Abstract
Rice (Oryza sativa) glutelins are synthesized on the endoplasmic reticulum as larger precursors, which are then transported via the Golgi to the protein storage vacuole (PSV), where they are processed into acidic and basic subunits. Three independent glutelin precursor mutant4 (glup4) rice lines, which accumulated elevated levels of proglutelin over the wild type, were identified as loss-of-function mutants of Rab5a, the small GTPase involved in vesicular membrane transport. In addition to the plasma membrane, Rab5a colocalizes with glutelins on the Golgi apparatus, Golgi-derived dense vesicles, and the PSV, suggesting that Rab5a participates in the transport of the proglutelin from the Golgi to the PSV. This spatial distribution pattern was dramatically altered in the glup4 mutants. Numerous smaller protein bodies containing glutelin and α-globulin were evident, and the proteins were secreted extracellularly. Moreover, all three independent glup4 allelic lines displayed the novel appearance of a large dilated, structurally complex paramural body containing proglutelins, α-globulins, membrane biomarkers for the Golgi apparatus, prevacuolar compartment, PSV, and the endoplasmic reticulum luminal chaperones BiP and protein disulfide isomerase as well as β-glucan. These results indicate that the formation of the paramural bodies in glup4 endosperm was due to a significant disruption of endocytosis and membrane vesicular transport by Rab5a loss of function. Overall, Rab5a is required not only for the intracellular transport of proglutelins from the Golgi to the PSV in rice endosperm but also in the maintenance of the general structural organization of the endomembrane system in developing rice seeds.
Developing plant seeds accumulate large quantities of storage proteins that are coded by two superfamilies of genes, globulins and prolamins (Shewry et al., 1995). Rice (Oryza sativa) is unique in that it accumulates major quantities of both storage protein types in the form of glutelins and prolamins as well as a third minor species, α-globulins (Tanaka et al., 1980; Krishnan and White, 1995). Glutelin, the dominant storage protein in rice, is homologous to leguminous 11S globulins and is packaged in a protein storage vacuole (PSV; Zhao et al., 1983). Unlike the saline-soluble 11S globulins, however, rice glutelins are only soluble in dilute acid and alkali solutions. During seed development, rice glutelin polypeptides are initially synthesized on the endoplasmic reticulum (ER) membrane as 57-kD proglutelin (Yamagata et al., 1982), which is then transported to the PSVs (Krishnan et al., 1986; Yamagata and Tanaka, 1986), where the precursors are cleaved to acidic and basic subunits (Yamagata et al., 1982). The rice α-globulin, a member of the prolamin superfamily, is also packaged in the PSV, where together with glutelins it forms PB-II (Yamagata et al., 1982).
Many storage proteins have been shown to be transported from the ER to PSVs via the Golgi apparatus (Chrispeels, 1983). In maturing bean (Phaseolus vulgaris) and pea (Pisum sativum) cotyledons, the storage proteins are concentrated early at the ends of the cis-Golgi cisternae and released as dense vesicles (Chrispeels, 1983; Hohl et al., 1996; Okita and Rogers, 1996). These dense vesicles are then transported to the PSV via an intermediate prevacuolar compartment (PVC). Golgi-associated dense vesicles containing glutelin and globulin are readily evident in developing rice endosperm cells, supporting a role for the Golgi apparatus in routing these storage proteins to the PSV (Krishnan et al., 1990, 1992). An alternative direct ER-to-PSV pathway bypassing the Golgi has also been proposed (Takahashi et al., 2005), although such PAC-like vesicles (Hara-Nishimura et al., 1998) have not been seen in other ultrastructural studies of rice protein body biogenesis (Ogawa et al., 1989; Krishnan et al., 1990, 1992; Takemoto et al., 2002). Hence, depending on the plant species, transport of storage proteins from the ER to PSVs occurs by vesicular transport by a Golgi-dependent and/or Golgi-independent pathway (Li et al., 2006).
Irrespective of the role of the Golgi, transport of storage proteins to the PSV requires a vacuolar sorting receptor (VSR) that targets the cargo to the PVC. BP-80 was identified based on its specific binding to the vacuolar peptide determinant NPIR motif. BP-80 is enriched in clathrin-coated vesicles, where it targets the thiol protease aleurain to the lytic vacuole, and is predominantly concentrated on the lytic PVCs in Arabidopsis (Arabidopsis thaliana) and tobacco (Nicotiana tabacum) BY-2 cells (Kirsch et al., 1994; Miao et al., 2006). Orthologous VSRs from Arabidopsis (AtVSR1) and pumpkin (Cucurbita sp.; PV72) have been suggested to play a role in storage protein transport to the PSV based on their secretion of storage proteins in knockout mutants and their presence on the PVC (Shimada et al., 1997, 2003; Otegui et al., 2006), although this relationship between VSR and storage protein targeting may be indirect (Zouhar et al., 2009) A second receptor type, RMR, interacts with the vacuolar signal determinants of the barley (Hordeum vulgare) lectin, bean phaseolin, and tobacco chitinase (Park et al., 2007) and is responsible for the formation of dense vesicles formed at the Golgi (Hinz et al., 2007). Interestingly, whereas AtVSR1 has been suggested to be recycled by a retromer complex from the PVC to the Golgi (Shimada et al., 2006), RMR is cotransported with the storage protein cargo to the PSV (Hinz et al., 2007).
In addition to VSRs or RMR, targeting of the storage protein-containing vesicles to the PVC and PSV requires the Rab family of small GTPases, which regulate and specify vesicular trafficking (Woollard and Moore, 2008). The best-studied small GTPase and the most relevant to storage protein transport to the PSV is Rab5. The small GTP-binding protein Rab5 was previously localized on early endosomes and on the cytoplasmic face of the plasma membrane (Gorvel et al., 1991). Arabidopsis has three Rab5 orthologs, RAB5F2a/RHA1, RAB5F2b/ARA7, and RAB5F1/ARA6, which are located on punctate structures labeled with FM4-64, a tracer of endocytosis (Nielsen et al., 2008). The RAB5F proteins have been implicated in the trafficking of soluble proteins to the vacuole based on the disruption of trafficking induced by the expression of dominant negative forms of this protein. Consistent with this role is that RAB5F2a/RHA1 is found on the PVC of Arabidopsis protoplasts (Sohn et al., 2003) while RAB5F2b/ARA7 is localized to the PVC and Golgi apparatus in tobacco leaf epidermal cells (Kotzer et al., 2004).
We have characterized eight independent rice mutants that accumulate large quantities of the 57-kD proglutelin polypeptide. The mutants were named endosperm storage protein mutant2 (esp2; Kumamaru et al., 1987, 1988) and glutelin precursor mutant1 (glup1) to glup7 (Kumamaru et al., 2007; Satoh-Cruz et al., 2010a; Ueda et al., 2010). Gene-gene interaction analyses of the eight mutant lines indicate that esp2 is the most epistatic while glup3 is the most hypostatic among the eight mutant genes (Ueda et al., 2010). This relationship suggests that the factors encoded by ESP2 and GLUP3 function upstream and downstream, respectively, of the other GLUP genes in the pathway of the synthesis, trafficking, and accumulation of proglutelin. This hypothesis is supported by studies that centered on the identification of the ESP2 and GLUP3 genes. ESP2 encodes PDIL1-1 (for protein disulfide isomerase-like 1-1), which catalyzes the formation of intramolecular disulfide bonds in the ER and serves as a chaperone (Takemoto et al., 2002; Satoh-Cruz et al., 2010a), while GLUP3 codes for a vacuolar processing enzyme, which proteolytically processes proglutelin into acidic and basic subunits within the PSV (Kumamaru et al., 2010). Based on these results, we hypothesized that the responsible factors of GLUP1-glup2-glup7 mutants and glup4-glup6-GLUP5 mutants participate in the intracellular trafficking of proglutelin from the ER to the Golgi apparatus and from the Golgi apparatus to the PSV, respectively (Ueda et al., 2010). The high-resolution mapping analysis of the glup4 gene suggested that GLUP4 encoded the small GTPase Rab5a, and it was hypothesized that Rab5a participated in the intracellular transport of proglutelin (Kumamaru et al., 2007; Satoh-Cruz et al., 2010b). The colocalization of Rab5a from rice with Golgi and PVC markers derived from other plants in Arabidopsis protoplasts (Wang et al., 2010) also supports this hypothesis. However, evidence for a direct role of Rab5a in proglutelin transport to the PSV has yet to be obtained.
In order to identify the role of Rab5a in the intracellular trafficking of the storage proteins in rice endosperm, we studied several glup4 mutant alleles. DNA sequence analysis showed that they all coded for Rab5a, the small GTPase involved in vesicular transport of storage proteins to the PSV. Mutations in Rab5a not only disrupt proglutelin trafficking but also mediate the appearance of large paramural body (PMB)-containing biomarkers for ER, Golgi, PVC, and PSV. Our results show that Rab5a plays multiple roles in the trafficking of proteins to the PSV and in the general maintenance of the endomembrane system in plants.
RESULTS
The glup4 Mutant Accumulates Large Amounts of Proglutelin
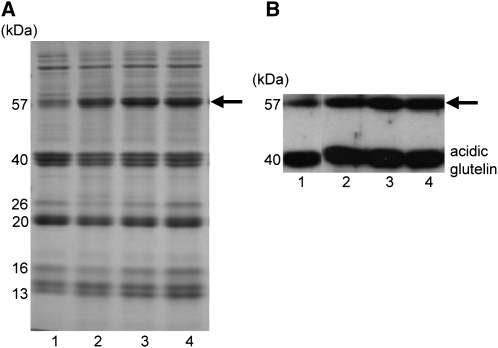
Figure 1A shows the profile of total seed protein from three independent glup4 mutant lines, EM425, EM956, and EM960, as well as the wild type. All three glup4 lines contained elevated amounts of the 57-kD proglutelin polypeptide and correspondingly reduced levels of the proteolytically processed acidic and basic glutelin subunits in comparison with the wild type. These corresponding changes in the levels of proglutelin and processed acidic and basic subunits between the wild type and glup4 mutants are readily discernible by immunoblot analysis of these proteins (Fig. 1B).
Figure 1.
Storage protein composition from seeds of wild-type and glup4 mutant lines. Seed protein extracts were separated on SDS-polyacrylamide gels (A) and then subjected to immunoblot analysis using anti-glutelin acidic subunit antibodies (B). Lane 1, The wild type; lane 2, EM425; lane 3, EM956; lane 4, EM960. EM425, EM956, and EM960 are glup4 allelic mutant lines. The arrows denote the 57-kD proglutelin polypeptide.
Sequence Analysis of Genomic DNA Encoding Rab5a in glup4 Lines
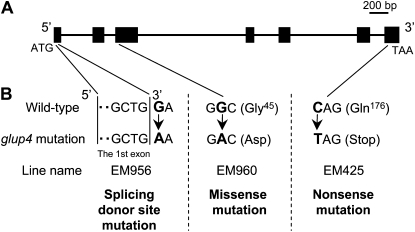
To determine whether GLUP4 was the Rab5a structural gene, genomic DNAs from the wild type and three glup4 alleles, EM425, EM956, and EM960, were sequenced (Fig. 2). Comparison with the wild-type gene sequences revealed that each of the three glup4 lines contained single nucleotide substitutions. Two of these point mutations mediated codon changes where Gly-45 and Gln-176 were replaced by Asp and a premature termination stop codon in EM960 and EM425, respectively. Gly-45 resides in a conserved peptide domain that binds specific effectors of Rab5a activity (Zhu et al., 2004), while the premature termination removes the prenylation residues required for membrane attachment of Rab5a. In EM956, a guanine base was replaced by an adenine at nucleotide positions 43, which was located at the splicing donor site of the first intron. This mutation was expected to disrupt intron splicing and lead to frame shifts or deletions in the mRNA (Brown, 1996). The presence of significant structural mutations in two out of the three glup4 allelic genes (EM425 and EM956) and the apparent loss of function in the third glup4 allele (EM960) indicates that the GLUP4 gene encodes the structural gene for the small GTPase Rab5a.
Figure 2.
Mutation sites of the Rab5a gene in three glup4 mutant lines. A, Structure of the GTPase Rab5a gene on chromosome 12 (Satoh-Cruz et al., 2010b). ATG and TAA indicate the initiation and termination codons, respectively. Black boxes indicate the positions of the seven exons. The Rab5a reading frame spans 612 bp and codes for 203 amino acids. B, Mutation sites in glup4 allelic lines. In EM956, a G→A mutation is situated at the splicing donor site of the first intron. EM960 contains a missense mutation resulting in an amino acid change located in the effector-binding site, while EM425 contains a nonsense mutation resulting in the premature termination of protein synthesis.
Immunoblot Analysis of Rab5a in Developing Seeds of the glup4 Mutant
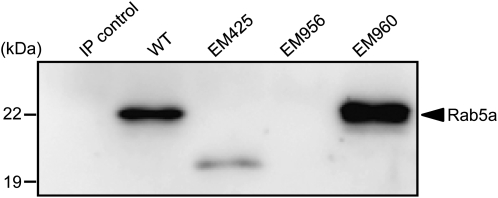
Immunoblot analysis of small samples (10–20 μL) of total protein extracts from developing or mature seeds failed to detect Rab5a, indicating that this small GTPase was present at very low levels. Therefore, immunoprecipitation (IP) experiments were carried out where Rab5a antibodies were incubated with 1 mL of seed extracts followed by immunoblot analysis of the proteins captured by the initial IP (Fig. 3). In EM960, which contains an amino acid substitution, the mutant Rab5a had the same molecular size as the wild type. In EM425, the size of the mutant Rab5a was much smaller than the wild type, a result consistent with the premature termination of translation mediated by the introduction of the nonsense mutation. The products from the Rab5a gene in EM425 and the wild type are calculated to be 19,309.94 and 22,146.12 D, respectively. The results of IP analysis coincided with the theoretical molecular size of Rab5a protein in EM425. In addition, its levels were considerably reduced compared with those of the wild type. This observation suggests that the truncated Rab5a, which lacked the C-terminal Cys residues needed for prenylation, was more susceptible to protein turnover than the wild type. Rab5a was not detected in EM956, indicating that the mutated splicing donor site of the first intron resulted in a nontranslatable RNA transcript produced by aberrant RNA splicing. The absence of Rab5a activity in EM956 and the loss of function in EM960 and EM425 (see “Discussion”) indicate that this small GTPase is responsible for the abnormal accumulation of proglutelin. Hereafter, we name the GLUP4 gene product as GLUP4-Rab5a.
Figure 3.
Immunoblot analysis of Rab5a in the various glup4 mutant lines. IP control, Without anti-Rab5a antibody; WT, wild type. Seed extracts (1 mL) were subjected to IP using anti-Rab5a antibody. The immunoprecipitates were then subjected to immunoblot analysis using anti-Rab5a antibody. Note that EM425 contains a truncated Rab5a (19.3 kD) consistent with the premature termination of synthesis of this protein by the introduction of a nonsense codon, while EM956 lacks the Rab5a polypeptide due to aberrant RNA splicing.
Expression Analysis of Other Rab5a Homologs
A database search (KOME: Knowledge-Based Oryza Molecular Biological Encyclopedia; http://cdna01.dna.affrc.go.jp/cDNA/) revealed the presence of three additional Rab5a homologs in rice (Supplemental Fig. S1). The expression profiles in seeds taken from the rice expression profile database (RiceXPro; http://ricexpro.dna.affrc.go.jp) for the homologs indicated that GLUP4-Rab5a (AK061116) has the highest expression level among the Rab5a homolog genes. The expression of the Rab5a homolog genes in developing seeds was confirmed by reverse transcription-PCR (Supplemental Fig. S2). GLUP4-Rab5a (AK061116) showed the highest expression level in seeds and was 5- to 10-fold higher than the other Rab5a genes. Hence, GLUP4-Rab5a is the major form expressed during seed development. Supplemental Figure S3 shows a phylogenetic tree indicating the relationship between GLUP4-Rab5a and the homologs in rice (Supplemental Fig. S1) and Arabidopsis (Nielsen et al., 2008).
Protein Body Formation in glup4
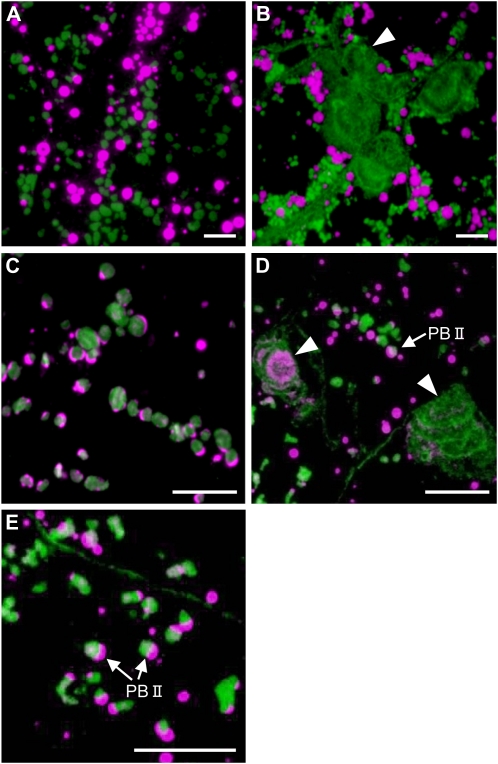
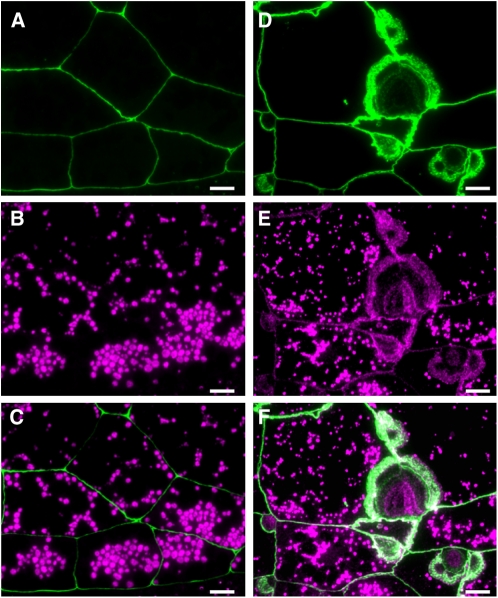
To determine the basis of the elevated levels of proglutelin in the various glup4 lines, immunofluorescence microscopy studies were carried out. Developing rice endosperm exhibits two types of protein bodies, PB-I and PB-II. PB-I contains prolamins, whereas PB-II contains glutelins and α-globulins. In the wild type and the glup4 mutant, PB-I and PB-II labeled by prolamin and glutelin antibodies (Fig. 4, A and B) or PB-II labeled by α-globulin and glutelin antibodies (Fig. 4, C–E) were observed. In addition, the glup4 mutant contained one or more large distended structures containing granules labeled with glutelin antibodies located adjacent to the cell wall (Fig. 4B; Supplemental Fig. S4). As all three glup4 lines contained this dilated structure, its formation (Supplemental Fig. S4) is due to the loss of function of Rab5a.
Figure 4.
Immunofluorescence microscopy of protein bodies in the wild type and the glup4 mutant, EM956. A and C, The wild type. B, D, and E, glup4 (EM956). Secondary antibodies labeled with rhodamine (magenta) and fluorescein isothiocyanate (FITC; green) were used to visualize the reaction of prolamin and glutelin antibodies, respectively, in A and B. Secondary antibodies labeled with rhodamine and FITC were used to detect antigen recognized by anti-α-globulin and anti-glutelin, respectively, in C, D, and E. Arrowheads indicate the presence of the novel large distended structures containing proglutelin and α-globulin. In many instances, the large distended structures are clustered in neighboring cells (B). Glutelins are present in overlapping layers (B and D). Also note that the distribution of proglutelins and globulin within the large distended structures is spatially distinct (D). Bars = 10 μm.
In wild-type cells, glutelins and α-globulins are packaged together in PSVs, although their distribution is stratified in this organelle, with glutelins located in the bulky crystalloid and α-globulins located at the peripheral, amorphous matrix regions (Fig. 4C). The novel distended structures in glup4 endosperm were labeled not only by glutelin antibodies but also by α-globulin antibodies (Fig. 4D). In glup4, the distribution of glutelin and α-globulin within PSVs appears normal, although the PSVs are much smaller than that seen in the wild type (Fig. 4E). While the size of PB-II in the wild type increased gradually during endosperm development, the size of PB-II in all glup4 lines changed very little between 1 and 3 weeks of development (Supplemental Fig. S4). These observations indicate that the normal transport of proglutelin as well as α-globulin to PB-II (PSV) is disrupted by the loss of function of Rab5a in glup4 mutants and that proglutelin accumulates in large distended structures adjoining the cell surface. The presence of small PB-IIs under the absence of GLUP4-Rab5a in glup4 seeds suggest that the other Rab5a species would partially participate in the intracellular transport of storage proteins to the PSV.
Transmission Electron Microscopy of glup4 Mutant Endosperm Indicates That the Large Distended Structures Are PMBs
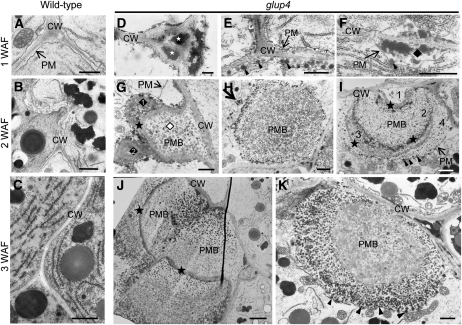
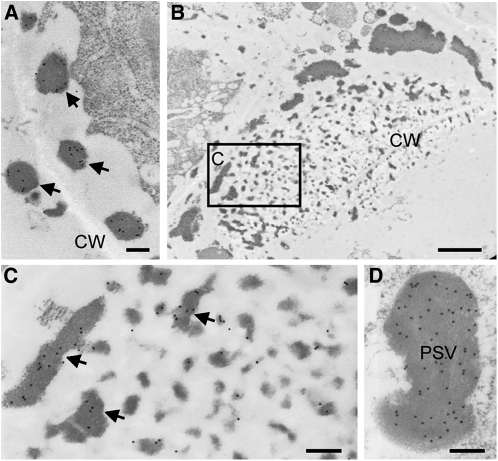
To obtain additional insights on this novel large distended structure in glup4 endosperm, the ultrastructure of developing glup4 and wild-type seeds was observed by transmission electron microscopy (Fig. 5). In young glup4 endosperm at 1 week after flowering (WAF), electron-dense materials (indicated by stars) and granules (indicated by arrowheads) were routinely detected in the extracellular space (Fig. 5D) and the paramural space between the plasma membrane and the cell wall (Fig. 5, E and F), respectively. Such materials and granules were not observed in wild-type endosperm (Fig. 5A). The electron-dense granules in glup4 were labeled by glutelin antibodies (Fig. 6A), indicating that significant amounts of proglutelin were not transported to its normal deposition site, PB-II, but instead were secreted extracellularly.
Figure 5.
Electron micrographs depicting secretory granules and the ultrastructure of PMBs in developing endosperms of glup4. A to C depict sections of developing wild-type endosperm at 1 WAF, 2 WAF, and 3 WAF, respectively. Corresponding sections from glup4 (EM956) are shown in D to F, G to I, and J and K. White stars in D denote the electron-dense materials in the extracellular space, while the arrowheads in E indicate electron-dense granules in the space between the plasma membrane and the cell wall. F shows the presence of a newly formed PMB located between the plasma membrane (arrow) and the cell wall containing electron-dense granules (diamond). White and black diamonds show the distinct inclusions within PMBs in G. In several instances, internal inclusions of the PMBs are surrounded by cell wall or cell wall-like structures (stars in G, I, and J). Arrows in H shows the dense vesicle close to the PMB in cytoplasm. Arrowheads in E, F, I, and K indicate the granules within PMBs. CW, Cell wall; PM, plasma membrane. Bars = 500 nm in A, D, E, and F; 1 μm in B, C, G to I, and K; and 2 μm in J.
Figure 6.
Immunolocalization of glutelin to the electron-dense granules and the PMBs in glup4 (EM956). A, The secreted electron-dense granules located between the cell wall (CW) and the plasma membrane are depicted with arrows. B, Image of PMBs close to the cell wall. C, Enlarged image of the area enclosed by the box in B. D, Enlarged image of PSV. Gold particles (15 nm) indicate the reaction of glutelin antibody. Bars = 1 μm in B and 200 nm in A, C, and D.
The large distended structures containing numerous small granules were readily observed in 2-WAF endosperm. As the large distended structures exist between the plasma membrane and cell wall, we will refer to these structures as PMBs. PMBs were first recognized as invaginations of the plasma membrane containing membranous or vesicular structures (Marchant and Robards, 1968). Interestingly, PMBs were observed in Arabidopsis embryos of the vps9 mutation, which codes for a defective GTP/GDP exchange factor of Rab5 GTPase (Goh et al., 2007).
Ultrastructural analysis showed that the PMBs were nonuniform in appearance. In Figure 5G, the PMB was composed of three separate inclusions: a larger inclusion (indicated by the white diamond) surrounded by a thick cell wall-like material continuous with the cell wall and two smaller adjoining inclusions (indicated by diamonds) enclosed by the plasma membrane and cell wall-like material. The upper inclusion body (diamond 1) appears to contain lamellar membrane structures with electron-dense granules on its surface, while the lower inclusion (diamond 2) was more uniform in appearance but also contained peripherally localized electron-dense granules. Cell wall-like material was also observed associated with other PMBs. The PMB in Figure 5I appears to contain four separate overlapping inclusions, with the inner three covered with cell wall-like layers (indicated by stars). The innermost and smallest layer (1) is surrounded by thick cell wall and contains very little granular material. The next two layers (2 and 3) are surrounded by thinner layers of cell wall-like material and contain numerous granules. The outermost layer (4) is surrounded by the plasma membrane and contains granules of varying electron density (indicated by arrowheads). By contrast to the structures depicted in Figure 5, G and I, the PMB in Figure 5H appears to be devoid of cell wall-like material and contains numerous granules with electron-dense types located on the periphery. Cytoplasm-localized electron-dense granules were also clustered near the PMB (Fig. 5H, arrow). The granules associated with the PMB were labeled with glutelin antibodies (Fig. 6, B and C).
In 3-WAF glup4 endosperm, the PMBs were much larger than those observed in younger endosperm, indicating that they continued to grow during development (Fig. 5, J and K). In Figure 5K, PMBs contained an outer layer of numerous small electron-dense vesicles (arrowheads) that surrounded a central core containing amorphous granules of lighter electron density. Interestingly, the secreted, cell wall-associated electron-dense granules observed in 1- or 2-WAF endosperm were absent in 3-WAF glup4 mutant seeds (Fig. 5E), suggesting that the PMBs were formed by the accumulation of secreted granules. Overall, these results indicate that a significant amount of proglutelin is not transported to its normal deposition site, PB-II, but is secreted extracellularly to form the PMBs and that GLUP4-Rab5a is required for the intracellular trafficking of proglutelins and α-globulin to PSVs.
Colocalization of Proglutelin and Rab5a
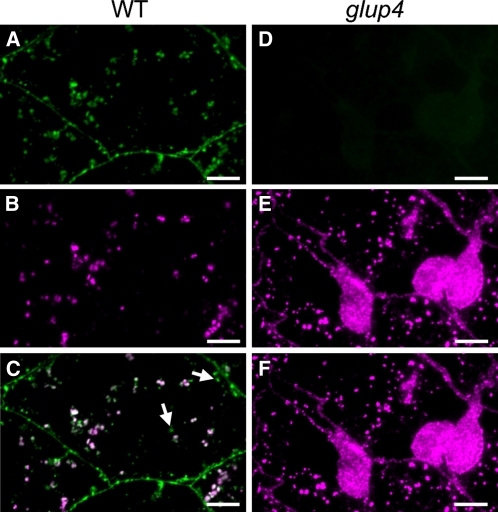
To obtain further insight on the function of GLUP4-Rab5a in the intracellular transportation of proglutelin, immunofluorescence and immunoelectron microscopy studies were performed using glutelin and Rab5a antibodies. When viewed by immunofluorescence, Rab5a is distributed to the plasma membrane and as cytoplasmic granules within wild-type endosperm cells (Fig. 7A). Glutelin is observed as cytoplasmic granules (Fig. 7B), with a significant number colocalizing with Rab5a (Fig. 7C). Immunofluorescence microscopy of glup4 (EM956) seed shows no detectable Rab5a at the plasma membrane or cytoplasm (Fig. 7D), which confirms the absence of Rab5a by immunoblot analysis (Fig. 3). These results demonstrate the specificity of the antibody for GLUP4-Rab5a.
Figure 7.
Subcellular localization of Rab5a in 2-WAF developing wild-type and glup4 (EM956) seeds by immunofluorescence microscopy. A to C, The wild type (WT). D to F, glup4 (EM956). Secondary antibodies labeled with FITC (green) and rhodamine (magenta) were used to visualize the reaction of Rab5a in A and D and glutelin antibodies in B and E, respectively. C and F are the merged images of A/B and D/E, respectively. Arrows indicate the granules showing only Rab5a signal. Bars = 10 μm.
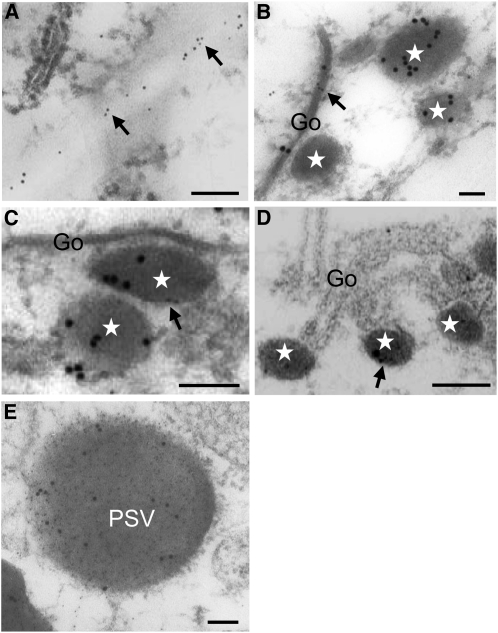
The localization of Rab5a in wild-type endosperm was observed also by immunoelectron microscopy (Fig. 8). Consistent with immunofluorescence analysis, Rab5a is distributed to the cell surface (Fig. 8A). The Golgi apparatus (Fig. 8B), dense vesicles derived from the Golgi apparatus (Fig. 8, B–D), and PSV (Fig. 8E) contained Rab5a as well as glutelin. The colocalization of Rab5a and glutelin in the Golgi apparatus, the dense vesicles, and the PSVs supports a role for GLUP4-Rab5a in the transport of proglutelin from the Golgi apparatus to PSVs.
Figure 8.
Subcellular localization of Rab5a in 2-WAF developing wild-type seeds by immunoelectron microscopy. The distribution of Rab5a and glutelins is denoted by 5-nm (denoted by arrows) and 15-nm gold particles, respectively, in A to D. The distribution of glutelin and Rab5a in normal PSVs is denoted by 5-nm and 15-nm gold particles, respectively, in E. Stars indicate the Golgi-derived dense vesicles. CW, Cell wall; Go, Golgi apparatus. Bars = 100 nm.
Characterization of PMBs in glup4 Endosperm
Distribution of Golgi, PVC, and PSV Markers
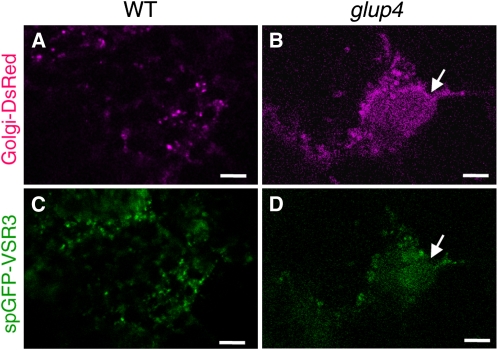
Available evidence indicates that Rab5a functions in vesicular transport between the Golgi and the lytic vacuole via the PVC (Sohn et al., 2003; Kotzer et al., 2004). In an effort to identify the cellular basis for the origin of the PMB in the glup4 mutant, we analyzed the microscopic distribution of biomarkers for various components of the endomembrane system using either fluorescently tagged proteins or by immunofluorescence. A glup4 mutant was transformed with gene constructs coding for Golgi-DsRed (contains β-1,4-galactosyltransferase; Clontech) or spGFP-VSR3 (VSR1 homolog; Shimada et al., 2003), biomarker membrane proteins of the Golgi apparatus and the PVC (Tse et al., 2004), respectively. Confocal laser microscopic analysis of developing endosperm sections from transgenic plants showed that Golgi-DsRed and spGFP-VSR3 were observed as small fluorescent foci distributed throughout the developing endosperm cell in the wild type (Fig. 9, A and C). In glup4 mutants, these proteins were present not only as small foci but were also associated with the PMB (Fig. 9, B and D). The distribution of PVC markers to the PMB is also supported by immunoelectron microscopy studies using antibodies to PV72, which has been shown to be located in the PVC in transgenic BY-2 cells (Miao et al., 2006). Similar to wild-type cells, PV72 is distributed to the PSV PB-II in glup4. In addition, it is also distributed to the PMB in glup4 endosperm (Supplemental Fig. S5). These observations indicate that the normal distribution of Golgi and the PVC markers was disrupted and mislocalized by Rab5a loss of function.
Figure 9.
The distribution of Golgi-DsRed (A and B) and spGFP-VSR3 (C and D) in the wild type and glup4 (EM425). A and C, The wild type (WT). B and D, glup4 (EM425). Transgenic wild-type and glup4 lines expressing Golgi-DsRed (β-galactosyltransferase) or spGFP-VSR3 were generated, and the distribution of native fluorescence generated by the reporters was noted by confocal microscopy. Note that the PMBs (arrows) contain significant amounts of those membrane proteins that are markers for the Golgi and PVC. Bars = 10 μm. [See online article for color version of this figure.]
As mentioned above, the PMBs in the glup4 mutant contain glutelin and α-globulin, which is normally deposited in PSV. Therefore, we analyzed the distribution of α-TiP, the PSV-specific, membrane-associated water intrinsic protein by immunofluorescence microscopy. Supplemental Figure S6, A to C, shows the normal distribution pattern of α-TiP and glutelin within the PSV in the wild type. In glup4, α-TiP is also distributed throughout the PMBs (Supplemental Fig. S6, D–F). Hence, the membrane vesicular trafficking pathway from the Golgi to the PSV is significantly distorted in glup4.
Distribution of ER Chaperones
As reported earlier, BiP and PDI are asymmetrically distributed on the cortical ER, where BiP is enriched on the ER around the protein bodies (PB-I) containing the prolamin while PDIL1-1 is not readily detected in PB-I and appears restricted to the cisternal ER (Muench et al., 1997; Satoh-Cruz et al., 2010a; Onda et al., 2011; Supplemental Fig. S7F). Hence, we analyzed the distribution patterns of BiP and PDIL1-1 in glup4 endosperm. Both of these ER lumenal chaperones were readily observed on the surface of the PMB, although with different spatial distribution patterns (Supplemental Fig. S7, A–C). PDI is located on the inner layer of the PMB surface boundary, while BiP is distributed on an outer layer (Supplemental Fig. S7, D and E). This asymmetric distribution of these chaperones within the PMB is more readily seen in sections viewed near the top of this novel organelle (Supplemental Fig. S7E). This asymmetric distribution of BiP and PDI is more obvious when using red-green colors, as depicted in Supplemental Movie S1, indicating that both luminal chaperons were also secreted. The cortical ER network appears disrupted, as the normal reticulate network seen in the wild type (Supplemental Fig. S7F) is never observed.
When malfolded proteins accumulate in the ER lumen, the unfolded protein response is generated where molecular chaperones such as BiP are induced (Okushima et al., 2002). In the esp2 mutant, which accumulates the proglutelin within the ER, BiP levels are significantly elevated (Takemoto et al., 2002). However, glup4 endosperm contains normal levels of BiP (Ueda et al., 2010). These results suggest that glup4 mutations dramatically alter the ER architecture without inducing an unfolded protein response.
Accumulation of β-Glucan within the PMBs in glup4
Cell wall-like materials were observed in PMBs in glup4 endosperm (Fig. 5). In order to define the nature and distribution of this cell wall-like material associated with the PMBs in glup4 seeds, we performed double-labeling experiments with antibodies against glutelin and (1,3;1,4)-β-glucan or by staining of the β-glucan by calcofluor white. In the wild type, cell walls reacted with the β-glucan antibody (Fig. 10, A and C) and were readily stained by calcofluor white (Supplemental Fig. S8, A and C). In the glup4 line, the β-glucan is readily evident associated with the PMBs (Fig. 10, D and F; Supplemental Fig. S8, D and F). β-Glucan is distributed on the periphery of the PMBs, as suggested by its location relative to glutelin associated with these organelles. The presence of β-glucan suggests that the PMBs in glup4 are modified cell wall appositions, which are typically produced at the sites of fungal penetration through the cell wall (An et al., 2006).
Figure 10.
Immunofluorescense microscopy showing the localization of β-glucan in developing seeds (3 WAF). A to C, The wild type. D to F, glup4 (EM956). Secondary antibodies labeled with FITC (green) and rhodamine (magenta) were used to visualize the reaction of β-glucan in A and D and glutelin antibodies in B and E, respectively. C and F are merged images of A/B and D/E, respectively. Bars = 10 μm.
DISCUSSION
During seed development, rice glutelin polypeptides are initially synthesized on the ER membrane as a 57-kD precursor (Yamagata et al., 1982), which is then transported to the Golgi apparatus and finally to the PSVs (Krishnan et al., 1986; Yamagata and Tanaka, 1986), where the proglutelin is cleaved to acidic and basic subunits (Yamagata et al., 1982). The deposition of glutelins together with α-globulins within the PSV forms PB-II. We have identified various mutants that accumulate high amounts of proglutelins. Two of these mutants, esp2 and glup3, were previously characterized and contain genetic defects in PDIL1-1 and the vacuolar processing enzyme, which function within the ER and PSV, respectively (Takemoto et al., 2002; Kumamaru et al., 2010; Satoh-Cruz et al., 2010a). Genetic interactive analysis is consistent with the view that the remaining glup mutants are likely defective in an intermediate process between one of the initial steps catalyzed by PDIL1-1 in the ER and the final proteolytic processing of proglutelin in the PSV. It was hypothesized that the GLUP4 protein participated in the intracellular trafficking of proglutelin from the Golgi apparatus to the PSV (Ueda et al., 2010). In this study, we show that glup4 expresses a defective Rab5a, the small GTPase that controls a myriad of endocytosis-related processes in animals and plants (Nielsen et al., 2008).
Three glup4 alleles, two of which expressed protein while the third is a null mutation, were identified in this study (Fig. 2). EM960 contains a missense mutation resulting in Gly-45 being replaced by Asp (Fig. 2). This mutation lies in the switch 1 region of the conserved effector-binding domain (Zhu et al., 2004). A second allele, EM425, contains a nonsense mutation resulting in a truncated Rab5a missing the C-terminal 28 residues and, hence, lacks the prenylated Cys residues that facilitate the membrane attachment of Rab5a (van der Bliek, 2005). Hence, these loss-of-function glup4 alleles show that Rab5a requires the modulation of GTPase activity by one or more specific effectors in association with membranes. Moreover, these studies also indicate that the Rab5a antibodies are highly specific, as only a single polypeptide band is evident in the wild type and EM960 and a single truncated polypeptide band in EM425. This antibody specificity is also supported by the absence of Rab5a in immunoblots of EM956 and when EM956 endosperm sections are examined by immunofluorescence using Rab5a antibodies (Fig. 7, compare A and D).
Several studies in plant cells have demonstrated the function of Rab proteins in intracellular trafficking. Arabidopsis Rha1, a Rab5 homolog, and Rab1 play critical roles in the trafficking of soluble cargo protein such as sporamin from the PVC to the central vacuole (Sohn et al., 2003) and transport between the ER and the Golgi apparatus (Batoko et al., 2000), respectively. In animals, Rab5 was first found to control early endosome dynamics but later was found to be involved in several endocytosis-associated processes, including signal transduction. Unlike the case in animal systems, the plant Rab5 orthologs are located on a late endosome compartment, the PVC and Golgi (Sohn et al., 2003; Bolte et al., 2004; Kotzer et al., 2004; Lee et al., 2004; Haas et al., 2007). The Arabidopsis Rab5, RHA1/AtRabF2a, colocalizes with VSR, a marker of the PVC (Sohn et al., 2003; Lee et al., 2004), while a second Arabidopsis Rab5, ARA7/AtRabF2b, localizes to the PVC and Golgi apparatus in tobacco leaf epidermal cells (Kotzer et al., 2004). Wang et al. (2010) reported that the rice Rab5a colocalizes with the PVC marker, VSR2, in Arabidopsis protoplasts. Here, we present direct evidence on the location of GLUP4-Rab5a in developing rice endosperm. In addition to the plasma membrane, this small GTPase is observed on the Golgi apparatus, the Golgi-derived dense vesicles, and the PSV (Fig. 8), the latter observation being, to our knowledge, the first demonstration for the localization of Rab5a protein to this organelle. The loss of function of Rab5a results in the secretion of glutelin and α-globulin to the extracellular space. These observations support a role for GLUP4-Rab5a in the transport of proglutelins from the Golgi apparatus to the PSV. As large amounts of glutelin and α-globulin storage proteins are rapidly transported and accumulated into PSVs during the ripening stage of seed development, Rab5a protein is indispensable for effective transport of these storage proteins. Therefore, the loss of Rab5a function disrupts storage protein transport to the PSV, resulting in secretion of these proteins to the extracellular space, which ultimately leads to the formation of PMBs.
The secretion of soluble cargo as a result of the expression of a variant Rab5 has been demonstrated in other plant systems. These include the expression of dominant negative Rab5 mutants in Arabidopsis (Sohn et al., 2003) or tobacco cells (Kotzer et al., 2004). Likewise, secretion of soluble cargo can also occur if retrograde trafficking between the Golgi and PSV is disturbed. Mutations in VPS29 and VPS35, components of a retromer complex that is responsible for the recycling VSR1 from the PVC to the Golgi complex, result in the secretion of soluble cargo (Shimada et al., 2003, 2006; Yamazaki et al., 2008). Disruptions in vesicular transport between the Golgi and PSV result in rerouting the cargo to an alternative secretion pathway.
The Rab5a-mediated disruption in anterograde transport to the PSV is also reflected in the mislocalization of membrane proteins of the Golgi, PVC, and PSV to the PMB. These biomarkers are β-galactosyltransferase (Golgi), VSR3 and PV72 (PVC), and α-TIP (PSV). Hence, loss of function of Rab5a mediates a dramatic alteration in membrane trafficking in the endomembrane system of developing rice endosperm. The lumenal chaperones, BiP and PDI, are secreted and localized to the PMB (Supplemental Fig. S7). In wild-type endosperm, small amounts of BiP manage to escape the ER retrieval system at the Golgi and are transported to the PSV (Takahashi et al., 2005). Hence, the secretion of BiP in glup4-rab5a mutant seeds follows the same pathway as glutelin. The loss of Rab5a significantly alters the ER. Under light microscopy, the tubular-cisternal network of ER membranes with associated prolamin-containing PB-I was not readily observed (Supplemental Fig. S7). It is not clear whether the tubular network has been structurally altered or whether it is more fragile in glup4 mutants and is disrupted during preparation of the tissue sections for microscopy.
The PMB is surrounded by the lumenal markers BiP and PDI. Interestingly, as evident in the wild type, these lumenal chaperones are asymmetrically distributed on these ER membranes. A confocal microscopic image depicting the top section of the PMB suggests that the spatial distributions of BiP and PDI are distinct, with PDI concentrated within a core region surrounded by BiP (Supplemental Fig. S7). Although further studies are clearly required to discern the spatial arrangement of the ER markers on the PMB, it is apparent that Rab5a is required for maintaining the normal network of ER membranes.
In wild-type endosperm, glutelin RNAs are targeted to the cisternal ER, whereas prolamin RNAs are localized on the PB-ER that bind the prolamin protein bodies (Choi et al., 2000). In addition to reorganizing the endomembrane system, the loss of Rab5a function results in the mislocalization of glutelin RNAs from the cisternal ER to the PB-ER as well as to the PMBs (Doroshenk et al., 2010). By contrast, prolamin RNAs are correctly targeted to the PB-ER in glup4-rab5a. Hence, in developing rice endosperm, GLUP4-Rab5a is involved in multiple processes ranging from intracellular transport between the Golgi and PSV to maintenance of the ER network and targeting of glutelin RNAs. The role of Rab5a in this latter process indicates the involvement of membrane vesicles in RNA transport and localization.
A conspicuous phenotype of glup4-rab5a endosperm is the presence of large dilated PMBs. Although Wang et al. (2010) also reported similar structures, which they referred to as “vesicle-filled structures,” the nature of this novel organelle was not studied in the single Rab5a knockout mutant they identified. Our light and electron microscopy analyses showed that the PMBs exhibit a very complex structure containing multiple inclusions. Inclusions located next to the cell wall were usually covered with β-glucan, while adjoining inclusions positioned within the cytoplasm were bound by the plasma membrane and cell wall-like material (Fig. 5G). The presence of β-glucan on inclusions closest to the cell surface indicates that their formation is initiated extracellularly.
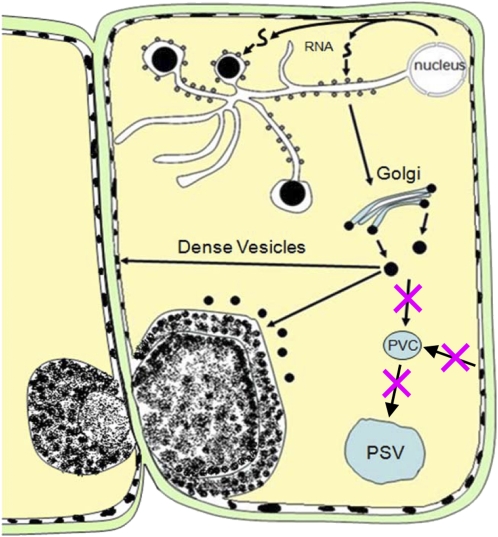
Figure 11 depicts a model that accounts for the cellular events that are likely responsible for PMB formation. Mislocalized glutelin RNAs are translated on the PB-ER, where the protein is exported to the Golgi apparatus, where it is packaged into dense vesicles (Doroshenk et al., 2010). Because of the loss of Rab5a activity in the glup4 mutant, most dense vesicles are not transported to the PSV but are secreted as electron-dense granules to the extracellular space. These granules are prominent in young endosperm cells but are apparently displaced to the growing PMB at later stages (Fig. 5). The PMBs not only contain secreted proglutelins and α-globulins but also membrane markers of the Golgi, PVC, and PSV, although membranes or membrane remnants are not readily detected. The distribution patterns of these storage proteins are not uniform but are spatially partitioned into separate areas of the PMB. This initial PMB is then covered with β-glucan, a feature that distinguishes this rice endosperm type from the ones observed in Arabidopsis (Goh et al., 2007). PMBs are typically seen as simple cytoplasmic invaginations bound by the plasma membrane. The basis for the involvement of β-glucan synthesis in rice endosperm PMBs is unclear, but it is interesting that in many instances, two or more of the PMBs are located on neighboring cells (Figs. 4B and 5J), a spatial arrangement suggesting that they are formed at or close to the plasmodesmata. The plasmodesma contains callose in the cell wall adjacent to the neck regions that participates in regulating the aperture of this intercellular pore. The formation of the PMBs at or near the plasmodesmata as well as the general disruption in endomembrane organization may induce callose synthesis as the PMB develops (Supplemental Fig. S8).
Figure 11.
A model for the formation of PMBs in the glup4 mutation. In the wild type, glutelin RNAs are translated in the cisternal ER, where the protein is exported to the Golgi apparatus, where it is packaged into dense vesicles. These dense vesicles are then transported to the PSV. The loss of Rab5a activity disrupts several processes, beginning with the mislocalization of glutelin RNAs to the ER that bound the prolamin protein bodies. Glutelin-containing dense vesicles are unable to merge with the PSV but secrete extracellularly. These granules accumulate between the plasma membrane and the cell wall, where they are partially endocytosed to form the PMBs. The layered appearance of the PMBs may be due to the sequential accumulation of cell wall material and the direct transport of electron-dense granules into the PMBs from the cytoplasm. [See online article for color version of this figure.]
In addition to the β-glucan-bound inclusions, the PMBs also contain inclusions bound solely by the plasma membrane. These inclusions are also filled with granules that are more electron dense than those seen in the β-glucan-associated inclusions (Fig. 5, G–I and K). Similarly sized, electron-dense granules are also evident in the cytoplasm located near or at the surface of the PMB (Fig. 5, H and I, respectively). The growth of the PMB during seed development and the close spatial relationship between the electron-dense granules on the PMB periphery and the cytoplasm suggest that these electron-dense granules are secreted directly into the PMB. Hence, PMBs are formed by a combination of secreted granules that are partially endocytosed, which may occur in early endosperm development, and the direct secretion of electron-dense granules at subsequent stages. It is speculated that this dual pathway would account for the appearance of multiple inclusions of the PMBs, some covered with cell wall-like material while others are bound only by the plasma membrane and contain electron-dense granules.
Although this study is directed at the major changes in trafficking and endomembrane organization induced by a loss of function of GLUP4-Rab5a in developing rice endosperm cells, this small GTPase is also essential for optimal plant growth. The glup4 mutant lines grow much slower than the wild type and take longer to flower (data not shown). Previous studies have shown that the rice GLUP4-Rab5a (AK061116) is required for normal nutrient-regulated root growth (Wang et al., 2002). Consistent with this view, GLUP4-Rab5a (AK061116) had the highest expression in root with weaker expression in shoots, flowers, and immature grains (refer to RiceXPro).
MATERIALS AND METHODS
Materials
The rice (Oryza sativa) glup4 mutant lines EM425, EM956, and EM960, induced by N-methyl-N-nitrosourea mutagenesis (Satoh et al., 2010) and characterized by their accumulation of substantial amounts of the 57-kD proglutelin (Satoh-Cruz et al., 2010b; Ueda et al., 2010), were used in these experiments. The full-length cDNA clone for OsVSR3 (AK072667) was obtained from the National Institute of Agrobiological Sciences in Tsukuba, Japan. pAcGFP1-Golgi and pDsRed-Monomer-N1 were purchased from Clontech. Rice plants were grown in the field or in a transgenic glasshouse at the Faculty of Agriculture, Kyushu University, and developing seeds, 5 to 28 d after flowering, were isolated and used in biochemical and microscopic analyses.
SDS-PAGE and Western-Blot Analysis
Proteins were extracted from seeds using 0.125 m Tris-HCl, 4% SDS, 4 m urea, and 5% β-mercaptoethanol, pH 6.8 (Ushijima et al., 2011). The proteins were resolved by SDS-PAGE, electrotransferred to nitrocellulose membranes, and then incubated for 1 h with Tris-buffered saline (TBS), 10 mm Tris-HCl, 0.15 m NaCl, 5% skim milk, pH 7.5, and primary antibody (1:1,000 dilution). After primary antibody binding, the blot was washed three times with TBS containing 0.05% Tween 20 (TBST) and then incubated with TBS containing 5% skim milk and the secondary antibody (1:2,500 dilution). The blot was washed three times with TBST and incubated with the enhanced chemiluminescence detection kit (GE Healthcare). It was then placed against x-ray film for detection of the bound primary antibody.
DNA Sequencing Analysis
DNA sequencing analysis was performed as described previously (Kumamaru et al., 2010). Total genomic DNA from the leaves of glup4 lines and the wild type was obtained using the cetyltrimethylammonium bromide method (Murray and Thompson, 1980). DNA sequence was determined using an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). DNA sequence analysis was performed using EditView 1.0.1 and AutoAssembler 2.1. Comparisons between wild-type and mutant sequences were performed using ClustalW of the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/top-e.html).
IP
Freshly harvested middevelopment rice seeds were dehulled, and then 1 g was extracted in 1.5 mL of IP buffer (20 mm Tris-HCl, pH 7.5, 75 mm NaCl, 1 mm EDTA, and 0.1% Nonidet P-40). The extract was then centrifuged for 10 min at 20,000g, and the supernatant was removed to a new tube and subjected to additional centrifugation using the same conditions. Protein A resin, which had previously been incubated overnight with either 1× phosphate-buffered saline alone (no antibody control) or anti-Rab5a antibody, was washed three times with 1 mL of IP buffer prior to the addition of the clarified seed extract prepared as described above. Extracts and protein A resins were incubated for 2 h prior to washing five times with 1 mL of IP buffer. The protein A resins were then boiled in SDS-containing extraction buffer and subjected to western-blot analysis using Rab5a antibody at a dilution of 1:1,000.
Plasmid Construction and Rice Transformation
AcGFP1-Golgi (Clontech) contains a segment of human β-1,4-galactosyltransferase and localizes at the trans-medial region of the Golgi apparatus (Miao et al., 2006). The GFP of the fusion protein was replaced with DsRed-Monomer (Clontech), generating Golgi-DsRed. The C-terminal region of OsVSR3 containing the membrane-spanning domain was amplified by PCR with primers 5′-ATAGGATCCAGCAAAGTTGCTTCTTCGTC-3′ and 5′-TATCTCGAGCCCAGTTAAGCTTGCTGCAA-3′ using a full-length cDNA clone (AK072667) as a template. The PCR product was then inserted downstream of the spGFP gene (Kawagoe et al., 2005), generating spGFP-VSR3. The two fusion genes were expressed under the control of the rice β-TIP promoter (Onda et al., 2009). The binary vector containing Golgi-DsRed and spGFP-VSR3 was constructed by the Gateway system (Invitrogen) as described previously (Onda et al., 2009). Transformation of rice was performed as described previously (Kawagoe et al., 2005).
Microscopic Analysis
Laser-scanning confocal microscopic analysis was performed as described previously (Onda et al., 2009). Immunocytochemical and fluorescence microscopic analyses were performed as described previously (Kumamaru et al., 2010). Samples were treated with antibodies raised against glutelin basic subunit (1:5,000), 14-kD prolamin (1:1,000), α-globulin (1:5,000), and α-TiP (1:1,000). Red fluorescent images were converted to magenta with Adobe Photoshop. Sections were examined with a transmission electron microscope at 80 kV.
Antibodies
Seed storage proteins were separated by SDS-PAGE, and individual bands were excised and solubilized by preparative electrophoresis. Antibodies to β-glutelin were then raised in mice. Antibodies against α-globulin and α-TiP were raised in rabbits. Antibody against 14-kD prolamin was generated as described previously (Nagamine et al., 2011). Antibodies against BiP and PDI were generated as described previously (Satoh-Cruz et al., 2010a). Antibodies against the recombinant expressed rice Rab5a were raised in rabbits. Antibody against (1,3;1,4)-β-glucan was purchased from Biosupplies Australia.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AK061116.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression analysis of Rab5a homologs in rice seed.
Supplemental Figure S2. Reverse transcription-PCR analysis of Rab5a homologs in developing rice seed.
Supplemental Figure S3. A phylogenetic tree of Rab5 proteins of rice and Arabidopsis.
Supplemental Figure S4. Immunofluorescence microscopy of protein bodies and the novel PMB structures in various glup4 allelic lines.
Supplemental Figure S5. Immunoelectron localization of the PVC marker PV72.
Supplemental Figure S6. Subcellular localization of α-TiP by immunofluorescence microscopy in 2-week-old developing glup4 endosperm.
Supplemental Figure S7. Subcellular localization of BiP and PDI in 2-week-old developing glup4 endosperm.
Supplemental Figure S8. Confocal microscopic images showing the PMB stained by calcofluor white.
Supplemental Movie S1. Localization of BiP and PDIL1-1 in developing endosperm of the glup4 mutant at 2 WAF.
Acknowledgments
We thank Dr. K. Yoshida (Taisei Co.) for valuable suggestions. The expression profile of Rab5a was obtained from the Rice Expression Profile Database (RiceXPro; http://ricexpro.dna.affrc.go.jp).
References
- An Q, Hückelhoven R, Kogel KH, van Bel AJ. (2006) Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell Microbiol 8: 1009–1019 [DOI] [PubMed] [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I. (2000) A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Brown S, Satiat-Jeunemaitre B. (2004) The N-myristoylated Rab-GTPase m-Rabmc is involved in post-Golgi trafficking events to the lytic vacuole in plant cells. J Cell Sci 117: 943–954 [DOI] [PubMed] [Google Scholar]
- Brown JW. (1996) Arabidopsis intron mutations and pre-mRNA splicing. Plant J 10: 771–780 [DOI] [PubMed] [Google Scholar]
- Choi SB, Wang C, Muench DG, Ozawa K, Franceschi VR, Wu Y, Okita TW. (2000) Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 407: 765–767 [DOI] [PubMed] [Google Scholar]
- Chrispeels MJ. (1983) The Golgi apparatus mediates the transport of phytohemagglutinin to the protein bodies in bean cotyledon. Planta 158: 140–151 [DOI] [PubMed] [Google Scholar]
- Doroshenk KA, Crofts AJ, Washida H, Satoh-Cruz M, Crofts N, Okita TW, Morris RT, Wyrick JJ, Fukuda M, Kumamaru T, et al. (2010) Characterization of the rice glup4 mutant suggests a role for the small GTPase Rab5 in the biosynthesis of carbon and nitrogen storage reserves in developing endosperm. Breed Sci 60: 556–567 [Google Scholar]
- Goh T, Uchida W, Arakawa S, Ito E, Dainobu T, Ebine K, Takeuchi M, Sato K, Ueda T, Nakano A. (2007) VPS9a, the common activator for two distinct types of Rab5 GTPases, is essential for the development of Arabidopsis thaliana. Plant Cell 19: 3504–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorvel JP, Chavrier P, Zerial M, Gruenberg J. (1991) rab5 controls early endosome fusion in vitro. Cell 64: 915–925 [DOI] [PubMed] [Google Scholar]
- Haas TJ, Sliwinski MK, Martínez DE, Preuss M, Ebine K, Ueda T, Nielsen E, Odorizzi G, Otegui MS. (2007) The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell 19: 1295–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I, Shimada T, Hatano K, Takeuchi Y, Nishimura M. (1998) Transport of storage proteins to protein storage vacuoles is mediated by large precursor-accumulating vesicles. Plant Cell 10: 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz G, Colanesi S, Hillmer S, Rogers JC, Robinson DG. (2007) Localization of vacuolar transport receptors and cargo proteins in the Golgi apparatus of developing Arabidopsis embryos. Traffic 8: 1452–1464 [DOI] [PubMed] [Google Scholar]
- Hohl I, Robinson DG, Chrispeels MJ, Hinz G. (1996) Transport of storage proteins to the vacuole is mediated by vesicles without a clathrin coat. J Cell Sci 109: 2539–2550 [DOI] [PubMed] [Google Scholar]
- Kawagoe Y, Suzuki K, Tasaki M, Yasuda H, Akagi K, Katoh E, Nishizawa NK, Ogawa M, Takaiwa F. (2005) The critical role of disulfide bond formation in protein sorting in the endosperm of rice. Plant Cell 17: 1141–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch T, Paris N, Butler JM, Beevers L, Rogers JC. (1994) Purification and initial characterization of a potential plant vacuolar targeting receptor. Proc Natl Acad Sci USA 91: 3403–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzer AM, Brandizzi F, Neumann U, Paris N, Moore I, Hawes C. (2004) AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J Cell Sci 117: 6377–6389 [DOI] [PubMed] [Google Scholar]
- Krishnan HB, Franceschi VR, Okita TW. (1986) Immunochemical studies on the role of the Golgi complex in protein-body formation in rice seeds. Planta 169: 471–480 [DOI] [PubMed] [Google Scholar]
- Krishnan HB, White JA. (1995) Morphometric analysis of rice seed protein bodies (implication for a significant contribution of prolamine to the total protein content of rice endosperm). Plant Physiol 109: 1491–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HB, White JA, Pueppke SG. (1990) Immunocytochemical evidence for the involvement of the Golgi apparatus in the transport of the vacuolar protein, gamma-secalin, in rye (Secale cereale) endosperm. Ceral Chem. 67: 360–366 [Google Scholar]
- Krishnan HB, White JA, Pueppke SG. (1992) Characterization and location of rice (Oryza sativa L.) seed globulins. Plant Sci 81: 1–11 [Google Scholar]
- Kumamaru T, Ogawa M, Satoh H, Okita TW. (2007) Protein body biogenesis in cereal endosperms. Olsen OA, , Endosperm: Development and Molecular Biology, Vol 8. Springer-Verlag, Berlin, pp 141–158 [Google Scholar]
- Kumamaru T, Satoh H, Iwata N, Omura T, Ogawa M. (1987) Mutants for rice storage proteins. III. Genetic analysis of mutants for storage proteins of protein bodies in the starchy endosperm. Jpn J Genet 62: 333–339 [DOI] [PubMed] [Google Scholar]
- Kumamaru T, Satoh H, Iwata N, Omura T, Ogawa M, Tanaka K. (1988) Mutants for rice storage proteins. 1. Screening of mutants for rice storage proteins of protein bodies in the starchy endosperm. Theor Appl Genet 76: 11–16 [DOI] [PubMed] [Google Scholar]
- Kumamaru T, Uemura Y, Inoue Y, Takemoto Y, Siddiqui SU, Ogawa M, Hara-Nishimura I, Satoh H. (2010) Vacuolar processing enzyme plays an essential role in the crystalline structure of glutelin in rice seed. Plant Cell Physiol 51: 38–46 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Sohn EJ, Lee MH, Hwang I. (2004) The Arabidopsis rab5 homologs rha1 and ara7 localize to the prevacuolar compartment. Plant Cell Physiol 45: 1211–1220 [DOI] [PubMed] [Google Scholar]
- Li L, Shimada T, Takahashi H, Ueda H, Fukao Y, Kondo M, Nishimura M, Hara-Nishimura I. (2006) MAIGO2 is involved in exit of seed storage proteins from the endoplasmic reticulum in Arabidopsis thaliana. Plant Cell 18: 3535–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant R, Robards AW. (1968) Membrane systems associated with the plasmalemma of plant cells. Ann Bot (Lond) 32: 457–471 [Google Scholar]
- Miao Y, Yan PK, Kim H, Hwang I, Jiang L. (2006) Localization of green fluorescent protein fusions with the seven Arabidopsis vacuolar sorting receptors to prevacuolar compartments in tobacco BY-2 cells. Plant Physiol 142: 945–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muench DG, Wu Y, Zhang Y, Li X, Boston RS, Okita TW. (1997) Molecular cloning, expression and subcellular localization of a BiP homolog from rice endosperm tissue. Plant Cell Physiol 38: 404–412 [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine A, Matsusaka H, Ushijima T, Kawagoe Y, Ogawa M, Okita TW, Kumamaru T. (2011) A role for the cysteine-rich 10 kDa prolamin in protein body I formation in rice. Plant Cell Physiol 52: 1003–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E, Cheung AY, Ueda T. (2008) The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiol 147: 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Kumamaru T, Satoh H, Omura T, Park T, Shintaku K, Baba K. (1989) Mutants for rice storage proteins. 2. Isolation and characterization of protein bodies from rice mutants. Theor Appl Genet 78: 305–310 [DOI] [PubMed] [Google Scholar]
- Okita TW, Rogers JC. (1996) Compartmentation of proteins in the endomembrane system of plant cells. Annu Rev Plant Physiol Plant Mol Biol 47: 327–350 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Koizumi N, Yamaguchi Y, Kimata Y, Kohno K, Sano H. (2002) Isolation and characterization of a putative transducer of endoplasmic reticulum stress in Oryza sativa. Plant Cell Physiol 43: 532–539 [DOI] [PubMed] [Google Scholar]
- Onda Y, Kumamaru T, Kawagoe Y. (2009) ER membrane-localized oxidoreductase Ero1 is required for disulfide bond formation in the rice endosperm. Proc Natl Acad Sci USA 106: 14156–14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onda Y, Nagamine A, Sakurai M, Kumamaru T, Ogawa M, Kawagoe Y. (2011) Distinct roles of protein disulfide isomerase and P5 sulfhydryl oxidoreductases in multiple pathways for oxidation of structurally diverse storage proteins in rice. Plant Cell 23: 210–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Herder R, Schulze J, Jung R, Staehelin LA. (2006) The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies. Plant Cell 18: 2567–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Mohammed O, Rogers JC. (2007) Golgi-mediated vacuolar sorting in plant cells: RMR proteins are sorting receptors for the protein aggregation/membrane internalization pathway. Plant Sci 172: 728–745 [Google Scholar]
- Satoh H, Matsusaka H, Kumamaru T. (2010) Use of N-methyl-N-nitrosourea treatment of fertilized egg cells for saturation mutagenesis of rice. Breed Sci 60: 475–485 [Google Scholar]
- Satoh-Cruz M, Crofts AJ, Takemoto-Kuno Y, Sugino A, Washida H, Crofts N, Okita TW, Ogawa M, Satoh H, Kumamaru T. (2010a) Protein disulfide isomerase like 1-1 participates in the maturation of proglutelin within the endoplasmic reticulum in rice endosperm. Plant Cell Physiol 51: 1581–1593 [DOI] [PubMed] [Google Scholar]
- Satoh-Cruz M, Fukuda M, Ogawa M, Satoh H, Kumamaru T. (2010b) Glup4 gene encodes small GTPase, Rab5a in rice. Rice Genet Newsl 25: 48–49 [Google Scholar]
- Shewry PR, Napier JA, Tatham AS. (1995) Seed storage proteins: structures and biosynthesis. Plant Cell 7: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Fuji K, Tamura K, Kondo M, Nishimura M, Hara-Nishimura I. (2003) Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 16095–16100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Koumoto Y, Li L, Yamazaki M, Kondo M, Nishimura M, Hara-Nishimura I. (2006) AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol 47: 1187–1194 [DOI] [PubMed] [Google Scholar]
- Shimada T, Kuroyanagi M, Nishimura M, Hara-Nishimura I. (1997) A pumpkin 72-kDa membrane protein of precursor-accumulating vesicles has characteristics of a vacuolar sorting receptor. Plant Cell Physiol 38: 1414–1420 [DOI] [PubMed] [Google Scholar]
- Sohn EJ, Kim ES, Zhao M, Kim SJ, Kim H, Kim YW, Lee YJ, Hillmer S, Sohn U, Jiang L, et al. (2003) Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell 15: 1057–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Saito Y, Kitagawa T, Morita S, Masumura T, Tanaka K. (2005) A novel vesicle derived directly from endoplasmic reticulum is involved in the transport of vacuolar storage proteins in rice endosperm. Plant Cell Physiol 46: 245–249 [DOI] [PubMed] [Google Scholar]
- Takemoto Y, Coughlan SJ, Okita TW, Satoh H, Ogawa M, Kumamaru T. (2002) The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol 128: 1212–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Sugimoto T, Ogawa M, Kasai Z. (1980) Isolation and characterization of two types of protein bodies in the rice endosperm. Agric Biol Chem 44: 1633–1639 [Google Scholar]
- Tse YC, Mo B, Hillmer S, Zhao M, Lo SW, Robinson DG, Jiang L. (2004) Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16: 672–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y, Satoh-Cruz M, Matsusaka H, Takemoto-Kuno Y, Fukuda M, Okita TW, Ogawa M, Satoh H, Kumamaru T. (2010) Gene-gene interactions between mutants that accumulate abnormally high amounts of proglutelin in rice seed. Breed Sci 60: 568–574 [Google Scholar]
- Ushijima T, Matsusaka H, Jikuya H, Ogawa M, Satoh H, Kumamaru T. (2011) Genetic analysis of cysteine-poor prolamin polypeptides reduced in the endosperm of the rice esp1 mutant. Plant Sci 181: 125–131 [DOI] [PubMed] [Google Scholar]
- van der Bliek AM. (2005) A sixth sense for Rab5. Nat Cell Biol 7: 548–550 [DOI] [PubMed] [Google Scholar]
- Wang X, Xia M, Chen Q, Wu Z, Wu P. (2002) Identification of a new small GTP-binding protein gene OsRab5a, genomic organization, and expression pattern analysis during nitrate supply and early nutrient starvation in rice (Oryza sativa L.) root. Plant Sci 163: 273–280 [Google Scholar]
- Wang Y, Ren Y, Liu X, Jiang L, Chen L, Han X, Jin M, Liu S, Liu F, Lv J, et al. (2010) OsRab5a regulates endomembrane organization and storage protein trafficking in rice endosperm cells. Plant J 64: 812–824 [DOI] [PubMed] [Google Scholar]
- Woollard AA, Moore I. (2008) The functions of Rab GTPases in plant membrane traffic. Curr Opin Plant Biol 11: 610–619 [DOI] [PubMed] [Google Scholar]
- Yamagata H, Sugimoto T, Tanaka K, Kasai Z. (1982) Biosynthesis of storage proteins in developing rice seeds. Plant Physiol 70: 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata H, Tanaka K. (1986) The site of synthesis and accumulation of storage proteins. Plant Cell Physiol 27: 135–145 [Google Scholar]
- Yamazaki M, Shimada T, Takahashi H, Tamura K, Kondo M, Nishimura M, Hara-Nishimura I. (2008) Arabidopsis VPS35, a retromer component, is required for vacuolar protein sorting and involved in plant growth and leaf senescence. Plant Cell Physiol 49: 142–156 [DOI] [PubMed] [Google Scholar]
- Zhao W-M, Gatehouse JA, Boulter D. (1983) The purification and partial amino acid sequence of a polypeptide from the glutelin fraction of rice grains: homology to pea legumin. FEBS Lett 162: 96–102 [Google Scholar]
- Zhu G, Zhai P, Liu J, Terzyan S, Li G, Zhang XC. (2004) Structural basis of Rab5-Rabaptin5 interaction in endocytosis. Nat Struct Mol Biol 11: 975–983 [DOI] [PubMed] [Google Scholar]
- Zouhar J, Rojo E, Bassham DC. (2009) AtVPS45 is a positive regulator of the SYP41/SYP61/VTI12 SNARE complex involved in trafficking of vacuolar cargo. Plant Physiol 149: 1668–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]