Abstract
The phytohormone auxin is a key regulator of plant growth and development that exerts its functions through F-box receptors. Arabidopsis (Arabidopsis thaliana) has four partially redundant of these receptors that comprise the TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX1 auxin receptor (TAAR) clade. Recent studies have shown that the microRNA miR393 regulates the expression of different sets of TAAR genes following pathogen infection or nitrate treatment. Here we report that miR393 helps regulate auxin-related development of leaves. We found that AtMIR393B is the predominant source for miR393 in all aerial organs and that miR393 down-regulates all four TAAR genes by guiding the cleavage of their mRNAs. A mutant unable to produce miR393 shows developmental abnormalities of leaves and cotyledons reminiscent of enhanced auxin perception by TAARs. Interestingly, miR393 initiates the biogenesis of secondary siRNAs from the transcripts of at least two of the four TAAR genes. Our results indicate that these siRNAs, which we call siTAARs, help regulate the expression of TAAR genes as well as several unrelated genes by guiding the cleavage of their mRNAs. Thus, miR393 and possibly siTAARs regulate auxin perception and certain auxin-related aspects of leaf development.
Auxins have a central role in plant growth, development, and responses to the environment. They are perceived by the four partially redundant auxin receptors TRANSPORT INHIBITOR RESPONSE1 (TIR1), AUXIN SIGNALING F-BOX1 (AFB1), AFB2, and AFB3 (Mockaitis and Estelle, 2008). These proteins are members of the TIR1/AFB2 clade of auxin receptors (TAARs) in the AFB family of plant F-box proteins (Mockaitis and Estelle, 2008). TAARs function as a component of SKP/CULLIN/F-BOX-ubiquitin ligase complexes that target members of the AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) transcriptional repressor protein family to proteasome-dependent degradation. Degradation of these AUX/IAA proteins releases the TOPLESS transcriptional corepressor and allows specific AUXIN RESPONSE FACTOR (ARF) transcription factors to act at the promoters of primary auxin-responsive genes to activate their transcription.
The microRNA (miRNA) miR393 has been implicated in down-regulating the expression of TAAR genes in Arabidopsis (Arabidopsis thaliana). In leaves, enhanced innate immunity in response to bacterial infection involves miR393-guided cleavage of TIR1, AFB2, and AFB3 transcripts and repression of AFB1 transcription (Navarro et al., 2006). In roots, response to nitrate involves miR393-guided cleavage of AFB3 mRNAs, but not mRNAs encoded by the other TAAR genes (Vidal et al., 2010).
Here we report that miR393 regulates some auxin-dependent developmental processes. We found that miR393 in aerial parts of the plant is encoded predominantly by AtMIR393B and regulates the expression of all four TAAR genes by guiding the cleavage of their mRNAs. Leaves and cotyledons of mutants unable to produce miR393 exhibit abnormalities expected for enhanced auxin perception by TAARs. Interestingly, unlike most miRNAs, miR393-guided cleavages also lead to the production of detectable amounts of secondary siRNAs from the transcripts of at least two of the four TAAR genes through a pathway similar to the canonical ta-siRNA pathway. We provide evidence that these secondary siRNAs, which we call siTAARs, regulate the expression of all four TAAR genes and several unrelated genes by guiding the cleavage of their mRNAs. Our results show that miR393 regulates auxin-dependent leaf development at the level of auxin perception and that this regulation involves a complex network of siTAARs.
RESULTS
The Gene AtMIR393B Is Required for miR393 Accumulation in Aerial Parts of Arabidopsis Plants
miR393 is potentially encoded by two genes, AtMIR393A (At2g39885) and AtMIR393B (At3g55734; Sunkar and Zhu, 2004). To examine the origin and function of miR393 in development, we characterized a T-DNA insertion mutant of AtMIR393B and verified that homozygous mir393b-1 plants were deficient in producing the corresponding primary transcript (Supplemental Fig. S1, A and B). Wild-type plants accumulated high but variable levels of miR393 in all aerial organs tested and very low levels in roots (Fig. 1A; Supplemental Fig. S1C). The high levels of miR393 accumulation in aerial organs were reduced by up to 100-fold in miR393b-1 mutant; whereas, the low levels in roots were not affected (Fig. 1A; Supplemental Fig. S1C). Thus, miR393 is developmentally regulated and arises primarily from AtMIR393B in aerial organs.
Figure 1.
Expression pattern and developmental roles of miR393. A, RNA-blot hybridization of RNA prepared from roots, leaves, stems, inflorescences (Inflor), and siliques of 50-d-old plants. Probed RNAs are indicated on the left. % Signal, The percent signal detected for mir393b-1 relative to wild-type Col-0 after both are normalized relative to signals for the unrelated miR171; RA, organ-specific accumulation of miR393 relative to leaves is normalized to the ethidium bromide (EtBr) loading standard. B, Top, Typical rosettes of Col-0 and mir393b-1 plants grown in short-day conditions for 28 d. B, Bottom, Abaxial view of leaves from 28-d-old Col-0 and mir393b-1 plants. The mir393b-1 mutant shows a greater number of leaves, more leaf elongation, and more leaf epinasty than wild-type Col-0. Additional views and higher magnification views are shown in Supplemental Figure S2.
The Gene AtMIR393B Is Required for Proper Leaf Development and Auxin-Regulated Cotyledon Epinasty
mir393b-1 plants differed phenotypically from wild type in leaf growth and morphology (Fig. 1B; Supplemental Fig. S2). Table I shows that relative to wild type, mir393b-1 plants had a greater numbers of leaves. These leaves exhibited an elongated, highly epinastic phenotype typical for auxin hypersensitivity (Boerjan et al., 1995), and were prone to early senescence. These results show that miR393, while not crucial for plant development, is required for normal leaf morphology and proper timing of senescence.
Table I. Phenotypic traits of Col-0 and mir393b-1 plants.
| Traits Analyzed | Mean Value ± sem | Pa | |
| Col-0 | mir393b-1 | ||
| 28-d-old plants | |||
| Rosette leaves per plant (n = 9) | 15.9 ± 0.5 | 17.9 ± 0.9 | 0.031 |
| Leaf blade length/width (n = 131) | 1.5 ± 0.03 | 1.6 ± 0.03 | 0.0007 |
| Leaf epinastyb (mm; n = 20) | 1.7 ± 0.1 | 3.4 ± 0.1 | 0.0001 |
| 74-d-old plants | |||
| Rosette leaves per plant (n = 26) | 55.4 ± 8.7 | 60.3 ± 9.6 | 0.029 |
| Senescent leaves per plantc (n = 26) | 3.6 ± 2.0 | 5.5 ± 2.2 | 0.0009 |
One-tailed distribution t test.
Measured by the vertical distance between the adaxial leaf side and the leaf margin.
Diagnosed by the appearance of chlorosis. Senescent leaves appeared more precociously in mir393b-1 plants than in Col-0 plants.
Cotyledon epinasty is a well-documented auxin-hypersensitivity response that depends on TAAR-mediated auxin perception (Hayashi et al., 2009). This prompted us to compare the incidence of extreme cotyledon epinasty of wild-type and mir393b-1 plants. When grown on standard medium, a high and significantly greater fraction of mir393b-1 mutants (79%; Fisher’s exact test, P < 2E-9) than wild-type plants (6.4%) exhibited the extreme cotyledon epinasty phenotype (Fig. 2). Moreover, this incidence was decreased by increasing the concentration of 1-N-naphthylphthalamic acid (NPA), an inhibitor of auxin transport and auxin responses (Fig. 2; Scanlon, 2003). These observations establish that the cotyledon epinasty response to auxin is regulated by miR393.
Figure 2.
Seedlings deficient for TAAR-derived secondary siRNAs exhibit an auxin-hypersensitive response. A, Representative images illustrating the normal range of cotyledon epinasty (left) and the extreme cotyledon epinasty typical of the auxin-hypersensitive response (right; Hayashi et al., 2009). B, The incidence of the auxin-hypersensitive response in populations of Col-0 (black bars), ago7-1 (right cross-hatched bars), mir393b-1 (white bars), and dcl4-2 (gray bars). Seedlings were grown on media containing the concentration of NPA indicated and harvested 4 d after germination. P values (two-sided Fisher’s exact test) for significant differences between pairs are indicated; NS, P > 0.05.
miR393 Guides the Cleavage and Down-Regulation of TAAR mRNAs
Approximately 60% to 70% of the 5′ RNA ligase-mediated (RLM)-RACE sites we detected for each TAAR transcript were at the positions expected for miR393-guided cleavage, showing that all members of the TAAR clade are targets of miR393 (Fig. 3; Supplemental Fig. S3). Further, we compared TAAR mRNA levels in wild type with that of mir393b-1 and dcl1-9 mutants, which are both deficient in miR393 accumulation (Fig. 4, A and C). Figure 5B shows that there was a positively correlated (P < 0.01) up-regulation in both mutants by 1.4- to 2.6-fold in the order TIR1 < AFB2 < AFB3 < AFB1. Together, these results show that miR393, produced by DCL1, is required for the down-regulation of all members of the TAAR clade in leaves.
Figure 3.
Fine mapping of cleavage sites in TAAR transcripts. Shown are the positions of cleavage sites detected in mRNAs encoded by TIR1 (At3g62980), AFB1 (At4g03190), AFB2 (At3g26810), and AFB3 (At1g12820). Exons (boxes), introns (thin lines), untranslated regions (thick lines), and start of transcription (broken arrow) are indicated. T-DNA insertions (white triangles) with their orientation from right to left border (arrowheads) are shown for afb2-3 (SALK_137151) and afb3-5 (SALK_016356) mutants. The cleavage sites identified by 5′ RLM-RACE for each gene are indicated by vertical arrows and the fraction of clones obtained is indicated above. Cleavage sites guided by the miRNA miR393 are in bold. The exact positions of the cleavage sites identified on the sequence of TAAR transcripts are shown in Supplemental Figure S3.
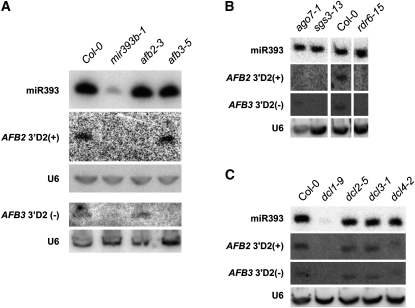
Figure 4.
Genetic requirements for accumulation of AFB2- and AFB3-derived siRNAs. Shown is RNA-blot hybridization using probes for miR393, AFB2 3′D2(+), AFB3 3′D2(−), and U6 RNA as a loading standard. Comparison of Col-0 with mir393b-1, afb2-3, and afb3-5 (A); ago7-1, sgs3-13, and rdr6-15 (B); and dcl1-9, dcl2-5, dcl3-1, and dcl4-2 (C). Similar results were obtained in at least two independent experiments. The differences observed in the ago7-1 sample are due to unequal loading. The ta-siRNAs from three TAS families, the miRNA that initiate their biogenesis, and three additional miRNAs were also probed as controls (Supplemental Figs. S1D and S5).
Figure 5.
Accumulation of TAAR mRNAs in mutants affecting siTAAR production. A, Comparison of AFB2 transcript accumulation in Col-0, mir393b-1, afb2-3, afb3-5, rdr6-14, sgs3-13, and ago7-1 by northern-blot hybridization. Ethidium bromide (EtBr)-stained 25S rRNAs serve as a loading standard. B, Real-time reverse transcription-qPCR of mRNAs encoded by TIR1 (white bars), AFB1 (black bars), AFB2 (horizontal cross-hatched bars), and AFB3 (gray bars) in dcl1-9, dcl4-2, mir393b-1, afb2-3, and afb3-5. mRNA contents are expressed as fold wild type relative to the ACTIN2 standard. Error bars, sem for two or three measurements of total RNA obtained from pools of at least 20 plants; *, significantly different from wild type (P < 0.05, t test of means). The primers used span the miR393 complementary sites.
Small RNAs Specifically Arise from AFB2 and AFB3 mRNAs
Unexpectedly, 30% to 40% of the cleavage sites we identified were not at positions anticipated for miR393 sites. These sites were distributed along the 3′ ends of the transcripts downstream of the miR393 binding sites (Fig. 3; Supplemental Fig. S3). Earlier deep sequencing studies identified low-abundance Arabidopsis sRNAs matching each TAAR mRNAs downstream of their miR393 binding sites that appeared rather imprecisely phased (Supplemental Table S1; Qi et al., 2005; Axtell et al., 2006; Howell et al., 2007). Thus, the cleavage sites we identified by 5′ RLM-RACE suggest that, in addition to miR393, these low-abundant sRNAs might contribute to regulating the expression of TAAR genes.
To examine the expression of sRNA representing each TAAR, we chose those most abundant sRNAs as judged from their reported number of sequencing reads (Qi et al., 2005; Axtell et al., 2006; Howell et al., 2007). The selected sRNAs were named by their origin and phase following the nomenclature of ta-siRNAs (Supplemental Fig. S4A; Allen et al., 2005). Although weakly abundant, AFB2 3′D2(+) (14 reads) and AFB3 3′D2(−) (17 reads) were detected in rosette leaves but not in inflorescences by RNA-blot hybridization (Supplemental Fig. S4B). No signals however were detected for TIR1 3′D9(+) (one read) or for AFB1 3′D14(+) (one read) in either leaves or in flowers (Supplemental Fig. S4B).
We could also identify the source of these TAAR-derived sRNAs in leaves by comparing their accumulation relative to wild type in the two T-DNA insertion mutants, afb2-3, which accumulates 5-fold less AFB2 mRNAs, and afb3-5, which accumulates 3-fold less AFB3 mRNAs relative to wild type (Figs. 4 and 5B). A weak signal for AFB2 3′D2(+) was consistently detected in afb3-5 and wild type but not in afb2-3. Conversely, a very weak signal for AFB3 3′D2(−) was consistently detected in afb2-3 and wild type, but not in afb3-5 (Fig. 4A). RNA-blot hybridization of miRNAs and ta-siRNAs unrelated to TAAR sRNAs gave similar signal strengths in the two deficiency mutants and in wild type (Supplemental Fig. S1D). Thus, the AFB2 3′D2(+) and AFB3 3′D2(−) signals we detected are specifically derived from AFB2 and AFB3 mRNAs, respectively.
siTAARs Are Secondary siRNAs Whose Biogenesis Depend on miR393
Arabidopsis ta-siRNAs are secondary siRNAs arising from noncoding transcripts that depend on DCL1, DCL4, RDR6, and SGS3 proteins for their biogenesis (Voinnet, 2009; Vazquez et al., 2010). No accumulation of AFB2 3′D2(+) and AFB3 3′D2(−) was detectable in the deficiency mutants rdr6-15 and sgs3-13; whereas, their accumulation in the deficiency mutants rdr1-1, rdr2-2, rdr3a-2, rdr3b-2, and rdr3c-1 was comparable to that of wild type (Fig. 4B; Supplemental Fig. S5). Thus, SGS3 and RDR6, but not the other members of the RDR family, are required for the accumulation of TAAR-derived sRNAs. Like the known ta-siRNAs from three different TAS families, the accumulation of TAAR sRNAs was blocked in dcl1-9, was shifted from the 21- to the 22-nt form in dcl4-2, and was unaffected in dcl2-5 and dcl3-1 mutants (Fig. 4, B and C; Supplemental Fig. S5). Of the known ta-siRNAs, only those derived from TAS3 require AGO7 for their biogenesis (Peragine et al., 2004; Vazquez et al., 2004b; Yoshikawa et al., 2005; Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006). In an experiment using both types of ta-siRNA as controls, we consistently found weak signals for AFB2 3′D2(+) and AFB3 3′D2(−) in the ago7-1 deficiency mutant (Fig. 4B; Supplemental Fig. S5). Therefore, production of TAAR sRNAs does not fully require AGO7.
ta-siRNAs depend on miRNA-guided cleavage to initiate their biogenesis and set their phasing (Allen et al., 2005). Recent studies have shown that the 22-nt rather than the 21-nt forms of miRNAs are important for the initiation of siRNA production from single-hit target transcripts (Chen et al., 2010; Cuperus et al., 2010). Because miR393 is 22-nt long, we tested whether the accumulation of TAAR sRNAs depended on miR393. Neither AFB2 3′D2(+) or AFB3 3′D2(−) were detected in the mir393b-1 mutant (Fig. 4A), showing that their biogenesis depends on the function of miR393. These findings, the phased arrangement of TAAR sRNAs downstream of the miR393 binding site (Qi et al., 2005; Axtell et al., 2006; Howell et al., 2007), and the requirement for RDR6, SGS3, DCL1, and DCL4, but not AGO7, for their biogenesis show that these TAAR sRNAs, which we refer to as siTAARs, are secondary siRNAs generated from coding transcripts using components of the canonical ta-siRNA pathway.
siTAARs Guide the Cleavage and Down-Regulation of TAAR mRNAs
Several predicted siTAAR target sites are shared among the different TAAR transcripts, suggesting that siTAARs might target both the mRNAs from which they were derived as well as from mRNAs encoded by different members of the TAAR clade. If siTAARs act in trans, then mutants deficient in production of sRNAs for one member of the clade should exhibit increased mRNA levels of the other members. We tested this hypothesis by comparing the accumulation of the four TAAR mRNAs in wild type and in mutants deficient in the production of either AFB2- or AFB3-derived siRNAs, and, using the dcl4-2 mutant as a positive control. As predicted by our hypothesis, the accumulation of AFB1 and AFB3 mRNAs relative to wild type was increased 1.5-fold in afb2-3; and, accumulation of AFB1 and AFB2 mRNAs was increased 1.5- and 2.3-fold in afb3-5 (Fig. 5B). Moreover, these increases were similar to that observed in the dcl4-2 mutant. Interestingly, the accumulation of TIR1 mRNAs was not affected in any of these mutants.
Although we cannot rule out the contribution of indirect transcriptional effects, the finding of siTAAR-guided cleavage sites in all TAAR mRNAs, the up-regulation of TAAR transcripts in dcl4-2, and the similar levels of up-regulation of TAAR transcripts in mutants deficient in sources of siTAARs are consistent with the hypothesis that siTAARs are involved in both autoregulation and transregulation of TAAR mRNAs. This conclusion and the genetic requirement for biogenesis of siTAARs were confirmed by the overaccumulation of AFB2 transcripts, the only TAAR transcripts detectable by northern-blot analysis, in afb3-5, rdr6-14, sgs3-13, but not in ago7-1 mutants (Fig. 5A). Interestingly, TIR1 mRNA levels were not increased either in the secondary siRNA-deficient dcl4-2 mutants or in the siTAAR-deficient mutants (Fig. 5B). Nevertheless, the cleavage sites we identified in the TIR1 mRNA are indicative of siRNA regulation, implying that miR393-guided cleavage is sufficient for proper regulation of TIR1 mRNAs in the siTAAR-deficient backgrounds. We cannot, however, rule out the possibility that siTAAR-guided cleavages also contribute to TIR1 mRNA regulation in specific tissues or cell types that might not be detected by our analyses done at the whole organ level.
siTAARs Also Regulate Targets Unrelated to the TAAR Clade
We identified additional siTAAR targets by screening the genome-scale parallel analysis of RNA ends (PARE) dataset comprising sRNAs and their guided cleavage sites in target RNAs (German et al., 2008). Two genes in this dataset had matches exactly at the positions expected for guided cleavage by siTAARs, namely, At5g24650 targeted by AFB2 3′D1(+)/AFB3 3′D1(+) and At3g18850, encoding the putative lysophosphatidyl acyltransferase LPAT5, targeted by AFB2 3′D6(+)/AFB3 3′D6(+) (Fig. 6A). Relative to wild type, the mRNA levels of these putative targets were 1.7- to 3.9-fold higher in dcl1-9 and mir393b-1 mutants deficient in miR393 accumulation and 1.3- to 2.4-fold higher in dcl4-2, afb2-3, and afb3-5 mutants deficient in siTAARs accumulation (Fig. 6B). These results together with the positions of the cleavage sites identified by PARE support the hypothesis that siTAARs can target genes outside of the TAAR clade.
Figure 6.
Regulation in trans of mRNAs unrelated to TAAR mRNAs by siTAARs. A, Sites in the coding regions of At5g24650 and LPAT5 (At3g18850) for cleavage guided by the siTAARs AFB2 3′D1(+), AFB3 3′D1(+), AFB3 3′D6(+), and AFB2 3′D6(+). Perfect (I) and wooble (o) base pairing are indicated. The cleavage sites (vertical arrows) and number of sequence reads obtained in the PARE dataset by German et al. (2008) are indicated. The ΔG values for target-sRNA pairs were calculated using the DINAMelt server. B, Real-time reverse transcription-qPCR of At5g24650 (white bars) and LPAT5 (black bars) mRNAs in mutants affected in siTAAR production. mRNA contents are expressed as fold wild type relative to the ACTIN2 standard. Error bars, sem for two or three measurements of total RNA obtained from pools of at least 20 plants; *, significantly different from wild type (P < 0.05, t test of means). The primers used span the siTAAR complementary sites.
siTAARs Are Likely Contributors to Auxin-Related Developmental Regulation
We examined the possibility that siTAARs contribute to miR393-mediated developmental regulation by analyzing the degree of cotyledon epinasty of dcl4-2 mutants known to be deficient in secondary siRNA production (Fig. 2; Gasciolli et al., 2005; Yoshikawa et al., 2005; Howell et al., 2007). The fraction of dcl4-2 plants (64%) showing the extreme cotyledon epinasty expected for hypersensitive responses to auxin was significantly greater (Fisher’s exact test, P < 5E-7) than that of wild type. In contrast, the ago7-1 mutant, which is deficient in ta-siARFs but not in siTAARs production, exhibited the same low incidence of epinasty as wild type. Although indirect, these important results and the fact that the dcl4-2 and mir393b-1 mutants show comparable, large increases in incidence of cotyledon epinasty, which is blocked by NPA in both cases, support the role of siTAARs in miR393-mediated regulation.
DISCUSSION
Our studies establish that miR393 is required to regulate the expression of all four TAAR genes, auxin perception homeostasis, and some aspects of auxin-dependent plant development. The mir393b-1 mutants exhibit only mild developmental defects; thus, we speculate that other pathways such as the ta-siARF or the ASYMMETRIC LEAF pathways (Adenot et al., 2006; Garcia et al., 2006; Xu et al., 2006) might act redundantly with miR393 to specify leaf development. Alternatively, the remaining traces of miR393 generated from AtMIR393A might be sufficient for proper plant development.
Our data show that miR393 generated from AtMIR393B is necessary to trigger the production of the siTAARs secondary siRNAs from at least two of the TAAR transcripts. Our data support the working hypothesis that both miR393 and siTAARs contribute to regulate the expression of TAAR genes and to regulate the auxin-related forms of leaf development we have examined (Fig. 7). The regulatory model we propose is based on three lines of evidence; namely, RACE identification of target cleavage sites; direct measurement of target mRNA levels in informative mutants; and, target predictions for known sRNAs in public deep-sequencing datasets. We show that miR393 targets all members of the TAAR family in Arabidopsis leaves. These targets, in turn, generate specific patterns of secondary siRNAs that act in a cascade on other TAAR mRNAs. Although the evidences for the function of siTAARs in the regulation of TAAR genes’ expression are rather indirect, their function also explains the paradoxical observation that artificial TAAR:GUS targets disrupted in their miR393 target site still show normal posttranscriptional regulation (Parry et al., 2009). Our model predicts that siTAARs generated from intact, endogenous TAAR mRNAs should regulate these artificial targets normally.
Figure 7.
Model for functions of siTAARs in auxin-mediated regulation. Shown are proposed sources and targets for siRNAs in the TAAR clade. Interaction of miR393 with cognate TAAR mRNA targets and initiates production of secondary siRNAs from the 3′ fragments of cleaved TAAR mRNAs. These siRNAs, in turn, target other TAAR mRNAs. The evidence for the pathways is indicated: Superscript 1, Identification of sRNAs by deep sequencing and in silico target prediction; superscript 2, RLM-RACE verification of target cleavage and measurements of target mRNA levels in informative mutants affected in the biogenesis of siTAARs; superscript 3, both 1 and 2. Note that only few TIR1- and AFB1-derived sRNAs have been identified and that assay of their transregulation will require identification of appropriate siRNA-deficient mutants.
Several of the regulatory links within the siTAAR pathway remain to be established. One problem is that siTAARs are of extremely low abundance and several may have escaped detection by deep sequencing. Another problem is that mutants lacking TIR1- or AFB1-siRNAs are not available and, thus, it was not yet possible to elucidate connections to the TIR1 and AFB1 nodes. It is also unclear if the 22-nt form of miR393 pairs preferentially to AFB2 and AFB3 mRNAs to trigger siTAAR biogenesis more efficiently than for TIR1 and AFB1 mRNAs. Finally, we have not as yet been able to distinguish between a shared network functioning in most cells in the tissues sampled and the distribution of subcomponents of the network in specific cell types.
Studies with single- and high-order TAAR mutants and expression studies have shown that TAAR family members have overlapping, partially divergent functions in auxin homeostasis and plant development as well as specific expression patterns (Dharmasiri et al., 2005; Parry et al., 2009). This suggests that the siTAAR network might have evolved to supplement, amplify, and stabilize the coordinated down-regulation by miR393 to ensure that proper TAAR steady-state levels and auxin signaling homeostasis are maintained throughout plant development. In Arabidopsis, TIR1 and AFB1 are paralogs that appear to have functionally diverged from AFB2 and AFB3 (Dharmasiri et al., 2005). It is tempting to speculate that the siTAAR network specific to the AFB2 and AFB3 node has evolved to ensure proper homeostasis of auxin perception independently from TIR1 and AFB1 (Fig. 7).
Another possible function is suggested by analogy to the ta-siARF network. Our comparison of leaf and cotyledon epinasty has shown that siTAARs and ta-siARFs have distinct as well as overlapping auxin-dependent functions. ta-siARFs act in a noncell autonomous fashion to regulate ARF2-, ARF3-, and ARF4-mRNAs, and, hence, abaxial leaf identity and juvenility (Chitwood et al., 2009; Schwab et al., 2009; Pulido and Laufs, 2010). We speculate that siTAARs might also act noncell autonomously, but upstream of ARF2/3/4, at the level of auxin perception. Finally, we found that siTAARs can target genes outside of the TAAR clade. Our database searches identified 314 additional potential targets for the 75 unique siTAARs (Supplemental Table S1). Relative to the Arabidopsis genome, this dataset was significantly enriched (Fisher’s exact test, P = 1.9E-3) by 2.5-fold in genes representing auxin-related GO terms. These findings raise the intriguing possibility that siTAARs have a more general function in coordinating a wide range of auxin-related processes.
CONCLUSION
In conclusion, miR393 regulates auxin signaling and auxin-mediated functions in Arabidopsis at several levels. We show that AtMIR393B is developmentally regulated: it is the major source of miR393 in aerial parts of the plant and expressed weakly or not at all in roots. Thus, there are at least three classes of TAAR gene regulation: (1) cleavage of all four TAAR mRNAs in developing leaves guided by miR393 originating from AtMIR393B; (2) cleavage of TIR1, AFB2, and AFB3 mRNAs and transcriptional regulation of AFB1 in bacteria-infected leaves guided by miR393 originating from AtMIR393A (Navarro et al., 2006); and, (3) miR393-guided cleavage of only AFB3 mRNAs in the response of roots to nitrate (Vidal et al., 2010). The siTAAR network we identified could help coordinate the expression of the TAAR gene family and might even act downstream of auxin perception to regulate auxin-dependent gene expression.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Unless stated otherwise, Arabidopsis (Arabidopsis thaliana) plants were grown in a greenhouse as described previously (Vazquez et al., 2008). Mutants used in this study are all in the Columbia-0 (Col-0) ecotype. The T-DNA insertion mutants dcl2-5 (SALK_123586), dcl3-1 (SALK_005512), dcl4-2 (GK-160G05), ago7-1 (SALK_037458), sgs3-13 (SALK_N039005), rdr1-1 (SAIL_672_F11), rdr2-2 (SALK_059661), and rdr6-15 (SAIL_617_H07) have been described previously (Peragine et al., 2004; Xie et al., 2004; Allen et al., 2005; Vazquez et al., 2008) and were all identified independently from the SALK collection. The T-DNA mutant dcl1-9 (N3828) in the Landsberg erecta background was introgressed into Col-0 by five successive backcrosses. To obtain homozygous lines of the mutants rdr3a-1 (SALK_036925), rdr3b-1 (SALK_143708), rdr3c-1 (SALK_008516), afb2-3 (SALK_137151), afb3-5 (SALK_016356), and mir393b-1 (SALK_018966), the presence of the T-DNA insert was determined by PCR using the primer LBb1 located at the left border of the T-DNA and gene-specific primers. Homozygous plants were then identified by using the gene-specific primers and primers located on the other side of the T-DNA insertion. The primers used are listed in Supplemental Table S2. Homozygosis for the T-DNA insert was confirmed for at least eight progeny.
Leaf and Growth Measurements
Plants were grown in soil in the greenhouse. For growth measurements, plants were raised in a Sanyo growth chamber (Brouwer AG) under short-day conditions (10 h light 21°C/14 h dark 18°C). For studies of cotyledon epinasty, seeds were surface sterilized for 10 min in 1.5% (w/v) sodium hypochlorite/70% (v/v) ethanol, rinsed three times with 100% (w/v) ethanol, air dried overnight in flow hood, and then sown on Murashige and Skoog solid medium supplemented as indicated with NPA (Fluka) in dimethyl sulfoxide or dimethyl sulfoxide alone. The seeds were stratified for 4 d at 4°C, transferred to short-days conditions, and 26 to 40 plants were scored for cotyledon epinasty 4 d later as described (Hayashi et al., 2009).
RNA Preparation and RNA Analysis
Unless stated otherwise, total RNAs were extracted from rosette leaves of 3-week-old plants using Trizol. Conventional northern blots were prepared by blotting 15 μg of total RNAs onto Hybond N+ (Amersham) membranes. Probes were prepared by random priming of PCR products using the Radprime kit (Invitrogen). Purification of sRNAs, preparation of sRNA blots with 20 μg of sRNAs (100 μg for the bottom section of Fig. 4A), labeling of oligonucleotide probes, and hybridizations of blots were done as described previously (Vazquez et al., 2008). The sequences of probes and primers are listed in Supplemental Table S2.
Modified 5′ RLM-RACE to Determine Cleavage Sites
Modified 5′ RLM-RACE was performed as described previously (Vazquez et al., 2004a). Two rounds of amplifications of TIR1, AFB1, AFB2, and AFB3 cDNAs were made using the GeneRacer 5′ primer and nested gene-specific primers. Amplification products without size selection on gel were cloned randomly, using a TOPO TA cloning kit (Invitrogen). Forty to 42 independent clones were sequenced for each gene.
Real-Time Quantitative Reverse Transcription-PCR
Synthesis of cDNAs was done from 2 μg total RNAs using the Superscript III cDNA synthesis kit (Invitrogen). Real-time quantitative (q)PCR were performed using power SYBR green PCR master mix (ABI) and analyzed as previously described (Vaucheret et al., 2004).The primer pairs used are listed in Supplemental Table S2. The mean from triplicate determinations of each transcript level was normalized to the corresponding mean for ACTIN2. For semiquantitative final-point PCR measurements of pri-miR393B and ACTIN2, PCR reactions were performed under standard PCR conditions using the primer pairs listed in Supplemental Table S2.
Bioinformatic Prediction of Small RNA Targets
siTAARs identified in public sRNA datasets (Qi et al., 2005; Axtell et al., 2006; Howell et al., 2007) were used for analysis with psRNATarget Web server against TAIR7 library (http://bioinfo3.noble.org/miRU2/) with default settings (Zhang, 2005).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. mir393b-1, afb2-3, and afb3-5 mutants accumulate normal levels of unrelated miRNAs and ta-siRNAs.
Supplemental Figure S2. Abnormalities in the development of mir393b-1 leaves.
Supplemental Figure S3. Fine mapping of cleavage sites in TAAR transcripts.
Supplemental Figure S4. Phased sRNAs potentially deriving from AFB2 and AFB3 mRNAs are detected in Arabidopsis leaf tissue sample.
Supplemental Figure S5. Genetic requirements for accumulation of trans-acting siRNAs and of the microRNA that initiates their biogenesis.
Supplemental Figure Table S1. siTAARs in public data sets and their predicted targets.
Supplemental Figure Table S2. Sequences of oligonucleotides and primers used in this study.
Acknowledgments
We thank Thomas Boller for critical comments, and Christian Körner, Andres Wiemken, and Thomas Boller for providing laboratory space at the Botanical Institute of the University of Basel. Author contributions: A.S.-A. and F.V. designed research; A.S.-A., D.W., E.A.-B., and F.V. performed research; C.K. and J.A. contributed analytic tools; A.S.-A., D.W., F.M., and F.V. analyzed data; F.M. and F.V. wrote the article.
References
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouché N, Gasciolli V, Vaucheret H. (2006) DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol 16: 927–932 [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. (2006) A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Van Montagu M, Inzé D. (1995) Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7: 1405–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH. (2010) 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA 107: 15269–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Nogueira FT, Howell MD, Montgomery TA, Carrington JC, Timmermans MC. (2009) Pattern formation via small RNA mobility. Genes Dev 23: 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC. (2010) Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol 17: 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16: 939–944 [DOI] [PubMed] [Google Scholar]
- Garcia D, Collier SA, Byrne ME, Martienssen RA. (2006) Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol 16: 933–938 [DOI] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. (2005) Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 15: 1494–1500 [DOI] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26: 941–946 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Kamio S, Oono Y, Townsend LB, Nozaki H. (2009) Toyocamycin specifically inhibits auxin signaling mediated by SCFTIR1 pathway. Phytochemistry 70: 190–197 [DOI] [PubMed] [Google Scholar]
- Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC. (2007) Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M. (2008) Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24: 55–80 [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M. (2009) Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci USA 106: 22540–22545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido A, Laufs P. (2010) Co-ordination of developmental processes by small RNAs during leaf development. J Exp Bot 61: 1277–1291 [DOI] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ. (2005) Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19: 421–428 [DOI] [PubMed] [Google Scholar]
- Scanlon MJ. (2003) The polar auxin transport inhibitor N-1-naphthylphthalamic acid disrupts leaf initiation, KNOX protein regulation, and formation of leaf margins in maize. Plant Physiol 133: 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Maizel A, Ruiz-Ferrer V, Garcia D, Bayer M, Crespi M, Voinnet O, Martienssen RA. (2009) Endogenous TasiRNAs mediate non-cell autonomous effects on gene regulation in Arabidopsis thaliana. PLoS ONE 4: e5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK. (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crété P, Bartel DP. (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Blevins T, Ailhas J, Boller T, Meins F., Jr (2008) Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res 36: 6429–6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crété P, Vaucheret H. (2004a) The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14: 346–351 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Legrand S, Windels D. (2010) The biosynthetic pathways and biological scopes of plant small RNAs. Trends Plant Sci 15: 337–345 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crété P. (2004b) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16: 69–79 [DOI] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutierrez RA. (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Yang L, Pi L, Liu Q, Ling Q, Wang H, Poethig RS, Huang H. (2006) Genetic interaction between the AS1-AS2 and RDR6-SGS3-AGO7 pathways for leaf morphogenesis. Plant Cell Physiol 47: 853–863 [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2005) miRU: an automated plant miRNA target prediction server. Nucleic Acids Res 33: W701–W704 [DOI] [PMC free article] [PubMed] [Google Scholar]