mass spectrometry to study conformational changes within the endopolygalacturonase II-homogalacturonan-polygalacturonase inhibiting protein system. Biochemistry 41: 10225–10233 [DOI] [PubMed] [Google Scholar]
- Koch MH, Vachette P, Svergun DI. (2003) Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q Rev Biophys 36: 147–227 [DOI] [PubMed] [Google Scholar]
- Komjanc M, Festi S, Rizzotti L, Cattivelli L, Cervone F, De Lorenzo G. (1999) A leucine-rich repeat receptor-like protein kinase (LRPKm1) gene is induced in Malus x domestica by Venturia inaequalis infection and salicylic acid treatment. Plant Mol Biol 40: 945–957 [DOI] [PubMed] [Google Scholar]
- Kozin MB, Svergun DI. (2001) Automated matching of high- and low-resolution structural models. J Appl Cryst 34: 33–41 [Google Scholar]
- Leckie F, Mattei B, Capodicasa C, Hemmings A, Nuss L, Aracri B, De Lorenzo G, Cervone F. (1999) The specificity of polygalacturonase-inhibiting protein (PGIP): a single amino acid substitution in the solvent-exposed beta-strand/beta-turn region of the leucine-rich repeats (LRRs) confers a new recognition capability. EMBO J 18: 2352–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggio C, Galantini L, Konarev PV, Pavel NV. (2009) Urea-induced denaturation process on defatted human serum albumin and in the presence of palmitic acid. J Phys Chem B 113: 12590–12602 [DOI] [PubMed] [Google Scholar]
- Lionetti V, Francocci F, Ferrari S, Volpi C, Bellincampi D, Galletti R, D’Ovidio R, De Lorenzo G, Cervone F. (2010) Engineering the cell wall by reducing de-methyl-esterified homogalacturonan improves saccharification of plant tissues for bioconversion. Proc Natl Acad Sci USA 107: 616–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipfert J, Doniach S. (2007) Small-angle x-ray scattering from RNA, proteins, and protein complexes. Annu Rev Biophys Biomol Struct 36: 307–327 [DOI] [PubMed] [Google Scholar]
- Manfredini C, Sicilia F, Ferrari S, Pontiggia D, Salvi G, Caprari C, Lorito M, De Lorenzo G. (2005) Polygalacturonase inhibiting protein 2 of Phaseolus vulgaris inhibits BcPG1, a polygalacturonase of Botrytis cinerea important for pathogenicity, and protects transgenic plants from infection. Physiol Mol Plant Pathol 67: 108–115 [Google Scholar]
- Mariotti L, Casasoli M, Migheli Q, Balmas V, Caprari C, De Lorenzo G. (2008) Reclassification of Fusarium verticillioides (syn. F. moniliforme) strain FC-10 as F. phyllophilum. Mycol Res 112: 1010–1011 [PubMed] [Google Scholar]
- Mertens HDT, Svergun DI. (2010) Structural characterization of proteins and complexes using small-angle x-ray solution scattering. J Struct Biol 172: 128–141 [DOI] [PubMed] [Google Scholar]
- Misas-Villamil JC, van der Hoorn RAL. (2008) Enzyme-inhibitor interactions at the plant-pathogen interface. Curr Opin Plant Biol 11: 380–388 [DOI] [PubMed] [Google Scholar]
- Nogales A, García C, Pérez J, Callow P, Ezquerra TA, González-Rodríguez J. (2010) Three-dimensional model of human platelet integrin alphaIIb beta3 in solution obtained by small angle neutron scattering. J Biol Chem 285: 1023–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orthaber D, Bergmann A, Glatter O. (2000) SAXS experiments on absolute scale with Kratky systems using water as a secondary standard. J Appl Cryst 33: 218–225 [Google Scholar]
- Payan F, Leone P, Porciero S, Furniss C, Tahir T, Williamson G, Durand A, Manzanares P, Gilbert HJ, Juge N, et al. (2004) The dual nature of the wheat xylanase protein inhibitor XIP-I: structural basis for the inhibition of family 10 and family 11 xylanases. J Biol Chem 279: 36029–36037 [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI. (2005) Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J 89: 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons C, D’Abramo M, Svergun DI, Orozco M, Bernadó P, Fernández-Recio J. (2010) Structural characterization of protein-protein complexes by integrating computational docking with small-angle scattering data. J Mol Biol 403: 217–230 [DOI] [PubMed] [Google Scholar]
- Powell AL, van Kan J, ten Have A, Visser J, Greve LC, Bennett AB, Labavitch JM. (2000) Transgenic expression of pear PGIP in tomato limits fungal colonization. Mol Plant Microbe Interact 13: 942–950 [DOI] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA. (2007) X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys 40: 191–285 [DOI] [PubMed] [Google Scholar]
- Raiola A, Camardella L, Giovane A, Mattei B, De Lorenzo G, Cervone F, Bellincampi D. (2004) Two Arabidopsis thaliana genes encode functional pectin methylesterase inhibitors. FEBS Lett 557: 199–203 [DOI] [PubMed] [Google Scholar]
- Segel DJ, Fink AL, Hodgson KO, Doniach S. (1998) Protein denaturation: a small-angle x-ray scattering study of the ensemble of unfolded states of cytochrome c. Biochemistry 37: 12443–12451 [DOI] [PubMed] [Google Scholar]
- Sicilia F, Fernandez-Recio J, Caprari C, De Lorenzo G, Tsernoglou D, Cervone F, Federici L. (2005) The polygalacturonase-inhibiting protein PGIP2 of Phaseolus vulgaris has evolved a mixed mode of inhibition of endopolygalacturonase PG1 of Botrytis cinerea. Plant Physiol 139: 1380–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev V, Eyal E, Gerzon S, Potapov V, Babor M, Prilusky J, Edelman M. (2005) SPACE: a suite of tools for protein structure prediction and analysis based on complementarity and environment. Nucleic Acids Res (Web Server issue) 33: W39–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli F, Mariotti L, Mattei B, Salvi G, Cervone F, Caprari C. (2009) Three aspartic acid residues of polygalacturonase-inhibiting protein (PGIP) from Phaseolus vulgaris are critical for inhibition of Fusarium phyllophilum PG. Plant Biol (Stuttg) 11: 738–743 [DOI] [PubMed] [Google Scholar]
- Stabinger H, Kratky O. (1978) A new technique for the measurement of the absolute intensity of x-ray small angle scattering: the moving slit method. Makromol Chem 179: 1655–1659 [Google Scholar]
- Svergun DI. (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J 76: 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Barberato C, Koch MH. (1995) CRYSOL—a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst 28: 768–773 [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MH. (2001) Determination of domain structure of proteins from x-ray solution scattering. Biophys J 80: 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabarani G, Thépaut M, Stroebel D, Ebel C, Vivès C, Vachette P, Durand D, Fieschi F. (2009) DC-SIGN neck domain is a pH-sensor controlling oligomerization: SAXS and hydrodynamic studies of extracellular domain. J Biol Chem 284: 21229–21240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J Appl Cryst 36: 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuppini A, Navazio L, Sella L, Castiglioni C, Favaron F, Mariani P. (2005) An endopolygalacturonase from Sclerotinia sclerotiorum induces calcium-mediated signaling and programmed cell death in soybean cells. Mol Plant Microbe Interact 18: 849–855 [DOI] [PubMed] [Google Scholar]