Abstract
4-Coumarate:coenzyme A ligase (4CL; EC 6.2.1.12) is a key enzyme in the phenylpropanoid metabolic pathways for monolignol and flavonoid biosynthesis. 4CL has been much studied in dicotyledons, but its function is not completely understood in monocotyledons, which display a different monolignol composition and phenylpropanoid profile. In this study, five members of the 4CL gene family in the rice (Oryza sativa) genome were cloned and analyzed. Biochemical characterization of the 4CL recombinant proteins revealed that the rice 4CL isoforms displayed different substrate specificities and catalytic turnover rates. Among them, Os4CL3 exhibited the highest turnover rate. No apparent tissue-specific expression of the five 4CLs was observed, but significant differences in their expression levels were detected. The rank in order of transcript abundance was Os4CL3 > Os4CL5 > Os4CL1 > Os4CL4 > Os4CL2. Suppression of Os4CL3 expression resulted in significant lignin reduction, shorter plant growth, and other morphological changes. The 4CL-suppressed transgenics also displayed decreased panicle fertility, which may be attributed to abnormal anther development as a result of disrupted lignin synthesis. This study demonstrates that the rice 4CLs exhibit different in vitro catalytic properties from those in dicots and that 4CL-mediated metabolism in vivo may play important roles in regulating a broad range of biological events over the course of rice growth and development.
In phenylpropanoid metabolism, 4-coumarate:coenzyme A ligase (4CL) mediates the activation of a number of hydroxycinnamic acids for the biosynthesis of monolignols and other phenolic secondary metabolites in higher plants (Lozoya et al., 1988; Allina et al., 1998; Hu et al., 1998; Ehlting et al., 1999; Lindermayr et al., 2002; Hamberger and Hahlbrock, 2004). The 4CL genes exist in plants as a family with multiple members. The 4CL family has four members in Arabidopsis (Arabidopsis thaliana; Ehlting et al., 1999), five members in rice (Oryza sativa), and four members in the moss Physcomitrella patens (Silber et al., 2008). In the aspen (Populus trichocarpa) genome, 17 genes are found to share sequence similarity with known 4CLs (Souza et al., 2008; Shi et al., 2010). Of these, five genes are classified as 4CL genes and the rest are classified as 4CL-like genes due to their different structures (Souza et al., 2008).
In gymnosperms and dicotyledonous angiosperms, the catalytic properties of 4CL isoforms have been studied extensively. 4CL isoforms with different substrate affinities may direct metabolic flux through different pathways to synthesize a variety of phenolic compounds, such as different monolignols, flavonoids, isoflavonoids, coumarins, suberin, and wall-bound phenolics (Boerjan et al., 2003; Naoumkina et al., 2010). Differences in substrate specificity can be attributed to variations in the sequences of 4CL isoforms or to posttranslational modification. Protein structure analysis revealed that both hydrophobicity and the size of the 4CL substrate-binding pocket play key roles in regulating substrate accessibility (Stuible et al., 2000; Stuible and Kombrink, 2001; Schneider et al., 2003; Hu et al., 2010).
4CLs can be classified into two distinct groups in dicots, type I and type II (Hu et al., 1998; Ehlting et al., 1999). Disruption of 4CL expression in vivo has demonstrated that type I 4CLs in dicots play a crucial role in regulating lignin accumulation while type II 4CLs impact the metabolism of other phenolic compounds (Kajita et al., 1996, 1997; Lee et al., 1997; Hu et al., 1998; Li et al., 2003a; Wagner et al., 2009). In aspen, for example, Pt4CL1, which is a type I 4CL, is involved in monolignol formation, while Pt4CL2, a type II 4CL, is involved in the synthesis of other phenolic compounds (Hu et al., 1998; Harding et al., 2002). Generally, the same type of 4CLs across different species of dicots share a high degree of sequence identity and can be grouped together into the same phylogenetic clade.
Monocotyledons contain different monolignol compositions and different wall-bound phenolic compounds compared with dicots (Hatfield et al., 2008, 2009). The genome of rice, a model species for monocots, contains five 4CL genes. However, the enzymatic properties and functions of 4CLs and how they catalyze various hydroxycinnamyl reactions in rice have been little studied. This paper presents a characterization of the 4CL gene family in japonica rice and shows that 4CLs play a role in regulating a broad range of biological events throughout the life cycle of rice.
RESULTS
Analysis of 4CLs in the Rice Genome
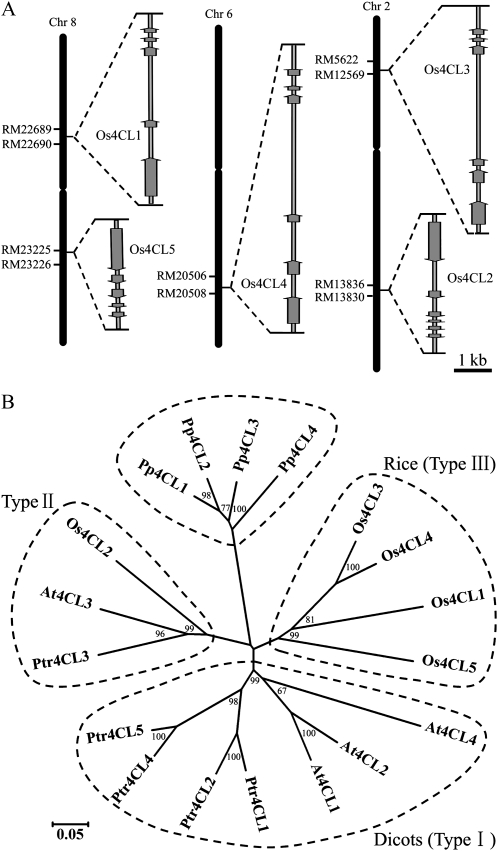
Five 4CLs are known in the rice genome (http://rice.plantbiology.msu.edu/). In accordance with the rice 4CL gene numbering system adopted in a previous study (Souza et al., 2008), these genes were named Os4CL1, Os4CL2, Os4CL3, Os4CL4, and Os4CL5. The five 4CLs are located on three different chromosomes (Fig. 1A). Expression of these five genes was confirmed by the isolation of their corresponding cDNAs from various tissues. The five cDNAs, found to be identical to their sequences in the rice genome database, were predicted to code for a family of proteins that includes Os4CL1 with 565 amino acids, Os4CL2 with 570 amino acids, Os4CL3 with 555 amino acids, Os4CL4 with 560 amino acids, and Os4CL5 with 540 amino acids (Supplemental Fig. S1). The identity between the 4CL protein sequences varied from 58% to 85% (Supplemental Table S1). Among them, 4CL4 and 4CL3 are the most similar to each other in sequence, with an identity of 85%. Phylogenetic analysis revealed that the 4CL genes from dicots, rice, and moss form separate clades (Fig. 1B). 4CLs in dicots can be divided into type I and type II. In rice, Os4CL2 belongs to the same clade as type II 4CLs in dicots, while the rest of the 4CLs (Os4CL1/3/4/5) belong to a separate cluster, hereafter named type III, which is distinct from the lignin-associated type I 4CLs found in dicots.
Figure 1.
Gene structure and phylogenetic analysis of the rice 4CL family. A, Schematic structure of rice 4CL genes on chromosomes. Black bars represent the chromosomes. Exons are represented as dark gray arrows, and introns are indicated by light gray lines between exons. The lengths of exons and introns are indicated by the scale bar. The five 4CL gene loci are as follows: Os4CL1, Os08g14760; Os4CL2, Os02g46970; Os4CL3, Os02g08100; Os4CL4, Os06g44620; Os4CL5, Os08g34790. B, Unrooted phylogenetic tree of 4CL isoforms from four representative species was constructed with bootstrap values after 1,000 trials. The scale bar corresponds to 0.05 amino acid substitutions per position in the sequence. 4CLs from dicot species were clustered in two clades, classified as type I and type II 4CLs. Four 4CLs from rice and the moss P. patens are grouped in separate clusters. Plant species are as follows: Os, Oryza sativa; At, Arabidopsis thaliana; Ptr, Populus trichocarpa; Pp, Physcomitrella patens.
Catalytic Properties of Rice 4CLs
As the sequences of the five rice 4CLs are rather different, they may display distinct enzymatic properties toward different hydroxycinnamate substrates. Thus, we produced recombinant proteins of the 4CLs to characterize their catalytic properties. Five hydroxycinnamic acid derivatives, cinnamate, 4-coumarate, caffeate, ferulate, and sinapate, were used to analyze the enzymatic activity of the 4CLs. The substrate preferences and enzyme turnover rates of the rice 4CL isoforms are summarized in Table I. Os4CL1 displayed a very low turnover rate. Os4CL2 was found to show a strong preference for ferulate, while Os4CL3 and Os4CL4, whose sequences are the most similar, showed a high affinity for 4-coumarate and ferulate. Os4CL5 clearly preferred 4-coumarate and ferulate while exhibiting low activity toward other hydroxycinnamic acid substrates. Os4CL5 was also able to convert sinapate to its corresponding CoA ester, a reaction that is rarely observed in 4CL catalysis. Overall, ferulate was a major substrate used by all five rice 4CLs. In addition to different substrate affinities and specificities (expressed in Km and Kenz), substantial differences in the catalytic turnover rates of rice 4CLs (expressed in Kcat) were also detected (Table I). Os4CL3 had the highest turnover rate while Os4CL1 had the lowest.
Table I. Enzyme activities of recombinant rice 4CLs.
The Km and Vmax of recombinant Os4CL1, Os4CL2, Os4CL3, Os4CL4, and Os4CL5 proteins were determined from a Lineweaver-Burk plot. The values represent means and se of three replicates. NC, No conversion.
| Enzyme | Substrate | Km | Vmax | Kcat | Kenz = Kcat/Km |
| μm | pkat mg−1 | min−1 | m−1 s−1 | ||
| Os4CL1 | Cinnamate | 9.4 ± 0.8 | 100 ± 8 | 0.37 | 663 |
| 4-Coumarate | 11.9 ± 0.2 | 130 ± 1 | 0.49 | 681 | |
| Caffeate | 29.3 ± 6.5 | 140 ± 31 | 0.52 | 296 | |
| Ferulate | 8.3 ± 0.4 | 110 ± 5 | 0.41 | 824 | |
| Sinapate | NC | NC | NC | NC | |
| Os4CL2 | Cinnamate | 21.7 ± 1.1 | 299 ± 15 | 1.13 | 867 |
| 4-Coumarate | 16.8 ± 0.7 | 629 ± 25 | 2.37 | 2,351 | |
| Caffeate | 27.6 ± 0.9 | 708 ± 29 | 2.67 | 1,612 | |
| Ferulate | 2.2 ± 0.1 | 613 ± 22 | 2.31 | 17,295 | |
| Sinapate | NC | NC | NC | NC | |
| Os4CL3 | Cinnamate | 28.2 ± 1.2 | 3,010 ± 132 | 10.92 | 6,458 |
| 4-Coumarate | 4.9 ± 0.5 | 4,700 ± 470 | 17.10 | 58,521 | |
| Caffeate | 10.9 ± 2.1 | 4,210 ± 813 | 15.30 | 23,330 | |
| Ferulate | 3.5 ± 0.2 | 4,930 ± 239 | 17.94 | 84,943 | |
| Sinapate | NC | NC | NC | NC | |
| Os4CL4 | Cinnamate | 15.7 ± 0.8 | 350 ± 18 | 1.28 | 1,361 |
| 4-Coumarate | 3.9 ± 0.3 | 770 ± 50 | 2.82 | 12,176 | |
| Caffeate | 5.8 ± 0.8 | 590 ± 82 | 2.16 | 6,206 | |
| Ferulate | 4.6 ± 0.6 | 520 ± 71 | 1.92 | 6,956 | |
| Sinapate | NC | NC | NC | NC | |
| Os4CL5 | Cinnamate | 54.4 ± 0.8 | 300 ± 4 | 1.02 | 312 |
| 4-Coumarate | 10.3 ± 1.5 | 830 ± 120 | 2.88 | 4,660 | |
| Caffeate | 26.1 ± 0.9 | 240 ± 8 | 0.84 | 535 | |
| Ferulate | 6.9 ± 0.1 | 590 ± 1 | 2.04 | 4,871 | |
| Sinapate | 58.9 ± 5.9 | 920 ± 90 | 3.18 | 899 |
In dicots, 4CLs usually display high activity toward 4-coumarate and caffeate but little or no activity toward ferulate (Hu et al., 1998; Ehlting et al., 1999; Lindermayr et al., 2002; Hamberger and Hahlbrock, 2004). The observed differences in catalytic properties between 4CLs in monocots and dicots may be the result of sequence variations in the region of the substrate-binding pocket. Crystal structure analysis identified Lys-303 as one of the key residues responsible for determining substrate specificity in the binding pocket of 4CL1 in Populus tomentosa (Hu et al., 2010). In contrast, the corresponding residue in the sequences of the five rice 4CLs is Ile, Leu, Met, Leu, and Met, respectively (Supplemental Fig. S1), rather different from the Lys residue at that position.
Expression Profile of 4CLs in Rice
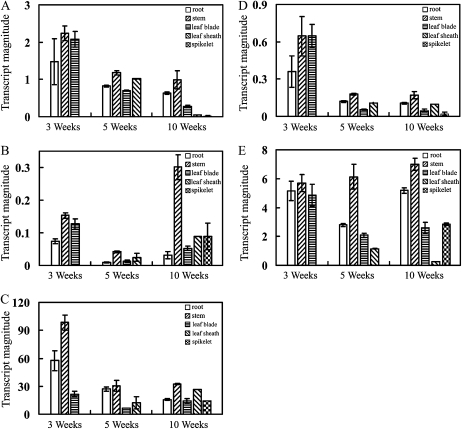
To further investigate how multiple 4CLs are involved in the development of various tissues, transcripts from the five 4CLs were quantitatively measured by real-time PCR using gene-specific primers (Supplemental Table S2). Total RNA was isolated from the roots, stems, leaf blades, leaf sheaths, and young panicles of rice plants at the age of 3 weeks, 5 weeks, and 10 weeks. Transcripts from the five rice 4CLs were detected across the range of collected tissues (Fig. 2) but with significant differences in their magnitude of expression. The Os4CL3 transcript was the most abundantly expressed, followed by Os4CL5 and Os4CL1, while the Os4CL2 transcript was the least abundant. These results indicated that all five 4CLs are expressed at quantitatively different levels over the course of plant growth but without any apparent tissue specificity.
Figure 2.
Expression profiles of rice 4CLs over the course of growth. Total RNAs were isolated from roots, culms, leaf blades, leaf sheaths, and panicles of rice plants at 3 weeks, 5 weeks, and 10 weeks old. Transcripts of rice 4CLs were determined using quantitative reverse transcription-PCR analysis. The rice ACTIN1 gene was used as a reference for normalization. A, Os4CL1. B, Os4CL2. C, Os4CL3. D, Os4CL4. E, Os4CL5. The results are means ± se of independent triplicate assays.
Localization of Os4CL3 Expression in Rice
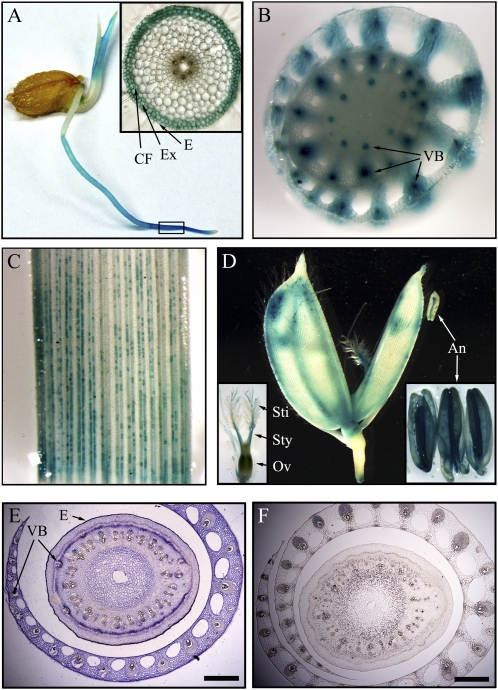
Os4CL3 had the fastest turnover rate in enzymatic catalysis and was also the most strongly expressed of the rice 4CLs during growth. To examine the expression of Os4CL3 in more detail in relation to specific cell types, an analysis of promoter activity and in situ localization was carried out. The promoter region of the Os4CL3 gene was cloned and the promoter-GUS fusion was transferred into rice plants to analyze promoter activity. Promoter-GUS activity was examined in the stem, root, leaf, and flower of the transgenic plants (Fig. 3). Os4CL3 promoter activity was detected in the exodermis and epidermis cells of root (Fig. 3A) and stem vascular cells (Fig. 3B). In leaves, activity was detected in developing vascular bundle cells as well as in parenchyma cells (Fig. 3C). In flowers, the promoter was active in the lemma, palea, stamens, and pistil (Fig. 3D).
Figure 3.
Activity determination of the Os4CL3 promoter and in situ localization of Os4CL3 expression. Localization of Os4CL3 expression was investigated through promoter-GUS assays and in situ hybridization. A, GUS staining of the promoter-GUS activity in a seedling. The inset shows a cross section of the root area indicated in the box. B, GUS staining of a cross section of stem. C, GUS staining of a leaf blade. D, GUS staining of a flower. The insets show pistils and anthers. E and F, In situ hybridization of stem cross sections (6-week-old plants) with an Os4CL3-specifc antisense probe (E) and with an Os4CL3-specific sense probe (F). An, Anther; CF, cortical fiber; E, epidermis; Ex, exodermis; Ov, ovary; Sti, stigma; Sty, style; VB, vascular bundle. Bars = 500 μm.
Cell type expression of rice Os4CL3 was further confirmed by in situ localization. Rice stems (6 weeks old) were sectioned and hybridized with specific Os4CL3 probes. Os4CL3 expression was detected in developing vascular bundle cells, which were undergoing strong lignification, as well as stem epidermis cells and sheath parenchyma cells, in which no lignin was being synthesized (Fig. 3E). Lignin is typically synthesized in the thickened secondary wall. Os4CL3 was found to be expressed in thickening vascular cells and nonthickening parenchyma cells throughout rice growth. This result suggests that depending on the cell type, Os4CL3 may play a role in the synthesis of lignin as well as other phenolic compounds.
Morphological Characteristics of Os4CL3-Suppressed Plants
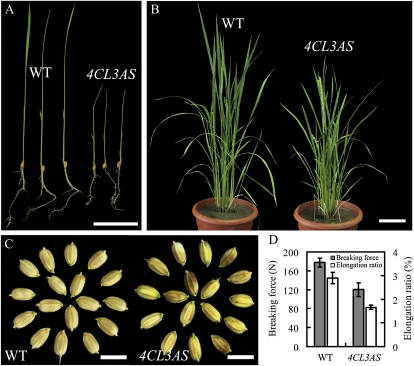
We transformed rice with the Os4CL3 antisense gene in order to suppress its expression. A significantly lower rate of plantlet induction (approximately 2%) from the transformed calli was observed compared with the typical rate (usually approximately 18% in our laboratory) of other gene transformations. Many of the first generation of Os4CL3 antisense transgenic lines also showed reduced plant height. About 5% of the plants were severely dwarfed or unable to undergo normal booting.
A total of 10 independent transgenic rice lines were selected and grown individually until the third generation (T3) to obtain homozygous transgenic plants. The homozygous transgenic plants were characterized for phenotypic changes at all stages of growth and development (Table II). At the 10-d seedling stage, both the root length and height of transgenic seedlings was reduced by approximately 50%. At maturity, the height of transgenic plants also showed significant differences compared with the wild type. Overall, antisense transgenics were shorter than wild-type plants (Fig. 4, A and B). However, a number of other agronomic traits, such as heading date, tiller number, blade length of flag leaf, blade width of flag leaf, and leaf index of flag leaf, in the transgenics were unchanged. Tensile strength, expressed either as breaking force or elongation ratio, was also significantly reduced in Os4CL3-suppressed plants (Fig. 4D). This change likely reflects alterations to the cell wall as a result of reduced lignification.
Table II. Morphologic analysis of the Os4CL3-suppressed transgenic rice.
Values are means ± se of 10 biological replicates. The second leaf was the first one under the flag leaf. Leaf index represents the ratio of leaf blade length to leaf blade width. Single asterisk indicates that the difference between the wild-type and transgenic lines is statistically significant at P < 0.05 by t test. Double asterisks indicate that the difference between the wild-type and transgenic lines is statistically significant at P < 0.01 by t test. DAS, Day after seeding.
| Trait | Wild Type | 4CL3AS |
| Plant height (cm) | 82.0 ± 6.4 | 65.5 ± 8.5** |
| Heading date (DAS) | 73.6 ± 3.0 | 73.2 ± 5.1 |
| Tiller no. | 9.5 ± 0.76 | 9.7 ± 1.0 |
| Flag leaf length (cm) | 23.3 ± 3.7 | 22.3 ± 4.7 |
| Flag leaf width (cm) | 1.18 ± 0.09 | 1.11 ± 0.03 |
| Leaf index of flag leaf | 21.5 ± 2.46 | 22.1 ± 4.9 |
| Second leaf length (cm) | 40.2 ± 5.8 | 41.5 ± 6.7 |
| Second leaf width (cm) | 1.01 ± 0.06 | 1.0 ± 0.06 |
| Leaf index of second leaf | 41.5 ± 3.7 | 42.9 ± 5.3 |
| Panicle length (cm) | 17.1 ± 0.42 | 16.1 ± 0.59 |
| Grain length (mm) | 7.01 ± 0.24 | 7.03 ± 0.26 |
| Grain width (mm) | 3.48 ± 0.35 | 3.49 ± 0.32 |
| 1,000-grain weight (g) | 23.9 ± 0.4 | 24.3 ± 0.4* |
| Fertility rate (%) | 95.3 ± 3.2 | 75.6 ± 6.7** |
Figure 4.
Phenotype of the Os4CL3-suppressed transgenic rice. Wild-type (WT) and Os4CL3-suppressed transgenic (Os4CL3AS) rice were grown in a phytotron. Morphology was systematically recorded from seedling to mature stages. A, Ten-day-old plants. B, Seventy-day-old plants. C, Mature rice grain. D, Measurement of the tensile strength of a rice stem. N represents the mechanical force unit in Newton. Error bars represent se of triple sample measurements (P < 0.01, by t test). Bars = 50 mm in A, 10 cm in B, and 7 mm in C.
Analysis of Lignin in Os4CL3-Suppressed Rice
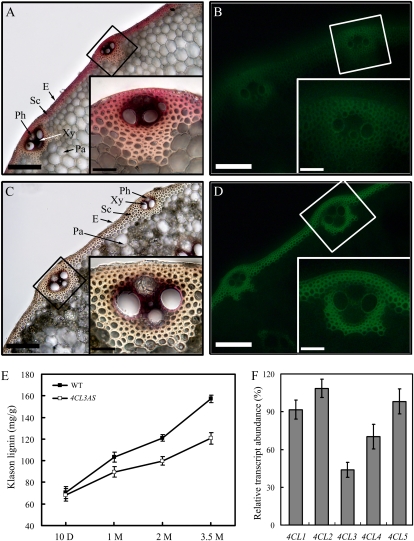
Previous studies in dicots have demonstrated that suppression of 4CL expression results in lower lignin content in cell walls (Kajita et al., 1996; Lee et al., 1997; Hu et al., 1999; Li et al., 2003a). To examine the cell wall in Os4CL3-suppressed transgenic rice, cross sections of culms were histochemically stained for lignin and cellulose. An overall reduction in lignin and increase in cellulose expression was observed in the walls of sclerenchyma cells (Fig. 5, A–D). The lignin content of the transgenic plants was also closely examined at different stages of plant growth. At 10 d, lignin content was low in both transgenic and wild-type plants. Lignin content gradually increased from 10-d-old seedlings to mature plants (about 100 d) in the wild type. The rate of increase was much slower in Os4CL3-suppressed transgenics (Fig. 5E) during growth, overall resulting in plants with lower lignin content.
Figure 5.
Lignin analysis and 4CL expression in transgenic rice. Rice culms at 3 months of age were sectioned and stained for lignin with phloroglucinol-HCl and for cellulose with calcofluor. A, Lignin staining in wild-type rice. B, Cellulose staining in wild-type rice. C, Lignin staining in Os4CL3AS transgenics. D, Cellulose staining in Os4CL3AS transgenics. Insets show closeup images of the vascular cells indicated in each box. Bars = 100 μm, and bars in insets = 50 μm. E, Klason lignin content in the course of growth from 10 d to 3.5 months. F, Transcripts from five 4CLs were measured in wild-type and Os4CL3AS transgenic plants at 2 weeks of age. After normalization using ACTIN1 as a reference, relative transcript abundance in the Os4CL3AS transgenics was calculated as a percentage of that in the wild type. Error bars represent se of three independent replicates. E, Epidermis; Pa, parenchyma cells; Ph, phloem; Sc, sclerenchyma cells; WT, wild type; Xy, xylem.
The link between lignin reduction and 4CL expression was further established by examining the expression of all five 4CL genes in Os4CL3-suppressed transgenics. Expression of the endogenous Os4CL3 in 2-week-old transgenics was approximately 50% of that in the wild type (Fig. 5F). Expression of Os4CL4 was also suppressed by about 30%, potentially because Os4CL3 and Os4CL4 share a relatively high degree of sequence identity with each other. Expression of the other 4CLs did not show significant suppression in the transgenics.
To test whether the suppression of Os4CL3 affected monolignol composition in rice, lignin was examined for the presence ρ-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) subunits (Supplemental Fig. S2; Supplemental Table S3). The three main monolignol subunits detected in wild-type rice were as follows: G units with about 70%, S units with about 20%, and H units with about 10% (Table III). A slightly different subunit ratio of approximately 75% G units, approximately 17% S units, and approximately 8% H units was detected in the lignin-reduced plants. Meanwhile, the total amount of detected monomers per Klason lignin unit in the transgenics was 963 μmol g−1 Klason lignin compared with 1,220 μmol g−1 in the wild type. The reduced thioacidolysis monomer yields, coupled with reduced S and increased G in the monomer composition, point to a more condensed lignin structure in Os4CL3-suppressed transgenic rice.
Table III. Determination of lignin monomer composition in rice plants.
The values represent means and se of three replicate determinations. Asterisks indicate that the difference of total H + G + S content between wild-type and transgenic plants is statistically significant at P < 0.01 by t test.
| Sample | Monomer Composition | Total H + G + S | Monomer Composition | Total H + G + S | Ratio of Monomer Composition | ||||||
| H | G | S | H | G | S | H | G | S | |||
| μmol g−1 dry wt | μmol g−1 Klason lignin | % | |||||||||
| Wild type | 16 ± 2 | 133 ± 10 | 38 ± 4 | 187 ± 15 | 122 ± 10 | 856 ± 64 | 242 ± 23 | 1,220 ± 94 | 10 | 70 | 20 |
| 4CL3AS | 10 ± 1 | 100 ± 10 | 22 ± 2 | 132 ± 13* | 77 ± 4 | 724 ± 65 | 162 ± 15 | 963 ± 84 | 8 | 75 | 17 |
Other Phenolic Compounds in 4CL Transgenic Rice
In addition to lignin, 4CLs are associated with the biosynthesis of other phenolic compounds. To understand how 4CL suppression disturbed the flux of phenolic intermediates in rice, we screened a group of intermediates, which were likely to be 4CL substrates, in both soluble and wall-bound forms in rice stem, leaf, booting panicles, and heading panicles. The phenolic compounds screened included 4-coumarate, caffeate, ferulate, 5-hydroxyferulate, and sinapate. In wild-type rice, 4-coumarate and ferulate were the two main compounds detected, while caffeate, 5-hydroxyferulate, and sinapate were not detected. Down-regulation of Os4CL3 clearly impacted the accumulation of 4-coumarate and ferulate bound to the cell wall and soluble inside the cell (Table IV). The amounts of both wall-bound and soluble 4-coumarate and ferulate were much higher in stem, leaf, booting panicles, and heading panicles of the transgenic plants, indicating that down-regulation of Os4CL3 has a significant impact on the metabolic routes of 4-coumarate and ferulate, which could be the two major hydroxycinnamates used by rice 4CLs in vivo.
Table IV. Content of free and wall-bound phenolics in various rice tissues.
The values represent means and se of three replicates.
| Phenolics | 4-Coumarate | Ferulate | |||||||
| Stems | Leaves | Booting Panicles | Heading Panicles | Stems | Leaves | Booting Panicles | Heading Panicles | ||
| μg g−1 dry wt | |||||||||
| Free | Wild type | 66 ± 7 | 52 ± 6 | 14 ± 1 | 16 ± 2 | 182 ± 19 | 162 ± 17 | 33 ± 2 | 37 ± 4 |
| 4CL3AS | 73 ± 8 | 74 ± 7 | 22 ± 3 | 27 ± 4 | 223 ± 24 | 234 ± 22 | 43 ± 3 | 59 ± 5 | |
| Wall bound | Wild type | 505 ± 55 | 380 ± 39 | 168 ± 17 | 180 ± 16 | 857 ± 88 | 870 ± 86 | 544 ± 43 | 553 ± 43 |
| 4CL3AS | 569 ± 52 | 547 ± 43 | 201 ± 20 | 214 ± 19 | 1,019 ± 81 | 1,199 ± 97 | 596 ± 51 | 601 ± 58 | |
Abnormal Anther Development in Os4CL3-Suppressed Rice
The fertility rate of the Os4CL3-suppressed transgenics was significantly reduced to about 75% compared with 95% in the control. The rice grain of the transgenics was also dotted with colored spots (Fig. 4C), although the panicle length, seed length, seed width, and 1,000-grain weight were not impacted. These results suggested that 4CL-mediated metabolism may play a significant role in seed formation in rice.
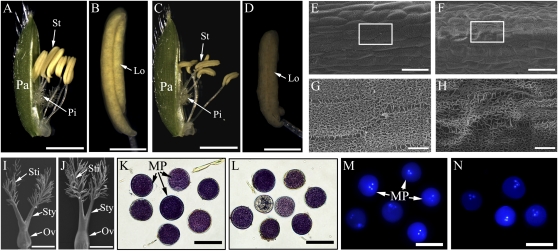
A systematic examination of the morphology of the flowering and reproductive structures in T3 homozygous transgenic plants led to the following observations (Fig. 6). First, microscopic analysis revealed that the anthers in the transgenic plants were shorter in length (Fig. 6, A–D). Second, the surface of the anther was wrinkled and contained larger pores (Fig. 6, E–H). However, no distinguishable differences were observed in the morphology of the stigma, style, and ovary between transgenic and wild-type plants (Fig. 6, I and J). The viability of pollen from the Os4CL3-suppressed plants was also not affected (Fig. 6, K–N). These results suggest that Os4CL3 suppression may affect fertility through impacting anther development.
Figure 6.
Microscopic analysis of rice flower morphology at the heading stage. Morphology of flowers from the wild type and Os4CL3AS transgenics was characterized. A, Spikelet of the wild type. Bar = 2 mm. B, Anther of the wild type. Bar = 500 μm. C, Spikelet of 4CL3AS. Bar = 2 mm. D, Anther of 4CL3AS. Bar = 500 μm. E and G, Scanning electron microscopy (SEM) images of the anther surface of the wild type. Bars = 50 μm in E and 10 μm in G. F and H, SEM images of the anther surface of 4CL3AS. Bars = 50 μm in F and 10 μm in H. I and J, SEM images of pistils of the wild type (I) and 4CL3AS (J). Bars = 500 μm. K and L, 3-(4,5-Dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide staining of pollen grains from the wild type (K) and 4CL3AS (L). Bars = 50 μm. M and N, DAPI staining of pollen grains from the wild type (M) and 4CL3AS (N). Bars = 50 μm. An, Anther; Lo, locule; MP, mature pollen; Ov, ovary; Pa, palea; Pi, pistil; St, stamen; Sti, stigma; Sty, style.
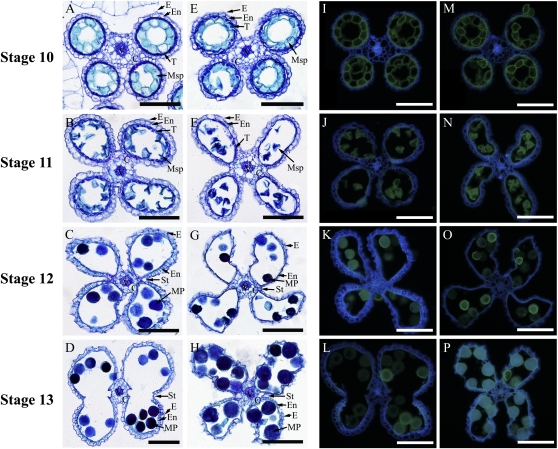
Rice anther development can be divided into 14 stages (Zhang and Wilson, 2009). We examined the cellular morphology of anther development stage by stage. Before stage 10, no noticeable morphological changes in the anthers of transgenic rice were observed (Fig. 7, A, E, I, and M). Beginning from stage 11 in normal development, the anther undergoes a degeneration of the middle and tapetum layers, formation of the falcate pollen grains in locules, and a characteristic U-shaped cell wall thickening in the endothecium layer (Zhang and Wilson, 2009). However, at stage 11, in contrast to the regular round locules that developed in the wild type (Fig. 7, B and J), the locules in Os4CL3AS transgenic plants were distorted (Fig. 7, F and N). At stage 12, while the endothecium layer thickens and becomes lignified in the wild-type anthers (Fig. 7, C and K), the thickening and lignification of the endothecium in Os4CL3AS transgenics were suppressed (Fig. 7, G and O). At stage 13, anther dehiscence, which is a critical process for pollination, occurred in the wild type (Fig. 7, D and L) but was seriously affected in Os4CL3AS transgenics (Fig. 7, H and P).
Figure 7.
Anther development in rice from stage 10 to 13. Rice anthers were transverse sectioned. The sections were observed with toluidine blue staining and under UV illumination. Four stages of anther development from the wild-type and Os4CL3AS transgenics were compared. In A to H, images were photographed with toluidine blue staining; in I to P, images were photographed under UV illumination. A to D and I to L, Anther development in the wild type from stage 10 to 13 with toluidine blue staining (A–D) and under UV illumination (I–L). E to H and M to P, Anther development in Os4CL3AS transgenics from stage 10 to 13 with toluidine blue staining (E–H) and under UV illumination (M–P). At stage 10, both the wild type (A and I) and Os4CL3AS (E and M) did not show obvious morphological differences in anther structures. At stage 11, the anther wall middle layer and endothecium degenerated, and typical falcate pollen grains were formed (B and J) and distorted locules in Os4CL3AS transgenic plants were observed (E and N). At stage 12, the endothecium underwent drastic secondary thickening and lignification, as indicated in striated U-shaped bands (C) and strong lignin UV illumination (K) in the wild-type anthers, whereas the endothecium secondary thickening and lignification were affected in Os4CL3AS transgenics (G and O). At stage 13, the anther dehiscence occurred in the wild type (D and L) but not in the abnormal anthers of Os4CL3AS transgenics (H and P). C, Connective tissue; E, epidermis; En, endothecium; MP, mature pollen; Msp, microspore; St, stomium; T, tapetum; V, vascular bundle. Bars = 100 μm.
DISCUSSION
Divergent Evolution of the Lignin-Associated 4CL Genes in Monocot Plants
4CLs have been extensively investigated in dicots and gymnosperms for their role in the biosynthesis of a variety of secondary metabolites, including lignin and flavonoids (Lozoya et al., 1988; Allina et al., 1998; Hu et al., 1998; Ehlting et al., 1999; Osakabe et al., 1999; Li et al., 2000; Harding et al., 2002; Lindermayr et al., 2002; Hamberger and Hahlbrock, 2004). In dicots, 4CLs can be classified into type I 4CLs, which are associated with lignin accumulation, and type II 4CLs, which are involved in the metabolism of other phenonic compounds (Hu et al., 1998; Ehlting et al., 1999; Harding et al., 2002). Sequence analysis revealed that rice Os4CL2 is classified in the same group as type II 4CLs, but the other four 4CLs in the rice genome belonged to a third phylogenetic group (type III) distinct from that of type II 4CLs in dicots. The high degree of sequence identity between 4CLs from rice and other monocot species, including perennial ryegrass (Lolium perenne), maize (Zea mays), and wheat (Triticum aestivum), found in GenBank databases (data not shown) indicates that monocot 4CLs likely share many similarities.
The presence of a third clade of 4CLs in monocots suggests that the 4CL genes evolved independently in the two groups of plants after the separation between monocots and dicots. Monocots, which branched off from dicots about 140 to 150 million years ago (Chaw et al., 2004), exhibit different anatomical, physiological, and chemical properties from dicots. G and S units are the major monomers of lignin in dicots, while a considerable amount of H units in addition to G and S units are found in monocot lignin (Boerjan et al., 2003; Hatfield et al., 2008, 2009). Differences in the composition of monolignols may be attributable to the divergent evolution of lignin-associated 4CL genes in monocots versus dicots.
The range of substrates used by 4CLs varies within and between plant species. Dicot 4CLs generally display a preference toward 4-coumarate and caffeate in vitro, while the five rice 4CLs we studied demonstrated a preference for 4-coumarate and ferulate but not caffeate. The rice 4CLs also showed activity toward cinnamate, a reaction that is rarely reported in the literature. 4CL substrate specificity is determined by its structure. 4CLs have highly conserved AMP- and hydroxycinnamate-binding pockets. Recently, the crystal structure of a 4CL from P. tomentosa revealed that type I and type II 4CLs featured different residues in the region of the substrate (hydroxycinnamate)-binding pocket (Hu et al., 2010). Differences in the substrate-binding pocket of type III rice 4CLs suggest that the 4CLs may have evolved to accommodate the utilization of different substrates in rice. Given the role of 4CLs in the synthesis of a variety of secondary phenolics, the affinity of 4CLs for different substrates could impact the physiological roles that 4CLs play in monocot versus dicot plants.
Conversion of Ferulate by 4CL Is a Main Metabolic Step of Lignin Biosynthesis in Rice
Suppression of Os4CL3 expression led to lignin reduction in vascular wall-thickened cells in rice and significantly affected the metabolism of soluble and wall-bound phenolic compounds. In particular, increased levels of 4-coumarate and ferulate accumulated in the transgenics in both soluble and wall-bound forms, suggesting that both may be intermediates converted by Os4CL3 for lignin synthesis in vivo. Consistently, 4-coumarate and ferulate are also preferred substrates for Os4CL3 during in vitro assays. However, previous studies showed that ρ-coumarate 3-hydroxylase (C3′H) is unable to directly use 4-coumarate as a substrate to produce caffeate. Instead, C3′H is thought to act in conjunction with ρ-hydroxycinnamoyl-CoA shikimate hydroxycinnamoyltransferase (HCT) to convert 4-coumarate to caffeoyl-CoA for lignin biosynthesis (Boerjan et al., 2003). Thus, 4-coumarate has been proposed as a main substrate of 4CL for G and S monolignol biosynthesis in dicots. Based on the HCT/C3′H pathways, ferulate may not be a substrate used by 4CL to synthesize G and S lignin units in dicots, and 4CL suppression should not cause ferulate accumulation. This is supported by several studies. For example, in Medicago truncatula, down-regulation of the monolignol pathway enzymes did not dramatically alter ferulate content (Chen et al., 2006). Suppression of 4CL in Pinus radiata caused a depletion of G units in lignin and at the same time increased flavonoids and condensed tannins in wood (Wagner et al., 2009). In monocots, ferulate and 4-coumarate are abundant intermediates for cell wall formation (de O Buanafina, 2009). Significant increase of ferulate accumulation in the Os4CL3-suppressed transgenic rice demonstrated that ferulate, in addition to 4-coumarate, is also a major 4CL substrate in vivo. Thus, 4CL conversion through a ferulate intermediate is a likely main pathway by which lignin is synthesized in rice.
Rice 4CL3 Plays a Broad Range of Roles in a Variety of Processes during Growth
In addition to lignin reduction, suppression of Os4CL3 expression also resulted in a series of morphological changes in rice, including lower efficiency of plantlet induction, retarded growth, weaker mechanical strength, spotted grains, and reduced fertility. These changes contrast with what is typically observed in dicots, where suppression of the lignin-associated type I 4CL expression results in significant lignin reduction but has little impact on plant growth properties. For example, suppression of 4CL expression in Arabidopsis leads to alteration in lignin subunit composition but no changes in growth properties, such as plant size, morphology, and fertility (Lee et al., 1997). Transgenic tobacco (Nicotiana tabacum) with depressed 4CL activities do not display morphological differences compared with the wild type (Kajita et al., 1996). In previous studies, morphological changes of lignin reduction were found to be related to tissue-specific expression of 4CLs and growth conditions. When the lignin-associated 4CL is constitutively suppressed in Populus tremuloides, the transgenic plants contain less lignin and show a faster growth in the greenhouse (Hu et al., 1999). Also with P. tremuloides, the xylem-specific suppression of the lignin-associated 4CL results in lignin reduction but no morphological differences relative to the control during the early growth stage (Li et al., 2003a). However, field evaluation of the transgenic plants indicates that the growth is affected in the 4CL-sppressed poplar when grown in the field for multiple years (Voelker et al., 2010). This suggests that the impact of lignin reduction on plant growth may be dependent on the severity of environmental conditions, indicating a role of lignin in defending against environmental stress. The effect of lignin reduction on morphological changes may also be associated with the plant’s anatomical structures. Thus, different classes of plants with major physiological and anatomical differences would react differently to the impact of lignin reduction. For example, suppression of 4CL in P. radiata, a gymnosperm, resulted in lignin reduction and more severe morphological changes compared with angiosperms (Wagner et al., 2009). Lignin reduction in P. radiata led to the collapse of tracheid cells, which has both water conduction and structural support functions (Wagner et al., 2009).
The extensive morphological changes observed in rice in response to Os4CL3 suppression support the suggestion that 4CLs play a role in a broader range of functions in monocots than in dicots. The broad role of 4CLs in monocots is supported by their expression pattern. Generally, lignin-associated 4CLs are expressed in a tissue-specific manner in dicots but are expressed across various tissues in monocots without apparent specificity. In our study, the Os4CL3 gene was found to be expressed in both wall-thickened vascular cells and parenchyma cells. Consistent with this expression pattern, the weakness in mechanical strength observed may reflect lower lignin content in the culms of the transgenics, while spotted grains may be a consequence of disruptions of phenolic metabolism in grain epidermis as a result of Os4CL3 suppression.
Previously, abnormal anther dehiscence has been reported in the mutants of cinnamoyl-coenzyme A reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD; Thevenin et al., 2011). Male sterility was also observed in the Arabidopsis triple mutant ccc (for cad-c, cad-d, and ccr1), which is unable to undergo anther dehiscence and fails to normally release pollen (Thevenin et al., 2011).
However, decreased fertility has not been investigated in other studies of 4CL suppression in plants. Our results indicated that abnormal anther development may be responsible for the lower fertility observed in transgenic rice. Rice is normally self-pollinating, and timely anther dehiscence is critical for pollination to occur effectively (Steiner-Lange et al., 2003; Mitsuda et al., 2005). U-shaped cell wall thickening is required for the generation of tension stress necessary for anther dehiscence (Steiner-Lange et al., 2003; Mitsuda et al., 2005). Disruption in cell wall thickening of the endothecium cells in 4CL-suppressed rice, which prevents the timely release of pollen, could cause the fertility decrease observed in Os4CL3-suppressed transgenic rice (Steiner-Lange et al., 2003; Mitsuda et al., 2005).
Overall, we found that the rice 4CLs display different catalytic properties compared with those in dicots and have a major impact on growth and physical properties of rice. By analyzing Os4CL3 as a key gene in the phenylpropanoid pathway, we have also identified a potential target for the engineering of lignin biosynthesis and mechanical strength in rice.
MATERIALS AND METHODS
Plant Material
Rice material used in this study was Oryza sativa japonica ‘Zhonghua 11’. The rice was grown in a phytotron under a 12-h photoperiod, 28°C day/22°C night temperature regime, and photon flux density of 200 to 250 μmol m−2 s−1.
Sequence Analysis
Five putative 4CL gene sequences were retrieved from the rice genome database (http://rice.plantbiology.msu.edu/). Sequences were analyzed using the BioEdit program version 7.0.0 (http://www.mbio.ncsu.edu/BioEdit/BioEdit.html) and the MEGA program version 3.1 (http://www.megasoftware.net/mega.html). Primers (4CL1F, 5′-TGTCACTGACAGATGGGGTCC-3′; 4CL1R, 5′-CACCAGCAAGAACAATAAACAGG-3′; 4CL2F, 5′-TTCCCACCACCTCCTCCCACTCT-3′; 4CL2R, 5′-CCCAACTGGCAACAACACGTACAAA-3′; 4CL3F, 5′-GCCGTCTCCTCGTGTAAC-3′; 4CL3R, 5′-TTGGCCTTAGCTGCTTTT-3′; 4CL4F, 5′-CATCAGTCAAGAGCGGAAGA-3′; 4CL4R, 5′-CACATAACGGGCATACAAAC-3′; 4CL5F, 5′-CGGAGAAGAAGAATGGGTTCGTTG-3′; 4CL5R, 5′-TAGCTCATGCCAGTTTCATCAGCAC-3′) were designed to flank the full coding region and used for cloning of the five rice 4CL cDNAs from the total RNA isolated from leaf tissue. The cloned 4CL cDNAs were sequenced with a perfect match to the sequences in the databases, confirming their identities.
Gene Expression Analysis
Total RNA was extracted from various tissues at different growth stages using the Trizol method following the manufacturer’s instructions (Invitrogen). Total RNA was treated with RNase-free DNase I to remove DNA contamination. Then, 2 μg of total RNA was reverse transcribed into cDNA using the ReverTra Ace qPCR RT Kit (Toyobo). Gene expression was assayed by quantitative PCR using the cDNA equivalent of 100 ng of total RNA. Gene-specific primers (Supplemental Table S2) were designed for PCR amplification. Quantitative PCR assays were performed with a MyiQ real-time PCR detection system (Bio-Rad) and SYBR Green real-time PCR Master Mix Plus (Toyobo). The gene expression data were normalized using the rice ACTIN1 gene (Os03g0718100) as a reference.
Rice 4CL Recombinant Protein Expression and Enzyme Property Analysis
4CL cDNAs with full coding regions (Os4CL1, 1,692 bp; Os4CL2, 1,707 bp; Os4CL3, 1,662 bp; Os4CL4, 1,677 bp; and Os4CL5, 1,617 bp) were subcloned into pET26b(+) vector and fused with a His tag at the C terminus (Invitrogen). After the sequence was confirmed, the construct was transformed into Escherichia coli BL21 (DE3) strain for recombinant protein expression and purification according to previous methods (Li et al., 2000). The purified protein was stored in 50 mm Tris-HCl buffer (pH 7.5) containing 14 mm β-mercaptoethanol and 30% glycerol at −20°C until enzyme activity analysis.
Enzyme activity was assayed following previous methods with minor modifications (Knobloch and Hahlbrock, 1975). Briefly, the reaction mixture of 200 μL contained 50 mm Tris-HCl (pH 7.5), 5 mm ATP, 5 mm MgC12, 0.2 mm CoA, 5 μg of purified recombinant 4CL protein (boiled protein was used as the control), and varying concentrations of substrates (20, 40, 100, 300, and 600 μm).The change in absorbance caused by CoA-ester formation was monitored at 311 nm for cinnamic acid, 333 nm for 4-coumaric acid, 346 nm for caffeic acid, 345 nm for ferulic acid, and 352 nm for sinapic acid. Extinction coefficients of the ester products were used to calculate enzyme activities according to previously established methods (Lee et al., 1997). Km (the concentration of substrate that leads to half-maximal velocity), Kcat (the maximum no. of molecules of substrate that an enzyme can convert to product per catalytic site per unit of time), Kenz (Kcat/Km), and Vmax (the maximum initial velocity of the enzyme-catalyzed reaction under the given conditions) values for recombinant 4CL1, 4CL2, 4CL3, 4CL4, and 4CL5 were determined as described (Li et al., 2000).
Transformation of Rice with Suppression of 4CL3 Expression
To suppress 4CL3 expression in rice plants, a rice 4CL3 cDNA fragment was cloned into a binary pHB vector in antisense orientation under the control of the 35S promoter (Mao et al., 2005). After the sequence of the construct was confirmed, it was mobilized into Agrobacterium tumefaciens strain EHA105. The Agrobacterium-mediated rice transformation was carried out according to Hiei et al. (1994). The transformants (named 4CL3AS) were selected in hygromycin and confirmed by PCR.
Analysis of the Os4CL3 Promoter
About 2.2 kb of the Os4CL3 promoter region was amplified by PCR from rice genomic DNA using two primers (4CL3P-F, 5′-CAAATCCAAAATTCCAAGTGC-3′; 4CL3P-R, 5′-TGTTGTCGATCTCGATGTCC-3′). After the amplified fragment was sequenced, the confirmed promoter fragment was subcloned into a pCAMBIA1301 vector upstream of uidA (GUS; CAMBIA). The construct was then transferred into Agrobacterium for rice transformation. The transgenic rice was analyzed for GUS expression. For the GUS assay, the transgenic rice was prepared and sectioned. The assay material was treated with acetone for 10 min at 4°C, washed three times with 100 mm NaPO4 buffer (pH 7.0), and incubated with a staining solution [100 mm NaPO4 (pH 7.0), 10 mm EDTA, 2 mm 5-bromo-4-chloro-3-indolyl-β-GlcA, 5 mm K4Fe(CN)6, 5 mm K3Fe(CN)6, and 0.2% Triton X-100] for 20 min to 3 h at 37°C. The reaction was stopped, and chlorophyll was extracted using 75% ethanol.
In Situ Hybridization and Histochemical Staining
An Os4CL3-specific fragment (206 bp) was amplified using two primers (Supplemental Table S2) and used as a probe for in situ hybridization. Digoxigenin-labeled antisense and sense probes were synthesized with T7 and SP6 RNA polymerase (Roche), respectively. Probes were shortened to 75- to 200-bp fragments by limited carbonate hydrolysis and quantified as described (Hejátko et al., 2006). The specificity and efficiency of the probes were confirmed by dot-blot analysis. Rice basal culm internodes (6 weeks old) were embedded in Paraplast (Sigma-Aldrich), cut into 8-μm-thick sections, and mounted onto precharged slides. Tissue fixation, embedding, hybridization, and signal detection were carried out as described (Harding et al., 2002). The slides were observed with a microscope and photographed.
For histochemical analysis, phloroglucinol-HCl staining was performed as described (Weng et al., 2010). Culms of 3-month-old rice plants were sectioned with a razor blade. Sections were stained with 1% phloroglucinol (w/v) in 12% HCl for 5 min and immediately observed with a light microscope. FAA (5% formaldehyde, 10% acetic acid, and 50% ethanol)-fixed culms and paraffin-embedded material were sectioned (9 μm thickness). Cellulose was stained with a 0.005% aqueous solution of calcofluor (fluorescent brightener 28; Sigma) for 2 min and visualized with an Olympus BX51 fluorescence microscope with a GFP filter according to Li et al. (2003b).
Microscopic Analysis of Flower Development
For scanning electron microscopy, flowering organs from wild-type and 4CL3AS lines were collected at different stages of development, fixed in FAA overnight, and dehydrated through serial concentrations of ethanol from 50% to 100%. Immediately after critical point drying, mounting, and gold coating, specimens were examined with a scanning electron microscope (JSM-5610LV; JEOL) at an accelerating voltage of 10 to 20 kV in low-vacuum mode.
To obtain cross sections of developing anthers, young panicles were fixed in FAA overnight, dehydrated through serial concentrations of ethanol from 50% to 100%, and then embedded. Samples were sectioned to 10 μm, mounted on glass slides, heat fixed, and stained with toluidine blue for bright-field microscopy. For UV microscopy, slides were directly photographed under UV illumination after paraffin was removed. Pollen viability was determined by 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide staining (Khatun and Flowers, 1995) and 4′,6-diamidino-2-phenylindole (DAPI) staining (Ishiguro et al., 2010). DAPI-stained pollen grains were observed by fluorescence microscopy under UV illumination.
Determination of Lignin Content and Composition
Klason lignin content was determined as described (Hatfield et al., 1994). In brief, 1 g (W1) of prepared sample was placed in a 50-mL conical flask. After reaction with 15 mL of 12 m H2SO4 at 30°C for 60 min, the sample was transferred to a 500-mL conical flask and diluted with 435 mL of water to 0.4 m H2SO4. After being autoclaved (121°C) for 60 min, the nonhydrolyzed residue was collected by filtration through a Fibertec P2 crucible, washed with hot water until acid free, and dried for 2 h at 130°C. The dried sample was cooled in a desiccator and weighed (W2). Then, the residue was ashed at 525°C for at least 3 h in a muffle furnace and weighed (W3) after being cooled to room temperature in a desiccator. Klason lignin content was determined using the following formula: Klason lignin (%) = (W2 − W3) × 100/W1.
Lignin subunits were analyzed by thioacidolysis as described with some modifications (Rolando et al., 1992). About 30 mg of sample powder (3-month-old rice plants) was added to a glass vial with Teflon-lined screw cap containing fresh thioacidolysis reagent (30 mL; 0.2 m boron trifluoride diethyl etherate in dioxane/ethanethiol, 8.75:1 [v/v]). After being dry sealed with a flow of nitrogen gas, the vial was placed in an oven at 100°C for 4 h with occasional shaking. After the reaction was stopped and cooled on ice, thioacidolytic products were transferred to a beaker for pH adjustment (to pH 3–4) with 0.4 m sodium bicarbonate and the addition of internal standard tetracosane. The organic phase was extracted using dichloromethane (30 mL × 3) through a separating funnel, pooled, and dehydrated with anhydrous sodium sulfate overnight. The solvent was removed by rotary evaporation at 40°C. The residue was redissolved in 1 mL of dichloromethane with some anhydrous sodium sulfate and stored in a desiccator at 4°C. A total of 20 μL of the resuspended sample was sylilated by combining with 50 μL of pyridine and 130 μL of N,O-bis-(trimethylsilyl)-trifluoroacetamide (99:1). After incubation for 40 min at 60°C, 3 μL of silylated product was analyzed by gas chromatography-mass spectrometry (GC-MS). An Agilent 6890 instrument, fitted with an Agilent 5975 inert mass selective detector and an Agilent HP-5MS column (30 mm × 0.25 mm × 0.25 μm film thickness) was used for analysis. Helium was used as the carrier gas at 1 mL min−1, and detector temperatures were set to 250°C. Monomers were identified by relative retention times and characteristic mass spectrum ions for H, G, and S monomers. The monomer abundance was quantified using tetracosane as an internal standard (Rolando et al., 1992).
Determination of Soluble and Wall-Bound Phenolics
Wall-bound phenolic extraction and analysis were carried out as described (Sosulski et al., 1982; Gou et al., 2009) with some modifications. Briefly, the leaves and stems (2 months old) were ground in liquid nitrogen and dried at 45°C for 2 d. A total of 0.5 g of fine-powder sample was extracted with 50 mL of methanol:acetone:water (7:7:6) for 24 h in a 37°C shaker at 120 rpm. Extracted residues were washed three times with 15 mL of water each time and subjected to alkaline hydrolysis for 16 h in 40 mL of 2 n NaOH in a 37°C shaker at 120 rpm. After centrifugation, the supernatant was collected, acidified to pH 3.0 with 6 n HC1, and extracted three times with an equal volume of water-saturated ethyl acetate. The organic phases were pooled, and the solvent was removed by rotary evaporation at 40°C. The residue was dissolved in 2 mL of methanol for HPLC-MS analysis.
Soluble phenolics were extracted with 40 mL of 0.1 m HCl in a 37°C shaker at 120 rpm for 16 h from 0.5 g of oven-dried ground powder. The supernatant was collected and further extracted three times with water-saturated ethyl acetate. The ethyl acetate extracts were then dried by rotary evaporation at 40°C and redissolved in 1 mL of methanol for HPLC-MS analysis.
HPLC-MS (Agilent 1200 series HPLC 6500 Series Accurate-Mass Quadrupole Time-of-Flight LC/MS device) was used for the detection and determination of soluble and wall-bound phenolics. A total of 3 μL of the prepared sample was injected into a Zorbax column (Eclips XDB-C18, 4.6 × 50 mm, 1.8 μm) and resolved in 0.1% acetic acid with increasing acetonitrile (at a constant rate of 0.2 mL min−1) for a gradient at 10% from 0 to 5 min, 15% from 5 to 20 min, 40% from 20 to 25 min, and 40% from 25 to 30 min. UV absorption was monitored at 214, 280, and 310 nm with a diode-array detector. MS analysis was carried out with a tandem linked LC/MSD Trap XCT system equipped with an electrospray ionization source (Agilent). Total ion chromatograms (90–1,500 mass-to-charge ratio) was acquired under negative mode with the following parameters: nebulizer pressure, 40 psiψ; drying gas flow, 9 L min−1; drying gas temperature, 350°C; and electrospray ionization voltage cap, 3,500 V. The MS spectrum of each particular peak was extracted for characterization of the resolved metabolites. Quantification of the phenolics was based on the detected area of UV absorbance against the standard curves, which were generated using authentic 4-coumarate and ferulate under the same conditions.
Measurement of Physical Properties
Breaking force and extensibility of 3-month-old rice culms were measured using a material tester (Instron 5867). The breaking force was measured as the force required for breaking a culm segment. Extensibility was expressed as a ratio of the distance extended before breaking versus the length of the tested sample. Culm of the third internode was cut into the same width and length and used for immediate measurement.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Os4CL1 (NM_001067888, Os08g14760), Os4CL2 (NM_001054354, Os02g46970), Os4CL3 (NM_001052604, Os02g08100), Os4CL4 (NM_001064787, Os06g44620), and Os4CL5 (NM_001068470, Os08g34790).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequences of rice 4CLs were aligned with a representative type I 4CL, P. tomentosa 4CL1, and a type II 4CL, Arabidopsis 4CL3.
Supplemental Figure S2. GC-MS analysis of thioacidolysis products from rice plants.
Supplemental Table S1. Amino acid sequence similarities of rice 4CLs compared with those of Arabidopsis, poplar, and moss.
Supplemental Table S2. Primers used in this study.
Supplemental Table S3. Main products obtained by thioacidolysis of various phenolic compounds in wild-type and 4CL3AS transgenic rice.
Acknowledgments
We thank Dr. Hongxuan Lin for assistance with microtome sectioning, Dr. Yining Liu for LC-(quadrupole time-of-flight)-MS analysis, Mr. Wenli Hu for GC-MS analysis, and Mr. Xiaoyan Gao for scanning electron microscopy. The vector pHB was provided by Dr. Hongquan Yang.
References
- Allina SM, Pri-Hadash A, Theilmann DA, Ellis BE, Douglas CJ. (1998) 4-Coumarate:coenzyme A ligase in hybrid poplar: properties of native enzymes, cDNA cloning, and analysis of recombinant enzymes. Plant Physiol 116: 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Chaw SM, Chang CC, Chen HL, Li WH. (2004) Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol 58: 424–441 [DOI] [PubMed] [Google Scholar]
- Chen F, Srinivasa Reddy MS, Temple S, Jackson L, Shadle G, Dixon RA. (2006) Multi-site genetic modulation of monolignol biosynthesis suggests new routes for formation of syringyl lignin and wall-bound ferulic acid in alfalfa (Medicago sativa L.). Plant J 48: 113–124 [DOI] [PubMed] [Google Scholar]
- de O Buanafina MM. (2009) Feruloylation in grasses: current and future perspectives. Mol Plant 2: 861–872 [DOI] [PubMed] [Google Scholar]
- Ehlting J, Büttner D, Wang Q, Douglas CJ, Somssich IE, Kombrink E. (1999) Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J 19: 9–20 [DOI] [PubMed] [Google Scholar]
- Gou JY, Yu XH, Liu CJ. (2009) A hydroxycinnamoyltransferase responsible for synthesizing suberin aromatics in Arabidopsis. Proc Natl Acad Sci USA 106: 18855–18860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberger B, Hahlbrock K. (2004) The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc Natl Acad Sci USA 101: 2209–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SA, Leshkevich J, Chiang VL, Tsai CJ. (2002) Differential substrate inhibition couples kinetically distinct 4-coumarate:coenzyme A ligases with spatially distinct metabolic roles in quaking aspen. Plant Physiol 128: 428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield R, Ralph J, Grabber JH. (2008) A potential role for sinapyl p-coumarate as a radical transfer mechanism in grass lignin formation. Planta 228: 919–928 [DOI] [PubMed] [Google Scholar]
- Hatfield RD, Jung HJG, Ralph J, Buxton DR, Weimer PJ. (1994) A comparison of the insoluble residues produced by the Klason lignin and acid detergent lignin procedures. J Sci Food Agric 65: 51–58 [Google Scholar]
- Hatfield RD, Marita JM, Frost K, Grabber J, Ralph J, Lu F, Kim H. (2009) Grass lignin acylation: p-coumaroyl transferase activity and cell wall characteristics of C3 and C4 grasses. Planta 229: 1253–1267 [DOI] [PubMed] [Google Scholar]
- Hejátko J, Blilou I, Brewer PB, Friml J, Scheres B, Benková E. (2006) In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat Protoc 1: 1939–1946 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hu WJ, Harding SA, Lung J, Popko JL, Ralph J, Stokke DD, Tsai CJ, Chiang VL. (1999) Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol 17: 808–812 [DOI] [PubMed] [Google Scholar]
- Hu WJ, Kawaoka A, Tsai CJ, Lung J, Osakabe K, Ebinuma H, Chiang VL. (1998) Compartmentalized expression of two structurally and functionally distinct 4-coumarate:CoA ligase genes in aspen (Populus tremuloides). Proc Natl Acad Sci USA 95: 5407–5412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Gai Y, Yin L, Wang X, Feng C, Feng L, Li D, Jiang XN, Wang DC. (2010) Crystal structures of a Populus tomentosa 4-coumarate:CoA ligase shed light on its enzymatic mechanisms. Plant Cell 22: 3093–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S, Nishimori Y, Yamada M, Saito H, Suzuki T, Nakagawa T, Miyake H, Okada K, Nakamura K. (2010) The Arabidopsis FLAKY POLLEN1 gene encodes a 3-hydroxy-3-methylglutaryl-coenzyme A synthase required for development of tapetum-specific organelles and fertility of pollen grains. Plant Cell Physiol 51: 896–911 [DOI] [PubMed] [Google Scholar]
- Kajita S, Hishiyama S, Tomimura Y, Katayama Y, Omori S. (1997) Structural characterization of modified lignin in transgenic tobacco plants in which the activity of 4-coumarate:coenzyme A ligase is depressed. Plant Physiol 114: 871–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita S, Katayama Y, Omori S. (1996) Alterations in the biosynthesis of lignin in transgenic plants with chimeric genes for 4-coumarate:coenzyme A ligase. Plant Cell Physiol 37: 957–965 [DOI] [PubMed] [Google Scholar]
- Khatun S, Flowers TJ. (1995) The estimation of pollen viability in rice. J Exp Bot 46: 151–154 [Google Scholar]
- Knobloch KH, Hahlbrock K. (1975) Isoenzymes of p-coumarate:CoA ligase from cell suspension cultures of Glycine max. Eur J Biochem 52: 311–320 [DOI] [PubMed] [Google Scholar]
- Lee D, Meyer K, Chapple C, Douglas CJ. (1997) Antisense suppression of 4-coumarate:coenzyme A ligase activity in Arabidopsis leads to altered lignin subunit composition. Plant Cell 9: 1985–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Popko JL, Umezawa T, Chiang VL. (2000) 5-Hydroxyconiferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation, a new view of monolignol biosynthesis in angiosperms. J Biol Chem 275: 6537–6545 [DOI] [PubMed] [Google Scholar]
- Li L, Zhou Y, Cheng X, Sun J, Marita JM, Ralph J, Chiang VL. (2003a) Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc Natl Acad Sci USA 100: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Qian Q, Zhou YH, Yan MX, Sun L, Zhang M, Fu ZM, Wang YH, Han B, Pang XM, et al. (2003b) BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15: 2020–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C, Möllers B, Fliegmann J, Uhlmann A, Lottspeich F, Meimberg H, Ebel J. (2002) Divergent members of a soybean (Glycine max L.) 4-coumarate:coenzyme A ligase gene family. Eur J Biochem 269: 1304–1315 [DOI] [PubMed] [Google Scholar]
- Lozoya E, Hoffmann H, Douglas C, Schulz W, Scheel D, Hahlbrock K. (1988) Primary structures and catalytic properties of isoenzymes encoded by the two 4-coumarate:CoA ligase genes in parsley. Eur J Biochem 176: 661–667 [DOI] [PubMed] [Google Scholar]
- Mao J, Zhang YC, Sang Y, Li QH, Yang HQ. (2005) A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc Natl Acad Sci USA 102: 12270–12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17: 2993–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoumkina MA, Zhao Q, Gallego-Giraldo L, Dai X, Zhao PX, Dixon RA. (2010) Genome-wide analysis of phenylpropanoid defence pathways. Mol Plant Pathol 11: 829–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe K, Tsao CC, Li L, Popko JL, Umezawa T, Carraway DT, Smeltzer RH, Joshi CP, Chiang VL. (1999) Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc Natl Acad Sci USA 96: 8955–8960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolando C, Monties B, La Pierre C. (1992) Thioacidolysis. Lin S, Dence C, , Methods in Lignin Chemistry. Springer-Verlag, Berlin, pp 334–349 [Google Scholar]
- Schneider K, Hövel K, Witzel K, Hamberger B, Schomburg D, Kombrink E, Stuible HP. (2003) The substrate specificity-determining amino acid code of 4-coumarate:CoA ligase. Proc Natl Acad Sci USA 100: 8601–8606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R, Sun YH, Li Q, Heber S, Sederoff R, Chiang VL. (2010) Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol 51: 144–163 [DOI] [PubMed] [Google Scholar]
- Silber MV, Meimberg H, Ebel J. (2008) Identification of a 4-coumarate:CoA ligase gene family in the moss, Physcomitrella patens. Phytochemistry 69: 2449–2456 [DOI] [PubMed] [Google Scholar]
- Sosulski F, Krygier K, Hogge L. (1982) Free, esterified, and insoluble-bound phenolic-acids. 3. Composition of phenolic-acids in cereal and potato flours. J Agric Food Chem 30: 337–340 [Google Scholar]
- Souza CdeA, Barbazuk B, Ralph SG, Bohlmann J, Hamberger B, Douglas CJ. (2008) Genome-wide analysis of a land plant-specific acyl:coenzyme A synthetase (ACS) gene family in Arabidopsis, poplar, rice and Physcomitrella. New Phytol 179: 987–1003 [DOI] [PubMed] [Google Scholar]
- Steiner-Lange S, Unte US, Eckstein L, Yang CY, Wilson ZA, Schmelzer E, Dekker K, Saedler H. (2003) Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J 34: 519–528 [DOI] [PubMed] [Google Scholar]
- Stuible H, Büttner D, Ehlting J, Hahlbrock K, Kombrink E. (2000) Mutational analysis of 4-coumarate:CoA ligase identifies functionally important amino acids and verifies its close relationship to other adenylate-forming enzymes. FEBS Lett 467: 117–122 [DOI] [PubMed] [Google Scholar]
- Stuible HP, Kombrink E. (2001) Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase allows the rational design of mutant enzymes with new catalytic properties. J Biol Chem 276: 26893–26897 [DOI] [PubMed] [Google Scholar]
- Thevenin J, Pollet B, Letarnec B, Saulnier L, Gissot L, Maia-Grondard A, Lapierre C, Jouanin L. (2011) The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol Plant 4: 70–82 [DOI] [PubMed] [Google Scholar]
- Voelker SL, Lachenbruch B, Meinzer FC, Jourdes M, Ki C, Patten AM, Davin LB, Lewis NG, Tuskan GA, Gunter L, et al. (2010) Antisense down-regulation of 4CL expression alters lignification, tree growth, and saccharification potential of field-grown poplar. Plant Physiol 154: 874–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Donaldson L, Kim H, Phillips L, Flint H, Steward D, Torr K, Koch G, Schmitt U, Ralph J. (2009) Suppression of 4-coumarate-CoA ligase in the coniferous gymnosperm Pinus radiata. Plant Physiol 149: 370–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng JK, Akiyama T, Bonawitz ND, Li X, Ralph J, Chapple C. (2010) Convergent evolution of syringyl lignin biosynthesis via distinct pathways in the lycophyte Selaginella and flowering plants. Plant Cell 22: 1033–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DB, Wilson ZA. (2009) Stamen specification and anther development in rice. Chin Sci Bull 54: 2342–2353 [Google Scholar]