Abstract
Abscisic acid-, stress-, and ripening-induced (ASR) proteins were first described about 15 years ago as accumulating to high levels during plant developmental processes and in response to diverse stresses. Currently, the effects of ASRs on water deficit tolerance and the ways in which their physiological and biochemical functions lead to this stress tolerance remain poorly understood. Here, we characterized the ASR gene family from maize (Zea mays), which contains nine paralogous genes, and showed that maize ASR1 (ZmASR1) was encoded by one of the most highly expressed paralogs. Ectopic expression of ZmASR1 had a large overall impact on maize yield that was maintained under water-limited stress conditions in the field. Comparative transcriptomic and proteomic analyses of wild-type and ZmASR1-overexpressing leaves led to the identification of three transcripts and 16 proteins up- or down-regulated by ZmASR1. The majority of them were involved in primary and/or cellular metabolic processes, including branched-chain amino acid (BCAA) biosynthesis. Metabolomic and transcript analyses further indicated that ZmASR1-overexpressing plants showed a decrease in BCAA compounds and changes in BCAA-related gene expression in comparison with wild-type plants. Interestingly, within-group correlation matrix analysis revealed a close link between 13 decreased metabolites in ZmASR1-overexpressing leaves, including two BCAAs. Among these 13 metabolites, six were previously shown to be negatively correlated to biomass, suggesting that ZmASR1-dependent regulation of these 13 metabolites might contribute to regulate leaf growth, resulting in improvement in kernel yield.
Since one-third of the world’s food is produced on irrigated land (Food and Agriculture Organization of the United Nations, 2008), the likely impacts of recurrent heat waves and water stress episodes on global food production are numerous. In the European heat wave of 2003, crop production was reduced by around 30% (Ciais et al., 2005). In maize (Zea mays), the most significant reductions in end-of-season kernel yields are observed when water deficit occurs during the flowering stage, either just before floral initiation or immediately after pollination, because of the impact of water deficit on ovary and young kernel abortions (Claassen, 1970; Boyer and Westgate, 2004). Water deficit during the vegetative growth phase also leads to reductions in overall productivity through reductions in kernel numbers (Boyer and Westgate, 2004). Finally, late-stage water deficit during the kernel-filling period can lead to reductions in yield, depending upon the dry matter reserves in the plant (McPherson and Boyer, 1977).
Trait-based approaches considering maize drought avoidance and dehydration tolerance mechanisms have been relatively slow to progress, as judged by the adoption of improved varieties (Salekdeh et al., 2009). Transgenic maize plants expressing the betA gene from Escherichia coli encoding choline dehydrogenase, a key enzyme in the biosynthesis of Gly betaine from choline, showed enhanced Gly betaine accumulation, resulting in greater kernel yield after drought stress in field tests (Quan et al., 2004). Expression of a maize CAAT box transcription factor, Z. mays nuclear factor Y B subunit 2 (ZmNF-YB2), has also been shown to confer drought tolerance and enhanced photosynthetic capacity under drought stress, with improvements in kernel yield in maize (Nelson et al., 2007). Additionally, two members of a family of bacterial RNA chaperones, cold shock protein A (CspA) from E. coli and CspB from Bacillus subtilis, were shown to confer vegetative tolerance and improved end-of-season kernel yield under water-limiting conditions in maize (Castiglioni et al., 2008). These studies clearly demonstrate that maize plants are amenable to improved water deficit tolerance through multiple mechanisms of action.
Having specific target traits and genes can markedly accelerate progress through the marker-assisted selection of parents and progeny in early generations (Tuberosa and Salvi, 2006). We used a proteomics strategy, with the use of a recombinant inbred line population derived from the cross between the maize inbred lines MBS847 and F2, differing in yield under water deprivation conditions, to identify candidate proteins associated with water stress response in maize (de Vienne et al., 1999). The protein quantity profiling of all individuals in the segregating population made it possible to treat the quantity of each protein as a quantitative trait and to identify a quantitative trait locus (QTL) for protein quantity, hereafter named protein quantity locus (PQL) according to Damerval et al. (1994). When the PQL colocated with a QTL related to water deficit response, a causal relationship could be inferred between the protein level and the trait variation. Furthermore, when the gene encoding the protein also colocated, it could be seen as a candidate gene. Among the identified candidate genes was a member of the abscisic acid-, stress-, and ripening-induced (ASR) gene family, ZmASR1, which colocated with its PQL and QTLs for leaf senescence and anthesis-silking interval (de Vienne et al., 1999; Jeanneau et al., 2002). These associations were validated by the observation that transgenic maize lines misexpressing ZmASR1 showed significant changes in leaf senescence under mild water deficit conditions in the field (Jeanneau et al., 2002). Interestingly, transgenic Arabidopsis (Arabidopsis thaliana) plants overexpressing an ASR gene from Lilium longiflorum (LLA23) or banana (Musa paradisiaca; MpASR) exhibited enhanced capacity to survive in water-limited conditions (Yang et al., 2005; Liu et al., 2010). Overexpression of the tomato (Solanum lycopersicum) ASR1 gene (SlASR1) in tobacco (Nicotiana tabacum) plants also resulted in decreased rates of water loss (Kalifa et al., 2004b). Together, these results suggest that water deficit tolerance is mediated at least in part through ASR proteins.
The ASR proteins are highly charged, hydrophilic, and low-Mr proteins that are widely present in the plant kingdom, except in the Brassicaceae family (for review, see Carrari et al., 2004; Battaglia et al., 2008). Since their discovery more than a decade ago, they have been described as accumulating to high levels during plant developmental processes, such as fruit ripening, pollen maturation, and Glc metabolism (Iusem et al., 1993; Wang et al., 1998; Cakir et al., 2003; Frankel et al., 2007; Yang et al., 2008), and in response to diverse stresses, including water deficit, salt, cold, limited light, and, lately, pathogen attack (Schneider et al., 1997; Riccardi et al., 1998; Vaidyanathan et al., 1999; Huang et al., 2000; Maskin et al., 2001; Kalifa et al., 2004b; Liu et al., 2010; Philippe et al., 2010; Henry et al., 2011). Nevertheless, it is noteworthy that their exact function remains enigmatic. Indeed, the molecular mechanisms underlying the biological roles of the ASR proteins cannot be deduced simply by sequence homology with other known proteins. SlASR1 and the grape (Vitis vinifera) ASR protein, VvMSA (for maturation-, stress-, and abscisic acid-induced protein), have been shown to possess DNA-binding activity (Cakir et al., 2003; Kalifa et al., 2004a; Rom et al., 2006; Maskin et al., 2007; Shkolnik and Bar-Zvi, 2008). On the basis of such findings, these ASR proteins have been proposed to regulate the transcription of sugar- and abiotic stress-regulated genes in fruit and vegetative tissues, respectively (Cakir et al., 2003; Kalifa et al., 2004b; Frankel et al., 2007; Saumonneau et al., 2008). However, a dual function of LLA23 and SlASR1 proteins as a regulator and a protective molecule upon water deficit has also been proposed (Yang et al., 2005, 2008; Konrad and Bar-Zvi, 2008).
To validate further the candidate gene ZmASR1 and gain insight into its function, we characterized the ASR gene family of maize. We showed that ZmASR1 was one of the most highly expressed ASR genes in maize and found evidence that its expression had a large overall impact on maize vegetative productivity and yield that was maintained under water-limited stress conditions in the field. We identified 25 genes that were either transcriptionally or posttranscriptionally regulated by ZmASR1, of which seven were involved in branched-chain amino acid (BCAA) biosynthesis. Our studies further revealed a close link between 13 decreased metabolites in ZmASR1-overexpressing leaves, including two BCAAs, and support the idea that ZmASR1-dependent regulation of BCAA biosynthetic genes might contribute to yield improvement.
RESULTS
ZmASR1 Belongs to the Maize ASR Gene Family, Which Consists of at Least Nine Members
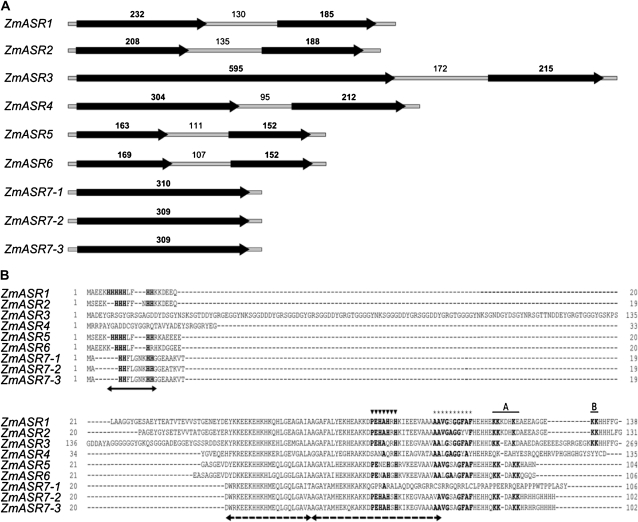
To identify ZmASR genes, we first analyzed the public databases for the presence of putative ASR coding sequences in maize using the ZmASR1 protein sequence as the query (Riccardi et al., 1998). The search was then extended to maize genomic contigs (The Institute for Genomic Research), high-throughput genomic sequences, and the recently released maize genomic sequence (Schnable et al., 2009), which were matched with the EST sequences and used to design specific primers for DNA and cDNA amplification in maize lines used for mapping analysis and transgenesis. This led to the identification of nine ZmASR genes in the maize genome (Fig. 1; Supplemental Tables S1 and S2), including the published gene Bss1 (for bundle sheath strands-specific gene 1; Furumoto et al., 2000), renamed hereafter ZmASR4. ZmASR1 to ZmASR7-3 mapped to five of the 10 maize chromosomes, with chromosomes 2 and 10 harboring two loci each and chromosome 3 harboring three loci (Supplemental Table S1). ZmASR7-1 and ZmASR7-2 were separated by 3 kb, whereas ZmASR7-3 was separated from this cluster by approximately 100 kb. When the nucleotide sequences of the nine ZmASR cDNAs were aligned, sequence identity values of 15% to 97% were observed between the individual maize genes (Supplemental Table S3). All ZmASR genes possessed a conserved structure composed of two exons and one intron, except ZmASR7-1, ZmASR7-2, and ZmASR7-3, whose first and second exons were fused (Fig. 1A). This fusion between exon 1 and exon 2 was also found in sorghum (Sorghum bicolor) SbASR6 and SbASR7, suggesting that it took place before the divergence of those two species (Supplemental Table S4). In general, the length of exons 1 and 2 was conserved (approximately 200 bp), with the exception of exon 1 of ZmASR3, which was three times longer, like its barley (Hordeum vulgare), sorghum, and rice (Oryza sativa) counterparts (Supplemental Table S4).
Figure 1.
Gene structure of ZmASR genes. A, Schematic drawing of the exon/intron structure of ZmASR genes. Black arrows and thin gray bars indicate exons (boldface numbers) and introns (lightface numbers), respectively, the sizes of which are in bp. B, Alignment of the deduced amino acid sequences of ZmASR genes. Arrows denote the two highly conserved regions of ASR proteins: a small N-terminal consensus of approximately 18 to 20 amino acids containing a stretch of six His residues (solid arrow) and a large C-terminal region containing two ABA/WDS signatures (dashed arrows). Amino acid positions determined to be the Zn2+-dependent DNA-binding activity domain and a sequence possibly hindering DNA binding of the SlASR1 protein (Rom et al., 2006) are marked with triangles and stars, respectively. Amino acid positions determined to be the A and B regions of the bipartite nuclear localization signal of the LLA23 protein (Wang et al., 2005) are overlined. Amino acids identical to LLA23 or SlASR1 proteins are shown in boldface.
The maize ASR loci encoded small proteins of 102 to 269 amino acid long that met the hydrophilin criteria (Garay-Arroyo et al., 2000), in that their GRAVY index was between −1.13 and −1.37 and 8% to 28% of their amino acid residues are Gly, and were predicted to be “natively unfolded” in solution, given their enrichment in disorder-promoting charged amino acids (Supplemental Table S4; Goldgur et al., 2007). They all contained the two ASR-specific conserved motifs (e.g. a small N-terminal consensus of 18–20 amino acids containing a typical stretch of six His residues and a large C-terminal region of at least 80 amino acids containing two abscisic acid [ABA]/water deficit stress [WDS] signatures; Canel et al., 1995; Padmanabhan et al., 1997), except ZmASR3 and ZmASR4, which did not possess the N-terminal His-rich domain (Fig. 1B; Supplemental Table S4). The first exon of ZmASR3 encoded an additional domain that was not conserved in other ZmASR proteins. Additionally, ZmASR7-1 presented a single nucleotide insertion in a position corresponding to amino acid 57 within the second ABA/WDS signature, leading to a frameshift and consequently to a unique and maize-specific deduced amino acid sequence at the C terminus (Fig. 1B). With the exception of ZmASR4 and ZmASR5, ZmASR proteins contained a sequence motif at the C terminus (Fig. 1B; Supplemental Table S4), presenting highest identity (71%–86%) with the Zn2+-dependent DNA-binding activity domain of the SlASR1 protein (Rom et al., 2006). Close examination of the C-terminal extremity of ZmASR1, ZmASR2, and ZmASR3 revealed the presence of a region structurally similar to the bipartite nuclear localization signal of LLA23 (Fig. 1B; Supplemental Table S4; Wang et al., 2005), while five other ZmASR proteins presented only the first motif (A in Fig. 1B) but lacked the second motif (B in Fig. 1B).
To assess the diversity of the ASR gene family beyond maize, a phylogenetic tree was constructed that included the nine maize genes and 91 related genes from representatives of different groups of Spermatophyta found in the data banks (Supplemental Fig. S1). In the case of rice, we adopted the nomenclature published during the time course of this study (Philippe et al., 2010). Maximum likelihood and Bayesian reconstruction methods were congruent and confirmed the previously established division of plant ASR proteins into three distinct clades corresponding to conifer, eudicot, and monocot sequences (Supplemental Fig. S1; Frankel et al., 2007). By adding all available maize sequences, we showed that within monocots, the Commelinid sequences form a monophyletic group, with Poaceae sequences falling into two distinct clades, the I-1/I-2/I-3 clade and the II-1/II-2 clade (Supplemental Fig. S1). Within the I-1/I-2/I-3 clade, ZmASR1, ZmASR2, ZmASR3, and ZmASR4 were related to SbASR1, SbASR2, SbASR3, and OsASR6, respectively (Supplemental Fig. S1). Within the II-1/II-2 clade, ZmASR5 and ZmASR6 were related to SbASR5 and SbASR4, respectively, while ZmASR7-1, ZmASR7-2, and ZmASR7-3 appeared as three recent paralogs that were related to SbASR7 (Supplemental Fig. S1).
Response of ZmASR Transcript Levels to Water Deficit and Water Stress-Related Treatments
To investigate the role of the different ZmASR genes in the maize plant and under water stress conditions, quantitative reverse transcription (qRT)-PCR was used to monitor their transcript levels in leaf and kernel tissues at different developmental stages, the growth of both tissues being very sensitive to water deficit, and root tissue, which partly maintains growth under water deficit (Westgate and Steudle, 1985). Transcript levels of ZmASR6, ZmASR7-2, and ZmASR7-3 were not quantified because it was not possible to design gene-specific primers of sufficient quality for qRT-PCR. The relative abundance of the other six ZmASR amplicons could be compared, as we showed that the PCR efficiency for each gene was very similar (see “Materials and Methods”).
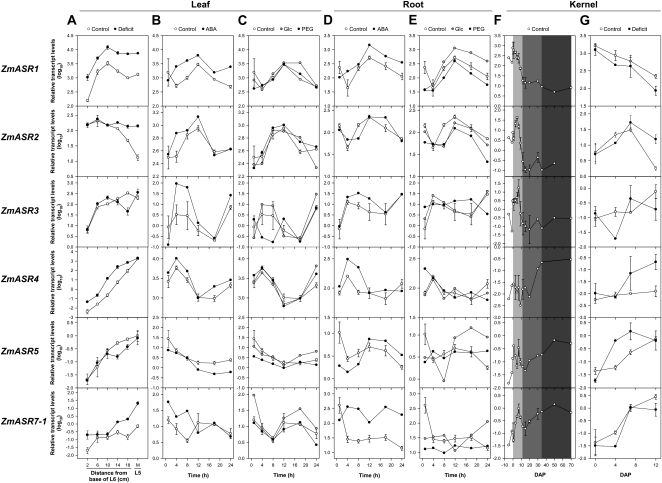
Under well-watered conditions, all the ZmASR genes tested were transcribed in leaf, kernel, and root tissues (Fig. 2). In leaves, the expression of ZmASR1 displayed a bell-shaped expression pattern (Fig. 2A), peaking at 10 cm from the leaf insertion point (e.g. beyond the growing zone, which has a constant length of 7–8 cm in maize; Tardieu et al., 2000). The transcript levels of ZmASR3, ZmASR4, ZmASR5, and ZmASR7-1 increased along the leaf, while the transcript levels of ZmASR2 were rather stable along the segments from 2 to 10 cm and decreased in the mature part of the leaf (Fig. 2A). In unfertilized and fertilized caryopses, the transcript levels of ZmASR1, ZmASR2, and ZmASR3 displayed a bell-shaped expression pattern, which peaked at 0, 7, and 9 d after pollination (DAP), respectively, and remained rather stable after 12 DAP (Fig. 2, F and G). The transcript levels of ZmASR4 increased moderately during the filling stage, whereas expression of ZmASR5 and ZmASR7-1 peaked at 7 DAP and, after a drop at 12 DAP, remained rather stable throughout development (Fig. 2, F and G). Under water deficit conditions, statistically significant transcript level increases were observed for ZmASR1, ZmASR2, ZmASR4, and ZmASR7-1 in leaves (Fig. 2A) but only for ZmASR4 in kernels (Fig. 2G).
Figure 2.
Transcript levels of ZmASR genes in leaf, root, and kernel tissues. A, ZmASR transcript levels in different leaf sections of B73 plants grown under well-watered (control) or water deficit (deficit) conditions. L5, Mature leaf 5; L6, growing leaf 6; M, middle of the fifth leaf blade. B to E, ZmASR transcript levels in leaves (B and C) and roots (D and E) from 10-d-old F2 plantlets supplied in continuous light during 24 h with culture medium (control) complemented with 3.7 μm ABA (B and D), 1% (w/v) Glc (C and E), or 7% (w/v) PEG 8000 (C and E). F and G, ZmASR transcript levels in developing kernels from A188 (F) or MBS847 (G) plants grown under well-watered (control) or water deficit (deficit; water interruption 7 d before pollination) conditions. The light gray, gray, and dark gray boxes indicate the early, filling, and desiccation phases of the maize kernel, respectively. qRT-PCR was performed on total RNA of the indicated tissues using gene-specific primers (Supplemental Table S2). Transcript levels were normalized against the stable endogenous ZmGRP2 gene and shown relative to ZmASR5 transcript levels in the middle of the fifth leaf blade in control conditions. Values represent means of three biological replicates ± se.
For plants, the responses to water deficit include both ABA-dependent and ABA-independent pathways (Nakashima et al., 2009). Moreover, ABA induces some ASR genes in a range of plant species, including tomato, tobacco, grape, banana, sugarcane (Saccharum officinarum), and rice (Rossi et al., 1998; Sugiharto et al., 2002; Cakir et al., 2003; Takasaki et al., 2008; Henry et al., 2011). Sugars such as Glc also exhibit interactions with ABA in controlling seedling development (Rolland et al., 2006), and grape and tomato ASR proteins may be involved in Glc metabolism (Cakir et al., 2003; Frankel et al., 2007). Additionally, polyethylene glycol (PEG)-induced water stress increases tomato, potato (Solanum tuberosum), and sugarcane ASR transcript levels (Amitai-Zeigerson et al., 1995; Huang et al., 2000; Doczi et al., 2002; Sugiharto et al., 2002). Therefore, we also studied ZmASR transcript levels in response to exogenous ABA (3.7 μm), Glc (1%), and PEG (7%) treatments. Transcript levels of ZmASR1, ZmASR2, ZmASR3, and ZmASR4 were up-regulated between 4 and 18 h in control leaves and roots, while those of ZmASR5 were slightly down-regulated after 4 h and up-regulated at 12 h in control leaves and roots, respectively (Fig. 2, B and D). Comparatively, transcript levels of ZmASR7-1 were up-regulated at 12 h and stable in control leaves and roots (Fig. 2, B and D). These expression data might be due to the memory of a circadian cycle. In response to ABA treatment, we found that in leaves, transcript levels of ZmASR5 were down-regulated, while those of ZmASR1, ZmASR3, and ZmASR4 were up-regulated (Fig. 2B). In roots, up-regulation by ABA appeared a common feature to ZmASR1, ZmASR2, ZmASR4, and ZmASR7-1 (Fig. 2D). Supplying Glc barely altered ZmASR1 transcript levels in leaves (Fig. 2C). In contrast, in roots, up-regulation by Glc appeared a common feature to ZmASR1 and ZmASR5 (Fig. 2E). This Glc response was unlikely to be due to an osmotic effect, since the PEG treatment, at an equal osmotic potential, had no effect on both ZmASR1 and ZmASR5 expression (Fig. 2E). Nevertheless, supplying PEG decreased ZmASR5 transcript levels in leaves as well as ZmASR2 and ZmASR7-1 transcript levels in roots (Fig. 2, C and E).
ZmASR1 Is the Primary Detectable ZmASR Protein Responding to Water Deficit in Leaves
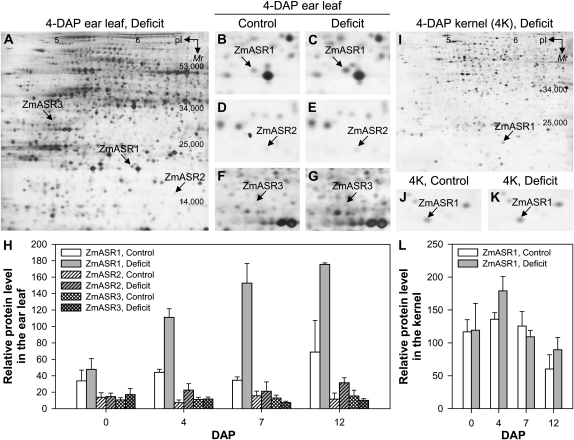
Expression at the transcript level is not always reflected at the protein level, so we combined separation of soluble proteins by two-dimensional gel electrophoresis (2-DE) with quantitative analysis of silver-stained gels and identification by liquid chromatography-tandem mass spectrometry (LC-MS/MS) to investigate the protein expression pattern of the ZmASR proteins in ear leaf and kernel tissues harvested at different DAPs from plants subjected to water deprivation 7 d before pollination. Our 2-DE condition allowed the analysis of five ZmASRs (ZmASR1, ZmASR2, ZmASR3, ZmASR4, and ZmASR5), the pI of the remaining ZmASRs (ZmASR6, ZmASR7-1, ZmASR7-2, and ZmASR7-3) being higher than that of the immobilized pH gradient strips used. 2-DE analysis revealed ZmASR1, ZmASR2, and ZmASR3 in ear leaves (Fig. 3, A–G) and only ZmASR1 in kernels (Fig. 3, I–K). A gel-free analysis, which has the advantage of being independent of the protein pI, gave the same results (data not shown). Interestingly, ZmASR1, encoded by one of the most highly expressed ZmASR genes, was the most abundant detected ZmASR isoform (Fig. 3H). Under well-watered conditions, its expression was rather stable during the early phase in kernels (Fig. 3H), paralleling the slight decrease observed at the transcript levels only after 7 DAP (Fig. 2G). Under water deficit conditions, its expression gradually increased in ear leaves (Fig. 3H), while it did not show any significant change in kernels (Fig. 3L). In contrast, ZmASR2 showed a relatively moderate increase in ear leaves in response to water deficit, while ZmASR3 did not show any change (Fig. 3H). These data suggest that ZmASR1 encodes the major ASR isoform for water stress responses in maize.
Figure 3.
Expression profile of ZmASR1, ZmASR2, and ZmASR3 proteins in ear leaf and kernel tissues. 2-DE gels of ZmASR1, ZmASR2, and ZmASR3 proteins extracted from 0-, 4-, 7-, and 12-DAP ear leaf (150 μg of protein per strip) or kernel (30 μg of protein per strip) of MBS847 plants daily irrigated (control) or subjected to water deprivation (deficit) 7 d before pollination are shown. A to G, 2-DE gel images of 4-DAP ear leaf in well-watered (B, D, and F) and water deficit (A, C, E, and G) conditions with the identified ZmASR1, ZmASR2, and ZmASR3 isoforms. H, Quantification of ZmASR1, ZmASR2, and ZmASR3 isoforms in the ear leaf. I to K, 2-DE gel images of 4-DAP kernel (4K) in well-watered (J) and water deficit (I and K) conditions with the identified ZmASR1 isoform. L, Quantification of the ZmASR1 isoform in the kernel. Values represent means of three biological replicates ± se.
Ectopic Expression of ZmASR1 Maintains Kernel Yield under Water-Limited Conditions
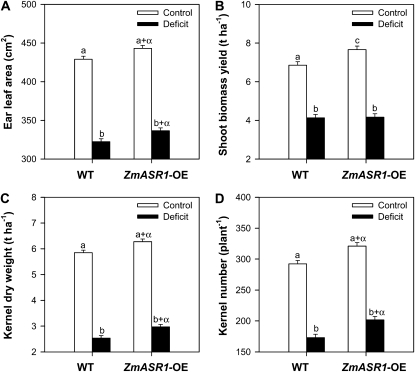
To address ZmASR1 function, transgenic maize plants overexpressing the ZmASR1 coding sequence (ZmASR1-OE) under the control of the constitutive cassava vein mosaic virus promoter (Verdaguer et al., 1998) were generated. After selection and regeneration, followed by two backcrosses with the maize line F2, nine independent hemizygous lines that carried a unique T-DNA insertion and expressed ZmASR1 protein (data not shown) and their wild-type sister lines were selected and evaluated in a field site under well-watered and water-limited conditions (see “Materials and Methods”; Supplemental Fig. S2, A and B). qRT-PCR experiments on leaf 11 (below the ear leaf) 5 d before silk emergence showed that transgenic lines expressed ZmASR1 about 13- and 4-fold more than wild-type lines under well-watered and water-limited conditions, respectively (Supplemental Fig. S2C). An across-event analysis demonstrated that ZmASR1-OE plants exhibited a significant 3% and 4% increase in ear leaf area relative to wild-type plants under both well-watered and water-limited stress conditions, respectively, and a significant 12% increase in shoot biomass yield in well-watered conditions (Fig. 4, A and B; Supplemental Table S5). ZmASR1-OE plants also demonstrated an increase (P < 0.10) in dry leaf weight and total chlorophyll content under both well-watered and water-limited stress conditions (Supplemental Table S5). Mean yield of wild-type plants at the water deficit block was 2.5 tons ha−1, representing an approximately 43% reduction in yield compared with wild-type plants under well-watered conditions (Fig. 4C; Supplemental Table S5). Yield averages of ZmASR1-OE plants as a group were significantly greater than in wild-type plants, by 7% and 17% under well-watered and water-limited conditions, respectively (Fig. 4, C and D; Supplemental Table S5). Together, these results provided evidence that ectopic expression of ZmASR1 improved and maintained maize kernel yield under well-watered and water-limited conditions in the field.
Figure 4.
ZmASR1-OE plants demonstrate improved vegetative and reproductive performance under water-limiting conditions. Ear leaf area (A), shoot biomass yield (B), kernel dry weight (C), and kernel number (D) values of wild-type (WT) and ZmASR1-OE plants are shown for groupings of nine individual ZmASR1 events. Values are means ± se (n = 52–60). When two samples show different letters above the bar, the difference between them is significant (normal letters, P < 0.05). When both genotype and treatment effects are significant, a, b, a+α, and b+α are indicated (see “Materials and Methods”; and Supplemental Table S5).
Transcriptomic, Proteomic, and Metabolic Adjustments in ZmASR1-OE Plants
To obtain additional cues with regard to ZmASR1 function, we used the 46K array constructed by the Maize Oligonucleotide Array Project (http://www.maizearray.org) followed by qRT-PCR experiments, 2-DE, and gas chromatography coupled to time-of-flight MS analysis to contrast the transcriptome, the proteome, and the metabolome, respectively, of ZmASR1-OE leaves with that of wild-type leaves 5 d before silk emergence under well-watered and water deficit conditions.
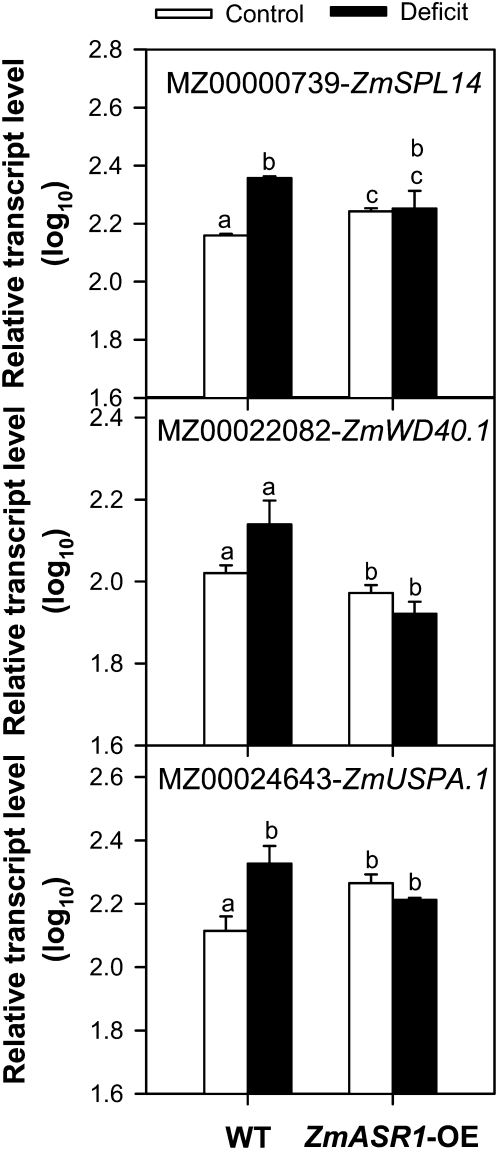
A first gene list of 16 differentially expressed genes was established from the transcriptome comparison between ZmASR1-OE and wild-type leaves under both treatment conditions based on P < 0.01 by two-way ANOVA and the criterion of a 1.5-fold change in transcript abundance. To confirm the differential expression, qRT-PCR experiments were performed based on the same samples that had been used for the initial microarray analysis. Because it was not possible to design gene-specific primers of sufficient quality for qRT-PCR for five target genes, the list was shortened to 11 candidates. Only three of the 11 potential target genes were significantly affected by ZmASR1-OE with opposite effects: (1) up-regulation of MZ00000739 and MZ00024643 by ZmASR1-OE under well-watered conditions; (2) down-regulation of MZ00022082 by ZmASR1-OE under both well-watered and water deficit conditions (Fig. 5; Supplemental Table S2). MZ00000739 (ZmSPL14) likely encoded a nuclear SQUAMOSA promoter-binding protein (SBP) box transcription factor of the plant-specific subfamily IIa, most closely related to AtSPL14 from Arabidopsis (Stone et al., 2005; Guo et al., 2008). MZ00022082 encoded a protein with unknown function carrying a WD40 repeat-like-containing domain and was accordingly hereafter named ZmWD40.1. Finally, MZ00024643 shared highest identity with At5g14680 (75%) and At3g01520 (71%) sequences encoding Arabidopsis vacuolar USPA (for universal stress protein A of E. coli) domain proteins (Kerk et al., 2003; Endler et al., 2006) and was accordingly hereafter named ZmUSPA.1.
Figure 5.
Transcript levels of confirmed ZmASR1 target genes identified by microarray. qRT-PCR was performed on total RNA prepared from the 11th leaves of wild-type (WT) and ZmASR1-OE plants that had been used for the initial microarray analysis using gene-specific primers (Supplemental Table S2). Transcript levels were normalized against the stable endogenous ZmGRP2 gene and shown relative to ZmBCAT2 transcript levels in the 11th leaf in well-watered (control) conditions. Values represent means of biological duplicates ± se. When two samples show different letters above the bar, the difference between them is significant (normal letters, P < 0.05).
The proteome comparison revealed 22 protein spots that were consistently and significantly more abundant (seven spots) or less abundant (15 spots) in ZmASR1-OE leaves than in wild-type leaves in one or both treatment conditions (Table I). Subsequent analyses by LC-MS/MS led to the identification of 16 of them and showed that they were encoded by 15 distinct genes (Table I; Supplemental Table S6). These comprised five up-regulated proteins, annotated as class II aspartyl-tRNA synthetase, β-d-glucosidase, spermidine synthase 1, glucan endo-1,3-β-glucosidase 5, and NAD-dependent epimerase/dehydratase, and 11 down-regulated proteins, annotated as trigger factor-like protein, adenylosuccinate synthetase (two spots encoded by the same gene), plastid transcriptionally active 16, 3-isopropylmalate dehydrogenase (IPMDH; two isoforms, ZmIPMDH1 and ZmIPMDH2), pyruvate dehydrogenase subunit E1β, sedoheptulose-1,7-bisphosphatase, PSII stability/assembly factor, C2-domain containing protein, and thiazole biosynthetic enzyme 1-1. Interestingly, none of the corresponding 15 genes generated expression data with significant P value thresholds in the transcriptomic analysis (data not shown).
Table I.
Proteins that showed significant changes in ZmASR1-OE leaves compared with wild-type leaves
| Spot IDa | Trend in ZmASR1-OE | Ratio of ZmASR1-OE to the Wild Typeb | ANOVA Tablec | Maize Gene Modeld | Annotatione | Class | |||
| Control | Deficit | ZmASR1-OE | Deficit | Interaction | |||||
| s0732 | Down | 0.99 | 0.34 | NR | NR | 0.007 | NI | ND | ND |
| s1021 | Up | 1.30 | 2.22 | 0.020 | 0.752 | 0.071 | GRMZM2G019121 | Class II aspartyl-tRNA synthetase (AspRS) | Amino acid activation |
| s1202 | Up | 2.94 | 1.74 | 0.020 | 0.420 | 0.561 | GRMZM2G008247 | β-d-Glucosidase (GLU) | Carbohydrate metabolism |
| GRMZM2G034152 | Flavin-containing amine oxidase | ||||||||
| s1388 | Down | 0.50 | 0.55 | 0.006 | 0.304 | 0.578 | GRMZM2G127393 | Trigger factor-like protein | Protein folding |
| s1406 | Down | 0.13 | 0.73 | NR | NR | 0.027 | GRMZM2G123204 | Adenylosuccinate synthetase (AdSS) | Purine biosynthesis |
| GRMZM2G020446 | Diaminopimelate decarboxylase | ||||||||
| s1422 | Down | 0.42 | 0.98 | NR | NR | 0.031 | GRMZM2G123204 | Adenylosuccinate synthetase (AdSS) | Purine biosynthesis |
| s1442 | Down | 0.26 | 0.56 | 0.015 | 0.491 | 0.118 | NI | ND | ND |
| s1444 | Down | 0.66 | 0.74 | 0.005 | 0.289 | 0.686 | GRMZM2G449496 | Plastid transcriptionally active 16 (PTAC16) | RNA regulation |
| s1612 | Down | 0.37 | 0.86 | NR | NR | 0.001 | GRMZM2G104613 | 3-Isopropylmalate dehydrogenase 2 (IPMDH2) | Leu biosynthesis |
| GRMZM2G803490 | 3-Isopropylmalate dehydrogenase 1 (IPMDH1) | ||||||||
| s1641 | Down | 0.48 | 0.88 | NR | NR | 0.026 | GRMZM2G803490 | 3-Isopropylmalate dehydrogenase 1 (IPMDH1) | Leu biosynthesis |
| s1886 | Down | 0.68 | 0.65 | 0.019 | 0.248 | 0.953 | GRMZM2G097226 | Pyruvate dehydrogenase subunit E1β (PDH-E1β) | Glycolysis and tricarboxylic acid cycle |
| s1904 | Down | 0.70 | 0.63 | 0.007 | 0.017 | 0.859 | AC147602.5_FGP004 | Sedoheptulose-1,7-bisphosphatase (SBPase) | Calvin cycle |
| s1913 | Down | 1.14 | 0.68 | NR | NR | 0.001 | GRMZM2G102838 | PSII stability/assembly factor (HCF136) | Protein assembly |
| s2020 | Down | 0.99 | 0.50 | NR | NR | 0.010 | AC210204.3_FGP002 | C2 domain-containing protein | Stress |
| s2141 | Down | 0.30 | 0.68 | 0.005 | 0.035 | 0.150 | NI | ND | ND |
| s2210 | Down | 0.71 | 0.38 | 0.019 | 0.215 | 0.258 | GRMZM2G018375 | Thiazole biosynthetic enzyme 1-1 (THI1) | Thiamine biosynthesis |
| s2275 | Up | 1.82 | 1.23 | 0.050 | 0.013 | 0.055 | GRMZM2G064163 | Spermidine synthase 1 | Polyamine biosynthesis |
| s2316 | Up | 1.56 | 1.29 | 0.011 | 0.028 | 0.133 | GRMZM2G078566 | Glucan endo-1,3-β-glucosidase 5 | Carbohydrate metabolism |
| s2322 | Up | 1.75 | 1.34 | 0.005 | 0.436 | 0.108 | NI | ND | ND |
| s2460 | Down | 0.82 | 0.63 | 0.035 | 0.023 | 0.112 | NI | ND | ND |
| s2528 | Up | 1.54 | 1.21 | 0.020 | 0.778 | 0.244 | GRMZM2G068244 | NAD-dependent epimerase/dehydratase | Coenzyme binding |
| s3353 | Up | 1.85 | 1.32 | 0.020 | 0.306 | 0.170 | NI | ND | ND |
Identification number of the corresponding protein spot on the two-dimensional reference map.
The expression values are reported relative to the wild-type samples in the same culture condition (n = 2).
Proteins were categorized as ZmASR1-OE (boldface text), deficit (underlined text), and interaction (italicized text) as follows: ZmASR1-OE, proteins that showed significant (P < 0.05) changes in ZmASR1-OE leaves compared with wild-type leaves when the additive model could be retained; deficit, proteins that showed significant (P < 0.05) changes under water deficit conditions compared with well-watered conditions when the additive model could be retained; interaction, proteins that showed significant (P < 0.05) changes in ZmASR1-OE leaves compared with wild-type leaves by the Bonferroni method when the additive model could not be retained. Experimental details are described in “Materials and Methods.” NR, Not relevant.
Maize genome release 5a.59 of November 2010 (http://www.maizesequence.org). NI, Not identified by LC-MS/MS.
Manually improved annotation from SwissProt, GenBank, Trembl, and InterPro databases. Annotations in italicized text were eliminated based on the absence of correspondence between the theoretical Mr and the observed Mr and/or the protein abundance index (see Supplemental Table S6). ND, Not determined.
Nontargeted metabolite profiling identified 84 unique compounds (Supplemental Table S7). Among them, 20 were consistently and significantly affected by ZmASR1-OE with opposite effects: increase in lactate and urea in water deficit conditions and decrease in 18 other metabolites, including 10 amino acids (Ile, Leu, Val, Phe, Trp, Asn, Gln, Pro, Ala, and Gly), two of their derivatives or potential precursors (benzoate and citramalate), and three sugars (galactinol, Glc, and Suc), in both well-watered and water deficit conditions (Table II).
Table II.
Metabolites that showed significant changes in ZmASR1-OE leaves compared with wild-type leaves
| Pathway | Metabolite | Trend in ZmASR1-OE | Ratio of ZmASR1-OE to the Wild Typea | ANOVA Tableb | |||
| Control | Deficit | ZmASR1-OE | Deficit | Interaction | |||
| BCAAs | Ile | Down | −0.11 | −0.15 | 0.010 | 0.014 | 0.573 |
| Leu | Down | −0.08 | −0.14 | 0.007 | 0.002 | 0.284 | |
| Val | Down | −0.09 | −0.11 | 0.010 | 0.143 | 0.790 | |
| Aromatic amino acids | Phe | Down | −0.06 | −0.12 | 0.011 | 0.001 | 0.189 |
| Trp | Down | −0.11 | −0.16 | 0.016 | 0.014 | 0.583 | |
| Glu family | Asn | Down | −0.13 | −0.20 | 0.044 | 0.170 | 0.624 |
| Gln | Down | −0.07 | −0.12 | 0.037 | 0.006 | 0.550 | |
| Pro | Down | −0.06 | −0.08 | 0.032 | 0.003 | 0.783 | |
| RFO | Galactinol | Down | −0.13 | −0.08 | 0.010 | 0.000 | 0.415 |
| Saccharides | Glc | Down | −0.17 | −0.08 | 0.004 | 0.000 | 0.221 |
| Suc | Down | −0.14 | −0.08 | 0.037 | 0.277 | 0.505 | |
| γ-Aminobutyrate shunt | Ala | Down | −0.08 | −0.09 | 0.024 | 0.012 | 0.856 |
| Others | trans-Aconitate | Down | −0.22 | −0.10 | 0.021 | 0.183 | 0.280 |
| Benzoate | Down | −0.09 | −0.11 | 0.039 | 0.173 | 0.833 | |
| trans-Caffeoylquinate | Down | −0.13 | −0.16 | 0.033 | 0.078 | 0.781 | |
| Citramalate | Down | −0.10 | −0.19 | 0.040 | 0.005 | 0.434 | |
| Gly | Down | −0.11 | −0.11 | 0.005 | 0.005 | 0.968 | |
| Lactate | Up | 0.31 | 1.81 | NR | NR | 0.045 | |
| Monomethylphosphate | Down | −0.16 | −0.21 | 0.004 | 0.020 | 0.551 | |
| Urea | Up | 0.37 | 1.23 | NR | NR | 0.042 | |
Values (log10) are reported relative to the wild-type samples (n = 2).
Metabolites were categorized as ZmASR1-OE (boldface text), deficit (underlined text), and interaction (italicized text) as follows: ZmASR1-OE, metabolites that showed significant (P < 0.05) changes in ZmASR1-OE leaves compared with wild-type leaves when the additive model could be retained; deficit, metabolites that showed significant (P < 0.05) changes under water deficit conditions compared with well-watered conditions when the additive model could be retained; interaction, metabolites that showed significant (P < 0.05) changes in ZmASR1-OE leaves compared with wild-type leaves under water deficit conditions by the Bonferroni method when the additive model could not be retained. Experimental details are described in “Materials and Methods.” NR, Not relevant.
ZmASR1-OE Influences the Expression of Additional BCAA- and Pro-Related Target Genes and a Specific Combination of Metabolites
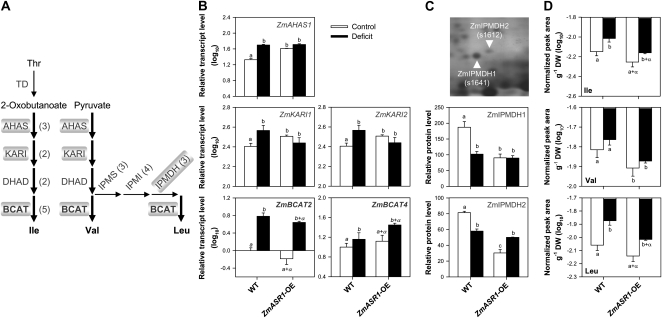
Since the decrease in the three BCAAs Ile, Leu, and Val was in reasonable agreement with that of the ZmIPMDH1 and ZmIPMDH2 isoforms involved in Leu biosynthesis (Fig. 6), we hypothesized that ZmASR1 could regulate the transcription of key genes involved in drought-responsive pathways that were modified by ZmASR1-OE. Therefore, we identified those genes in the maize genome based on sequence homology to Arabidopsis genes and examined the expression level of 32 of them in wild-type and ZmASR1-OE leaves by qRT-PCR experiments (Fig. 6; Supplemental Fig. S3; Supplemental Table S2; The Arabidopsis Information Resource and AraCyc; Fang et al., 1992; Taji et al., 2002; Martin et al., 2006; Less and Galili, 2008; Urano et al., 2009). We did not find evidence for transcriptional regulation of ZmIPMDH1 and ZmIPMDH2/3 genes, in agreement with the transcriptomic analysis (Supplemental Fig. S3). However, three other BCAA-related transcripts, one for acetohydroxyacid synthase (ZmAHAS1) and two for ketolacid reductoisomerase (ZmKAR1 and ZmKARI2), showed significant increases in ZmASR1-OE leaves in comparison with wild-type leaves under well-watered conditions (Fig. 6). Furthermore, one transcript for branched-chain aminotransferase (ZmBCAT4; BCAA biosynthesis) and two transcripts for Δ1-pyrroline-5-carboxylate synthetase (ZmP5CS2 and ZmP5CS3; Pro biosynthesis) were up-regulated under both treatment conditions, while another branched-chain aminotransferase paralog (ZmBCAT2) was down-regulated in ZmASR1-OE leaves under both treatment conditions (P < 0.10; Fig. 6; Supplemental Fig. S3). These seven biosynthetic genes were induced in response to water deficit, but only one, ZmBCAT2, had an expression pattern consistent with the metabolic changes in ZmASR1-OE leaves compared with wild-type leaves (Fig. 6).
Figure 6.
ZmASR1-OE influences BCAA-related gene expression and BCAA composition. A, The BCAA biosynthetic pathway. Substrates and products are in lightface and boldface text, respectively. Enzymes are in gray, and the number of isoforms identified in maize is indicated in parentheses. The enzymes in boldface are the rate-limiting enzymes during the water deficit response. Thick arrows indicate common steps in BCAA biosynthesis. Enzymes in boxes are ZmASR1 targets. B to D, Relative transcript (B), protein (C), and metabolite (D) levels in the 11th leaves of wild-type (WT) and ZmASR1-OE plants according to Figure 5, Table I, and Table II. Values represent means of biological duplicates ± se. When two samples show different letters above the bar, the difference between them is significant (roman letters, P < 0.05; italic letters, P < 0.10). When both genotype and treatment effects are significant, a, b, a+α, and b+α are indicated (see “Materials and Methods”). AHAS, Acetohydroxyacid synthase; DHAD, dihydroxyacid dehydratase; DW, dry weight; IPMI, isopropylmalate isomerase; IPMS, isopropylmalate synthase; TD, Thr deaminase.
To provide further insight into the impact of ZmASR1-OE on the transcriptome, proteome, and metabolome of the maize leaf, a within-group correlation matrix analysis was performed. Among 10,153 pairwise correlations computed between 36 transcripts (ZmASR1, ZmSPL14, ZmWD40.1, ZmUSPA.1, and the 32 genes related to metabolic pathways modified by ZmASR1-OE), 22 protein spots, the total chlorophyll content, and 84 metabolites, we obtained 765, 170, and 23 individual significant correlations at P < 0.05, P < 0.01, and P < 0.001, respectively (Supplemental Tables S8 and S9). These numbers were nearly twice that expected under the null hypothesis (Supplemental Table S9). Furthermore, the number of positive and negative individual significant correlations did not follow a symmetric binomial distribution (Supplemental Table S9). Similar differences were observed for the transcript-transcript, metabolite-metabolite, and transcript-metabolite submatrices, suggesting that the individual significant correlations obtained for these submatrices were most likely valuable (Supplemental Table S9). The analysis of the metabolite-metabolite submatrix revealed that 13 of the 18 metabolites that were less abundant in ZmASR1-OE leaves than in wild-type leaves showed high positive correlation to each other (Table III). Interestingly, six of them were previously reported to be negatively correlated to biomass in Arabidopsis (Table III; Meyer et al., 2007; Sulpice et al., 2009). Furthermore, six others showed high positive correlation with metabolites negatively correlated to biomass in Arabidopsis (Table III; Meyer et al., 2007; Sulpice et al., 2009). The analysis of the transcript-metabolite submatrix showed that ZmASR1 target transcripts involved in BCAA biosynthesis were positively correlated with each other and/or BCAAs and revealed a close link between six ZmASR1 target transcripts (ZmBCAT4, ZmKARI2, ZmP5CS2, ZmP5CS3, ZmUSPA.1, and ZmWD40.1) and the specific combination of 13 decreased metabolites (Supplemental Tables S10 and S11).
Table III.
Pairwise correlations of ZmASR1 target metabolites against decreased and/or biomass-related metabolites
| Pathway | Parameter xa | Trend in ZmASR1-OE | Parameter yb | Correlation (x,y)c |
| Glu family | Gln | Down | Leu | −0.8897 |
| BCAAs | Leu | Down | Gln | −0.8897 |
| Ile | Down | Leu | 0.9708 | |
| Val | Down | Phe | 0.9188 | |
| Pro | 0.9935 | |||
| Aromatic amino acids | Phe | Down | Val | 0.9188 |
| Pro | 0.9319 | |||
| Glu family | Pro | Down | Phe | 0.9319 |
| Val | 0.9935 | |||
| RFO | Galactinol | Down | Benzoate | 0.9598 |
| Citrate | 0.9371 | |||
| Monomethylphosphate | 0.8921 | |||
| Raffinose | 0.8893 | |||
| Succinate | 0.9280 | |||
| Threonate | 0.9378 | |||
| Saccharides | Glc | Down | trans-Aconitate | 0.9409 |
| Ala | 0.9330 | |||
| Ascorbate | 0.8875 | |||
| trans-Caffeoylquinate | 0.9264 | |||
| Citramalate | 0.9457 | |||
| Malate | 0.9564 | |||
| Suc | 0.9006 | |||
| Suc | Down | Ascorbate | 0.9692 | |
| trans-Caffeoylquinate | 0.9532 | |||
| Citrate | 0.9587 | |||
| Ethanolamine | 0.8896 | |||
| Glc | 0.9006 | |||
| Malate | 0.9466 | |||
| Orn | 0.9113 | |||
| γ-Aminobutyrate shunt | Ala | Down | trans-Aconitate | 0.9643 |
| Citramalate | 0.9656 | |||
| Glc | 0.9330 | |||
| Malate | 0.9579 | |||
| Others | trans-Aconitate | Down | Ala | 0.9643 |
| Citramalate | 0.9669 | |||
| Glc | 0.9409 | |||
| Malate | 0.9118 | |||
| Raffinose | 0.8973 | |||
| Benzoate | Down | Galactinol | 0.9598 | |
| Monomethylphosphate | 0.8797 | |||
| Raffinose | 0.8909 | |||
| Threonate | 0.8848 | |||
| trans-Caffeoylquinate | Down | Ascorbate | 0.9735 | |
| Ethanolamine | 0.9181 | |||
| Glc | 0.9264 | |||
| Malate | 0.8884 | |||
| Suc | 0.9532 | |||
| Citramalate | Down | trans-Aconitate | 0.9669 | |
| Ala | 0.9656 | |||
| Glc | 0.9457 | |||
| Malate | 0.8958 | |||
| Monomethylphosphate | Down | Benzoate | 0.8797 | |
| Galactinol | 0.8921 | |||
| Raffinose | 0.9420 |
Metabolites that showed significant changes in ZmASR1-OE leaves compared with wild-type leaves (P < 0.05).
Metabolites that showed significant decreases in ZmASR1-OE leaves compared with wild-type leaves and/or were correlated to biomass in Arabidopsis (Meyer et al., 2007; Sulpice et al., 2009). Boldface italic type and underlined lightface italic type distinguish metabolites negatively and positively correlated to biomass, respectively, in Arabidopsis (Meyer et al., 2007; Sulpice et al., 2009).
Correlations were calculated from residual data (P < 0.05). The original data are in Supplemental Table S8.
DISCUSSION
Expression of ASR Genes in Response to Water Deficit in Maize and Other Poaceae Species
Here, we identified nine ASR genes in the maize genome, making the ZmASR gene family the biggest ASR gene subfamily identified to date. Phylogenetic analyses indicated several duplication events after the divergence of Liliales from Commelinids, giving rise to five paralogous clades of ASR genes in the Poaceae family. Anchoring of the ASR genes on the maize physical map shows that all Poaceae sequences can be assigned to one of the Poaceae ancestor protochromosomes previously defined by Salse et al. (2008). Thus, sequences from the subclades I-1, I-2, I-3, II-1, and II-2 trace back to chromosomes A11/A12, A4, A1, A2/A4, and A1, respectively. Consequently, the data support the idea that the Poaceae intermediate ancestor with 12 protochromosomes (Murat et al., 2010) already had seven ASR genes. The sequence of events raising this small gene family in grasses might have taken place during the diploidization process following the Poaceae-specific whole-genome duplication (Murat et al., 2010). A larger sample of sequence data from monocots other than Poaceae might help to further elucidate this point.
Our gene-specific qRT-PCR expression data combined with 2-DE analysis and the recent work of Philippe et al. (2010) in rice strongly suggest that members of the Poaceae subclade I-1, including ZmASR1 and OsASR5, are the most prevalent ASR proteins in major plant tissues. It is noteworthy that recently released large-scale quantitative proteomics studies only revealed ZmASR1 and ZmASR2 in maize leaves (Friso et al., 2010; Majeran et al., 2010). In C4 Poaceae species, such as maize, a key component of mature leaves is the partitioning of photosynthetic processes between the bundle sheath and the mesophyll. Among the 25,800 transcripts (about 80% of the predicted maize transcriptome) that were revealed along the maize leaf gradient, Li et al. (2010) identified only five (ZmASR1, ZmASR2, ZmASR3, ZmASR4, and ZmASR5) of the nine ZmASR genes characterized in this study. It is striking that they all belong to the suite of genes (21%) that were determined to be differentially expressed between bundle sheath and mesophyll cells (Li et al., 2010). Thus, ZmASR4 showed enriched expression in bundle sheath cells, in agreement with a previous work that used differential screening analysis (Furumoto et al., 2000), while ZmASR1, ZmASR2, ZmASR3, and ZmASR5 showed enriched expression in mesophyll cells. Such a pattern of expression suggests that the ZmASR gene family shows subfunctionalization. Interestingly, almost all Poaceae sequences in subclades I-1, I-2, and II-2, including ZmASR1, ZmASR2, ZmASR3, ZmASR7-2, and ZmASR7-3, share a putative nuclear localization signal and substantial sequence identity with the Zn2+-dependent DNA-binding activity domain of LLA23 and SlASR1 proteins, raising the interesting possibility that the ASR proteins belonging to these subclades act as regulators modulating gene expression. Unlike the other members of the maize ASR gene family, ZmASR7-1, ZmASR7-2, and ZmASR7-3 do not contain any introns. Because ZmASR7-1 was expressed at a low level, whereas the expression of ZmASR7-2 and ZmASR7-3 was not determined by qRT-PCR, conventional RT-PCR was used to monitor the transcript levels of ZmASR7-2 and ZmASR7-3 and to rule out the possibility that these genes might be pseudogenes. Both transcripts were detected in leaf and root tissues, although at very low levels, regardless of the water stress-related conditions (Supplemental Fig. S4).
Additionally, our transcript and protein expression data confirmed and extended findings of up-regulation by water deficit of ZmASR1 in maize leaves (Riccardi et al., 1998, 2004). Indeed, we showed up-regulation of ZmASR1, ZmASR2, ZmASR4, and ZmASR7-1 by water deficit in leaves, although direct treatment with ABA in leaves induced elevations of ZmASR1, ZmASR3, and ZmASR4. Up-regulation by water deficit and ABA treatment was previously shown for the sugarcane and rice ZmASR1 orthologs SoDIP22 and OsASR5 (Sugiharto et al., 2002; Rabbani et al., 2003; Takasaki et al., 2008). These data indicate that members of the Poaceae subclade I-1, except ZmASR2, may play important roles in ABA-dependent pathways in the water deficit response in Poaceae leaves. In contrast, ZmASR2 and ZmASR7-1 are regulated by ABA-independent pathways under water deficit in mature leaves, as reported previously for potato DS2 genes (Silhavy et al., 1995; Doczi et al., 2002). It is worth noting that maize and rice (Philippe et al., 2010) ASR genes belonging to Poaceae subclades I-3 (ZmASR4 and OsASR6) and II-2 (ZmASR7-1 and OsASR1) were all up-regulated by water deficit in mature leaves. In contrast, maize and rice ASR genes belonging to Poaceae subclades I-2 (ZmASR3 and OsASR4) and II-1 (ZmASR5 and OsASR3) were not significantly altered and down-regulated by water deficit in mature leaves, respectively. Therefore, these data suggest that the ASR expression in response to water deficit represents an evolutionarily conserved regulatory mechanism in the Poaceae and several unrelated eudicot plants. A better understanding of the physiological roles of the maize ASR genes will require more exhaustive definition of their expression profiles in response to water deficit at the cellular level. It would also be important to determine whether these genes are affected by water deficit at the posttranslational level.
ZmASR1 Maintained Kernel Yield under Water Deficit Conditions through a Set of Low-Mr Metabolites Related to Growth Rate
ASR overexpression studies in plants are exemplified by works carried out on Solanoideae (Kalifa et al., 2004b; Frankel et al., 2007) and Arabidopsis, although the latter does not include any ASR homologous genes (Yang et al., 2005; Liu et al., 2010). Our study demonstrated that ZmASR1-OE improved the kernel yield of transgenic maize in the field under well-watered and water-limited conditions. The positive yield impact of ZmASR1 was both on kernel number and weight. These results were unexpected based on our previous PQL study, which did not reveal any QTL for maize yield and its component in the vicinity of the ZmASR1 chromosomal region (de Vienne et al., 1999; Jeanneau et al., 2002). Nevertheless, it may be pointed out that recently identified QTLs for ear length under water-stressed conditions (Lu et al., 2006) and female flowering time (Marino et al., 2009) are centered on the ZmASR1 locus. The finding that ZmASR1-OE plants demonstrated significant improvements in ear leaf area and shoot biomass yield, as well as increases in leaf dry weight and total chlorophyll content, implies that these improvements in vegetative productivity translated into improvements in reproductive performance and kernel yield. Enhanced vegetative tolerance under well-watered conditions is surprising, as we would have expected no significant differences, as described previously under mild water deficit conditions (Jeanneau et al., 2002). Nevertheless, transgenic maize plants overexpressing ZmNF-YB2 were darker green and flowered 1 to 3 d earlier than the control plants under conditions of ample water supply (Nelson et al., 2007). Additionally, enhanced vegetative tolerance and improved kernel yield under water deficit conditions were produced by the overexpression of ZmNF-YB2, betA, or Csp genes in maize (Quan et al., 2004; Nelson et al., 2007; Castiglioni et al., 2008). Continued evaluation of ZmASR1-OE across different hybrid genetic backgrounds adapted for the marketplace will be important ongoing work. Furthermore, testing the ability of other water deficit-inducible ZmASR genes for their ability to confer water deficit tolerance will further define the structure-function relationships and improve our understanding of the mode of action of the ZmASR proteins.
A noteworthy observation from our data is that 18 low-Mr metabolites decrease in mature ZmASR1-OE leaf. Of high relevance are 12 standard or related amino acids and three sugars: (1) Ala, which is synthesized from pyruvate; (2 and 3) Asn and Gln, which are the central regulators of carbon/nitrogen metabolism; (4) Gly, whose metabolism occurs by several different pathways, including Thr catabolism; (5–7) Ile (Thr-derived pathway) and the two other BCAAs, Leu and Val (pyruvate-derived pathway), that show the highest hydrophobicity among the proteinogenic amino acids and are accordingly the major constituents of transmembrane regions of membrane proteins; (8 and 9) Phe and Trp, whose synthesis initiates from chorismate and that serve in plants as precursors for a wide range of secondary metabolites having multiple functions; (10) Pro, which is synthesized from either Glu or Orn and has multiple functions in stress adaptation, recovery, and signaling; (11) benzoate, which derives from Phe and is a precursor to several important benzenoid compounds, including the defense signaling compound salicylic acid; (12) citramalate, which may serve as a precursor for Ile biosynthesis in plants as in microorganisms (de Kraker et al., 2007; Joshi et al., 2010; Less et al., 2010; Szabados and Savouré, 2010); (13) Suc, which is the major end product of photosynthesis and the major transport form of carbon from source and sink; (14) its cleavage product Glc, which enhances the expression of genes implied in carbohydrate metabolism (Koch, 1996; Trouverie et al., 2004); (15) galactinol (raffinose family oligosaccharide [RFO]), which plays a novel role in the protection of cellular metabolism, in particular, the photosynthesis of chloroplast, from oxidative damage caused by several types of abiotic stresses (Nishizawa et al., 2008). Our finding that ZmASR1-OE triggers an adjustment in the Glc level is in good agreement with the observation that a QTL for hexose content colocates with the ZmASR1 locus (Pelleschi et al., 2006) and a previous report in potato in which SlASR1 was overexpressed (Frankel et al., 2007). Additionally, the lower levels of Pro in ZmASR1-OE plants is in close agreement with previous reports in tobacco and potato plants in which SlASR1 was overexpressed and down-regulated, respectively (Kalifa et al., 2004b; Frankel et al., 2007). Lower levels of Phe, Val, and Suc had also been described for potato plants overexpressing SlASR1 (Frankel et al., 2007), although the remaining other decreased metabolites had never been identified before. Interestingly, among the 18 decreased metabolites, 13 were positively correlated to each other. Previous reports documented that biomass can be predicted by a specific metabolite status (Meyer et al., 2007; Sulpice et al., 2009). It is intriguing that among the 13 positively correlated metabolites, six (Ala, Phe, Val, Suc, benzoate, and citramalate) were previously shown to be negatively correlated to biomass (Meyer et al., 2007; Sulpice et al., 2009). It is also noteworthy that six others (transaconitate, transcaffeoylquinate, galactinol, Glc, monomethylphosphate, and Pro) showed positive correlation to metabolites negatively correlated to biomass (Meyer et al., 2007; Sulpice et al., 2009). Additionally, it was found that Pro levels accumulated under water deficit conditions were negatively correlated with maize yield (Udomprasert et al., 1999). Since ZmASR1-OE plants demonstrated improvement and maintenance in shoot biomass yield under well-watered and water-limited conditions, respectively, as well as decreases in this specific set of 13 metabolites under both treatment conditions, we hypothesize that ZmASR1 might negatively impact the level of these 13 metabolites, which would contribute to biomass yield and subsequently result in improvements and maintenance of kernel yield under well-watered and water deficit conditions, respectively.
Influence of ZmASR1 on the Expression of Genes Involved in BCAA Biosynthesis
What is the relationship between ZmASR1 and the level of these 13 metabolites that may impact on biomass and kernel yield? Our global comparative study led to the identification of 25 genes that were either transcriptionally (10 genes) or posttranscriptionally (16 proteins encoded by 15 distinct genes) up-regulated (eight transcripts and five proteins) or down-regulated (two transcripts and 11 proteins) by ZmASR1-OE in mature leaves. Three-quarters of them encoded proteins normally localized in plastids or accumulating in these organelles under osmotic stress and involved in primary metabolic pathways. Importantly, the microarray analysis and the additional qRT-PCR experiments revealed different target genes from the proteomic analysis. Therefore, the two approaches were complementary. It is quite evident that some genes with transcript changes in ZmASR1-OE leaves compared with wild-type leaves might not have been revealed by the proteomic analysis because of their low abundance. Nevertheless, none of the 16 identified proteins that showed substantial changes in abundance in ZmASR1-OE leaves compared with wild-type leaves were transcriptionally regulated in ZmASR1-OE leaves. This is not necessarily surprising, since it has been shown recently that posttranscriptional regulation can affect the protein abundance without affecting transcript levels (Böhmer and Schroeder, 2011). These data, therefore, strengthen the idea that ZmASR1, similar to other ASR proteins, may act both as a transcriptional regulator and a chaperone-like protein protector (Saumonneau et al., 2008; Urtasun et al., 2010). A synergistic relationship was found between the osmolyte Gly betaine and the SlASR1 protein, suggesting a combined mechanism of action of these two proteins in response to abiotic stresses (Konrad and Bar-Zvi, 2008). However, the enhanced Gly betaine accumulation in transgenic betA-overexpressing maize lines led to greater increases in total soluble sugars and free amino acids (Quan et al., 2004), in contrast to the metabolite effects we report in ZmASR1-OE plants. Thus, the mechanism of action of ZmASR1 is likely to be different from that of Gly betaine. Just recently, a wheat (Triticum aestivum) group 2 LEA protein named DHN-5, which is closely related to the maize LEA protein RAB17 and involved in salt and osmotic tolerance (Brini et al., 2007), has been shown to enhance the thermostability and activity of β-d-glucosidase (Brini et al., 2010), a target protein of ZmASR1. It is striking that β-d-glucosidase and 12 other ZmASR1 target genes belong to the list of established and potential thioredoxin (Trx) targets (He et al., 2009; Montrichard et al., 2009). Closer analysis shows that seven additional ZmASR1 target genes may contain Cys residues involved in disulfide bridges, making a total of 20 ZmASR1 target genes potentially linked to Trx. Thus, our results open the way to the very speculative hypothesis that ZmASR1 exhibits a chaperone-like activity on selectively redox-regulated proteins.
Among the established and potential Trx targets with expression changes in ZmASR1-OE plants, we found seven genes involved in BCAA biosynthesis. A unique feature of BCAA biosynthesis, which seems to occur exclusively in plastids, is that Val and Ile are formed in two parallel pathways using the same enzymes, namely AHAS, KARI, dihydroxyacid dehydratase, and BCAT, while Leu formation branches off from 2-oxoisovalerate, the last intermediates of the Val biosynthetic pathway, to follow a three-step chain elongation catalyzed by isopropylmalate synthase, isopropylmalate isomerase, and IPMDH, which ends with a transamination step catalyzed by a BCAT (Binder et al., 2007; Joshi et al., 2010). Decreases in ZmBCAT2 transcripts and ZmIPMDH1 and ZmIPMDH2 proteins in ZmASR1-OE leaves in comparison with wild-type leaves fit well with the finding that ZmASR1-OE triggers significant decreases in BCAAs. Furthermore, transcript levels of ZmAHAS1, ZmKARI1, ZmKARI2, and ZmBCAT4 were correlated with each other and/or the levels of the specific set of 13 decreased metabolites. Importantly, ZmKARI1 and ZmKARI2 showed enriched expression in mesophyll cells in comparison with bundle sheath cells (Majeran et al., 2005; Friso et al., 2010), similar to that was observed for ZmASR1 (Li et al., 2010). There are contradictory reports in the literature on the distribution of ZmIPMDH1 and ZmIPMDH2 across mesophyll and bundle sheath chloroplasts, with a 2-fold enrichment in either mesophyll (Majeran et al., 2005) or bundle sheath (Friso et al., 2010) chloroplasts. Distribution data for ZmAHAS1, ZmBCAT2, and ZmBCAT4 are as yet unavailable, although it is noteworthy that ZmBCAT1 and ZmBCAT3 showed enriched expression in mesophyll cells (Friso et al., 2010). Additionally, it may be noted that Arabidopsis Trx-m1 is the most effective Trx on AtIPMDH1 in comparison with Arabidopsis Trx-f1 or bacterial Trx (He et al., 2009). Its maize ortholog (Trx-m4) also showed enriched expression in mesophyll cells (Majeran et al., 2005; Friso et al., 2010). Taken together, our data demonstrate the causal relationship between the overexpression of ZmASR1 and genes involved in BCAA biosynthesis and support the idea that the improvement in biomass associated with ZmASR1-OE involves the coordinated transcriptional regulation of BCAA biosynthetic genes. The resolution of the other ZmASR1 target genes is severely hampered by the fact that only limited functional information is available on them. In this respect, a noteworthy observation is that Cañas et al. (2010) have just recently shown that increasing the expression of the enzyme P5CS may have a beneficial effect on yield. Additional experiments will be required to reveal the role of the other ZmASR1 gene targets in water deficit tolerance.
MATERIALS AND METHODS
Plant Materials and Cultures
Stress Treatments at the Vegetative Stage
To carry out the water deficit experiment, maize (Zea mays) B73 plants grown in a greenhouse (photoperiod of 16 h supplemented with artificial lighting, 25°C/18°C [day/night], and 60%/55% [day/night] relative humidity) with appropriate daily watering (50% soil water content) until the fifth leaf reached a 35-cm length (about 21 d on average) were deprived of water (WDS test) or daily irrigated (50% soil water content; control condition). The fifth and sixth leaves were harvested when growth of the sixth leaf stopped (0.2 cm or less; WDS test) or after 3 d of constant leaf elongation rate (control condition), 5 to 7 h after the onset of the light period. Four-centimeter-long fragments of the sixth leaves and a 10-cm-long fragment from the middle of the fifth leaf blade were immediately frozen in liquid nitrogen. For ABA, Glc, and PEG treatments, we used leaves and roots described in detail by Trouverie et al. (2004). All samples were ground in a 2-mL microcentrifuge tube containing both 5- and 7-mm steel beads for two to three series of 20 s using a TissueLyser II at 20 Hz (Qiagen) and stored at −80°C until analysis.
Stress Treatment at the Reproductive Stage
Tissues used to establish expression profiles during maize ovule and kernel development (−5 to 70 DAP) were described in detail by Massonneau et al. (2005). To carry out the water deficit experiment, maize MBS847 plants were grown in the greenhouse as described by Qin et al. (2004). Seven days before pollination, plants were deprived of water or irrigated daily. Sampling took place at 0, 4, 7, and 12 DAP, 5 to 7 h after the onset of the light period. The ears and 10-cm-long fragments from the bottom thirds of the corresponding axillary leaves were harvested from plants subjected to each treatment. Mature ovules and kernels were collected from the central part of the ears, separated from the adhering perianth and the pedicel, and stored in liquid nitrogen. All plant materials were ground in a mortar in liquid nitrogen and stored at −80°C until analysis.
Stress Treatment and Yield Trial in the Field
To evaluate the ZmASR1-OE plants in the field under limited water treatment, nine independently integrated ZmASR1-OE events were grown at Magneraud, France, in May 2005 using two randomized block designs (one to four rows per event; 45 plants per row in each block, transgene-positive and transgene-negative plants in each row identified by PCR during the late vegetative stage of development). One block was maintained in a well-watered condition and the other one placed under limited water supply using mobile devices preventing watering by rainfall (Supplemental Fig. S1A) and three sets of two Watermark probes dispatched at the top third, middle third, and bottom third of each block. For each location, there were two probes placed vertically in the soil, 30 and 60 cm below the surface, and connected to a data logger, allowing daily collection of soil water potential data. These data and growth stage of plants were integrated into the “grapher Watermark” software designed for a specific quality of soil, allowing the monitoring of the trial for signs of water deficit throughout the season. Consequently, the limiting-water condition was achieved at the late vegetative stage of development (e.g. from 19 d before silk emergence; Supplemental Fig. S1, B and C). Among the nine events, the three with strongest expression of the transgene (two to three rows per event in each block) were selected for further analysis. Two-centimeter-long fragments in the bottom third of the 11th leaves below the ear leaf were collected 57 d after sowing (5 d before silk emergence), 5 to 7 h after the onset of the light period, and immediately frozen in liquid nitrogen. They were then ground in a 2-mL microcentrifuge tube containing both 5- and 7-mm steel beads for two to four series of 20 s, using a TissueLyser II at 20 Hz (Qiagen). Biological duplicates were formed by dividing each row in two groups and pooling the three selected events, without changing the proportion of each row: samples included 46 plants in total, with 14 to 18 plants per event and two to three rows per event. All samples were stored at −80°C until analysis.
Plant Transformation
The plasmid used for the production of ZmASR1-OE plants contains the backbone of vector pSB12 (Komari et al., 1996), a Basta resistance cassette (rice Actin promoter and intron, Bar and Nos terminators) next to the right border, and the ZmASR1 cDNA of maize MBS847 (GenBank accession no. AX297905), under the control of the cassava vein mosaic virus promoter, next to the left border. Agrobacterium tumefaciens-mediated transformation of maize inbred line A188 was based on a published protocol (Ishida et al., 1996). For each transformation event, the number of T-DNA insertions was evaluated by Southern blot, the integrity of the transgene was verified by PCR, and the expression level of the transgene was evaluated by RT-PCR and 2-DE as described (Jeanneau et al., 2002; Depège-Fargeix et al., 2011). Seventeen independent transformation events were crossed with the maize inbred line F2, in which the ZmASR1 protein is not detected (Riccardi et al., 1998; Jeanneau et al., 2002). T1 events were then selected and backcrossed one more time with the maize inbred line F2. Among the 17 independently integrated T2 events, the nine with strongest expression of the transgene compared with the endogenous ZmASR1 gene based on 2-DE were evaluated in the field. In the T2 generation containing 50% transgenics, the wild-type plants were used as control plants.
Database Search, Gene Structure Determination, and Chromosomal Location of ZmASR Genes
To search for ZmASR1 homologs in maize, we used the ZmASR1 cDNA in BLAST analysis using EST assemblies in The Gene Index database (http://compbio.dfci.harvard.edu/tgi/plant.html), assemblies of genomic DNA fragments at The Plant Genomics Resources (http://blast.jcvi.org/tgi_maize/index.cgi), high-throughput genomic sequences from bacterial artificial chromosome clones at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/HTGS), and the recent release of the maize genome sequence (http://maizesequence.org). The members of the maize ASR gene family suggested from these combined in silico analyses were confirmed by sequencing of amplified cDNA and genomic DNA of maize B73 plus one to five additional maize inbred lines (Mo17, F2, F252, MBS847, and/or A188). They were then mapped on the intermated B73 × Mo17 (IBM) or F2 × F252 (LHRF) mapping panels using the REFMAP050110 (IBM_Gnp2004) or LHRF_Gnp2004 framework map, respectively (Falque et al., 2005), using gene-specific primers (Supplemental Table S2). Alternatively, BLAST analysis on MaizeSequence.org was used in the absence of polymorphism between the maize inbred lines that led to the IBP or LHRF mapping panels. Deduced amino acid sequences were annotated by BLAST against known ASR proteins at NCBI and query against InterPro (http://www.ebi.ac.uk/Tools/pfa/iprscan/). The ProtParam tool (Gasteigler et al., 2005; http://web.expasy.org/protparam) was also used to determine the physical and chemical properties of the deduced amino acid sequences.
Phylogenetic Analysis
Sequence alignments were managed using BioEdit version 7.0.0 (Hall, 1999) and visually refined on the basis of the amino acid sequences. Bayesian phylogenetic analysis was carried out using MrBayes version 3.1.1 (Huelsenbeck and Ronquist, 2001) with a general time-reversible model, with a proportion of invariable sites and a γ-distribution for site-specific rates and a partition according to the three codon positions on the four most conserved regions only (272-nucleotide alignment matrix). Three chains (two-heated) were run twice for 107 generations, with a sampling of the cold chain parameters every 100 generations and a burn-in of 25,000 samples. Convergence was followed with potential scale reduction factor and average sd of split frequencies. A majority rule consensus tree was built with posterior probabilities of nodes above 0.85 × 100 indicated.
qRT-PCR and RT-PCR Analyses
The total RNA isolation from maize tissues collected in the stress experiments (leaves, roots, mature ovules, and kernels) with the TRIzol Reagent (Invitrogen) and ethanol precipitation was carried out as described (Trouverie et al., 2004). Subsequent DNase treatment and DNase inactivation were carried out according to the instructions of the supplier (Ambion). Two micrograms of total RNA was reverse transcribed using random hexamers (Invitrogen), 100 units of SuperScript II (Invitrogen), and 40 units of recombinant Rnasin RNase inhibitor (Promega) in a final volume of 20 μL. For expression profiles during maize ovule and kernel development, the total RNA isolation with phenol/chloroform extraction and acetic acid/ethanol precipitation, as well as subsequent DNase treatment and RT, were carried out as described (Massonneau et al., 2005).
Primer sequences were designed using Primer Express 2.0 (Applied Biosystem). Subsequent qRT-PCR analysis was carried out as described (Capelle et al., 2010). The calibration step of the experiment checked for equivalent PCR efficiency of the different genes (to allow comparison and normalization). Standard curves (log of cDNA dilution versus cycle threshold) using serial 10-fold dilution of cDNA were built for each pair of selected primers, a 100% efficiency corresponding to a slope of −3.3 (Marino et al., 2003). Practically, only pairs of primers yielding a slope of −3.3 ± 0.1 were selected. The specificity of the amplification (checked by dissociation curve analysis, gel electrophoresis, and sequencing of the PCR product) and the use of appropriate control genes were also assessed. Normalization of the results was achieved using the Gly-rich RNA-binding protein2 (ZmGRP2) gene. After an initial test of five genes (actin, polyubiquitin, α-tubulin, H+-ATPase, and ZmGRP2) on the range of tissues and treatments we wished to compare, only ZmGRP2 was found to be stably expressed, in agreement with recent genome-wide observations in 55 tissues of maize B73 line (Sekhon et al., 2011). Conventional RT-PCR analysis was used to provide evidence that ZmASR7-2 and ZmASR7-3 genes are expressed. It was performed using the real-time PCR system and the SYBR Green PCR master mix as described (Capelle et al., 2010) to allow comparison with the qRT-PCR experiments. Products of the PCR were loaded on agarose gels and stained with ethidium bromide.
Microarray Analysis
Total RNA was extracted using the Nucleospin 8 RNA Extraction Kit (Macherey-Nagel) and in vitro amplified based on suggestions from the Maize Oligonucleotide Array Project (http://www.maizearray.org/files/cRNA_Target_Production_Using_RNA_Amplification.pdf). Hybridization was carried out as described (http://www.maizearray.org/files/Hybridization_Protocol_For_cRNA_Targets.pdf) using the 46K array constructed by the Maize Oligonucleotide Array Project. Quantile normalization of the raw data was carried out using the R version 2.4.1 software with a log10 transformation of the data initially performed. The criteria for the inclusion of a gene in the list of differentially expressed genes were a 1.5-fold increase or decrease and P < 0.01 by two-way ANOVA. The cDNA and genomic sequences corresponding to the 70-mers present on the microarray were established by BLAST of the 70-mers against the high-throughput genomic sequences database and the recent release of the maize genome sequence and regularly updated. Deduced amino acid sequences were annotated by BLAST against the Arabidopsis (Arabidopsis thaliana) genome at NCBI and screened for known conserved domains using the Center for Biological Sequence Analysis database (http://www.cbs.dtu.dk/services/).
Protein Extraction, Gel Staining, and 2-DE Analysis
Protein extraction with TCA/acetone precipitation was carried out as described (Méchin et al., 2007). Proteins were solubilized in UKS (for urea-K2CO3-sodium dodecyl sulfate) buffer (Méchin et al., 2007) and quantified using the 2-D Quant Kit (GE Healthcare Life Sciences). Isoelectrofocusing was carried out using 24-cm-long, pH 4 to 7 Immobiline Dry Strips (GE Healthcare Life Sciences) to which 50 μg (for protein quantification; 150 and 30 μg for ear leaf and kernel tissues, respectively) or 300 μg (for protein identification) of dissolved protein was added. Active rehydration and focusing were achieved at 20°C in a Protean IEF Cell (Bio-Rad) by increasing the voltage step by step from 50 to 10,000 V (13 h at 50 V, 0.5 h at 200 V, 0.5 h at 500 V, 1 h at 1,000 V, then increase to 10,000 V) and stopped when 84,000 Vh was reached. Strips were equilibrated as described (Görg et al., 1987) and sealed at the top of a 1-mm-thick two-dimensional gel (24 × 22 cm) with 1% (w/v) low-melting agarose in SDS electrophoresis buffer (25 mm Trizma base, 0.2 m Gly, and 0.1% [v/v] SDS). Separation of continuous gels (11% [w/v] acrylamide, 2.9% [w/v] piperazine diacrylamide) was performed at 14°C (20 V for 1 h, 140 V for 15 h, and 20 V for 2 h) in a Protean Plus Dodeca Cell Electrophoresis Chamber (Bio-Rad) until the bromphenol blue front reached the end of the gel. For protein quantification, a silver-staining procedure was performed as described (Méchin et al., 2003). Scanning was carried out at 300 dpi with a 16-bit grayscale pixel depth using an image scanner (Amersham Biosciences) and analyzed with Progenesis software (Nonlinear Dynamics) according to Zivy (2007). For protein identification, gels were stained in colloidal Coomassie blue according to Yan et al. (2000) and spots were excised for MS. Excised gels were restained with silver nitrate as described (Méchin et al., 2003) and compared with the nonexcised silver-stained gels.
LC-MS/MS and Protein Identification
Two-dimensional gel spot digestion and LC-MS/MS analysis were performed as described (Page et al., 2010). A database search was performed with X!Tandem (version 2010.01.01.4; http://www.thegpm.org/tandem/). Enzymatic cleavage was declared as a trypsin digestion with one possible miscleavage. Cys carboxyamidomethylation and Met oxidation were set to static and possible modifications, respectively. Precursor mass and fragment mass tolerance were 2.0 and 0.8, respectively. A refinement search was added with similar parameters, except that semitryptic peptide and possible N-terminal protein acetylation were searched. The maize genome sequence (release 4a.53; 53,764 entries), the UniProt database restricted to maize (http://www.uniprot.org; release 15.11; 43,694 entries), and a contaminant database including in particular trypsin and keratins were used. Only peptides with a E value smaller than 0.1 were reported. Identified proteins were filtered and grouped using the X!Tandem pipeline (http://pappso.inra.fr/bioinfo/xtandempipeline/) with the following criteria: (1) a minimum of two different peptides was required with an E value smaller than 0.05; (2) a protein E value (calculated as the product of unique peptide E values) smaller than 10−4 was required. In the case of identification with only two or three MS/MS spectra, similarity between the experimental and the theoretical MS/MS spectra was visually checked. To take redundancy into account, proteins with at least one peptide in common were grouped. This allowed grouping of proteins of similar function. Within each group, proteins with at least one specific peptide relative to other members of the group were reported as subgroups.
Chlorophyll Content Measurement and Metabolite Analysis
Total chlorophyll content was measured after extraction of leaf material into 80% acetone and assay at 663 and 645 nm as described (Arnon, 1949). Gas chromatography coupled to time-of-flight MS analysis was carried out as described (Noctor et al., 2007). Because automated peak integration was occasionally erroneous, integration was verified manually for each compound in all analyses.
Two-Way ANOVA and Within-Group Correlation Matrix Analysis
A log10 transformation of the data was initially performed when the se values showed that the se increased in proportion to the treatment. Either nontransformed or log10-transformed values were then used for two-way ANOVA using R software version 2.8.1 (http://cran.at.r-project.org). The two possible means models for two-way ANOVA are the additive model and the interaction model (Scheffé, 1999; Seltman, 2010). An F test for interactions was performed to determine whether the additive model could be retained. When the interaction model was needed, the Bonferroni method was applied for pairwise comparisons. The significance was placed at the 0.05 level.
In a multivariate ANOVA model, the within-group correlation matrix is calculated from the correlation matrix of residuals after regressing out the influence of the fixed effects by linear regression (Hair et al., 2010). Functions of the R software were used. We were unable to test the association between two variables independently of the others because we did not have enough data to determine partial correlation coefficients. Therefore, the null hypothesis of a pairwise correlation coefficient was evaluated by a test derived from an F test and extended to all correlation coefficients using the Bonferroni method. The significance was placed at the 0.05 level.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Majority rule consensus tree obtained using Bayesian interference analysis of 100 ASR sequences.
Supplemental Figure S2. Stress treatment and yield trial of ZmASR1-OE plants.
Supplemental Figure S3. Relative expression profile of selected genes encoding ZmASR1 targets and/or rate-limiting enzymes in leaves 11 of ZmASR1-OE and wild-type plants.
Supplemental Figure S4. Transcript levels of ZmASR7-1, ZmASR7-2, and ZmASR7-3 genes in leaf and root tissues.
Supplemental Table S1. ZmASR gene mapping.
Supplemental Table S2. Sequences of the primers used for mapping and qRT-PCR analyses.
Supplemental Table S3. Sequence identity between individual ZmASR genes.
Supplemental Table S4. Summary of exon lengths of various ASR genes from Poaceae and properties of their deduced amino acid sequences.
Supplemental Table S5. ZmASR1-OE plants maintain kernel yield under water-limited conditions.
Supplemental Table S6. Proteins that showed significant changes in ZmASR1-OE leaves compared with wild-type leaves, their annotated function, location, and quantification.
Supplemental Table S7. Unique metabolites identified in ZmASR1-OE and wild-type leaves.
Supplemental Table S8. Within-group correlation matrix.
Supplemental Table S9. Number of individual significant correlations for the whole matrix and its submatrices.
Supplemental Table S10. Pairwise correlations of ZmASR1 targets against BCAA-related ZmASR1 targets.
Supplemental Table S11. Pairwise correlations of ZmASR1 target transcripts against decreased and/or biomass-related metabolites.
Acknowledgments
We thank Ingrid Doridant and Carine Hécart for maize transformation and culture, Carine Remoué for help with physical mapping, the Biogemma transcriptomic team for the hybridization and analysis of the microarray, and Jean Coursol for guiding us through the statistical analyses. Caroline Lelarge-Trouverie is acknowledged for preliminary metabolomic analysis on ZmASR1-OE.
References
- Amitai-Zeigerson H, Scolnik PA, Bar-Zvi D. (1995) Tomato Asrl mRNA and protein are transiently expressed following salt stress, osmotic stress and treatment with abscisic acid. Plant Sci 110: 205–213 [Google Scholar]
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA. (2008) The enigmatic LEA proteins and other hydrophilins. Plant Physiol 148: 6–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S, Knill T, Schuster J. (2007) Branched-chain amino acid metabolism in higher plants. Physiol Plant 129: 68–78 [Google Scholar]
- Böhmer M, Schroeder JI. (2011) Quantitative transcriptomic analysis of abscisic acid-induced and reactive oxygen species-dependent expression changes and proteomic profiling in Arabidopsis suspension cells. Plant J 67: 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer JS, Westgate ME. (2004) Grain yields with limited water. J Exp Bot 55: 2385–2394 [DOI] [PubMed] [Google Scholar]
- Brini F, Hanin M, Lumbreras V, Amara I, Khoudi H, Hassairi A, Pagès M, Masmoudi K. (2007) Overexpression of wheat dehydrin DHN-5 enhances tolerance to salt and osmotic stress in Arabidopsis thaliana. Plant Cell Rep 26: 2017–2026 [DOI] [PubMed] [Google Scholar]
- Brini F, Saibi W, Amara I, Gargouri A, Masmoudi K, Hanin M. (2010) Wheat dehydrin DHN-5 exerts a heat-protective effect on β-glucosidase and glucose oxidase activities. Biosci Biotechnol Biochem 74: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Cakir B, Agasse A, Gaillard C, Saumonneau A, Delrot S, Atanassova R. (2003) A grape ASR protein involved in sugar and abscisic acid signaling. Plant Cell 15: 2165–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañas RA, Quilleré I, Lea PJ, Hirel B. (2010) Analysis of amino acid metabolism in the ear of maize mutants deficient in two cytosolic glutamine synthetase isoenzymes highlights the importance of asparagine for nitrogen translocation within sink organs. Plant Biotechnol J 8: 966–978 [DOI] [PubMed] [Google Scholar]
- Canel C, Bailey-Serres JN, Roose ML. (1995) Pummelo fruit transcript homologous to ripening-induced genes. Plant Physiol 108: 1323–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelle V, Remoué C, Moreau L, Reyss A, Mahé A, Massonneau A, Falque M, Charcosset A, Thévenot C, Rogowsky P, et al. (2010) QTLs and candidate genes for desiccation and abscisic acid content in maize kernels. BMC Plant Biol 10: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Fernie AR, Iusem ND. (2004) Heard it through the grapevine? ABA and sugar cross-talk: the ASR story. Trends Plant Sci 9: 57–59 [DOI] [PubMed] [Google Scholar]
- Castiglioni P, Warner D, Bensen RJ, Anstrom DC, Harrison J, Stoecker M, Abad M, Kumar G, Salvador S, D’Ordine R, et al. (2008) Bacterial RNA chaperones confer abiotic stress tolerance in plants and improved grain yield in maize under water-limited conditions. Plant Physiol 147: 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciais P, Reichstein M, Viovy N, Granier A, Ogée J, Allard V, Aubinet M, Buchmann N, Bernhofer C, Carrara A, et al. (2005) Europe-wide reduction in primary productivity caused by the heat and drought in 2003. Nature 437: 529–533 [DOI] [PubMed] [Google Scholar]
- Claassen HC. (1970) The determination of low levels of cobalt-60 in environmental waters by liquid scintillation counting. Anal Chim Acta 52: 229–235 [DOI] [PubMed] [Google Scholar]
- Damerval C, Maurice A, Josse JM, de Vienne D. (1994) Quantitative trait loci underlying gene product variation: a novel perspective for analyzing regulation of genome expression. Genetics 137: 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kraker JW, Luck K, Textor S, Tokuhisa JG, Gershenzon J. (2007) Two Arabidopsis genes (IPMS1 and IPMS2) encode isopropylmalate synthase, the branchpoint step in the biosynthesis of leucine. Plant Physiol 143: 970–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depège-Fargeix N, Javelle M, Chambrier P, Frangne N, Gerentes D, Perez P, Rogowsky PM, Vernoud V. (2011) Functional characterization of the HD-ZIP IV transcription factor OCL1 from maize. J Exp Bot 62: 293–305 [DOI] [PubMed] [Google Scholar]
- de Vienne D, Leonardi A, Damerval C, Zivy M. (1999) Genetics of proteome variation for QTL characterization: application to drought-stress response in maize. J Exp Bot 50: 303–309 [Google Scholar]
- Doczi R, Csanaki C, Banfalvi Z. (2002) Expression and promoter activity of the desiccation-specific Solanum tuberosum gene, StDS2. Plant Cell Environ 25: 1197–1203 [Google Scholar]
- Endler A, Meyer S, Schelbert S, Schneider T, Weschke W, Peters SW, Keller F, Baginsky S, Martinoia E, Schmidt UG. (2006) Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol 141: 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falque M, Décousset L, Dervins D, Jacob AM, Joets J, Martinant JP, Raffoux X, Ribière N, Ridel C, Samson D, et al. (2005) Linkage mapping of 1454 new maize candidate gene loci. Genetics 170: 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang LY, Gross PR, Chen CH, Lillis M. (1992) Sequence of two acetohydroxyacid synthase genes from Zea mays. Plant Mol Biol 18: 1185–1187 [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization of the United Nations (2008) Climate change, water and food security. http://www.fao.org/nr/water/docs/HLC08-FAOWater-E.pdf (February 26, 2008) [Google Scholar]
- Frankel N, Nunes-Nesi A, Balbo I, Mazuch J, Centeno D, Iusem ND, Fernie AR, Carrari F. (2007) ci21A/Asr1 expression influences glucose accumulation in potato tubers. Plant Mol Biol 63: 719–730 [DOI] [PubMed] [Google Scholar]
- Friso G, Majeran W, Huang M, Sun Q, van Wijk KJ. (2010) Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol 152: 1219–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumoto T, Hata S, Izui K. (2000) Isolation and characterization of cDNAs for differentially accumulated transcripts between mesophyll cells and bundle sheath strands of maize leaves. Plant Cell Physiol 41: 1200–1209 [DOI] [PubMed] [Google Scholar]
- Garay-Arroyo A, Colmenero-Flores JM, Garciarrubio A, Covarrubias AA. (2000) Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J Biol Chem 275: 5668–5674 [DOI] [PubMed] [Google Scholar]
- Gasteigler E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD,, Bairoch A. (2005) Protein identification and analysis tools on ExPASy server. Walker JM, , The Proteomics Protocols Handbook. Humana Press, New York, pp 571–607 [Google Scholar]
- Goldgur Y, Rom S, Ghirlando R, Shkolnik D, Shadrin N, Konrad Z, Bar-Zvi D. (2007) Desiccation and zinc binding induce transition of tomato abscisic acid stress ripening 1, a water stress- and salt stress-regulated plant-specific protein, from unfolded to folded state. Plant Physiol 143: 617–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görg A, Postel W, Weser J, Günther S, Strahler JR, Hanash SM, Somerlot L. (1987) Elimination of point streaking on silver stained two-dimensional gels by addition of iodoacetamide to the equilibration buffer. Electrophoresis 8: 122–124 [Google Scholar]
- Guo AY, Zhu QH, Gu X, Ge S, Yang J, Luo J. (2008) Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 418: 1–8 [DOI] [PubMed] [Google Scholar]
- Hair JF, Black WC, Babin BJ, Anderson RE. (2010) Multivariate Data Analysis, Ed 7. Prentice Hall, Upper Saddle River, NJ [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95–98 [Google Scholar]
- He Y, Mawhinney TP, Preuss ML, Schroeder AC, Chen B, Abraham L, Jez JM, Chen S. (2009) A redox-active isopropylmalate dehydrogenase functions in the biosynthesis of glucosinolates and leucine in Arabidopsis. Plant J 60: 679–690 [DOI] [PubMed] [Google Scholar]
- Henry IM, Carpentier SC, Pampurova S, Van Hoylandt A, Panis B, Swennen R, Remy S. (June 1, 2011) Structure and regulation of the Asr gene family in banana. Planta http://dx.doi.org/10.1007/s00425-011-1421-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JC, Lin SM, Wang CS. (2000) A pollen-specific and desiccation-associated transcript in Lilium longiflorum during development and stress. Plant Cell Physiol 41: 477–485 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Ishida Y, Saito H, Ohta S, Hiei Y, Komari T, Kumashiro T. (1996) High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat Biotechnol 14: 745–750 [DOI] [PubMed] [Google Scholar]
- Iusem ND, Bartholomew DM, Hitz WD, Scolnik PA. (1993) Tomato (Lycopersicon esculentum) transcript induced by water deficit and ripening. Plant Physiol 102: 1353–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanneau M, Gerentes D, Foueillassar X, Zivy M, Vidal J, Toppan A, Perez P. (2002) Improvement of drought tolerance in maize: towards the functional validation of the Zm-Asr1 gene and increase of water use efficiency by over-expressing C4-PEPC. Biochimie 84: 1127–1135 [DOI] [PubMed] [Google Scholar]
- Joshi V, Joung JG, Fei Z, Jander G. (2010) Interdependence of threonine, methionine and isoleucine metabolism in plants: accumulation and transcriptional regulation under abiotic stress. Amino Acids 39: 933–947 [DOI] [PubMed] [Google Scholar]
- Kalifa Y, Gilad A, Konrad Z, Zaccai M, Scolnik PA, Bar-Zvi D. (2004a) The water- and salt-stress-regulated Asr1 (abscisic acid stress ripening) gene encodes a zinc-dependent DNA-binding protein. Biochem J 381: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalifa Y, Perlson A, Gilad A, Konrad Z, Scolnik PA, Bar-Zvi D. (2004b) Over-expression of the water and salt stress-regulated Asr1 gene confers an increased salt tolerance. Plant Cell Environ 27: 1459–1468 [Google Scholar]
- Kerk D, Bulgrien J, Smith DW, Gribskov M. (2003) Arabidopsis proteins containing similarity to the universal stress protein domain of bacteria. Plant Physiol 131: 1209–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KE. (1996) Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 509–540 [DOI] [PubMed] [Google Scholar]
- Komari T, Hiei Y, Saito Y, Murai N, Kumashiro T. (1996) Vectors carrying two separate T-DNAs for co-transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J 10: 165–174 [DOI] [PubMed] [Google Scholar]
- Konrad Z, Bar-Zvi D. (2008) Synergism between the chaperone-like activity of the stress regulated ASR1 protein and the osmolyte glycine-betaine. Planta 227: 1213–1219 [DOI] [PubMed] [Google Scholar]
- Less H, Angelovici R, Tzin V, Galili G. (2010) Principal transcriptional regulation and genome-wide system interactions of the Asp-family and aromatic amino acid networks of amino acid metabolism in plants. Amino Acids 39: 1023–1028 [DOI] [PubMed] [Google Scholar]
- Less H, Galili G. (2008) Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses. Plant Physiol 147: 316–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al. (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Liu HY, Dai JR, Feng DR, Liu B, Wang HB, Wang JF. (2010) Characterization of a novel plantain Asr gene, MpAsr, that is regulated in response to infection of Fusarium oxysporum f. sp. cubense and abiotic stresses. J Integr Plant Biol 52: 315–323 [DOI] [PubMed] [Google Scholar]
- Lu G-H, Tang J-H, Yan F-B, Ma X-Q, Li J-S, Chen S-J, Ma J-C, Liu Z-X, L-Z E, Zhang Y-R, et al. (2006) Quantitative trait loci mapping of maize yield and its components under different water treatments at flowering time. J Integr Plant Biol 48: 1233–1243 [Google Scholar]
- Majeran W, Cai Y, Sun Q, van Wijk KJ. (2005) Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. Plant Cell 17: 3111–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Friso G, Ponnala L, Connolly B, Huang M, Reidel EJ, Zhang C, Asakura Y, Bhuiyan NH, Sun Q, et al. (2010) Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. Plant Cell 22: 3509–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino JH, Cook P, Miller KS. (2003) Accurate and statistically verified quantification of relative mRNA abundances using SYBR Green I and real-time RT-PCR. J Immunol Methods 283: 291–306 [DOI] [PubMed] [Google Scholar]
- Marino R, Ponnaiah M, Krajewski P, Frova C, Gianfranceschi L, Pè ME, Sari-Gorla M. (2009) Addressing drought tolerance in maize by transcriptional profiling and mapping. Mol Genet Genomics 281: 163–179 [DOI] [PubMed] [Google Scholar]
- Martin A, Lee J, Kichey T, Gerentes D, Zivy M, Tatout C, Dubois F, Balliau T, Valot B, Davanture M, et al. (2006) Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18: 3252–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskin L, Frankel N, Gudesblat G, Demergasso MJ, Pietrasanta LI, Iusem ND. (2007) Dimerization and DNA-binding of ASR1, a small hydrophilic protein abundant in plant tissues suffering from water loss. Biochem Biophys Res Commun 352: 831–835 [DOI] [PubMed] [Google Scholar]
- Maskin L, Gudesblat G, Moreno J, Carrari F, Frankel N, Sambade A, Rossi M, Iusem ND. (2001) Differential expression of the members of the Asr gene family in tomato (Lycopersicon esculentum). Plant Sci 161: 739–746 [Google Scholar]
- Massonneau A, Condamine P, Wisniewski JP, Zivy M, Rogowsky PM. (2005) Maize cystatins respond to developmental cues, cold stress and drought. Biochim Biophys Acta 1729: 186–199 [DOI] [PubMed] [Google Scholar]
- McPherson HG, Boyer J. (1977) Regulation of grain yield by photosynthesis in maize subjected to water deficiency. Agron J 69: 714–718 [Google Scholar]
- Méchin V, Consoli L, Le Guilloux M, Damerval C. (2003) An efficient solubilization buffer for plant proteins focused in immobilized pH gradients. Proteomics 3: 1299–1302 [DOI] [PubMed] [Google Scholar]
- Méchin V, Damerval C, Zivy M. (2007) Total protein extraction with TCA-acetone. Methods Mol Biol 355: 1–8 [DOI] [PubMed] [Google Scholar]
- Meyer RC, Steinfath M, Lisec J, Becher M, Witucka-Wall H, Törjék O, Fiehn O, Eckardt A, Willmitzer L, Selbig J, et al. (2007) The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 4759–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montrichard F, Alkhalfioui F, Yano H, Vensel WH, Hurkman WJ, Buchanan BB. (2009) Thioredoxin targets in plants: the first 30 years. J Proteomics 72: 452–474 [DOI] [PubMed] [Google Scholar]
- Murat F, Xu JH, Tannier E, Abrouk M, Guilhot N, Pont C, Messing J, Salse J. (2010) Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution. Genome Res 20: 1545–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K. (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149: 88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, Warner DC, Anstrom DC, Bensen RJ, Castiglioni PP, Donnarummo MG, et al. (2007) Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA 104: 16450–16455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Shigeoka S. (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Bergot G, Thominet D, Mauve C, Lelarge-Trouverie C, Prioul JL. (2007) A comparative study of amino acid analysis in leaf extracts by gas chromatography-time of flight-mass spectrometry and high performance liquid chromatography with fluorescence detection. Metabolomics 3: 161–174 [Google Scholar]
- Padmanabhan V, Dias DM, Newton RJ. (1997) Expression analysis of a gene family in loblolly pine (Pinus taeda L.) induced by water deficit stress. Plant Mol Biol 35: 801–807 [DOI] [PubMed] [Google Scholar]
- Page D, Gouble B, Valot B, Bouchet JP, Callot C, Kretzschmar A, Causse M, Renard CM, Faurobert M. (2010) Protective proteins are differentially expressed in tomato genotypes differing for their tolerance to low-temperature storage. Planta 232: 483–500 [DOI] [PubMed] [Google Scholar]
- Pelleschi S, Leonardi A, Rocher J-P, Cornic G, de Vienne D, Thevenot C, Prioul JL. (2006) Analysis of the relationships between growth, photosynthesis and carbohydrate metabolism using quantitative trait loci (QTLs) in young maize plants subjected to water deprivation. Mol Breed 17: 21–39 [Google Scholar]
- Philippe R, Courtois B, McNally KL, Mournet P, El-Malki R, Le Paslier MC, Fabre D, Billot C, Brunel D, Glaszmann JC, et al. (2010) Structure, allelic diversity and selection of Asr genes, candidate for drought tolerance, in Oryza sativa L. and wild relatives. Theor Appl Genet 121: 769–787 [DOI] [PubMed] [Google Scholar]
- Qin L, Trouverie J, Chateau-Joubert S, Simond-Côte E, Thevenot C, Prioul JL. (2004) Involvement of the Ivr2-invertase in the perianth during maize kernel development under water stress. Plant Sci 166: 371–379 [Google Scholar]
- Quan R, Shang M, Zhang H, Zhao Y, Zhang J. (2004) Engineering of enhanced glycine betaine synthesis improves drought tolerance in maize. Plant Biotechnol J 2: 477–486 [DOI] [PubMed] [Google Scholar]
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccardi F, Gazeau P, de Vienne D, Zivy M. (1998) Protein changes in response to progressive water deficit in maize: quantitative variation and polypeptide identification. Plant Physiol 117: 1253–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccardi F, Gazeau P, Jacquemot MP, Vincent D, Zivy M. (2004) Deciphering genetic variations of proteome responses to water deficit in maize leaves. Plant Physiol Biochem 42: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Rom S, Gilad A, Kalifa Y, Konrad Z, Karpasas MM, Goldgur Y, Bar-Zvi D. (2006) Mapping the DNA- and zinc-binding domains of ASR1 (abscisic acid stress ripening), an abiotic-stress regulated plant specific protein. Biochimie 88: 621–628 [DOI] [PubMed] [Google Scholar]
- Rossi M, Carrari F, Cabrera-Ponce JL, Vázquez-Rovere C, Herrera-Estrella L, Gudesblat G, Iusem ND. (1998) Analysis of an abscisic acid (ABA)-responsive gene promoter belonging to the Asr gene family from tomato in homologous and heterologous systems. Mol Gen Genet 258: 1–8 [DOI] [PubMed] [Google Scholar]
- Salekdeh GH, Reynolds M, Bennett J, Boyer J. (2009) Conceptual framework for drought phenotyping during molecular breeding. Trends Plant Sci 14: 488–496 [DOI] [PubMed] [Google Scholar]
- Salse J, Bolot S, Throude M, Jouffe V, Piegu B, Quraishi UM, Calcagno T, Cooke R, Delseny M, Feuillet C. (2008) Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 20: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saumonneau A, Agasse A, Bidoyen MT, Lallemand M, Cantereau A, Medici A, Laloi M, Atanassova R. (2008) Interaction of grape ASR proteins with a DREB transcription factor in the nucleus. FEBS Lett 582: 3281–3287 [DOI] [PubMed] [Google Scholar]
- Scheffé H. (1999) The Analysis of Variance. John Wiley & Sons, New York [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schneider A, Salamini F, Gebhardt C. (1997) Expression patterns and promoter activity of the cold-regulated gene ci21A of potato. Plant Physiol 113: 335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM. (2011) Genome-wide atlas of transcription during maize development. Plant J 66: 553–563 [DOI] [PubMed] [Google Scholar]
- Seltman HJ. (2010) Experimental design and analysis. http://www.stat.cmu.edu/˜hseltman/309/Book/Book.pdf (September 8, 2011)
- Shkolnik D, Bar-Zvi D. (2008) Tomato ASR1 abrogates the response to abscisic acid and glucose in Arabidopsis by competing with ABI4 for DNA binding. Plant Biotechnol J 6: 368–378 [DOI] [PubMed] [Google Scholar]
- Silhavy D, Hutvágner G, Barta E, Bánfalvi Z. (1995) Isolation and characterization of a water-stress-inducible cDNA clone from Solanum chacoense. Plant Mol Biol 27: 587–595 [DOI] [PubMed] [Google Scholar]
- Stone JM, Liang X, Nekl ER, Stiers JJ. (2005) Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J 41: 744–754 [DOI] [PubMed] [Google Scholar]
- Sugiharto B, Ermawati N, Mori H, Aoki K, Yonekura-Sakakibara K, Yamaya T, Sugiyama T, Sakakibara H. (2002) Identification and characterization of a gene encoding drought-inducible protein localizing in the bundle sheath cell of sugarcane. Plant Cell Physiol 43: 350–354 [DOI] [PubMed] [Google Scholar]
- Sulpice R, Pyl ET, Ishihara H, Trenkamp S, Steinfath M, Witucka-Wall H, Gibon Y, Usadel B, Poree F, Piques MC, et al. (2009) Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci USA 106: 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados L, Savouré A. (2010) Proline: a multifunctional amino acid. Trends Plant Sci 15: 89–97 [DOI] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29: 417–426 [DOI] [PubMed] [Google Scholar]
- Takasaki H, Mahmood T, Matsuoka M, Matsumoto H, Komatsu S. (2008) Identification and characterization of a gibberellin-regulated protein, which is ASR5, in the basal region of rice leaf sheaths. Mol Genet Genomics 279: 359–370 [DOI] [PubMed] [Google Scholar]
- Tardieu F, Reymond M, Hamard P, Granier C, Muller B. (2000) Spatial distributions of expansion rate, cell division rate and cell size in maize leaves: a synthesis of the effects of soil water status, evaporative demand and temperature. J Exp Bot 51: 1505–151411006302 [Google Scholar]
- Trouverie J, Chateau-Joubert S, Thévenot C, Jacquemot MP, Prioul JL. (2004) Regulation of vacuolar invertase by abscisic acid or glucose in leaves and roots from maize plantlets. Planta 219: 894–905 [DOI] [PubMed] [Google Scholar]
- Tuberosa R, Salvi S. (2006) Genomics-based approaches to improve drought tolerance of crops. Trends Plant Sci 11: 405–412 [DOI] [PubMed] [Google Scholar]
- Udomprasert N, Kijjanon J, Thiraporn R, Machuay A. (1999) Effects of water deficit at tasseling on proline and abscisic acid levels and yield of corn. Kasetsart J Nat Sci 33: 310–316 [Google Scholar]
- Urano K, Maruyama K, Ogata Y, Morishita Y, Takeda M, Sakurai N, Suzuki H, Saito K, Shibata D, Kobayashi M, et al. (2009) Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J 57: 1065–1078 [DOI] [PubMed] [Google Scholar]
- Urtasun N, Correa García S, Iusem ND, Bermúdez Moretti M. (2010) Predominantly cytoplasmic localization in yeast of ASR1, a non-receptor transcription factor from plants. Open Biochem J 4: 68–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan R, Kuruvilla S, Thomas G. (1999) Characterization and expression pattern of an abscisic acid and osmotic stress responsive gene from rice. Plant Sci 140: 21–30 [Google Scholar]
- Verdaguer N, Sevilla N, Valero ML, Stuart D, Brocchi E, Andreu D, Giralt E, Domingo E, Mateu MG, Fita I. (1998) A similar pattern of interaction for different antibodies with a major antigenic site of foot-and-mouth disease virus: implications for intratypic antigenic variation. J Virol 72: 739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CS, Liau YE, Huang JC, Wu TD, Su CC, Lin CH. (1998) Characterization of a desiccation-related protein in lily pollen during development and stress. Plant Cell Physiol 39: 1307–1314 [DOI] [PubMed] [Google Scholar]
- Wang HJ, Hsu CM, Jauh GY, Wang CS. (2005) A lily pollen ASR protein localizes to both cytoplasm and nuclei requiring a nuclear localization signal. Physiol Plant 123: 314–320 [Google Scholar]
- Westgate ME, Steudle E. (1985) Water transport in the midrib tissue of maize leaves: direct measurement of the propagation of changes in cell turgor across a plant tissue. Plant Physiol 78: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan JX, Wait R, Berkelman T, Harry RA, Westbrook JA, Wheeler CH, Dunn MJ. (2000) A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometry. Electrophoresis 21: 3666–3672 [DOI] [PubMed] [Google Scholar]
- Yang CY, Chen YC, Jauh GY, Wang CS. (2005) A lily ASR protein involves abscisic acid signaling and confers drought and salt resistance in Arabidopsis. Plant Physiol 139: 836–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY, Wu CH, Jauh GY, Huang JC, Lin CC, Wang CS. (2008) The LLA23 protein translocates into nuclei shortly before desiccation in developing pollen grains and regulates gene expression in Arabidopsis. Protoplasma 233: 241–254 [DOI] [PubMed] [Google Scholar]
- Zivy M. (2007) Quantitative analysis of 2D gels. Methods Mol Biol 355: 175–194 [DOI] [PubMed] [Google Scholar]