Abstract
Compounds of the terpenoid class play numerous roles in the interactions of plants with their environment, such as attracting pollinators and defending the plant against pests. We show here that the genome of cultivated tomato (Solanum lycopersicum) contains 44 terpene synthase (TPS) genes, including 29 that are functional or potentially functional. Of these 29 TPS genes, 26 were expressed in at least some organs or tissues of the plant. The enzymatic functions of eight of the TPS proteins were previously reported, and here we report the specific in vitro catalytic activity of 10 additional tomato terpene synthases. Many of the tomato TPS genes are found in clusters, notably on chromosomes 1, 2, 6, 8, and 10. All TPS family clades previously identified in angiosperms are also present in tomato. The largest clade of functional TPS genes found in tomato, with 12 members, is the TPS-a clade, and it appears to encode only sesquiterpene synthases, one of which is localized to the mitochondria, while the rest are likely cytosolic. A few additional sesquiterpene synthases are encoded by TPS-b clade genes. Some of the tomato sesquiterpene synthases use z,z-farnesyl diphosphate in vitro as well, or more efficiently than, the e,e-farnesyl diphosphate substrate. Genes encoding monoterpene synthases are also prevalent, and they fall into three clades: TPS-b, TPS-g, and TPS-e/f. With the exception of two enzymes involved in the synthesis of ent-kaurene, the precursor of gibberellins, no other tomato TPS genes could be demonstrated to encode diterpene synthases so far.
Terpenes constitute a large class of compounds that serve multiple roles in plants. They are made of a five-carbon isoprene-building unit. Some hormones, such as GAs, abscisic acid, strigolactones, and cytokinins, are made up in whole or in part by terpenes. A much larger class of terpenes belong to the class of compounds defined as specialized metabolites, those compounds whose synthesis is restricted to specific plant lineages and that serve specific physiological and ecological roles. Many thousands of such terpenes are known in the plant kingdom, with each plant species capable of synthesizing only a small fraction of this repertoire (Pichersky et al., 2006; Chen et al., 2011).
The enzymes that synthesize the backbone of the specialized monoterpenes (C10), sesquiterpenes (C15), and diterpenes (C20), as well as isoprene and the diterpene ent-kaurene, the precursor of GAs, belong to the structurally related terpene synthase (TPS) family (Bohlmann et al., 1998). At one extreme, the genome of the moss Physcomitrella patens has a single active TPS gene, encoding the bifunctional enzyme copalyl diphosphate synthase/kaurene synthase (CPS/KS; Hayashi et al., 2006). This protein is responsible for the synthesis of ent-kaurene from geranylgeranyl diphosphate (GGPP) via the intermediate copalyl diphosphate (CPP). The TPS gene families in other plant genomes examined range in size from 19 to 152 (Chen et al., 2011).
A recent classification of TPS genes divides them into seven clades: TPS-a, TPS-b, TPS-c, TPS-d, TPS-e/f, TPS-g, and TPS-h. TPS-d is a gymnosperm-specific clade, and TPS-h is thus far specific to the lycopod Selaginalla moellendorffii (Chen et al., 2011). TPS-a, TPS-b, and TPS-g are angiosperm-specific clades, with TPS-a containing mostly sesquiterpene and diterpene synthases and the TPS-b clade, together with the much smaller TPS-g clade, consisting mostly of monoterpene synthases. TPS-c is believed to be the ancestral clade, and it contains the gymnosperm and angiosperm CPS genes. TPS-e/f contains gymnosperm and angiosperm KS genes (in angiosperm and gymnosperm plants, ent-kaurene is also synthesized from GGPP via CPP in two steps, but the reactions are catalyzed by separate CPS and KS enzymes) and various other TPSs (see below).
To date, the TPS gene family has been studied most extensively in Arabidopsis (Arabidopsis thaliana) Columbia ecotype. About one-third of the proteins encoded by its 32 functional or potentially functional TPS genes (the genome also contains eight mutated TPS genes) have been assigned a biochemical function in the synthesis of monoterpenes, sesquiterpenes, and diterpenes in flowers, leaves, or roots (Sun and Kamiya, 1994; Yamaguchi et al., 1998; Bohlmann et al., 2000; Chen et al., 2003, 2004; Fäldt et al., 2003; Tholl et al., 2005; Herde et al., 2008). The roles of these terpenes range from pollinator attraction to defense against a range of biotic agents (Gershenzon and Dudareva, 2007). Most of the functional Arabidopsis TPS genes are expressed constitutively, whereas some are induced under specific stress conditions (Tholl et al., 2005; Herde et al., 2008; Huang et al., 2010).
While the genome sequences of several cultivated plants, such as rice (Oryza sativa), sorghum (Sorghum bicolor), and poplar (Populus trichocarpa), have been substantially determined (International Rice Genome Sequencing Project, 2005; Tuskan et al., 2006; Paterson et al., 2009), detailed analysis of the presence and role of terpenes in these plants has not been published. The flowers of all three of these species are wind pollinated, making it unlikely that terpenes contribute significantly to their pollination. However, recent analysis has shown that they have TPS gene families comparable in size to that of Arabidopsis (Chen et al., 2011), and it is likely that further investigation will uncover specific roles for many of their TPS genes. Grape (Vitis vinifera), on the other hand, has an extremely large TPS gene family with perhaps as many as 69 active TPS genes, commensurate with the known presence of a large variety of mostly monoterpenes in the flowers and fruits (Martin et al., 2010). However, little is known about the function of grape TPS genes outside the reproductive structures.
Tomato (Solanum lycopersicum) is a widely cultivated herbaceous crop plant whose genome was recently substantially determined by the International Tomato Genome Sequencing Consortium (Solgenomics.net). Tomato terpenes are found in large quantities in the glandular trichomes that are present on its leaves, stems, young fruits, and part of the flowers and are believed to act as defense compounds (van Schie et al., 2007; Schilmiller et al., 2009, 2010a; Kang et al., 2010). Insect herbivory as well as jasmonic acid (JA) treatment increase the production of some monoterpenes such as linalool as well as of (E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene (TMTT), a C16 homoterpene derived from oxidative cleavage of the diterpene geranyllinalool (Ament et al., 2004; Herde et al., 2008). However, in contrast to the fruits of many other species, where volatile monoterpenes and sesquiterpenes are a major determinant of flavor (Sharon-Asa et al., 2003; Aharoni et al., 2004; Pechous and Whitaker, 2004), the tomato fruit is mostly devoid of such volatile terpenes, although some aroma compounds are derived from the degradation of carotenoids (Lewinsohn et al., 2005; Goff and Klee, 2006; Davidovich-Rikanati et al., 2007).
Consistent with the hypothesis that terpenes are produced at low levels in nontrichome cells, TPS transcripts are not well represented in EST databases from various tissues (Schilmiller et al., 2009, 2010b; Dai et al., 2010). Mining these databases as well as homology-based gene isolation techniques have led to the identification in tomato and related species of several typical monoterpene and sesquiterpene synthases that use the transprenyl diphosphates geranyl diphosphate (GPP) and e,e-farnesyl diphosphate (trans-trans-FPP, usually referred to as FPP), respectively, as substrates (van der Hoeven et al., 2000; van Schie et al., 2007). In contrast, an unusual plastid-localized monoterpene synthase that is active in cultivated tomato trichomes and uses neryl diphosphate (NPP), the cis-isomer of GPP, to catalyze the formation of β-phellandrene and several other monoterpenes was also recently described, together with the gene encoding the enzyme neryl diphosphate synthase (NDPS1), responsible for the synthesis of NPP (Schilmiller et al., 2009). In total, however, the functions of only eight tomato TPS genes have been reported to date.
The determination of the sequence of the cultivated tomato genome has made it possible to characterize its TPS gene family and to study the roles of terpenes and terpene synthases in this important crop. The availability of closely related wild Solanum species allows us to test how domestication has affected the structure and function of TPS genes. We report here that with a total of 29 functional or potentially functional genes and 15 mutated genes found to date, the tomato TPS gene family is similar in size to that of Arabidopsis. As with Arabidopsis, the majority of the putatively functional TPS genes in tomato show some expression in one or more organs. However, the tomato genome has a significantly larger number of sesquiterpene synthases than Arabidopsis, and thus far the only identified diterpene synthases are those involved in GA biosynthesis.
RESULTS
Analysis of TPS Gene Models in the Tomato Genome
Searches (see “Materials and Methods”) of the tomato genomic sequence provided by the International Tomato Genome Sequencing Consortium (http://solgenomics.net/genomes/Solanum_lycopersicum/; the July 2011 release was the latest used in this study) identified multiple gene models with homology to TPS sequences (Table I; Supplemental Fig. S1). We designated these gene models as TPS1 through TPS44. As with any early-stage genome sequence releases, sequential tomato genome releases have presented revisions of earlier releases, and we have accordingly modified and improved our TPS gene models with each update. Such improvements have included the elimination of gene models recognized by the curators of the genome sequence or by us to be the same sequence assembled elsewhere in the genome or incorrectly assembled sequences. In our analysis, in some cases when the sequence of the gene from the first ATG codon to the termination codon was incomplete but cDNAs could be obtained (in this study or elsewhere) or sufficient genomic sequences were available, we amplified genomic DNA spanning the gaps and determined the sequence of the amplified fragments, thus completing the sequence of the exons and introns of the gene. Changes to the annotations of individual genes from our work are found in the Supplemental Figure S1.
Table I. A list of tomato terpene synthase genes, their characteristics, and the enzymes they encode.
| Chromosome | TPS | No. of Exons | Transcript Observed by RT-PCR | Full-Length cDNA Constructed | Main Product(s) of in Vitro Enzymatic Activity | Substrate |
| 1 | 1 | 7 | Yes | Noa | Mutant | – |
| 2 | 3(−)b | No | No | Mutant | – | |
| 3 | 7 | Yes | Yesc | Camphene/tricyclenec | GPP | |
| 4 (MTS2) | 7 | Yes | Yesd | β-Phellandrened | GPP | |
| 5 (MTS1) | 8 | Yes | Yesd | Linaloold | GPP | |
| 6 | 7 | No | No | Mutant | – | |
| 7 | 7 | Yes | Yesc | β-Myrcene/limonenec | GPP | |
| 8 | 7 | Yes | Yesc | 1,8-Cineolec | GPP | |
| 1,8-Cineolec | NPP | |||||
| 22 | 7 | No | No | Mutant | – | |
| 30 | (−)3(−)1 | No | No | Mutant | – | |
| 31 | 6 | Yes | Yese | Viridiflorenee | e,e-FPP | |
| 32 | 6 | Yes | Yesc | Viridiflorenec | e,e-FPP | |
| Unidentifiedc | z,z-FPP | |||||
| 33 | 6 | Yes | No | NDf | – | |
| 34 | (−)3 | Yes | No | Mutant | – | |
| 35 | 6 | Yes | No | ND | – | |
| 2 | 25 | 7 | No | No | ND | – |
| 26 | (−)4 | No | No | Mutant | – | |
| 27 | 7 | No | No | ND | – | |
| 38 | 7 | Yes | Yesc | α-Bergamotenec | e,e-FPP | |
| 4 | 28 | 7 | No | No | ND | – |
| 5 | 43 | 7 | No | No | Mutant | |
| 6 | 9 (Sst1) | 7 | Yes | Yesg | Germacreneg | e,e-FPP |
| 10 | 7 | Yes | No | ND | – | |
| 11 | 2(−)2(−) | No | No | Mutant | – | |
| 12 (CAHS) | 7 | Yes | Yesh | β-Caryophyllene/α-humuleneh | e,e-FPP | |
| 13 | 7 | Yes | Noa | Mutant | – | |
| 23 | (−)1 | No | No | Mutant | – | |
| 36 | 7 | Yes | Yesc | Unidentifiedc | z,z-FPP | |
| 40 (CPS1) | 15 | Yes | Yesi | Copalyl diphosphatei | GGPP | |
| 7 | 15 | 7 | Yes | Noa | Mutant | – |
| 16 | 7 | Yes | No | ND | – | |
| 24 (KS) | 15 | Yes | Yesc | ent-Kaurenec | CPP | |
| 8 | 18 | 14 | Yes | Yesc | ND | – |
| 19 | 14 | Yes | No | ND | – | |
| 20 (PHS1) | 14 | Yes | Yesj | β-Phellandrenej | NPP | |
| 21 | 14 | Yes | Yesc | ND | – | |
| 41 | 12 | Yes | Yesb | ND | – | |
| 9 | 14 | 7 | Yes | No | β-Bisabolenec | e,e-FPP |
| α-Bisabolenec | z,z-FPP | |||||
| 44 | (−)3(−) | No | No | Mutant | ||
| 10 | 37 | 7 | Yes | Yesc | Linalool | GPP |
| Nerolidolc | e,e-FPP | |||||
| 39 | 7 | Yes | Yesc | Linalool | GPP | |
| Nerolidolc | e,e-FPP | |||||
| 42 | (−)4(−) | No | No | Mutant | ||
| 12 | 17 | 7 | Yes | Yesc | Valencenee | e,e-FPP |
| 29 | (−)1(g)2(g)k | No | No | Mutant | – | |
| Total | 44 | 29 | 20 |
Aberrant splicing (Supplemental Fig. S1).
(−) indicates that the rest of the sequence of this mutant gene is not present in the genome, preceded and/or followed by the specified number of exons.
This work.
ND, Not determined.
g, A gap in the genomic sequence, preceded and/or followed by the specified number of exons.
As of July 2011, a total of 44 TPS gene models were identified. These 44 TPS gene models include 29 gene models that appear to have uncompromised open reading frames (TPS3, TPS4, TPS5, TPS7–TPS10, TPS12, TPS14, TPS16–TPS21, TPS24, TPS25, TPS27, TPS28, and TPS31–TPS41). In contrast, 15 TPS gene models (TPS1, TPS2, TPS6, TPS11, TPS13, TPS15, TPS22, TPS23, TPS26, TPS29, TPS30, TPS34, and TPS42–TPS44) appear to have mutations and deletions that would preclude the production of proteins of similar length and enzymatic activity (Table I; Supplemental Fig. S1). When a gene had only one or two mutations, we verified the mutations by isolating the affected genomic DNA by PCR and sequencing the PCR product and in some cases also by isolating cDNAs and demonstrating that they contained the mutations or were aberrantly spliced (Supplemental Fig. S1).
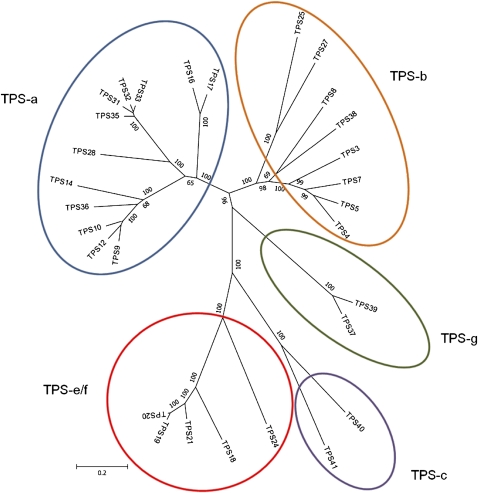
Phylogenetic analysis of the tomato TPS genes (Fig. 1; Supplemental Fig. S2) indicates that 19 belong to the TPS-a clade, including 12 functional or potentially functional genes (TPS9, TPS10, TPS12, TPS14, TPS16, TPS17, TPS28, TPS31, TPS32, TPS33, TPS35, and TPS36) and seven mutated genes (TPS11, TPS13, TPS15, TPS23, TPS29, TPS30, and TPS34). Another 13 TPS genes are in clade TPS-b, including eight functional or potentially functional ones (TPS3, TPS4, TPS5, TPS7, TPS8, TPS25, TPS27, and TPS38) and five mutated genes (TPS1, TPS2, TPS6, TPS22, and TPS26). The TPS-g clade includes two functional genes, TPS37 and TPS39, and two mutated genes, TPS42 and TPS43, while there are two functional or potentially functional genes in the TPS-c clade (TPS40 and TPS41) and one mutated gene (TPS44), and five functional or potentially functional genes (TPS18, TPS19, TPS20, TPS21, and TPS24) in the TPS-e/f clade (Fig. 1; Supplemental Fig. S2).
Figure 1.
Phylogenetic tree of the proteins encoded by the 29 functional or potentially functional TPS genes in the tomato genome.
A search of the Solanum Trichome Project (http://www.trichome.msu.edu/), the SOL Genomics Network (http://solgenomics.net/), and GenBank EST databases identified ESTs from 20 of the 29 TPS genes deemed potentially functional (Table II) as well as from the presumptive pseudogenes TPS1, TPS13, and TPS15 (Supplemental Fig. S1; see below). Reverse transcription (RT)-PCR analysis of different organs of growth chamber-grown plants with specific primers for all 29 potentially functional genes identified transcripts for 26 of these genes (Table II; Supplemental Fig. S3), with the exceptions being TPS25, TPS27, and TPS28 (the RT-PCR analysis was done with 40 cycles to identify rare transcripts and was not meant to be quantitative). The enzymatic activities of the proteins encoded by eight of the 29 potentially functional TPS genes were previously reported (Table I; see below). We obtained additional full-length cDNAs (Table I) and biochemically tested recombinant proteins obtained by expression in Escherichia coli with potential prenyl diphosphate substrates. As it had been shown that tomato produces NPP in the plastids, all putative plastid-localized terpene synthases were tested with GPP and NPP as well as GGPP. Since a gene encoding a protein with homology to NDPS1 but that does not appear to have a transit peptide is present in the tomato genome (see below), we tested putative cytosol-localized terpene synthases with both e,e-FPP and z,z-FPP substrates. Terpene synthases without a clear transit peptide were tested with all these substrates.
Table II. Detection of transcripts of the 29 functional or potentially functional terpene synthase genes in tomato by RT-PCR.
RT-PCR was performed in 40 cycles as described in “Materials and Methods” using the accession M82, except for TPS19 and TPS39, whose transcripts were searched for in other accessions as well (see below). The list of primers used in the RT-PCR experiments is given in Supplemental Table S1.
| Chromosome | TPS | Young Leaf | Young Leaf Trichome | Fully Expanded Leaf | Fully Expanded Leaf Trichome | Stem | Stem Trichome | Flower | Green Immature Fruit | Red Fruit | Root | ESTs in the Databases |
| 1 | TPS3 | − | − | − | − | + | + | − | + | − | + | No |
| TPS4 | − | − | − | − | + | − | − | + | − | + | Yes | |
| TPS5 | + | + | + | + | + | + | + | + | + | − | Yes | |
| TPS7 | + | − | − | − | − | − | + | + | − | − | Yes | |
| TPS8 | + | − | + | − | − | − | − | − | − | + | Yes | |
| TPS31 | − | + | − | − | − | + | + | − | − | + | Yes | |
| TPS32 | − | + | − | − | − | + | + | + | − | + | Yes | |
| TPS33 | − | — | − | − | − | + | − | + | + | + | No | |
| TPS35 | − | − | − | − | − | − | − | − | − | + | Yes | |
| 2 | TPS25 | − | − | − | − | − | − | − | − | − | − | No |
| TPS27 | − | − | − | − | − | − | − | − | − | − | No | |
| TPS38 | − | − | − | − | − | − | + | − | − | − | No | |
| 4 | TPS28 | − | − | − | − | − | − | − | − | − | − | No |
| 6 | TPS9 | + | + | + | + | + | + | + | + | − | + | Yes |
| TPS10 | + | + | + | − | − | + | + | + | − | − | No | |
| TPS12 | + | + | + | + | + | + | + | + | + | + | Yes | |
| TPS36 | − | − | − | − | − | − | − | + | + | − | No | |
| TPS40 | + | − | + | − | + | − | + | + | − | + | Yes | |
| 7 | TPS16 | + | + | + | + | − | + | + | − | − | − | Yes |
| TPS24 | − | + | − | − | − | + | − | + | − | + | Yes | |
| 8 | TPS18 | − | − | − | − | − | − | + | + | − | + | Yes |
| TPS19a | − | − | − | − | − | + | − | − | − | − | Yes | |
| TPS20 | + | + | + | + | + | + | + | + | + | + | Yes | |
| TPS21 | + | + | + | − | + | + | + | + | − | + | Yes | |
| TPS41 | + | + | + | + | + | + | + | − | − | − | Yes | |
| 9 | TPS14 | − | − | − | − | − | − | + | − | − | + | Yes |
| 10 | TPS37 | + | + | − | + | + | + | + | + | − | + | No |
| TPS39b | − | + | − | − | − | − | + | − | − | − | Yes | |
| 12 | TPS17 | + | + | + | + | + | + | + | + | + | + | Yes |
| Total | 29 | 12 | 14 | 11 | 7 | 11 | 16 | 18 | 16 | 6 | 17 | 20 |
No TPS19 transcripts were observed in accession M82; here, + indicates transcripts observed in the MP1 accession.
No TPS39 transcripts were observed in accession M82; here, + indicates transcripts observed in the Moneymaker accession.
Characterization of TPS-a Clade Genes
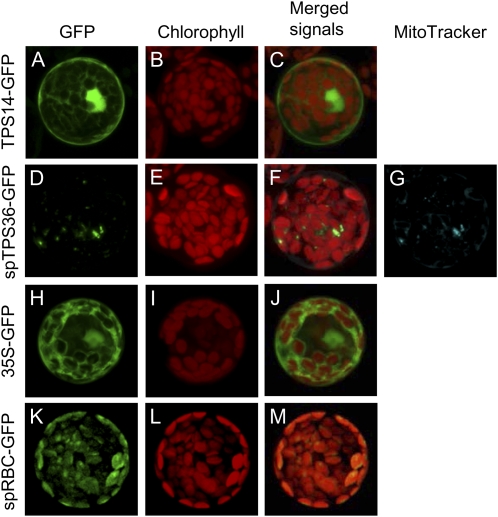
Ten of the 12 potentially functional tomato genes in this clade encode presumed cytosolic synthases, as they have 18 or fewer amino acids upstream of the conserved RP/R motif believed to be part of the mature protein and thus are likely to encode sesquiterpene synthases (Supplemental Fig. S4). In contrast, the N terminus of TPS14 extends 27 residues upstream of its RP motif, but this sequence includes three acidic amino acids, which is unusual for plastid transit peptides (Keegstra et al., 1989). TPS36, the remaining gene of this class, does appear to encode for a protein with a transit peptide (Supplemental Fig. S4). To determine the subcellular localization of the TPS14 and TPS36 proteins, we constructed gene fusions to the GFP gene and examined the localization of the fused proteins in Arabidopsis protoplast transient expression assays. TPS14-GFP fusion protein was localized to the cytosol (Fig. 2, A–C). Because cells expressing the full-length TPS36-GFP fusion protein showed no fluorescence, we fused the first 60 codons of TPS36 to GFP and observed green fluorescence that coincided with the location of the mitochondria rather than the chloroplast or the cytosol (Fig. 2, D–G).
Figure 2.
Subcellular localization of TPS14 and TPS36. The complete open reading frame of TPS14 (TPS14-GFP) and the first 60 codons of TPS36 (tpTPS36-GFP) were fused to a downstream GFP and transiently expressed in Arabidopsis leaf protoplasts. GFP fluorescence indicates the location of each fusion protein; the location of the chloroplasts was determined by chlorophyll autofluorescence (shown in red), and the location of mitochondria was determined by the fluorescence of MitoTracker Red dye (shown in artificial blue color to distinguish it from chlorophyll autofluorescence). The column labeled “Merged Signals” provides a view of all fluorescent signals obtained for this sample. A to C, TPS14-GFP. D to G, tpTPS36-GFP. H to J, Expression of a nonfused GFP showing cytosolic (and nuclear) localization, used here for control. K to M, Expression of a Rubisco-transit peptide-GFP fusion gene (Nagegowda et al., 2008) showing plastid localization of the protein, used here for control.
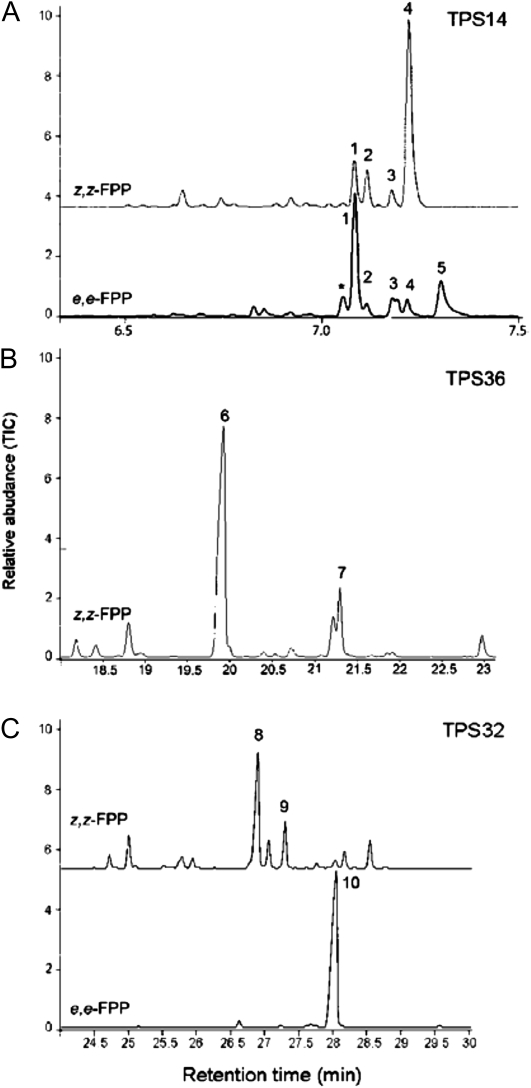
The functional or potentially functional genes in the tomato TPS-a clade can be divided into three subclades (Fig. 1). Four of the five members in the first subclade, TPS9, TPS10, TPS12, and TPS36, reside on chromosome 6, with TPS14 found on chromosome 9 (Table I; Fig. 3). Published data indicate that TPS9 (also called Sst1) catalyzes the formation of the sesquiterpene germacrene C from e,e-FPP in the VFNT variety (Colby et al., 1998), and the closely linked TPS12 catalyzes the formation of the sesquiterpenes β-caryophyllene and α-humulene (and was therefore named CAH synthase, or CAHS) from e,e-FPP in the M82 variety (Schilmiller et al., 2010b). These three sesquiterpenes are the major sesquiterpenes found in tomato leaf trichomes (van der Hoeven et al., 2000). Our in vitro assays indicated that TPS14 catalyzes the formation of several bisabolene isomers from either all-trans-e,e-FPP, and the same bisabolene isomers (but at different ratios) as well as nerolidol were produced by the enzyme with all-cis-z,z-FPP substrate (Fig. 4A). TPS36 shows the highest identity to the diterpene-producing cembratrienol synthases from Nicotiana sylvestris (Ennajdaoui et al., 2010). It encodes an enzyme that catalyzes the formation of an unidentified sesquiterpene from z,z-FPP (Fig. 4B) but has little or no activity with e,e-FPP, GPP, NPP, or GGPP.
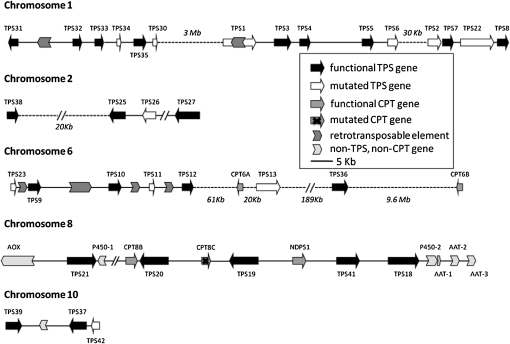
Figure 3.
The organization of TPS genes and other genes in clusters on chromosomes 1, 2, 6, 8, and 10. AAT, Alcohol acyltransferase; AOX, alcohol oxidase; CPT, confirmed or putative cis-prenyltransferase; P450, putative cytochrome P450 oxidoreductase. The P450-2 gene has a large insertion in the first exon, and the AAT-1 gene has multiple deletions and mutations.
Figure 4.
GC analysis of the products formed in the reactions catalyzed by TPS14, TPS36, and TPS32 with z,z-FPP and e,e-FPP as the substrates. The reaction products (peaks) were identified by comparison of their GC-MS profiles with authentic standards and NIST libraries: (1) β-bisabolene, (2) γ-bisabolene (cis), (3) γ-bisabolene (trans), (4) α-bisabolene (trans), (5) nerolidol, (6) unidentified, (7) unidentified, (8) viridiflorene, (9) unidentified, (10) unidentified. TPS14, TPS36, and TPS32 enzymatic products were analyzed on different columns with different programs (see “Materials and Methods”). GC analysis is shown for alternative substrates only when activity of the enzyme with such a substrate was more than 10% compared with the best substrate.
TPS16 and TPS17 define a separate subclade and are most similar to the previously characterized germacrene D synthases from poplar, grape, and kiwifruit (Actinidia deliciosa; Arimura et al., 2004; Lücker et al., 2004; Nieuwenhuizen et al., 2009). TPS17 was recently shown to encode a valencene synthase (Bleeker et al., 2011).
Four of the five members of the third TPS-a subclade are in close proximity to each other on chromosome 1 (TPS31, TPS32, TPS33, and TPS35; Figs. 1 and 3). The enzymes encoded by these genes are more similar to the sesquiterpene-forming vetispiradiene synthase from the solanaceous species Hyoscyamus muticus (Back and Chappell, 1995) and germacrene A synthase from Pogostemon cablin (Deguerry et al., 2006). TPS32 encodes an enzyme that catalyzes the formation of viridiflorene from e,e-FPP but can also use z,z-FPP to make several unidentified sesquiterpenes (Fig. 4C). TPS31 was also recently shown to make viridiflorene from e,e-FPP and to have no activity with z,z-FPP (Bleeker et al., 2011). TPS31, TPS32, TPS33, and TPS35 are unusual in that they are missing the sixth and final intron characteristic of TPS-a family members (Table I; Supplemental Fig. S5). In contrast, the less closely related TPS28 gene, which belongs to the same subclade but is present on chromosome 4, has an exon-intron structure more typical of other TPS-a members (Table I). Interestingly, TPS34, which is a mutated gene that is missing the first four exons and contains mutations in the remaining exons, is located between TPS33 and TPS35 and contains the intron equivalent to the sixth intron in TPS-a members. TPS30, another mutated gene in this cluster, has a deletion encompassing the region that would have contained this intron.
Expression of TPS-a Genes
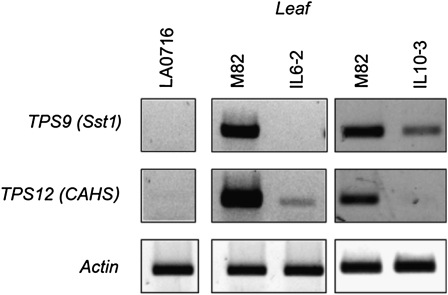
Evidence for mRNA accumulation was obtained for all genes encoding potentially functional TPS-a proteins, with the exception of TPS28 (Table II). As TPS9 (Sst1), encoding a germacrene C synthase, and TPS12 (CAHS), encoding β-caryophyllene and α-humulene synthase, have been shown to be the two TPS genes responsible for the majority of the sesquiterpenes found in the leaf trichomes (Colby et al., 1998; Schilmiller et al., 2010), we analyzed their expression in two chromosomal substitution lines, IL6-2 and IL10-3, reported to have low levels of sesquiterpenes in the leaves (Schilmiller et al., 2010a). IL6-2 contains a small segment of the Solanum pennellii chromosome 6, where TPS9 and TPS12 are located, and IL10-3 contains a segment from chromosome 10 of S. pennellii (Schilmiller et al., 2010a). RT-PCR analysis using primers for these two genes detected few or no transcripts in either S. pennellii LA0716 or IL6-2 (Fig. 5), in agreement with the absence of β-caryophyllene and α-humulene in LA0716 and IL6-2 trichome extracts. Surprisingly, TPS9 and TPS12 also showed much reduced expression in the isogenic line IL10-3 (Fig. 5). It is likely that genes involved in the regulation of these two genes are located on this segment of chromosome 10.
Figure 5.
RT-PCR analysis of transcripts of TPS9 and TPS12 in leaves of tomato (accession M82), S. pennellii (accession LA0716), and the introgression lines IL6-2 and IL10-3.
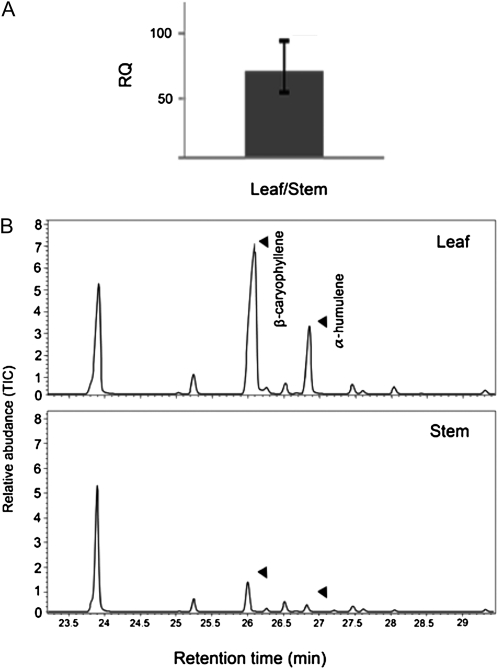
As previously shown based on EST representation in cDNA libraries (Schilmiller et al., 2010b), TPS12 is more highly expressed in type 6 leaf trichomes than in type 6 stem trichomes. Quantitative (q)RT-PCR results showed that accumulation of TPS12 transcript was approximately 80-fold higher in the leaf versus stem trichomes (Fig. 6A), consistent with the significantly higher concentration of β-caryophyllene and humulene detected in leaf compared with stem tissues (Fig. 6B).
Figure 6.
Comparison of transcript levels of TPS12 and β-caryophyllene and α-humulene levels in leaves and stems. A, qRT-PCR analysis of TPS12 transcripts. B, GC analysis of sesquiterpene volatiles detected in leaves and stems. RQ, Ratio of qRT-PCR results; TIC, total ion chromatograph.
Despite the presence of mutations in TPS13 and TPS15 that are predicted to prevent the production of functional protein, transcripts of these genes were found in various tissues. TPS13 contains an in-frame stop codon in exon 4 in addition to an 88-nucleotide insertion of plastidial genomic DNA in the coding region of exon 2 that is predicted to cause a frame shift. In TPS15, the end of the first exon and the following GT splice site is deleted; therefore, splicing is unlikely to occur properly. As expected, some cDNAs amplified by RT-PCR showed no splicing of intron 1 while others showed missplicing of the latter part of the intron, utilizing as the 5′ splicing border an alternative GT dinucleotide located in the middle of the intron (Supplemental Fig. S1).
Characterization of the TPS-b Clade
Genes in this clade typically encode monoterpene synthases. The tomato genome has eight potentially functional TPS-b genes (Fig. 1; Supplemental Fig. S6). Two of the TPS-b clade genes, TPS4 (MTS2) and TPS5 (MTS1), were previously demonstrated to encode monoterpene synthases that catalyze the formation of mostly β-phellandrene and linalool, respectively, from GPP (van Schie et al., 2007).
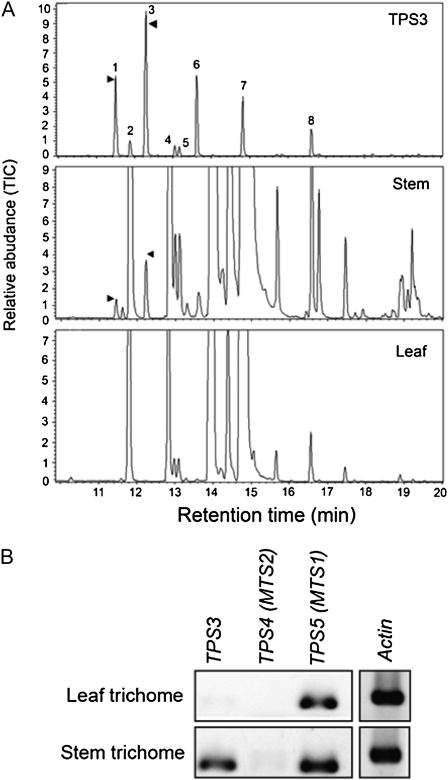
Sequences of TPS3, another potentially functional gene in the TPS-b clade, were not found in publicly available tomato EST databases including trichome ESTs. Nevertheless, RT-PCR analysis revealed the expression of this gene in the stem trichome, the green immature fruit, and the root tissue (Table II). TPS3 encodes a protein of 607 amino acids, including a putative N-terminal transit peptide of approximately 50 amino acids, with 60% and 58% identity to MTS2 (TPS4) and MTS1 (TPS5), respectively (Supplemental Fig. S6). The full-length TPS3 cDNA was expressed in E. coli, and the resulting protein was found to use only GPP as substrate to produce camphene and tricyclene (peaks 3 and 1, respectively, in the top panel in Fig. 7A) as the major products, as well as other monoterpenes. Consistent with the detection of TPS3 transcripts in stem but not in leaf tissue (Fig. 7B), camphene and tricyclene were detected in stem trichomes but not in leaf trichomes (middle and bottom panels, respectively, in Fig. 7A).
Figure 7.
Identification of the major products of TPS3 enzymatic activity in vitro as camphene and tricyclene, and correlation with the presence of these compounds and of transcripts of TPS3 in leaf and stem tissues of tomato. A, TPS in vitro activity with GPP as the substrate (top panel) and GC analysis of monoterpenes extracted from leaves and stems by SPME (bottom two panels). B, RT-PCR analysis of TPS3 transcripts in leaves and stems. The reaction products (peaks) were identified by comparison of their GC-MS profiles with authentic standards and NIST libraries: (1) tricyclene, (2) α-pinene, (3) camphene, (4) sabinene, (5) β-pinene, (6) β-myrcene, (7) limonene, (8) unidentified. TIC, Total ion chromatograph.
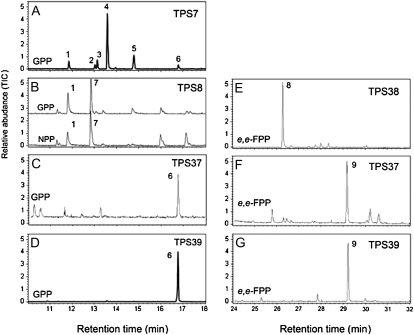
TPS7 is another member of the TPS-b family with the general features of a monoterpene synthase. It has a predicted open reading frame of 592 amino acids with a putative N-terminal plastid transit peptide of approximately 45 residues. TPS7 shows the highest amino acid identity with TPS5 (70%) and TPS4 (69%) and is primarily expressed in the flower and in the green immature fruit as well as in the leaf (Table II). Recombinant TPS7 catalyzed the formation of mainly β-myrcene and limonene, in addition to modest amounts of α-pinene and β-pinene from GPP (Fig. 8A). These monoterpenes are detected in the flower and green immature fruit as well as in the leaf, which is consistent with the expression of TPS7 in these tissues (Table II).
Figure 8.
GC analysis of the products formed in vitro by the enzymatic activities of TPS7 (A), TPS8 (B), TPS37 (C and F), TPS39 (D and G), and TPS38 (E). Enzymes were incubated with GPP, NPP, e,e-FPP, and z,z-FPP, and products were analyzed as described in “Materials and Methods.” Reaction products (peaks) were identified by comparison of their GC-MS profiles with authentic standards and NIST libraries: (1) α-pinene, (2) sabinene, (3) β-pinene, (4) β-myrcene, (5) limonene, (6) linalool, (7) 1,8-cineole, (8) α-bergamotene, (9) nerolidol. GC analysis is shown for alternative substrates only when activity of the enzyme with such a substrate was more than 10% compared with the best substrate. TIC, Total ion chromatograph.
Another member of the tomato TPS-b clade is the 597-residue TPS8 protein. A TPS8 cDNA was expressed in E. coli, and enzyme assays with whole cell extract showed that TPS8 catalyzed the formation of 1,8-cineole from both GPP and NPP (Fig. 8B). This activity is consistent with the observation that it has highest identity (77%) to a 1,8-cineole synthase from the Solanaceae species Nicotiana suaveolens. The TPS8 transcripts were found primarily in root but also in leaf tissue, but 1,8-cineole could not be detected in any of these tissues. Another unusual feature of the TPS8 protein is that its putative N-terminal transit peptide is unusually short and has an atypical amino acid composition, containing negatively charged residues (Supplemental Fig. S6). This is in contrast to the N. suaveolens 1,8-cineole synthase, which has a plastidic transit peptide of typical length and amino acid composition (Roeder et al., 2007).
TPS25, TPS27, and TPS38, at the base of the TPS-b clade, are located in a cluster on chromosome 2, together with the mutated gene TPS26 (Fig. 3; Supplemental Fig. S1). No transcripts for TPS25 or TPS27 were detected in any of the tissues studied (Table II). The proteins encoded by TPS25 and TPS27, both of which appear to have a very short N-terminal extension beyond the RR motif (ER in TPS25; Supplemental Fig. S6), show the highest similarity to β-ocimene/α-farnesene synthase from grape, which were reported to be able to use GPP or e,e-FPP as substrate to give the respective monoterpene or sesquiterpene in vitro (Martin et al., 2010). TPS38, the third potentially functional gene in this TPS-b subgroup, also appears to have no transit peptide, and it encodes an enzyme that uses e,e-FPP to make the sesquiterpene α-bergamotene (Fig. 8E). It is not active with GPP, NPP, or z,z-FPP.
The remaining five genes in the TPS-b clade appear to be mutated. TPS1 on chromosome 1 is interrupted by a copia long terminal repeat-like sequence inserted in intron 4 (Supplemental Fig. S1). In addition, the 3′ splice site of intron 4 is AC rather than AG, which should prevent this intron from being spliced correctly. As predicted, several cDNAs obtained by RT-PCR lacked exon 4, indicating that splicing occurred from the end of exon 3 to the beginning of exon 5. The remaining cDNAs were missing not only exon 4 but also the beginning part of exon 5, due to utilization of an alternative AG splice site within exon 5 (Supplemental Fig. S1). Despite altered splicing, TPS1 transcripts were observed in almost all of the tissues tested, with the exception of red fruit and root. The remaining pseudogenes have a variety of defects. TPS2 is predicted to include only the first three exons due to mutations that introduce stop codons in the reading frame (Supplemental Fig. S1). The tomato genome assembly revealed a TPS6 gene model with a deletion of approximately 80 nucleotides of exon 3 and a stop codon in the middle of exon 7 (Supplemental Fig. S1). Both mutations were confirmed by sequencing tomato genomic DNA. Neither ESTs nor transcripts were found for TPS6 in any of the tissues tested (Table II). Consistent with the lack of mRNA accumulation, the TPS22 gene model has a large insertion in exon 2 and numerous stop codons in the reading frames (Supplemental Fig. S1). The TPS26 gene model has only the last four exons (exons 4–7), and deletions in the last three of these are predicted to introduce frame shifts and a premature stop codon into the open reading frame (Supplemental Fig. S1).
Variation in Monoterpene Profiles between Tomato and S. pennellii and TPS-b Gene Expression
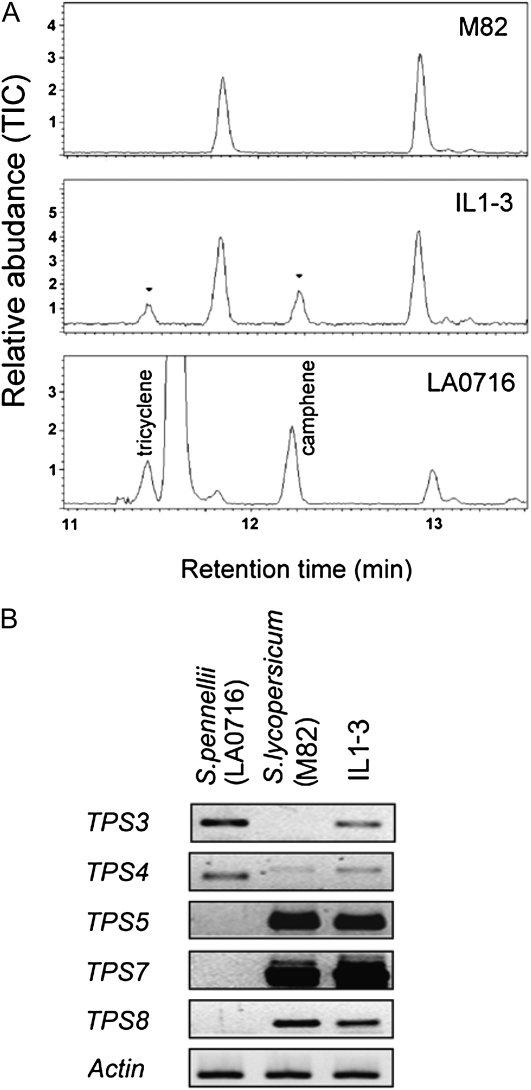
As described above, TPS3 is responsible for the synthesis of camphene and tricyclene in tomato stems, and neither TPS3 expression nor camphene and tricyclene are observed in tomato leaves. However, camphene and tricyclene are observed in leaves of the introgression line IL1-3, which has a large region of tomato chromosome 1 substituted by S. pennellii, as well as in leaves of S. pennellii LA7016 (Fig. 9A). Chromosome 1 contains the TPS-a genes TPS1, TPS3, TPS4, TPS5, TPS6, TPS2, TPS7, TPS22, and TPS8, with all of these genes closely linked to each other in this order (Fig. 3). To determine which TPS gene on chromosome 1 is responsible for the synthesis of camphene and tricyclene in leaves of IL1-3 and S. pennellii LA7016, we compared the expression profiles of the potentially functional genes in this cluster (TPS3, TPS4, TPS5, TPS7, and TPS8) in S. pennellii LA7016 and tomato. The result of this comparison (Fig. 9B) indicates that the expression of TPS3, but not of TPS4, TPS5, TPS7, or TPS8, correlates with the presence of camphene and tricyclene, consistent with the functional characterization of TPS3. It is likely that the S. pennellii chromosomal segment in IL1-3 contains TPS3 but not TPS4, TPS5, TPS7, and TPS8 (Fig. 3) and that the expression of TPS3 in leaves in this line is due to cis-regulatory elements.
Figure 9.
RT-PCR analysis of the expression of monoterpene synthase genes from the chromosome 1 gene cluster in leaves of tomato (accession M82), S. pennellii (accession LA0716), and IL1-3 plants, and comparison of the monoterpenes found in these leaves. A, GC analysis. B, RT-PCR analysis. TIC, Total ion chromatograph.
TPS37 and TPS39 Are the Two Functional Members of the TPS-g Clade in Tomato
The proteins encoded by TPS37 and TPS39 (Supplemental Fig. S6) belong to the TPS-g clade, and both catalyze the formation of linalool from GPP (Fig. 8, C and D) and nerolidol from e,e-FPP (Fig. 8, F and G) but had little or no activity with GGPP or cis-prenyldiphosphates. As with most members of the angiosperm TPS-g clade, TPS37 and TPS39 are characterized by the absence of the conserved RRX8W motif at the N terminus (Supplemental Fig. S1). They both have a short (approximately 30-residue) N-terminal extension from the putative mature N terminus, which in the case of TPS39 includes two negatively charged amino acids. Thus, it is not clear if these N-terminal extensions function as plastid-targeting sequences. TPS37 was expressed in leaf and stem trichomes (young and fully expanded leaf) as well as in flower and root tissues, while TPS39 transcripts were observed only in young leaves and in flowers (Table II). TPS37 and TPS39 are found near each other on chromosome 10, together with the mutated gene TPS42 (Fig. 3). A second mutated gene, TPS43, is found on chromosome 5. It includes an insertion in exon 1 and a deletion in exon 6, both of which cause shifts in the reading frame.
TPS40 and TPS41 Belong to the TPS-c Clade
TPS40 was previously reported to encode CPS, the plastidic enzyme that converts GGPP to CPP as part of GA biosynthesis (Rebers et al., 1999). TPS41, part of a cluster of TPS genes on chromosome 8 (Fig. 3), is similar to TPS40. However, it does not appear to encode a transit peptide (Supplemental Fig. S7). While its function is not known, it is expressed in several organs, leaves, stems, and flowers. TPS44 on chromosome 9 is a mutated gene related to TPS40 that contains only parts of exons 6, 7, and 8 (Supplemental Fig. S1).
TPS24, TPS18, TPS19, TPS20, and TPS21 Belong to the TPS-e/f Clade
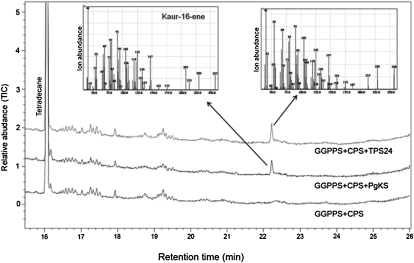
TPS24 is present on chromosome 7 and is most similar to KS enzymes from other species (Supplemental Figs. S1 and S8). A TPS24 cDNA was expressed in E. coli cells engineered to synthesize CPP (Cyr et al., 2007), and ent-kaur-16-ene was detected in these cells (Fig. 10). Thus, the tomato TPS24 gene encodes a bona fide ent-kaurene synthase.
Figure 10.
GC analysis demonstrating that TPS24 catalyzes the formation of ent-kaur-16-ene from CPP. E. coli cells containing the plasmid pGGeC, which carries the A. grandis GGPPS cDNA and the Arabidopsis CPS cDNA, and E. coli cells containing the pGGeC plasmid as well as a second plasmid with the P. glauca ent-kaurene synthase or the tomato TPS24 cDNA were used. The E. coli cells expressing TPS24 produced a peak that had the identical retention time and MS pattern as the peak produced by PgKS, which had previously been authenticated as ent-kaur16-ene (Keeling et al., 2010). TIC, Total ion chromatograph.
We had previously shown that the protein encoded by TPS20 (PHS1) is a plastidic enzyme that catalyzes the formation of mostly β-phellandrene as well as several other monoterpene products from NPP (Schilmiller et al., 2009). The available genome assembly indicates that TPS20 and the remaining tomato TPS-e/f genes, including TPS18, TPS19, and TPS21, are present in a cluster at the tip of chromosome 8. However, the sequence assembly in this region is poor, presumably due in part to these duplicated genes, and all these genes, or gene fragments, are presented on short scaffolds that are not physically linked. Amplifying genomic DNA followed by sequencing enabled us to close many gaps, but not all (Fig. 3; Supplemental Fig. S1). An additional complicating factor was the observation that sequence comparison of the full-length TPS19 cDNA and TPS20 cDNA indicated that the two are more than 98% identical, with most of the differences concentrated in the regions encoding the C terminus of the protein. Because of this high level of similarity, sequences of genomic fragments containing TPS19 and TPS20 could not be unambiguously assigned to either one of them, further explaining the difficulties that the genome assemblers have had in putting together this region. Nonetheless, by anchoring the beginnings and ends of TPS19 and the end of TPS20 to adjacent genes, we were able to determine that the two genes are arranged in tandem (Fig. 3). The proteins encoded by TPS18 and TPS21 have somewhat lower levels of sequence similarity to TPS20 (68% and 88% identity, respectively), and they are present on the periphery of this TPS gene cluster. As noted above, TPS41, the gene encoding a CPS-like protein, is also present on this cluster, upstream of TPS18. Moreover, NDPS1 and a second gene encoding a protein with 86% identity to NDPS1 (designated as CPT8B, for cis-prenyltransferase) are also found interspersed with the TPS genes (Fig. 3).
Expression of TPS20 was observed in most tissues, but TPS19 transcripts were detected only in stem trichomes (Table II). Transcripts of TPS18 and TPS21 were detected in flowers, roots, and green fruits, and TPS21 transcripts were also observed in leaves and stems (Table II). TPS18 and TPS21 cDNAs were expressed in E. coli, with no activity observed for either of these proteins with any of the substrates tested (GPP, NPP, e,e-FPP, z,z-FPP, GGPP, and CPP).
Variation in α-Phellandrene and β-Phellandrene Profiles and TPS20 Activity between Tomato and S. pennellii
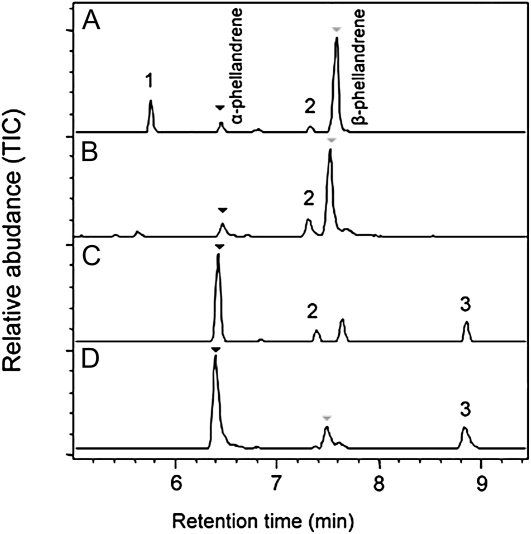
While β-phellandrene is the major product of TPS20 (PHS1) in tomato, trichomes of S. pennellii contain mostly α-phellandrene and considerably lower levels of β-phellandrene (Fig. 11; Schilmiller et al., 2009). The trichomes of IL8-1-1, the line that carries a segment of chromosome 8 from S. pennellii, have the same monoterpene profile as do the trichomes of S. pennellii, with α-phellandrene being the predominant compound (Fig. 11). To examine the basis for this difference, we obtained a full-length cDNA of the S. pennellii TPS20 ortholog. SpTPS20 encodes a protein that is 94% identical to the tomato TPS20 (PHS1; Supplemental Fig. S8). After expressing SpPHS1 cDNA in E. coli and obtaining a purified enzyme (see “Materials and Methods”), enzymatic assays showed that SpPHS1 uses NPP (but not GPP) to produce mostly α-phellandrene and significantly lower levels of β-phellandrene (Fig. 11). Kinetic analysis of the purified SpPHS1 showed a Km value for NPP of 9.7 μm and a turnover rate of 53 s−1 (compared with 9.1 μm and 4.1 s−1, respectively, for the SlPHS1; Schilmiller et al., 2009).
Figure 11.
Comparison of the enzymatic activities of the proteins encoded by TPS20 genes from tomato and S. pennellii. A, GC analysis of tomato (accession M82) trichome monoterpenes. B, GC analysis of the products of the in vitro reaction catalyzed by purified SlPHS1 with NPP as the substrate. C, GC analysis of S. pennellii (LA0716) trichome monoterpenes. D, GC analysis of the products of the in vitro reaction catalyzed by purified SpPHS1 with NPP as the substrate. Numbered peaks are as follows: (1) δ-2-carene, (2) limonene, (3) γ-terpinene. TIC, Total ion chromatograph.
JA Induces Several Terpene Synthases in Stem Trichomes
It was previously shown that transcript levels of TPS5 (MTS1; linalool synthase) in trichomes and emission of the homoterpene TMTT, derived from the diterpene geranyllinalool, in leaves both increased approximately 2-fold in cultivated tomato following JA treatment (Ament et al., 2006). This led us to test the effects of JA treatment on the expression of other TPS genes normally expressed in trichomes. After treatment with 1 mm JA for 24 h, cv Moneymaker RNA was extracted from isolated stem trichomes and expression profiling was performed with the Illumina GA II System. The results (Table III) confirmed that TPS5 is indeed induced by JA. In addition, the monoterpene synthases TPS3, TPS7, and TPS39 as well as the sesquiterpene synthase TPS31 were also induced.
Table III. JA induction of TPS genes in stem trichomes.
Tomato cv Moneymaker was used. For details of the experiment, see “Materials and Methods.”
| Chromosome | TPS Transcripts Detected | JA Induction |
| fold | ||
| 1 | 3 | 7 |
| 5 (MTS1) | 2.3 | |
| 7 | 3 | |
| 8 | – | |
| 31 | 4 | |
| 6 | 9 (SST1) | – |
| 12 (CAHS) | – | |
| 7 | 16 | – |
| 24 | – | |
| 8 | 19 | – |
| 20 (PHS1) | – | |
| 41 | – | |
| 10 | 39 | 2.2 |
| 12 | 17 | – |
| Total | 14 |
DISCUSSION
The Tomato TPS Gene Family Includes Members from All Known Angiosperm Clades But May Contain a Low Number of Diterpene Synthases
We show here that the genome of tomato contains at least 44 TPS genes, including 29 that are functional or potentially functional, and report the in vitro activity of 10 of the 29 TPS enzymes. Together with the eight terpene synthases whose activities were previously reported, the function of more than half of the tomato terpene synthases have now been characterized in vitro. These enzymes are involved in the synthesis of monoterpenes and sesquiterpenes as well as of ent-kaurene, the diterpene precursor of GAs (Colby et al., 1998; van Schie et al., 2007; Schilmiller et al., 2009, 2010b; this work).
In some cases, the products seen in the in vitro assay of a tomato TPS enzyme correspond well to terpenes found in the tissues in which the encoding gene is expressed, as is the case for TPS3 (camphene synthase) and TPS12 (CAHS). However, in several cases, we have not been able to correlate the in vitro enzyme products with metabolites extracted from plant tissues. For example, TPS8 catalyzes the formation of 1,8-cineole, but we have not yet detected this compound in leaves or roots, where TPS8 is expressed. This is not unusual, however; for example, a 1,8-cineole synthase is expressed in Arabidopsis roots (Chen et al., 2004), but detection of 1,8-cineole required an extremely sensitive method (Steeghs et al., 2004). The inability to detect the products of several other TPS enzymes, for example TPS14, TPS17, and TPS38, whose major products are γ-bisabolene, valencene, and α-bergamotene, respectively, in the tissues in which these genes are expressed may likewise be due to low levels of these compounds or further conversion of these compounds to metabolites not detected by gas chromatography-mass spectrometry (GC-MS). It is also important to note that because multiple TPS genes are often expressed in the same tissue, and many of these TPSs would have overlapping ranges of products, it is not straightforward to identify the specific contribution that each TPS enzyme makes to the profile of terpenes observed in that tissue.
The availability in tomato of NPP in plastids (Schilmiller et al., 2009), and possibly it and other prenyl diphosphates in the cytosol and other subcellular compartments of the cell (there are several additional genes encoding proteins with homology to NDPS1 in tomato; see below), further complicates the direct linking of a specific TPS with a specific product in a given tissue. So does the common observation that many monoterpene synthases will react in vitro with C15 prenyl diphosphates and that sesquiterpene synthases will react in vitro with C10 prenyl diphosphates (Schnee et al., 2002; Davidovich-Rikanati et al., 2008; Nieuwenhuizen et al., 2009; Jones et al., 2011), substrates that may or may not be available to them in planta. Thus, the biological significance of all these in vitro activities can only be fully determined by combining biochemical characterization with genetic manipulation.
It is also clear that not all observed terpenes in tomato could be traced to the activities of the already studied TPS enzymes. For example, the diterpene geranyllinalool and its derivative TMTT, a homoterpene, were emitted by tomato and geranyllinalool synthase enzymatic activity was demonstrated in tomato (Ament et al., 2006), yet the gene responsible for its formation has yet to be identified, perhaps because TMTT emission might be due to gene activities in leaf cells outside the trichomes.
The TPS genes and proteins of tomato fall into all known angiosperm TPS clades, TPS-a, TPS-b, TPS-e/f, TPS-c, and TPS-g (Fig. 1; Supplemental Fig. S2). Clade TPS-a, with the largest number of potentially functional genes (12, with seven enzymes characterized), appears to encode mostly cytosolic sesquiterpene synthases, with one protein, TPS36, present in the mitochondria, but its in vivo product is not yet known. TPS-a is an angiosperm-specific clade that typically contains the largest number of TPS genes in a given genome, and enzymes in this clade usually are sesquiterpene or diterpene synthases (Chen et al., 2011). In the Arabidopsis genome, 22 of the 32 TPS genes are in TPS-a. However, only four Arabidopsis genes in this clade encode cytosolic sesquiterpene synthases lacking a transit peptide (Aubourg et al., 2002). Most of the remaining 18 Arabidopsis TPS-a genes likely encode diterpene synthases that are targeted to the chloroplasts, but some are apparently targeted to the mitochondria, although the biochemical activities of none of these 18 enzymes have yet been reported (Aubourg et al., 2002; Chen et al., 2003; Tholl and Lee, 2011).
The eight potentially functional tomato TPS-b genes (six of which are functionally characterized) encode both monoterpene and sesquiterpene synthases. Most enzymes previously characterized from this angiosperm-specific TPS-b clade are monoterpene synthases. For example, all six Arabidopsis Columbia TPS-b clade enzymes are monoterpene synthases predicted to be localized in the plastids (Aubourg et al., 2002). However, in Arabidopsis ecotype Wassilewskija, an ortholog of a TPS-b protein that is lacking a transit peptide is found in the cytosol and it uses e,e-FPP to make farnesene, although it can use GPP in vitro to make ocimene (Huang et al., 2010). Similar TPS-b cytosolic sesquiterpene synthases have been reported in kiwifruit and apple (Malus domestica; Pechous and Whitaker, 2004; Nieuwenhuizen et al., 2009), among others. Tomato has three TPS-b genes, TPS25, TPS27, and TPS38, encoding proteins that appear to lack a transit peptide. We could detect the expression of only one of them, TPS38, and the protein it encodes reacted with e,e-FPP in vitro to make the sesquiterpene α-bergamotene (Fig. 8E).
Clade TPS-g contains two genes, TPS37 and TPS39, encoding similar enzymes with linalool/nerolidol synthase activity (Fig. 8, C, D, F, and G). Many angiosperms have one or just a few TPS genes that belong to this clade. These genes encode enzymes that catalyze the formation of acyclic monoterpenes such as linalool and β-myrcene from GPP, although some linalool synthases in TPS-g can also use e,e-FPP to make nerolidol, and the products they produce in planta depends on their subcellular localization (Chen et al., 2011). Based on sequence analysis alone, it is not clear if the TPS37 and TPS39 proteins possess a plastid-targeting transit peptide. Thus, the specific functions of TPS37 and TPS39 and their subcellular localization remain to be determined.
Clade TPS-c in plants contains the CPS gene involved in GA biosynthesis. In some species, additional CPS-like genes are found encoding plastid-localized enzymes that make precursors for other diterpenes, such as the labdane-type diterpenes in maize (Zea mays) and Cistus creticus (Falara et al., 2010; Chen et al., 2011). In tomato, the TPS-c clade includes CPS and a second gene, TPS41, which lacks a transit peptide and whose function is still unknown.
Clade TPS-e/f includes a group of plastid-localized enzymes with a large repertoire of biosynthetic activities. For example, the gymnosperm and angiosperm KS enzymes (including tomato TPS24) that use CPP as substrate to make the C20 diterpene ent-kaurene (the GA precursor) are in this clade. In addition, however, a Clarkia breweri linalool synthase is a TPS-e/f enzyme that uses GPP to produce linalool (Dudareva et al., 1996), and a TPS-e/f diterpene synthase from Arabidopsis uses GGPP to make geranyllinalool (Chen et al., 2011). We previously showed that tomato TPS20 in this clade is a monoterpene synthase that uses NPP to make mostly β-phellandrene (Schilmiller et al., 2009), and Sallaud et al. (2009) reported that the TPS20 ortholog in Solanum habrochaites, ShSBS, encodes an enzyme that uses z,z-FPP as the substrate to make the sesquiterpenes santalene and bergamotene, the predominant volatile terpenoids in the trichomes and also the precursors of the dominant terpenoid acids in the glands. The tomato genome contains three additional TPS genes in this clade, TPS18, TPS19, and TPS21, but their functions have yet to be determined.
Variation in the Expression of TPS Genes
Twenty of the 29 functional or potentially functional tomato TPS genes were represented by ESTs in publicly available databases. Our RT-PCR results with gene-specific primers for all functional TPS genes extended the number of detectable transcripts to 26 genes in at least one or another of the tomato tissues and organs tested, including trichomes, leaves, stems, flowers, green immature fruits, red fruits, and roots. No transcripts were detected for three of the nonmutated genes (TPS25, TPS27, and TPS28). Four TPS genes, TPS5, TPS9, TPS20, and TPS17, exhibited a broad expression profile with transcripts detected in at least nine out of the 10 tissues tested. On the other hand, there were several genes (e.g. TPS7, TPS8, TPS14, and TPS38) that were found expressed specifically in only one or just a few tissues (Table II). Overall, the organs from which the largest number of TPS genes are expressed are the root and the flower (with 17 and 18 genes, respectively), while leaves and stems bearing trichomes also show the expression of large numbers of TPS genes; only six and seven TPS genes were found to be expressed in red fruits and fully expanded leaves (Table II).
While expression of TPS genes in trichomes of various species was reported (Bertea et al., 2006; Schilmiller et al., 2009; Ennajdaoui et al., 2010; Falara et al., 2010), intraspecies variation of TPS gene expression in trichomes of different organs and differences between closely related species have not yet been examined in detail. Comparison between the expression patterns of the tomato TPS genes in trichomes isolated from leaf versus stem revealed both qualitative and quantitative differences consistent with the distinct volatile profiles of the two organs. For example, elevated levels of CAHS (TPS12) transcripts in the leaf trichome versus the stem trichome (Fig. 6) are consistent with the higher concentrations of β-caryophyllene and α-humulene and the higher amounts of CAHS protein in the leaf versus the stem (Schilmiller et al., 2010b). The expression of TPS12 as well as TPS9 (germacrene synthase) is lower in S. pennellii leaves compared with tomato leaves, consistent with the reported lower levels of β-caryophyllene, α-humulene, and germacrene in leaf trichomes of S. pennellii (Schilmiller et al., 2010b), and these differences in levels of expression appear to be controlled both at the loci encoding these genes and elsewhere in the genome (Fig. 5). We further showed that TPS3 encodes a camphene synthase that is expressed in the stem trichomes but not in leaf trichomes. Camphene and tricyclene, the enzymatic products of TPS3, were detected only in the stem (Fig. 7). Moreover, in the substitution line IL1-3, characterized by the presence of a segment of chromosome 1 from the wild species S. pennellii in the tomato background, the TPS3 ortholog is expressed in the leaf trichomes and camphene is detected as a leaf volatile (Fig. 9). These results indicate that differential TPS tissue-specific expression in Solanum species is in part responsible for the observed distinct intraspecific and interspecific chemical variability.
Since our general expression profiling by RT-PCR was done in tomato cv M82 while the measurements for JA inducibility in stem trichomes was done in tomato cv Moneymaker, we were also able to observe intraspecific variation in TPS gene expression. Transcripts of TPS10, TPS21, TPS32, and TPS33 were observed in stem trichomes of M82 plants but not in the stem trichomes of Moneymaker plants. On the other hand, transcript levels of TPS7 and TPS8 were detected in stem trichomes of Moneymaker plants but were not detected in the stem trichomes of M82. However, most of the TPS genes expressed in this tissue in one cultivar were also expressed in the other (Tables II and III).
Many TPS Genes in Tomato Are Arranged in Clusters
The majority of the TPS genes in the tomato genome are located in clusters on chromosomes 1, 2, 6, 8, and 10 (Fig. 3). The rest of the genes are located elsewhere on chromosomes 4, 5, 7, 9, 10, and 12, without another TPS gene nearby. Two clusters of TPS genes are localized on chromosome 1. The first cluster consists of nine TPS-b genes (TPS1, TPS3, TPS4, TPS5, TPS6, TPS2, TPS7, TPS22, and TPS8) and the second consists of six TPS-a genes (TPS31, TPS32, TPS34, TPS35, and TPS30), with most of the genes organized in the same orientation, the only exception being TPS31. A distance of approximately 3 Mb separates the two TPS gene clusters on chromosome 1.
Three intact TPS-b genes, TPS25, TPS27, and TPS38, are clustered on chromosome 2, together with a mutated gene, TPS26 (Fig. 3). These three genes are most similar in sequence to each other among the tomato TPS family, and the proteins they encode are also more similar to some cytosolic enzymes in species outside Solanum that use e,e-FPP to make sesquiterpenes such as farnesene than they are to the other tomato TPSs. We showed here that the TPS38 protein, which lacks a clear transit peptide, uses e,e-FPP to make α-bergamotene but cannot use GPP to synthesize monoterpenes (we did not detect the expression of TPS25 and TPS27 and therefore did not characterize the activity of the encoded enzymes). Because of their overall sequence divergence from the rest of the tomato TPS-b monoterpene synthases, which are located on a cluster on chromosome 1, it is likely that the initial duplication that gave rise to the progenitor of TPS25, TPS27, and TPS38 is ancient and may predate the origin of the Solanaceae. The apparent functional divergence of this group of genes from the TPS-b genes on chromosome 1 may also date to that time.
The TPS genes in the cluster on chromosome 6 are also found in the same orientation (Fig. 3). This cluster consists of the TPS-a genes TPS23, TPS9, TPS10, TPS11, and TPS12, and two other genes in this clade, TPS13 and TPS36, are within 100 kb (Fig. 3). Since TPS36 as well as TPS14 (whose gene is on chromosome 9) can use z,z-FPP as an in vitro substrate (Fig. 4), it is noteworthy that a gene encoding a cis-prenyltransferase-like protein, designated CPT6A, is present in this cluster.
Four of the TPS genes that cluster on chromosome 8 (Fig. 3) belong to the TPS-e/f subclade (TPS18, TPS19, TPS20, and TPS21), and the fifth one, TPS41, belongs to the TPS-c subclade. The positions of the TPS-e/f genes on the chromosome are not contiguous; they are interspersed with NDPS1, the gene encoding neryl diphosphate synthase (Schilmiller et al., 2009), as well as another cis-prenyltransferase-like protein, CPT8B. Also in the vicinity are two genes encoding putative cytochrome P450 proteins (one of them mutated), one gene encoding a putative alcohol oxidase, and three genes encoding putative acyltransferases of the BAHD family (one of them mutated). Of particular note is that TPS20 (PHS1) and NDPS1, which encode the enzymes that catalyze the formation of the substrate utilized by PHS1, are very close to each other, as previously predicted from published mapping studies (Schilmiller et al., 2009). TPS41 is related to the bona fide CPS of tomato (Rebers et al., 1999), but it lacks the characteristic DXDD motif of CPS enzymes and also appears to lack a transit peptide. While the specific enzymatic activities of the proteins encoded by TPS18, TPS19, and TPS21 are not yet known, it is possible that NDPS1 and TPS41 encode enzymes that provide substrates for these terpene synthases. Furthermore, the other genes in this cluster may encode enzymes that further modify the products of the enzymes encoded by the TPS genes in this cluster in a manner analogous to similar clusters of genes in specialized metabolism seen in other species (Qi et al., 2004; Swaminathan et al., 2009).
The clustering of TPS genes and the close association of TPS genes with genes encoding enzymes that could potentially provide substrates for TPS enzymes or modify terpenes once they are synthesized has been observed before. For example, 18 of the 40 Arabidopsis TPS genes are found in groups of two to five genes, while six GGPPS-like genes, five cytochrome P450 genes, and two glycosyltranferase genes are found in close proximity to some of these TPS genes (Aubourg et al., 2002). In contrast, in grape, which has a considerably larger TPS family, 129 out of 152 TPS genes are arranged in clusters of two to 45 genes, and only two of these TPS genes are found close to other putative terpenoid metabolism-related genes (a prenyltransferase and a cytochrome P450; Martin et al., 2010).
Structural Evolution of Tomato TPS Genes
The observations that in each TPS cluster, the genes of the cluster share a higher degree of identity with each other than with any other genes in the genome, and that in most cases they are also less similar to any TPS genes outside the Solanaceae, suggest that these clusters are the result of recent tandem duplications and possibly functional divergence. Gene losses also occur, and the 12 mutated TPS genes in the tomato genome bear witness to such stochastic events. Seven of the mutated genes have the full complement of exons but show small internal deletions and mutations. Another five mutated genes currently have only a few upstream or downstream exons (TPS23 only has the last exon). These nonfunctional genes may have undergone a major deletion, or they may have been the result of incomplete duplications. At any rate, they are clearly nonfunctional. Again, examples of mutated TPS genes as well as partially deleted/duplicated TPS genes have been observed in Arabidopsis and other species (Aubourg et al., 2002; Martin et al., 2010).
Loss and gain of introns are also evident in the tomato TPS gene family. While most tomato TPS-a genes bear seven introns, the monophyletic group of TPS31, TPS32, TPS33, and TPS35 on chromosome 1 (Fig. 1) contains only five introns, having lost the equivalent of the sixth intron in the rest of the TPS-a genes (Table I; Supplemental Figs. S1 and S5). In contrast, all but one of the TPS-b genes contain seven introns; TPS5 contains an additional intron that appears to be the result of an insertion of a DNA fragment, only part of which is spliced out to give the mature mRNA, thus causing an insertion of 22 amino acid residues in the middle of the protein (Table I; Supplemental Figs. S1, S5, and S6).
CONCLUSION
Ripe tomato fruits are mostly devoid of terpenoid flavor compounds other than those derived from the degradation of carotenoids (Goff and Klee, 2006; Mathieu et al., 2009). Extensive breeding programs that focused primarily on larger fruit yields may have decreased the amount of defensive terpenoids produced in the vegetative part of the plant, perhaps indirectly through the reduction in trichome density in cultivated tomatoes as compared with the density found in wild species (Blauth et al., 1998; Simmons et al., 2003). Until this work, there was scant evidence for the expression of tomato TPS genes in the EST databases. However, our systematic analysis of the tomato genome sequence indicates that the number of TPS genes in the genome is similar to that found in other genomes, and the proportion of TPS genes with mutations in the coding region, 15 out of 44, is only 2-fold higher than that found in Arabidopsis (eight out of 40; Aubourg et al., 2002). It is of interest to look at both differences in expression levels of TPS genes and biochemical activity of the encoded proteins among Solanum species to understand the effects of domestication on the various functions of the TPS family in the plant.
MATERIALS AND METHODS
Plant Growth and Conditions
Seeds of tomato (Solanum lycopersicum) and Solanum pennellii LA0716 were obtained from the Tomato Genetic Resource Center (http://tgrc/ucdavis.edu). Throughout this paper, when not specifically indicated, the tomato plants used were of cv M82. Seedlings were grown in Jiffy peat pots (Hummert International) in a controlled-growth chamber maintained for 16 h in the light (300 μE m−2 s−1; mixed cool-white and incandescent bulbs) at 28°C and 8 h in the dark at 20°C.
Identification of TPS Gene Models in the Tomato Genome
The prepublication draft assembly of the tomato genome that was made publicly available from the International Tomato Genome Sequencing Consortium facilitated the screening of the genomic scaffolds with known TPS sequences from the nonredundant protein database maintained by the National Center for Biotechnology Information. In particular, TPS sequences were used as a query sequence for TBLASTN searches of SOL Genomic Network databases. The FGENESH gene prediction tool (www.softberry.com) was used for initial annotation of the predicted tomato TPS genes. The gene models were further manually curated to accomplish a more accurate gene annotation considering available ESTs as well as cDNA sequences after PCR amplification using gene-specific primers (Supplemental Fig. S1). The tomato-derived cDNA sequences along with those of homolog genes from other species facilitated the prediction of exon-intron boundaries using the Sim4 nucleotide sequence alignment program (Florea et al., 1998). ChloroP was the prediction tool used to identify potential N-terminal plastid transit peptides in the predicted amino acid sequences corresponding to each TPS gene (Emanuelsson et al., 1999). Multiple sequence alignments were generated with the ClustalW program (Chenna et al., 2003). Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 4 (Tamura et al., 2007).
Gene Expression Analysis
Expression Analysis of Tomato TPS Genes Performed by RT-PCR
Total RNA was isolated with the RNAeasy kit (Qiagen), treated with the DNA-free kit (Ambion) to remove genomic DNA contamination, and used for first-strand cDNA synthesis using SuperScript II reverse transcriptase and oligo(dT)12–18 primer (Invitrogen) according to the manufacturer’s protocol. PCR amplification was performed with GoTaq Green Master Mix (Promega) using an initial denaturation step for 2 min at 94°C, followed by denaturation for 30 s at 94°C, annealing for 30 s at 68°C, extension for 30 s at 72°C, and a final extension of 5 min for 40 cycles to enable the detection of rare transcripts. Alignment of the tomato TPS cDNA sequences allowed the design of gene-specific primers for their PCR amplification, while actin was used as an internal control (Supplemental Table S1).
Expression Analysis of TPS12 Performed by qRT-PCR
Total trichomes were collected in liquid nitrogen from stems and leaves of growth chamber M82 plants. Total RNA extraction and first-strand cDNA synthesis were performed as mentioned above. The resulting cDNA was diluted 10-fold, and 1 μL was used as a template for PCR amplification in a 30-μL reaction using Power SYBR Green PCR master mix (Applied Biosystems) and gene-specific primers (Supplemental Table S1). Reactions were performed with the StepOnePlus Real-Time PCR System (Applied Biosystems) with the following cycles: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. A final dissociation step was performed to assess the quality of the amplified product. Relative expression levels of TPS12 in stem and leaf trichomes were calculated by using the relative quantification method normalized to the expression levels of tomato elongation factor 1 (GenBank accession no. X14449).
JA Treatment
Four-week-old Moneymaker C32 tomato plants were sprayed either with JA solution (1 mm JA + 0.05% SilwetL-77 in tap water) or with control solution (0.05% SilwetL-77 in tap water). Tissue was collected 30 min, 2 h, 8 h, and 24 h later. Stem trichomes were isolated by shaking in liquid N2, and subsequently, total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer’s instructions. Equal amounts of RNA from the different time points were pooled, creating the control and JA samples.
Expression Profiling Database Construction
mRNA was amplified and purified using the MessageAmp II aRNA Amplification kit (Applied Biosystems) according to the manufacturer’s instructions. Synthesis of cDNA was performed using the MessageAmp II aRNA Amplification kit (Applied Biosystems) according to the manufacturer’s instructions with modifications of the adapters to enable sequencing of 3′ cDNA ends. Shearing and ligation were carried out using standard Illumina paired-end adapters containing a specific sample identifier tag. Adapter-ligated cDNA fragments were column purified with the Qiaquick PCR purification kit (Qiagen). Expression profiling was performed using the Illumina GA II System. The GA reads were mapped to contigs obtained from a GS FLX Titanium run (454 Life Sciences, Roche Diagnostics) by Vertis Biotechnologie.
Isolation of Full-Length TPS cDNAs
The full-length cDNAs for TPS3 (lacking the transit peptide-coding region) and TPS8 were obtained by PCR amplification with gene-specific primers (Supplemental Table S1) based on genomic DNA sequence information. The cDNA used as template was synthesized from M82 stem trichome and root RNA. PCR amplification was performed with the KOD DNA polymerase (Novagen) and the following PCR conditions: 2 min at 95°C and then 20 s at 95°C, 10 s at 58°C, and 30 s at 68°C min for 30 cycles. The open reading frames of TPS3 and TPS8 were then inserted into the bacterial expression vector pEXP5-NT/TOPO (Invitrogen).
For TPS7, TPS38, and TPS39 genes, primers (Supplemental Table S1) were designed to amplify the entire open reading frame from cDNA prepared from flower tissue. For recombinant protein expression, a truncated version of TPS7 (beginning at amino acid 45) was amplified using primers T7F2/T7R2, digested with NheI/NotI, and ligated into the corresponding sites of pET28b. For TPS38, a modified version of the gene was synthesized (Genscript) and inserted between the NheI/NotI sites of pET28b. For TPS39, the open reading frame was amplified using primers T39F/T39R and transferred to pEXP5-NT/TOPO, according to the manufacturer’s instructions (Invitrogen).
The S. pennellii full-length PHS1 (TPS20) was amplified from leaf cDNA using primers based on SlPHS1 cDNA (Supplemental Table S1). The SpPHS1 open reading frame was then inserted into pEXP5-NT/TOPO (Invitrogen) to facilitate the expression of a recombinant version of the protein excluding the transit peptide and with insertion of an artificial initiation Met (at amino acid 45).
The entire open reading frame of TPS24 was amplified from cDNA prepared from M82 green fruit tissue using primers TPS24-F1 and TPS24-R1 (Supplemental Table S1). PCR amplification was performed with the KOD DNA polymerase (Novagen) for 2 min at 94°C followed by 40 cycles of 15 s at 94°C, 30 s at 55°C, and 2 min at 72°C, and then 10 min at 72°C. For recombinant protein expression, a truncated version of TPS24 (without the transit peptide-coding region) was amplified using primers TPS24-F2 and TPS24-R2 (Supplemental Table S1) and then ligated into the pEXP5-NT/TOPO (Invitrogen).
Protoplast Transformation and Confocal Microscopy
The open reading frame of TPS14 and a region corresponding to the first 180 nucleotides of TPS36 were amplified with primer pairs TPS14FOR/REV and T36F9/R4, respectively (Supplemental Table S1) and ligated between the SacI and BamHI sites of pSAT6a (Yu et al., 2010), creating an in-frame C-terminal fusion with GFP. Arabidopsis (Arabidopsis thaliana) mesophyll protoplasts were obtained using the “tape sandwich” method described by Wu et al. (2009). Approximately 25 μg of each GFP fusion construct was mobilized into protoplasts using the polyethylene glycol-mediated transfection procedure (http://genetics.mgh.harvard.edu/sheenweb). GFP fluorescence was visualized 16 to 30 h following transfection with a Leica SP5 laser scanning confocal microscope. A DM6000B microscope base was utilized to capture images using LAS AF version 2.4.1 build 6384 software, a double-dichroic 488/561 beam splitter, a 488-nm argon laser with 496- to 555-nm spectral detection for GFP, a 561-nm laser with 569- to 634-nm spectral detection for MitoTracker Red (Invitrogen), and chloroplast autofluorescence detection from both lasers at 651 to 800 nm.
Expression in Escherichia coli and TPS Enzyme Assays
TPS3 and TPS8
E. coli BL21-CodonPlus(DE3) cells (Stratagene) containing the plasmids pEXP5-NT-TOPO:TPS3 and pEXP5-NT-TOPO:TPS8 were grown in Luria-Bertani medium containing the appropriate antibiotics until optical density of the culture at 600 nm (OD600) reached 0.5 to 0.7 and then induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside (IPTG) at 18°C for 16 h. Cell pellets were resuspended in assay buffer containing 50 mm HEPES, 7.5 mm MgCl2, 100 mm KCl, 5 mm dithiothreitol, and 10% (v/v) glycerol, pH 7.0. The enzyme reactions contained 0.2 mL of E. coli crude protein extract in assay buffer (after sonication) and 20 μm prenyl diphosphate substrate. All commercially available prenyl-diphosphate substrates, including GPP, NPP, e,e-FPP, z,z-FPP, and GGPP, were used (Echelon Biosciences). Assay mixtures were incubated at 30°C for 1 h, and the resultant mixture was directly exposed to a polydimethylsiloxane solid-phase microextraction (SPME) fiber (Supelco) at 42°C for 15 min.
TPS7, TPS32, TPS38, and TPS39
Plasmids pET28b:TPS7, pEXP5-NT/TOPO:TPS32, pET28b:TPS38, and pEXP5-NT/TOPO:TPS39 were transferred into E. coli BL21-CodonPlus(DE3)-RIPL cells (Stratagene), which were grown at 37°C in Luria-Bertani medium containing the appropriate antibiotics. When the OD600 reached 0.6, IPTG was added to a final concentration of 1 mm and cells were transferred to 18°C and incubated for an additional 16 h. Cell pellets were resuspended in buffer A (50 mm MOPS, 5 mm MgCl2, 20 μm MnCl2, 5 mm dithiothreitol, and 10% glycerol [v/v], pH 7.0) and ruptured by sonication. Clarified crude extracts were desalted on PD-10 columns equilibrated with 100 mm sodium phosphate and 150 mm NaCl, pH 7.0, and recombinant His6-tagged proteins were partially purified via Ni2+ affinity chromatography, according to the manufacturer’s instructions (Qiagen). Partially purified recombinant proteins were desalted on PD-10 columns equilibrated with buffer A, and protein concentration was estimated by the method of Bradford (1976) using bovine serum albumin as the standard. Enzymatic assays were performed in buffer A containing 10 to 50 μg of protein and 40 μm of substrate in a volume of 250 μL. Reaction products were collected by SPME.
TPS24
E. coli C41 cells containing the plasmid pGGeC (Cyr et al., 2007), which carries the Abies grandis GGPPS gene and the maize (Zea mays) CPS gene, and E. coli cells containing the pGGeC plasmid as well as a second plasmid with the Picea glauca KS (Keeling et al., 2010) or the tomato TPS24 gene were grown until A600 reached 0.5 to 0.7 and then induced with 0.4 mm IPTG at 16°C for 16 h, after which 7.5 mL of the culture medium was extracted with 500 μL of hexane containing 100 ng μL−1 tetradecane, and 3 μL was injected for GC-MS. For obtaining ent-kaurene standard, E. coli cells carrying pGGeC were transformed with a plasmid containing a cDNA of PgKS, the gene encoding ent-kaurene synthase from P. glauca (Keeling et al., 2010), and grown under the same conditions as described above.
TPS14 Enzyme Assays
The enzymatic assays for TPS14 were performed as described by Bleeker et al. (2011).
SpPHS1 Enzyme Assays
The conditions used for product identification and kinetic studies were as described for SlPHS1 by Schilmiller et al. (2009).
GC-MS Analysis of Terpenes
GC-MS Tomato Tissue Chemical Analysis
Terpene standards were obtained from Sigma-Aldrich. For analysis of total trichome terpenes in M82, IL8-1-1, IL1-4, and LA0716 plants, leaflets from the second leaf after the newly emerging leaf of 3-week-old plants were dipped with gentle rocking for 1 min in 750 μL of methyl tert-butyl ether containing 10 ng μL−1 tetradecane internal standard. For type VI trichome analysis, approximately 200 type VI glands from 3-week-old plants (M82, IL8-1-1, and IL1-4) and greenhouse-grown plants (LA0716) were picked using a pulled Pasteur pipette into 100 μL of methyl tert-butyl ether containing tetradecane internal standard. Two microliters of the extract was injected into an EC-WAX column (Grace Davison; 30 m length, 0.25 μm film thickness, and 0.32 mm i.d.) on a GC17-A (Shimadzu) coupled to a QP-5000 GC-MS system. Injector temperature was 220°C and working on splitless mode. Interface temperature was 280°C. The temperature program was as follows: 44°C for 3.5 min, 5°C min−1 up to 200°C, 70°C min−1 up to 275°C, and hold for 1 min. In the case where SPME fiber was used for extraction, the tissue was exposed to the fiber in a 2-mL glass vial for 15 min at 42°C. The fiber was then injected in a Shimadzu QP-2010 GC-MS system fitted with an HP-5MS column (0.25 mm diameter, 30 m long, and 0.25 μm film thickness) at 250°C, and after a 2-min isothermal hold at 50°C, the column temperature was increased by 10°C min−1 to 275°C with a 10-min isothermal hold at 275°C.
GC-MS Analysis of TPS Enzymatic Assay Products
After enzyme assay incubations, the SPME fiber that was used to absorb the volatile terpenes produced was injected and analyzed on a Shimadzu QP-2010 GC-MS system fitted with an HP-5MS column (0.25 mm diameter, 30 m long, and 0.25 μm film thickness). Samples were injected onto the column at 250°C, and after a 2-min isothermal hold at 50°C, the column temperature was increased by 10°C min−1 to 275°C with a 10-min isothermal hold at 275°C.
For TPS14 enzymatic assays, terpenoid products were separated on a DB-5 column (10 m × 180 μm, 0.18 μm film thickness; Hewlett-Packard) in an 6890N gas chromatograph (Agilent) with a temperature program set to 40°C for 1.5 min, ramp to 250°C by 30°C min−1, with a 2.5-min isothermal hold at 250°C.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers JN408284, JN408285, JN408286, JN408287, JN408288, JN408289, JN412093, JN412092, JN412091, JN412090, JN412089, JN412088, JN412072, FJ797957, JN412071, JN412087, JN412086, JN412085, JN412084, JN412083, JN412082, JN412081, JN412080, JN412079, JN412078, JN412077, JN412076, JN412075, JN412074, and JN412073.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Annotated sequences of the tomato TPS genes.
Supplemental Figure S2. Phylogenetic tree of the proteins encoded by the 29 functional or potentially functional tomato TPS genes and “clade representative” protein sequences from other species.
Supplemental Figure S3. RT-PCR results with specific primers for the potentially functional TPS genes in tomato M82 and RNA from various tissues.
Supplemental Figure S4. Sequence alignment of the proteins encoded by the functional or potentially functional tomato TPS-a genes.
Supplemental Figure S5. Exon/intron structure of the tomato TPS genes.
Supplemental Figure S6. Sequence alignment of the proteins encoded by the functional or potentially functional tomato TPS-b and TPS-g genes.
Supplemental Figure S7. Sequence alignment of the proteins encoded by the functional or potentially functional tomato TPS-c genes.
Supplemental Figure S8. Sequence alignment of the proteins encoded by the functional or potentially functional tomato TPS-e/f genes.
Supplemental Table S1. Information about primers used in this work.
Acknowledgments
We thank Dr. Dani Zamir (Hebrew University) for providing seeds of the S. pennellii introgression lines, Dr. Reuben Peters (Iowa State University) for providing plasmid pGGeC, and Dr. Joerg Bohlmann (University of British Columbia) for providing plasmid carrying the PgKS cDNA. We also thank Dr. Natalia Dudareva (Purdue University) for the spRBC-GFP construct, Dr. Yoel Shiboleth (University of Michigan) for advice on protoplast transformation, and Greg Sobocinski (University of Michigan) for technical advice with confocal microscopy.
References
- Aharoni A, Giri AP, Verstappen FW, Bertea CM, Sevenier R, Sun Z, Jongsma MA, Schwab W, Bouwmeester HJ. (2004) Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 16: 3110–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ament K, Kant MR, Sabelis MW, Haring MA, Schuurink RC. (2004) Jasmonic acid is a key regulator of spider mite-induced volatile terpenoid and methyl salicylate emission in tomato. Plant Physiol 135: 2025–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ament K, Van Schie CC, Bouwmeester HJ, Haring MA, Schuurink RC. (2006) Induction of a leaf specific geranylgeranyl pyrophosphate synthase and emission of (E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene in tomato are dependent on both jasmonic acid and salicylic acid signaling pathways. Planta 224: 1197–1208 [DOI] [PubMed] [Google Scholar]
- Arimura G, Huber DP, Bohlmann J. (2004) Forest tent caterpillars (Malacosoma disstria) induce local and systemic diurnal emissions of terpenoid volatiles in hybrid poplar (Populus trichocarpa x deltoides): cDNA cloning, functional characterization, and patterns of gene expression of (−)-germacrene D synthase, PtdTPS1. Plant J 37: 603–616 [DOI] [PubMed] [Google Scholar]
- Aubourg S, Lecharny A, Bohlmann J. (2002) Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Genet Genomics 267: 730–745 [DOI] [PubMed] [Google Scholar]
- Back K, Chappell J. (1995) Cloning and bacterial expression of a sesquiterpene cyclase from Hyoscyamus muticus and its molecular comparison to related terpene cyclases. J Biol Chem 270: 7375–7381 [DOI] [PubMed] [Google Scholar]
- Bertea CM, Voster A, Verstappen FW, Maffei M, Beekwilder J, Bouwmeester HJ. (2006) Isoprenoid biosynthesis in Artemisia annua: cloning and heterologous expression of a germacrene A synthase from a glandular trichome cDNA library. Arch Biochem Biophys 448: 3–12 [DOI] [PubMed] [Google Scholar]
- Blauth SL, Churchill GA, Mutschler MA. (1998) Identification of quantitative trait loci associated with acylsugar accumulation using intraspecific populations of the wild tomato, Lycopersicon pennellii. Theor Appl Genet 96: 458–467 [DOI] [PubMed] [Google Scholar]
- Bleeker PM, Spyropoulou EA, Diergaarde PJ, Volpin H, De Both MTJ, Zerbe P, Bohlmann J, Falara V, Matsuba Y, Pichersky E, et al. (August 5, 2011) RNA-seq discovery, functional characterization, and comparison of sesquiterpene synthases from Solanum lycopersicum and Solanum habrochaites trichomes. Plant Mol Biol http://dx.doi.org/10.1007/s11103-011-9813-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlmann J, Martin D, Oldham NJ, Gershenzon J. (2000) Terpenoid secondary metabolism in Arabidopsis thaliana: cDNA cloning, characterization, and functional expression of a myrcene/(E)-beta-ocimene synthase. Arch Biochem Biophys 375: 261–269 [DOI] [PubMed] [Google Scholar]
- Bohlmann J, Meyer-Gauen G, Croteau R. (1998) Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA 95: 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Chen F, Ro DK, Petri J, Gershenzon J, Bohlmann J, Pichersky E, Tholl D. (2004) Characterization of a root-specific Arabidopsis terpene synthase responsible for the formation of the volatile monoterpene 1,8-cineole. Plant Physiol 135: 1956–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tholl D, Bohlmann J, Pichersky E. (2011) The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J 66: 212–229 [DOI] [PubMed] [Google Scholar]
- Chen F, Tholl D, D’Auria JC, Farooq A, Pichersky E, Gershenzon J. (2003) Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15: 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby SM, Crock J, Dowdle-Rizzo B, Lemaux PG, Croteau R. (1998) Germacrene C synthase from Lycopersicon esculentum cv. VFNT cherry tomato: cDNA isolation, characterization, and bacterial expression of the multiple product sesquiterpene cyclase. Proc Natl Acad Sci USA 95: 2216–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr A, Wilderman PR, Determan M, Peters RJ. (2007) A modular approach for facile biosynthesis of labdane-related diterpenes. J Am Chem Soc 129: 6684–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Wang G, Yang DS, Tang Y, Broun P, Marks MD, Sumner LW, Dixon RA, Zhao PX. (2010) TrichOME: a comparative omics database for plant trichomes. Plant Physiol 152: 44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich-Rikanati R, Lewinsohn E, Bar E, Iijima Y, Pichersky E, Sitrit Y. (2008) Overexpression of the lemon basil α-zingiberene synthase gene increases both mono- and sesquiterpene contents in tomato fruit. Plant J 56: 228–238 [DOI] [PubMed] [Google Scholar]
- Davidovich-Rikanati R, Sitrit Y, Tadmor Y, Iijima Y, Bilenko N, Bar E, Carmona B, Fallik E, Dudai N, Simon JE, et al. (2007) Enrichment of tomato flavor by diversion of the early plastidial terpenoid pathway. Nat Biotechnol 25: 899–901 [DOI] [PubMed] [Google Scholar]
- Deguerry F, Pastore L, Wu S, Clark A, Chappell J, Schalk M. (2006) The diverse sesquiterpene profile of patchouli, Pogostemon cablin, is correlated with a limited number of sesquiterpene synthases. Arch Biochem Biophys 454: 123–136 [DOI] [PubMed] [Google Scholar]
- Dudareva N, Cseke L, Blanc VM, Pichersky E. (1996) Evolution of floral scent in Clarkia: novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell 8: 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennajdaoui H, Vachon G, Giacalone C, Besse I, Sallaud C, Herzog M, Tissier A. (2010) Trichome specific expression of the tobacco (Nicotiana sylvestris) cembratrien-ol synthase genes is controlled by both activating and repressing cis-regions. Plant Mol Biol 73: 673–685 [DOI] [PubMed] [Google Scholar]
- Falara V, Pichersky E, Kanellis AK. (2010) A copal-8-ol diphosphate synthase from the angiosperm Cistus creticus subsp. creticus is a putative key enzyme for the formation of pharmacologically active, oxygen-containing labdane-type diterpenes. Plant Physiol 154: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fäldt J, Arimura G, Gershenzon J, Takabayashi J, Bohlmann J. (2003) Functional identification of AtTPS03 as (E)-beta-ocimene synthase: a monoterpene synthase catalyzing jasmonate- and wound-induced volatile formation in Arabidopsis thaliana. Planta 216: 745–751 [DOI] [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. (1998) A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res 8: 967–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon J, Dudareva N. (2007) The function of terpene natural products in the natural world. Nat Chem Biol 3: 408–414 [DOI] [PubMed] [Google Scholar]
- Goff SA, Klee HJ. (2006) Plant volatile compounds: sensory cues for health and nutritional value? Science 311: 815–819 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Kawaide H, Notomi M, Sakigi Y, Matsuo A, Nozaki H. (2006) Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Lett 580: 6175–6181 [DOI] [PubMed] [Google Scholar]
- Herde M, Gärtner K, Köllner TG, Fode B, Boland W, Gershenzon J, Gatz C, Tholl D. (2008) Identification and regulation of TPS04/GES, an Arabidopsis geranyllinalool synthase catalyzing the first step in the formation of the insect-induced volatile C16-homoterpene TMTT. Plant Cell 20: 1152–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Abel C, Sohrabi R, Petri J, Haupt I, Cosimano J, Gershenzon J, Tholl D. (2010) Variation of herbivore-induced volatile terpenes among Arabidopsis ecotypes depends on allelic differences and subcellular targeting of two terpene synthases, TPS02 and TPS03. Plant Physiol 153: 1293–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jones CG, Moniodis J, Zulak KG, Scaffidi A, Plummer JA, Ghisalberti EL, Barbour EL, Bohlmann J. (2011) Sandalwood fragrance biosynthesis involves sesquiterpene synthases of both the terpene synthase (TPS) a and TPS-b subfamilies, including santalene synthases. J Biol Chem 286: 17445–17454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Shi F, Jones AD, Marks MD, Howe GA. (2010) Distortion of trichome morphology by the hairless mutation of tomato affects leaf surface chemistry. J Exp Bot 61: 1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K, Olsen LJ, Theg SM. (1989) Chloroplastic precursors and their transport across the membrane. Annu Rev Plant Physiol Plant Mo1 Biol 40: 471–501 [Google Scholar]
- Keeling CI, Dullat HK, Yuen M, Ralph SG, Jancsik S, Bohlmann J. (2010) Identification and functional characterization of monofunctional ent-copalyl diphosphate and ent-kaurene synthases in white spruce reveal different patterns for diterpene synthase evolution for primary and secondary metabolism in gymnosperms. Plant Physiol 152: 1197–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinsohn E, Sitrit Y, Bar E, Azulay Y, Ibdah M, Meir A, Yosef E, Zamir D, Tadmor Y. (2005) Not just colors: carotenoid degradation as a link between pigmentation and aroma in tomato and watermelon fruit. Trends Food Sci Technol 16: 407–415 [Google Scholar]
- Lücker J, Bowen P, Bohlmann J. (2004) Vitis vinifera terpenoid cyclases: functional identification of two sesquiterpene synthase cDNAs encoding (+)-valencene synthase and (−)-germacrene D synthase and expression of mono- and sesquiterpene synthases in grapevine flowers and berries. Phytochemistry 65: 2649–2659 [DOI] [PubMed] [Google Scholar]
- Martin DM, Aubourg S, Schouwey MB, Daviet L, Schalk M, Toub O, Lund ST, Bohlmann J. (2010) Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol 10: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu S, Cin VD, Fei Z, Li H, Bliss P, Taylor MG, Klee HJ, Tieman DM. (2009) Flavour compounds in tomato fruits: identification of loci and potential pathways affecting volatile composition. J Exp Bot 60: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagegowda DA, Gutensohn M, Wilkerson CG, Dudareva N. (2008) Two nearly identical terpene synthases catalyze the formation of nerolidol and linalool in snapdragon flowers. Plant J 55: 224–239 [DOI] [PubMed] [Google Scholar]
- Nieuwenhuizen NJ, Wang MY, Matich AJ, Green SA, Chen X, Yauk YK, Beuning LL, Nagegowda DA, Dudareva N, Atkinson RG. (2009) Two terpene synthases are responsible for the major sesquiterpenes emitted from the flowers of kiwifruit (Actinidia deliciosa). J Exp Bot 60: 3203–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Pechous SW, Whitaker BD. (2004) Cloning and functional expression of an (E,E)-alpha-farnesene synthase cDNA from peel tissue of apple fruit. Planta 219: 84–94 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Noel JP, Dudareva N. (2006) Biosynthesis of plant volatiles: nature’s diversity and ingenuity. Science 311: 808–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Bakht S, Leggett M, Maxwell C, Melton R, Osbourn A. (2004) A gene cluster for secondary metabolism in oat: implications for the evolution of metabolic diversity in plants. Proc Natl Acad Sci USA 101: 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebers M, Kaneta T, Kawaide H, Yamaguchi S, Yang YY, Imai R, Sekimoto H, Kamiya Y. (1999) Regulation of gibberellin biosynthesis genes during flower and early fruit development of tomato. Plant J 17: 241–250 [DOI] [PubMed] [Google Scholar]
- Roeder S, Hartmann AM, Effmert U, Piechulla B. (2007) Regulation of simultaneous synthesis of floral scent terpenoids by the 1,8-cineole synthase of Nicotiana suaveolens. Plant Mol Biol 65: 107–124 [DOI] [PubMed] [Google Scholar]
- Sallaud C, Rontein D, Onillon S, Jabès F, Duffé P, Giacalone C, Thoraval S, Escoffier C, Herbette G, Leonhardt N, et al. (2009) A novel pathway for sesquiterpene biosynthesis from Z,Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell 21: 301–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A, Shi F, Kim J, Charbonneau AL, Holmes D, Jones AD, Last RL. (2010a) Mass spectrometry screening reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines. Plant J 62: 391–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Miner DP, Larson M, McDowell E, Gang DR, Wilkerson C, Last RL. (2010b) Studies of a biochemical factory: tomato trichome deep expressed sequence tag sequencing and proteomics. Plant Physiol 153: 1212–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Schauvinhold I, Larson M, Xu R, Charbonneau AL, Schmidt A, Wilkerson C, Last RL, Pichersky E. (2009) Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc Natl Acad Sci USA 106: 10865–10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnee C, Köllner TG, Gershenzon J, Degenhardt J. (2002) The maize gene terpene synthase 1 encodes a sesquiterpene synthase catalyzing the formation of (E)-β-farnesene, (E)-nerolidol, and (E,E)-farnesol after herbivore damage. Plant Physiol 130: 2049–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon-Asa L, Shalit M, Frydman A, Bar E, Holland D, Or E, Lavi U, Lewinsohn E, Eyal Y. (2003) Citrus fruit flavor and aroma biosynthesis: isolation, functional characterization, and developmental regulation of Cstps1, a key gene in the production of the sesquiterpene aroma compound valencene. Plant J 36: 664–674 [DOI] [PubMed] [Google Scholar]
- Simmons AT, Gurr GM, McGrath D, Nicol HI, Martin PM. (2003) Trichomes of Lycopersicon spp. and their effect on Myzus persicae (Sulzer) (Hemiptera: Aphididae). Aust J Entomol 42: 373–378 [Google Scholar]
- Steeghs M, Bais HP, de Gouw J, Goldan P, Kuster W, Northway M, Fall R, Vivanco JM. (2004) Proton-transfer-reaction mass spectrometry as a new tool for real time analysis of root-secreted volatile organic compounds in Arabidopsis. Plant Physiol 135: 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Kamiya Y. (1994) The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6: 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ. (2009) CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21: 3315–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tholl D, Chen F, Petri J, Gershenzon J, Pichersky E. (2005) Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J 42: 757–771 [DOI] [PubMed] [Google Scholar]
- Tholl D, Lee S. (2011) Terpene specialized metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0143, doi/10.1199/tab.0143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- van der Hoeven RS, Monforte AJ, Breeden D, Tanksley SD, Steffens JC. (2000) Genetic control and evolution of sesquiterpene biosynthesis in Lycopersicon esculentum and L. hirsutum. Plant Cell 12: 2283–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie CC, Haring MA, Schuurink RC. (2007) Tomato linalool synthase is induced in trichomes by jasmonic acid. Plant Mol Biol 64: 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Sun T, Kawaide H, Kamiya Y. (1998) The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol 116: 1271–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Nguyen TT, Guo Y, Schauvinhold I, Auldridge ME, Bhuiyan N, Ben-Israel I, Iijima Y, Fridman E, Noel JP, et al. (2010) Enzymatic functions of wild tomato methylketone synthases 1 and 2. Plant Physiol 154: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS. (2009) Tape-Arabidopsis sandwich: a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]