Abstract
Phytoplasmas are insect-transmitted bacterial plant pathogens that cause considerable damage to a diverse range of agricultural crops globally. Symptoms induced in infected plants suggest that these phytopathogens may modulate developmental processes within the plant host. We report herein that Aster Yellows phytoplasma strain Witches’ Broom (AY-WB) readily infects the model plant Arabidopsis (Arabidopsis thaliana) ecotype Columbia, inducing symptoms that are characteristic of phytoplasma infection, such as the production of green leaf-like flowers (virescence and phyllody) and increased formation of stems and branches (witches’ broom). We found that the majority of genes encoding secreted AY-WB proteins (SAPs), which are candidate effector proteins, are expressed in Arabidopsis and the AY-WB insect vector Macrosteles quadrilineatus (Hemiptera; Cicadellidae). To identify which of these effector proteins induce symptoms of phyllody and virescence, we individually expressed the effector genes in Arabidopsis. From this screen, we have identified a novel AY-WB effector protein, SAP54, that alters floral development, resulting in the production of leaf-like flowers that are similar to those produced by plants infected with this phytoplasma. This study offers novel insight into the effector profile of an insect-transmitted plant pathogen and reports to our knowledge the first example of a microbial pathogen effector protein that targets flower development in a host.
Phytoplasmas are insect-vectored plant pathogens that infect a range of high-value agricultural crops, including coconut (Cocos nucifera; Bonnot et al., 2010), grapevine (Vitis vinifera; Constable, 2009), apple (Malus domestica; Seemüller et al., 2010), maize (Zea mays; Jović et al., 2009), oilseed rape (Brassica napus; Wang and Hiruki, 2001), carrot (Daucus carota), cabbage (Brassica sp.), onion (Allium cepa), and other vegetables (Lee et al., 2003). These bacterial pathogens are members of the class Mollicutes, which have diverged from the gram-positive Firmicutes through the loss of a cell wall and a reduction in genome size (Hogenhout et al., 2008). Phytoplasmas are obligate parasites that survive and replicate intracellularly within both insect and plant hosts. Within a plant, phytoplasmas inhabit the phloem and are injected directly into the cytoplasm of phloem sieve cells via the feeding activity of an insect vector. Insects capable of transmitting phytoplasmas comprise members of the order Hemiptera, including sap-sucking leafhoppers, planthoppers, and psyllids.
Phytoplasmas have a plant host range that is in part dependent upon the feeding range of their insect vectors (Hogenhout et al., 2008). Aster Yellows phytoplasma strain Witches’ Broom (AY-WB; Candidatus Phytoplasma asteris) is vectored by the polyphagous aster leafhopper Macrosteles quadrilineatus, which readily transmits this pathogen to a wide range of plants, including members of the Solanaceae and Brassicaceae families (Sugio et al., 2011). Phytoplasmas such as AY-WB induce a variety of symptoms in plants that are indicative of an abnormal development of host tissues, including the formation of witches’ broom (a dense mass of shoots originating from a single point), phyllody (conversion of floral organs into leaves), virescence (green pigmentation of tissues such as flower petals), and bolting (growth of elongated stalks). We hypothesized that phytoplasmas secrete virulence proteins (effectors) to induce such alterations in phenotype of host plants (Hogenhout et al., 2008; Bai et al., 2009; Sugio et al., 2011).
Plant pathogens typically employ a range of effectors that enable these organisms to manipulate developmental processes within the host to the benefit of the pathogen (Hogenhout et al., 2009; Busch and Benfey, 2010). Xanthomonas species produce transcription activator-like effectors (such as AvrBs3) that modulate the expression of target plant genes to induce mesophyll cell hypertrophy in susceptible hosts, a process that is proposed to facilitate dispersal of the pathogen (Marois et al., 2002). The actinomycete Rhodococcus fascians secretes phytohormones (cytokinins) that induce ectopic expression of KNOTTED-like homeobox genes in Arabidopsis (Arabidopsis thaliana), thereby maintaining infected tissues in a meristematic state that provides beneficial metabolites to the pathogen (Depuydt et al., 2008). Plant parasitic nematodes secrete effectors into the host root, including CLAVATA3 (CLV3)/embryo surrounding region-like effector proteins that function as ligand mimics. These effectors interact with the CLV2/CORYNE receptor complex, an interaction that is important to the successful formation of nematode-induced syncytia, which act as feeding sites from which the parasites obtain nutrition (Replogle et al., 2011). However, effectors that modulate flower development have not yet been identified.
Gram-negative plant pathogens often employ a complex secretory apparatus (type three secretion systems) to introduce effector proteins into host cells to manipulate cellular processes to the benefit of the pathogen (Ghosh, 2004). As phytoplasmas inhabit the cytoplasm of the immature and mature sieve cells that constitute the phloem, these bacteria are intracellular parasites and hence do not require an elaborate secretion system for delivering effectors across the plant cell membranes. Rather, phytoplasmas secrete effectors directly into the host cytoplasm of sieve cells via the Sec-dependent protein translocation pathway, and the effectors then unload from the phloem to target other plant cells by symplastic transport (Bai et al., 2009; Hoshi et al., 2009; Sugio et al., 2011). Putative secreted proteins can be identified by the presence of a signal peptide (Bendtsen et al., 2004) that is cleaved during export to yield a mature protein (Kakizawa et al., 2004). Mining of the AY-WB genome sequence resulted in the identification of 56 secreted AY-WB proteins (SAPs) that we predict to be secreted into the host cytoplasm via the AY-WB Sec-dependent pathway (Bai et al., 2009). These secreted proteins are candidate effectors, as they are likely to interact with host components in the plant or insect cells.
A couple of phytoplasma effectors have recently been functionally characterized. SAP11 is one of the 56 AY-WB effectors, and it was shown to possess a bipartite nuclear localization signal that is involved in the targeting of SAP11 to plant cell nuclei in AY-WB-infected plants (Bai et al., 2009). More recent work provides evidence that SAP11 destabilizes class II CINCINNATA-related TCP (for TEOSINTE BRANCHED1, CYCLOIDEA, PCF) transcription factors, resulting in crinkled leaf and witches’ broom phenotypes and the down-regulation of LIPOXYGENASE2 expression and jasmonic acid production, which are involved in the Arabidopsis defense response to the principal AY-WB insect vector M. quadrilineatus (Sugio et al., 2011). An effector (Tengu) identified from Candidatus Phytoplasma asteris Onion Yellows phytoplasma strain M (OY-M) also induces developmental phenotypes resembling the witches’ broom symptoms of infected plants and is thought to interfere with plant auxin biosynthesis or signaling pathways (Hoshi et al., 2009).
The development of flowers follows a strict regulatory program in Arabidopsis and involves the “ABC model” for floral organ formation (Coen and Meyerowitz, 1991; Causier et al., 2010). This model proposes that flowers are formed via the activity of three classes of functions (i.e. A, B, and C functions), involving primarily members of the MADS box proteins, which act to confer the identity of floral organs. Summarized briefly, flowering signals in the Arabidopsis photoperiodic pathway converge with the floral meristem identity genes APETALA1 (AP1)/CAULIFLOWER and LEAFY, which encode transcription factors that act to establish a floral meristem. These transcription factors activate the expression of floral organ identity genes, which include AP2 and MADS box domain transcription factors that activate downstream pathways to form sepals, petals, stamens, and carpels. Organ identity specification is further conferred to these organs by E-class proteins (SEP proteins), which serve as bridging proteins that facilitate protein-protein interactions among A-, B-, and C-class members (Immink et al., 2010). AGAMOUS is a C-class protein that is involved in stamen and carpel development and in the termination of flower development. Various genes involved in floral organ formation are misregulated in phytoplasma-infected plants (Pracros et al., 2006; Cettul and Firrao, 2010; Himeno et al., 2011). We hypothesized that phytoplasmas produce specific effectors that interfere with floral organ formation (Sugio et al., 2011).
In this work, we report the results of a comprehensive screen to identify an AY-WB effector that induces the formation of the leaf-like flowers that are associated with many phytoplasma strains. We detected transcripts for the majority of the 56 AY-WB candidate effector genes, of which 20 genes are more highly expressed in Arabidopsis and 16 genes are more highly expressed in the insect vector M. quadrilineatus. We screened the T1 generation of stable transgenic Arabidopsis lines expressing 52 effector genes and identified SAP54, which modifies flower architecture in Arabidopsis. This paper illustrates how genome sequence data of uncultivable organisms such as phytoplasmas can be explored to identify and functionally characterize pathogen effector proteins.
RESULTS
AY-WB Infection of Arabidopsis Induces an Alteration of Host Phenotype
To better define the symptoms that AY-WB induces in Arabidopsis, we exposed 4-week-old Arabidopsis ecotype Columbia (Col-0) plants to noncarrier (healthy control) and AY-WB-carrying M. quadrilineatus to allow inoculation of the bacterial pathogen. Plants were grown under short-day (SD) or long-day (LD) photoperiods. In both SD and LD, early symptoms of infection were observed at approximately 14 d post inoculation (dpi), as indicated by an accumulation of anthocyanins spreading outward from the midvein of rosette leaves in infected plants (Supplemental Fig. S1). As this pigmentation was rarely observed in healthy plants, it is likely related to AY-WB infection. At 3 weeks post inoculation, plants showed evidence of yellowing within the center of the rosette. The infected plants showed several signs of hyponastic growth, such as low leaf angles, long petioles, pale green leaf colors, and narrow leaf blades (Supplemental Fig. S2). At later stages of infection, the growth of plants was also significantly impacted by AY-WB (Table I), with healthy plants attaining a greater mass and height in both SD and LD. Infected plants showed statistically significant differences in rosette diameter (Table I; Supplemental Fig. S3) and leaf length and width (Table I; Supplemental Fig. S1). Symptoms were visually more pronounced in plants grown in SD (Supplemental Fig. S3), and these plants did not bolt and failed to form flowers, whereas healthy plants bolted and flowered (greater than 1 cm bolt, 37.1 ± 0.8 dpi, SD photoperiod). Infected plants did bolt at LD but remained shorter than healthy plants (Fig. 1A; Supplemental Fig. S4). The formation of witches’ broom (an increased production of axillary stems) is a characteristic of AY-WB infection (Zhang et al., 2004), and infected Arabidopsis plants exhibited a similar phenotype (Fig. 1, B and C). We found that infected plants produce a significantly greater number of axillary stems at 3 and 4 weeks post inoculation; however, the number of stems did not differ significantly between healthy and infected plants at 5 weeks post inoculation (Supplemental Fig. S5). Furthermore, we noted that infected plants exhibited considerable variation in branch number between individuals (range of three to 14 branches originating from the main bolt), whereas healthy plants showed little variation (range of three to six branches).
Table I. Phenotypic analysis of AY-WB-infected Arabidopsis.
Values shown are means ± se; n ≥ 10 plants per treatment. All measurements were obtained 44 and 54 dpi for SD and LD, respectively, unless otherwise indicated. Differences between healthy and infected plants are statistically significant if indicated by one (P < 0.05) or two (P < 0.0001) asterisks, as determined by Student’s t test. Infected plants did not bolt in SD; thus, these characteristics were not assessed. NA, Not applicable.
| SD |
LD |
|||
| Measurement | Healthy | Infected | Healthy | Infected |
| Fresh shoot mass (g)a | 11.93 ± 0.98 | 0.84 ± 0.05** | 8.96 ± 0.65 | 0.63 ± 0.09** |
| Height (cm) | NA | NA | 36.50 ± 0.80 | 8.50 ± 1.16** |
| Rosette diameter (cm) | 17.20 ± 0.68 | 8.64 ± 0.34** | 12.47 ± 0.19 | 11.17 ± 0.54* |
| Length of rosette leaf (cm)b | 8.87 ± 0.20 | 4.40 ± 0.10** | 6.32 ± 0.10 | 5.70 ± 0.16* |
| Width of rosette leaf (cm)b | 2.93 ± 0.06 | 1.03 ± 0.02** | 2.02 ± 0.04 | 1.08 ± 0.03** |
| No. of axillary shootsc | NA | NA | 9.00 ± 0.42 | 10.53 ± 0.86 |
| No. of branches from main stemc | NA | NA | 4.35 ± 0.17 | 6.60 ± 0.72* |
| Onset of bolting (>1 cm bolt; dpi) | NA | NA | 20.15 ± 0.47 | 20.23 ± 0.34 |
Plant biomass includes all aboveground tissues (i.e. the rosette and inflorescence).
The length and width of the three largest rosette leaves were measured for each plant (n = 30).
Measurements were obtained at 35 dpi.
Figure 1.
Phytoplasma AY-WB alters the phenotype of infected Arabidopsis. A, Healthy (left) and infected (right) plants. Infected plants show evidence of stunted growth (reduced height) and witches’ broom phenotype (B and C). B, Close-up of the healthy plant depicted in A. C, Close-up of the infected plant depicted in A shows an increased number of axillary stems. Bars = 1 cm. D and F, Images of flowers from healthy Arabidopsis plants show white petals. E and G, Flowers from infected plants show evidence of virescence (green petals and stamens) and phyllody (leaf-like sepals and petals). Note the increased flower size of the infected plants. Bars = 1 mm.
While the onset of flowering did not differ between AY-WB-infected and healthy plants when grown in LD (Table I), infected plants exclusively produced green flowers (Fig. 1, E and G). In healthy Arabidopsis, petals never contain photosynthetic pigments and the floral organs do not produce trichomes on either petals or carpels; the sepals of healthy plants produce primarily simple (unbranched) trichomes. In contrast, in the majority of infected plants, one or several large green flowers would emerge from the main stem (Supplemental Fig. S6). The sepals and petals of these “leaf-like” flowers were green and contained branched (stellate) trichomes (Supplemental Fig. S6); we also observed abundant stellate trichomes upon the carpels of infected plants. The stamens of infected plants were virescent, appeared to be shorter than those of healthy plants, and did not produce pollen (Supplemental Fig. S6). In many flowers, one or more vegetative shoots would emerge from the center of the green flowers in lieu of carpels (Supplemental Fig. S6), consistent with a loss of floral meristem determinacy. None of the flowers formed from infected plants produced siliques prior to plant death; thus, infected plants grown in SD (which failed to bolt) and LD (which failed to form siliques) were sterile.
AY-WB Candidate Effectors Are Expressed in Insect and Plant Hosts
We previously identified 56 candidate effector genes using a bioinformatics approach (Bai et al., 2009). However, it has not been investigated whether these are genuine genes that are transcribed by AY-WB in vivo and whether gene expression is influenced by host background. Hence, we analyzed their expression in Arabidopsis and M. quadrilineatus.
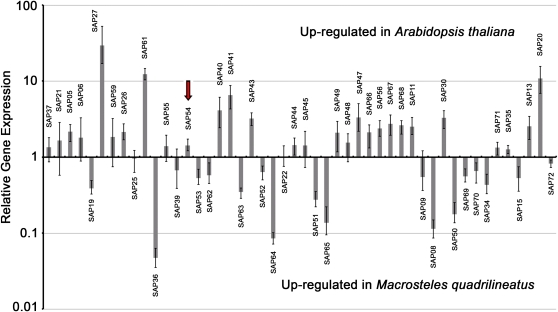
While most effector genes were transcribed in a plant host (Supplemental Fig. S7), gene expression was influenced by host background, with approximately half of the genes exhibiting higher transcript levels in plants and the other half in insects (Fig. 2; Supplemental Table S1). Specifically, the expression of 20 AY-WB candidate effector genes was significantly (P < 0.05) up-regulated in Arabidopsis compared with M. quadrilineatus. We have previously described the AY-WB effector protein SAP11, which is encoded within an operon-like region with four additional candidate effector genes (SAP56, SAP66, SAP67, and SAP68; Bai et al., 2009). We note that expression of all five genes is up-regulated in plants, consistent with the organization of these genes within an operon. Genes encoding 16 AY-WB candidate effectors were significantly up-regulated in insects compared with plants (Fig. 2). One of these effectors, SAP36, is encoded within a potential mobile unit (referred to as PMU1) whose expression we have previously shown to be up-regulated in insects, where PMU1 exists as an extrachromosomal circular element (Toruño et al., 2010).
Figure 2.
Expression of AY-WB genes encoding putative effector proteins. Phytoplasma gene expression was assessed in an infected model plant (Arabidopsis) and the AY-WB leafhopper vector (M. quadrilineatus). Gene expression was normalized against that of five control genes (replicative DNA helicase [AYWB_007], DNA gyrase subunit A [AYWB_254], pyruvate kinase [AYWB_434], 6-phosphofructokinase [AYWB_440], and docking protein FtsY [AYWB_064]). Relative gene expression was calculated by subtracting the ΔCt values of plant samples from the ΔCt values of insect samples to yield ΔΔCt, with data analysis performed by REST 2009 software (Qiagen and Pfaffl). SAP54 is indicated by the red arrow. Error bars represent se. [See online article for color version of this figure.]
In conclusion, the majority of the predicted effector genes are expressed in plants and insects, suggesting that they encode products that function in either host or in both hosts.
Screen of AY-WB Candidates Identifies an Effector That Affects Floral Development
To increase the likelihood of identifying the effector that targets floral development, we decided to generate stable transgenic Arabidopsis lines expressing all the AY-WB candidate effector genes. The genes were expressed under the control of the 35S cauliflower mosaic virus promoter. Fifty-two of the effector genes were successfully cloned and transformed into Arabidopsis Col-0, and the development of T1 transformants was observed to identify any plants that exhibited an abnormal phenotype. In this initial screen, we observed the growth and development of a minimum of 10 independent T1 transgenic lines per effector for the majority (48 of 52) of the candidate effectors (Supplemental Table S2).
Expression of a single effector called SAP54 (AYWB_224) resulted in severely altered flower morphology. Independent T1 transformant lines of Arabidopsis carrying the 35S::SAP54 transgene repeatedly formed flowers from which a secondary flower was generated in the fourth whorl in lieu of a carpel (flowers within flowers; Fig. 3, A and B), a phenotype that is associated with a loss of determinacy of the flower meristem. Furthermore, the petals produced by these flowers were often virescent (Fig. 3B), and the leaf-like sepals were covered in stellate trichomes (Fig. 3C). These phenotypic traits were also often observed in flowers produced by plants infected with AY-WB, suggesting that SAP54 plays a major role in modulating flower development in AY-WB-infected hosts.
Figure 3.
AY-WB effector SAP54 alters Arabidopsis flower development. A, Images of inflorescences of Col-0 (left) and 35S::SAP54 line 4 transgenic Arabidopsis (middle and right). Transgenic plants grown in SD (middle) and LD (right) photoperiods show indeterminate floral growth (flowers within flowers). Bar = 0.5 cm. B, Flowers from AY-WB-infected plants and 35S::SAP54 plants exhibit indeterminate flower phenotypes, leafy sepals, and green stamens. Two sepals, petals, and stamens were removed from the wild-type Col-0 flower (left) to show the carpel (ca). Arrowheads indicate shoots that emerge in lieu of carpels in flowers of AY-WB-infected plants (middle) and 35S::SAP54 transgenic line 4 (right). pe, Petals; se, sepals; st, stamens; tr, simple unbranched trichomes on sepals. Plants were grown in LD photoperiods. Bars = 0.1 cm. C, Close-up of green stamens and sepals with stellate trichomes (str), which are characteristic of leaves; sepals of wild-type Col-0 commonly have simple nonbranched trichomes (B, left). The arrowhead indicates stem in lieu of carpel. This plant was grown in a SD photoperiod. Bar = 0.1 cm. D, 35S::SAP54 plants have sepals (se) that remain attached to fruiting bodies, and siliques (si) are crinkled. Bar = 0.5 cm. E, Sepals attached to fruiting bodies of 35S::SAP54 line 1 flowers curl inward as opposed to those of 35S::SAP54 line 4, which curl outward. Furthermore, siliques (si) of 35S::SAP54 line 1 plants are less crinkled and stunted than those of line 4. Bars = 0.25 cm. F, AtSuc2::SAP54 plants that produce SAP54 in phloem tissue also show indeterminate leafy flowers. Stem is indicated with an arrowhead; fl, newly emerging flower bud from within the flower. Bar = 0.25 cm.
SAP54 Is an Effector That Alters Flower Development in Arabidopsis
The flower phenotypes of the SAP54-expressing plants were analyzed in greater detail and compared with the flowers of AY-WB-infected plants. The extent of the flower development defect caused by SAP54 was reflected in the observation that many T1 transformants were sterile or produced seeds that failed to germinate. However, we obtained two independent T3 lines (SAP54 lines 4 and 1) that expressed the gene encoding SAP54 (Supplemental Fig. S8) and were used in further experiments.
In noninfected wild-type Arabidopsis Col-0, four sepals enclose and protect four white petals, giving the flowers a characteristic cup-like shape (Fig. 3B). Six stamens are produced in the third whorl, and two fused carpels arise from the central fourth whorl to form the gynoecium, which houses the ovules. In comparison with the wild-type plants, the sepals of 35S::SAP54 line 4 were larger and did not enclose the petals but rather curled outward and away from the petals, a trait that was also observed in flowers of AY-WB-infected plants (Figs. 1, E and G, and 3B). Scanning electron microscopy revealed that the sepals of SAP54-expressing and infected plants are composed of irregular, interlocking cells that are more reminiscent of leaf pavement cells than the elongated cells that constitute the sepals of wild-type (healthy) Col-0 (Fig. 4A). The hypertrophic sepals of the transgenic line assumed an increasingly leaf-like appearance as the flower aged. White petals were occasionally present in newly opened flowers of 35S::SAP54 line 4 plants, but these either dropped from the plant or reverted to green leaf-like tissues as the flower matured. An increased number of stamens were produced in many flowers (often seven or eight stamens per flower instead of six) of plants grown in SD, and these occasionally produced trichomes (Supplemental Fig. S9). Frequently, these stamens were transformed into leaf-like organs via the outgrowth of vegetative tissue that resembled a leaf (Fig. 4B; Supplemental Fig. S9). In comparison, stamens of infected plants were virescent; however, these did not produce the leaf-like outgrowths observed of the transgenic line. Within the central whorl, one to as many as four new flowers was produced in lieu of carpels, indicative of a loss of floral determinacy (Fig. 3, A and B). In fact, this line yielded relatively few siliques that were invariably shorter in length than wild-type siliques, of wrinkled appearance, and with sepals still attached at the base (Fig. 3D). Furthermore, siliques were only produced by 35S::SAP54 line 4 at an advanced stage of growth (several weeks after wild-type siliques had shattered) and were restricted to the uppermost portion of the plant. Closer analyses of 35S::SAP54 line 4 flowers of plants grown in SD revealed that many secondary flowers emerging from the fourth whorl of flower-like structures had reiterations of sepals and stamens, which were both green in plants grown in SD (Fig. 5A). Furthermore, while these sepals were leaf like, as evidenced by the presence of stellate trichomes, they occasionally possessed carpelloid features, as indicated by the presence of stigmatic papillae along the distal edges of the sepals (Fig. 5, B and C).
Figure 4.
Cryoscanning electron microscopy images illustrating changes in flower phenotypes at the cellular and organ levels in AY-WB-infected plants and 35S:SAP54 lines. A, Examination of the adaxial surface of sepals from plants expressing SAP54 and plants infected with phytoplasma AY-WB reveals irregular and interlocking cells that resemble the pavement cells of a rosette leaf. Sepals of both 35S::SAP54 line 4 (above, SAP54-4) and 35S::SAP54 line 1 (not shown) are composed of these jagged cells. Bars = 100 μm. B, Stalks (arrowheads) and anthers (asterisks) of stamens of 35S::SAP54 line 4 plants are frequently transformed into leafy organs, whereas those of infected plants are virescent. Bars = 500 μm. C, Petals of 35S::SAP54 line 1 plants produce stellate trichomes (arrowheads) and show evidence of phyllody, consistent with phenotypes of these organs in AY-WB-infected plants. Bars = 200 μm. D, Carpels produced by SAP54-expressing transgenic lines and AY-WB-infected plants are bulbous and contain stellate trichomes. Bars = 200 μm.
Figure 5.
Secondary flowers of 35S::SAP54 line 4 exhibit abnormal development. A, Secondary flower bud from SAP54 line 4 shows reiterations of sepals and stamens. S indicates sepals; the asterisk indicates virescent anther. Bar = 200 μm. B, Flower bud emerging from the fourth whorl of a flower with carpelloid sepals bearing stigmatic papillae. This image is an enlargement of the boxed area in the inset. Bars = 100 μm and 200 μm (inset). C, Flower bud from SAP54 line 4 showing carpelloid sepals. Arrowheads indicate stigmatic papillae along the distal edge of sepals. Bar = 100 μm.
35S::SAP54 line 1 also produced abnormal flowers and siliques; however, the alteration in phenotype appeared to be less pronounced than that of 35S::SAP54 line 4. We also note that the phenotype observed in this line was more evident in early-arising flowers. Sepals did not curl away from petals, as observed in 35S::SAP54 line 4, although sepals frequently curled inward to form a dome in lieu of an open flower (Fig. 3E). Nonetheless, sepals produced from this line contained stellate trichomes and were composed of irregular and interlocking cells comparable to those observed in 35S::SAP54 line 4. Flowers originating from 35S::SAP54 line 1 produced white petals; however, these frequently reverted to green petals that contained both stomata and trichomes (Fig. 4C). Plants of this transgenic line did not exhibit a loss of floral determinacy (i.e. they did not form flowers within flowers); however, carpels produced by the earliest flowers were often bulbous and covered in stellate trichomes (Fig. 4D). Examination of the interior of the bulbous carpels revealed the presence of abnormal structures that resembled developing leaves or sepals (Supplemental Fig. S10).
In summary, these results provide evidence that SAP54 induces alterations in Arabidopsis flower phenotypes similar to those observed in AY-WB-infected plants and hence is likely to be responsible for symptom induction during AY-WB infection in plants.
Phloem-Expressed SAP54 Alters Floral Organ Identity
Phytoplasma are mainly restricted to the phloem; hence, the effectors, including SAP54, are likely to be released inside the phloem cells. These effectors may remain in the phloem or unload from the phloem to target other plant cells or organs (Bai et al., 2009; Hoshi et al., 2009). To determine if SAP54 also induces changes in flower development when produced in the plant phloem, we generated transgenic Arabidopsis plants that express SAP54 under the control of the phloem-specific AtSuc2 promoter (An et al., 2004; Corbesier et al., 2007; Mathieu et al., 2007). The AtSuc2::SAP54 transgenic lines showed the indeterminate leaf-like flowers similar to those observed in AY-WB-infected plants and 35S::SAP54 lines (Fig. 3F). Thus, SAP54 expressed in the phloem also alters flower development.
DISCUSSION
AY-WB was originally isolated from infected lettuce (Lactuca sativa), and this strain was also shown to infect China aster (Callistephus chinensis; Zhang et al., 2004). A comparison of the symptoms elicited by AY-WB in Arabidopsis, lettuce, and China aster reveals similarities in disease progression. Particularly striking are early-onset symptoms of hyponastic growth (abnormally upright leaves) and the late-arising symptoms of witches’ broom, phyllody, and virescence. These symptoms have been described in plants infected by a range of phytoplasmas (Firrao et al., 1996; Lee et al., 2004; Zhang et al., 2004; Naito et al., 2007; Zamharir and Mirabolfathi, 2011) and suggest that these phytopathogens alter the growth and development of their plant hosts. We have hypothesized that phytoplasmas such as AY-WB produce a range of proteinaceous effectors that are released into the cytoplasm of host cells to modulate developmental processes in a host-specific (i.e. regulated) manner (Hogenhout and Loria, 2008; Hogenhout et al., 2008; Bai et al., 2009; Toruño et al., 2010; Sugio et al., 2011). Having established that Arabidopsis is susceptible to AY-WB infection and exhibits symptoms that are characteristic of this phytoplasma strain, we set out to identify AY-WB effector(s) that induce the formation of the leaf-like flowers that are a hallmark of phytoplasma infection through the expression of candidate effectors in transgenic Arabidopsis.
We have expressed in Arabidopsis a total of 52 bacterial proteins that we had previously predicted to be secreted from AY-WB into a host cell (based upon the presence of a signal peptide; Bai et al., 2009). From this initial screen, we have identified one effector that induces obvious alterations in flower architecture upon expression in Arabidopsis. Plants expressing SAP54 produce abnormal leaf-like flowers that are reminiscent of symptoms of phyllody and virescence that are characteristic of AY-WB infection (Fig. 3). Indeed, the flowers produced by infected plants and SAP54-expressing lines of Arabidopsis are quite similar. For example, AY-WB-infected Arabidopsis plants frequently produce flowers that form shoots in lieu of carpels, as observed in one of the SAP54 transgenic lines. Flowers from infected plants are green, produce sepals, petals, and carpels with stellate trichomes, and have a leaf-like appearance, all attributes that are shared with plants that express the phytoplasma effector.
To the best of our knowledge, there is no pathogen effector identified to date that interferes with floral development; thus, our discovery of SAP54 is novel. SAP54 encodes a 10.7-kD protein having no homology with previously characterized proteins or domains. We examined available sequence data to identify homologs of this AY-WB effector and performed alignments to compare the proteins. SAP54 homologs are encoded in the genomes of at least three other phytoplasmas (Supplemental Fig. S11), two of which are members of the AY group of phytoplasmas (Onion Yellows phytoplasma strain OY-M and MD Aster Yellows; 16SrI-B), while Spiraea stunt phytoplasma (Candidatus Phytoplasma pruni; 16SrIII, X-disease group) is distantly related to the AY phytoplasmas. AY phytoplasmas (particularly members of the 16SrI-A and 16SrI-B subgroups) frequently elicit symptoms of abnormal floral development (Lee et al., 2004); thus, the presence of SAP54 homologs in representatives of this group is not surprising. Although we were unable to identify SAP54 homologs in a number of phytoplasmas known to influence flower architecture (e.g. stolbur phytoplasma; Pracros et al., 2006), our efforts were severely constrained by the limited availability of phytoplasma sequence data; thus, it cannot be concluded that these phytoplasmas do not encode a SAP54-like effector. Conversely, it is possible that these phytoplasmas employ different effectors to modulate flower development in their hosts.
The development of a flower is a highly regulated process that consists of four major stages: (1) initiating the transition from vegetative growth to reproductive growth (generation of the inflorescence meristem); (2) establishment of a floral meristem and its maintenance; (3) activation of floral organ identity genes and the formation of sepals, petals, stamens, and carpels; and (4) termination of the floral meristem (Coen and Meyerowitz, 1991; Sablowski, 2007). SAP54 expression in Arabidopsis strongly compromises flower organ identity and termination of the floral meristem (Figs. 3 and 4). The steps of flower development have been genetically dissected in Arabidopsis, providing a model system from which we may examine the mechanism by which SAP54 modulates floral development in future studies.
The complex phenotype exhibited by SAP54-expressing and AY-WB-infected plants recommends more than one host protein as a candidate that is targeted by SAP54. For example, the production of a secondary flower in place of a carpel (loss of fourth whorl determinacy) indicates that phytoplasmas might interfere in some way with the function of the C-class protein AGAMOUS, and there is evidence that the expression of an AGAMOUS ortholog is altered in tomato (Solanum lycopersicum) infected with stolbur phytoplasma (Pracros et al., 2006). We also notice striking similarities between flowers of AY-WB-infected plants and sep1 sep2 sep3/+ sep4 quadruple mutants (Ditta et al., 2004). Particularly, the sep quadruple mutants produce leaf-like flowers with sepals and carpels that contain stellate trichomes and in which many flowers also produce secondary flowers instead of carpels. One scenario is that SAP54 targets or impedes the function of the SEP proteins that are required for the formation of multiprotein complexes, thereby affecting the patterning of the developing flowers. An alternative hypothesis that does not preclude the first proposal is that SAP54 broadly targets members of the MADS box domain protein family, which constitute the largest and most important family of proteins to regulate flowering in Arabidopsis. This scenario is reminiscent of SAP11-mediated targeting of class II TCP transcription factors (Sugio et al., 2011). Of course, SAP54 might simply target one protein (e.g. SEP3) or a nonproteinaceous component (e.g. a microRNA). When expressed in the phloem, SAP54 also alters Arabidopsis flower development. Therefore, SAP54 may interact with its target in the phloem and/or unload from the phloem to target plant host factors in adjacent cells. The identification and localization of the host target of SAP54 and the mode of action by which this effector modulates flower development will be an interesting area of future research that may offer unique insights into flower development. Accordingly, studies to directly identify the host target of SAP54 are ongoing.
Little is known regarding gene regulation in the uncultivated phytoplasmas, and this study represents one of the first comprehensive surveys of phytoplasma gene expression. Toruño et al. (2010) examined the expression of genes encoded within the PMU1 region of the AY-WB genome, and we report herein the expression of an additional 56 effector genes. A subset of the examined genes was highly responsive to host background, being significantly up-regulated in either plants (20 SAPs) or insects (16 SAPs). It is likely that genes that exhibit a strong host-specific expression pattern encode proteins with functions relevant to the colonization of that host. However, it cannot be excluded that the proteins also have a function in the alternate host. For example, SAP11 (which is up-regulated in plants) is also present in great abundance in insect salivary glands (Bai et al., 2007); thus, it is likely that this protein is injected directly into the phloem by a vector insect while feeding. Consequently, SAP11 can be introduced into the phloem of a plant by two sources: the saliva of an infected insect and by way of secretion from the bacterial pathogen. Interestingly, the majority of candidate effector genes that are up-regulated in infected plants do not confer an obvious phenotype when expressed in transgenic Arabidopsis. These effectors may modulate processes (such as host defense) that do not yield a readily observable alteration in morphology. It is also possible that the activity of certain effectors is dependent upon interactions with other phytoplasma factors, which would not be available in a transgenic plant.
CONCLUSION
We have frequently hypothesized that phytoplasmas such as AY-WB produce effectors that manipulate the phenotype of their hosts to their own benefit (Hogenhout and Loria, 2008; Hogenhout et al., 2008; Bai et al., 2009; Sugio et al., 2011). The recent discovery and characterization of one such effector (SAP11) offered support to this hypothesis, and the identification of an additional effector (SAP54) that influences the phenotype of host flowers lends further credence. Nonetheless, the question of why phytoplasmas employ effectors that (for example) alter the development of flowers needs to be addressed. We have proposed several possibilities (Sugio et al., 2011). One hypothesis is that alterations in plant phenotype represent an attempt by the phytoplasmas to encourage insect activity (i.e. feeding and egg laying) as a means of facilitating phytoplasma dispersal in the environment. Phytoplasmas cannot survive outside of an insect or plant host, and thus the movement of phytoplasmas in natural (and agricultural) settings is dependent upon their insect vectors. Phloem-feeding insects such as leafhoppers may be attracted to plants that aggressively produce young, green, vegetative tissues. Effectors that increase the production of stems (such as SAP11) or produce leafy flowers (such as SAP54) may stimulate leafhopper feeding, increasing the frequency of phytoplasma acquisition by its vector. Another possibility is that interfering with flower development may extend the plant life span as annual plants die upon seed production. In this scenario, phytoplasmas that possess SAP54 or functionally similar effectors may have a competitive advantage against those that do not have these effectors. This study identifies a number of excellent candidates that likely play important roles in modulating host processes in both plants and insects, and we expect many fascinating discoveries relating to phytoplasma research in the near future.
MATERIALS AND METHODS
Analysis of AY-WB Symptoms in Arabidopsis
Prior to insect exposure, Arabidopsis (Arabidopsis thaliana) Col-0 plants were grown in a 10/14-h light/dark cycle at 22°C. To inoculate plants with AY-WB, trays of 10 4-week-old plants were exposed to 20 Macrosteles quadrilineatus leafhoppers (two leafhoppers per plant) and enclosed within perforated bags (300 × 400 mm). In this manner, 40 plants were exposed to noninfected leafhoppers (referred to as healthy plants), and an additional 40 plants were exposed to AY-WB-infected leafhoppers (infected plants). Immediately following the addition of insects, the bagged plants were randomly divided into two groups comprising equal numbers of healthy and infected plants. One group of plants (i.e. 20 healthy and 20 infected) was transferred to a growth room set at a 16/8-h light/dark cycle at 23°C/20°C (LD photoperiod). The second group of plants was grown in a chamber set at a 10/14-h light/dark cycle at 22°C/22°C (SD photoperiod). After 1 week, all insects were removed from the plants, which were then returned to their respective growth chambers for the duration of the experiment.
All plants exposed to AY-WB-infected insects exhibited marked symptoms of infection, including stunted growth and yellowing of developing rosette leaves within 3 weeks after inoculation. The length and width of leaves were assessed based upon measurements obtained from the three largest rosette leaves produced per plant for each plant in a group. Plant height was determined by measuring the height of the main bolt of a plant. Fresh shoot mass was determined after cutting off the roots and carefully removing any soil that adhered to the aboveground tissues. All measurements were obtained 44 and 54 dpi for SD and LD, respectively, unless otherwise indicated.
Expression Analysis of Phytoplasma Genes by Quantitative Reverse Transcription-PCR
Methods and materials used were as described previously (Toruño et al., 2010) with the following modifications. Five plant (Arabidopsis) and three insect (M. quadrilineatus) biological replicates were used to measure AY-WB gene expression in these respective hosts. As a means of generating material for expression studies in an insect host, three separate populations of M. quadrilineatus were generated by rearing noncarrier insects on AY-WB-infected China aster (Callistephus chinensis) to enable acquisition of the phytoplasma. After 4 weeks, insects were collected from each population for RNA extraction, and TaqMan reverse transcription (RT)-PCR (Christensen et al., 2004) was used to confirm the presence of AY-WB in these insects. A population of noncarrier insects was maintained on healthy oat plants (Avena sativa), and TaqMan RT-PCR was likewise performed to confirm the absence of AY-WB within these insects. To generate infected plants, five 3-week-old Arabidopsis Col-0 plants (grown in a 10/14-h light/dark cycle at 22°C) were exposed to AY-WB carrier M. quadrilineatus males (two leafhoppers per plant). At the same time, one 3-week-old plant was exposed to two noncarrier M. quadrilineatus males (healthy control). Following addition of the insects, all plants were transferred to a growth chamber (10/14-h light/dark cycle at 22°C) for the duration of the experiment. Leafhoppers were removed after 1 week, and plants were returned to the growth chamber and monitored for the development of symptoms of infection with AY-WB. Plants were harvested approximately 4 weeks after exposure to insects, and all five plants that were exposed to carrier insects exhibited early signs of phytoplasma infection. Roots were carefully trimmed from the plant rosette, which was immediately frozen in liquid nitrogen. Insect and plant material was homogenized and used for total RNA extraction. After DNaseI treatment, RNA was precipitated in ethanol and redissolved in water. DNaseI-treated samples were tested without a RT step to confirm the absence of significant amounts of contaminating genomic DNA. One microgram of total RNA was used for cDNA synthesis.
Primers were designed with Primer Express software (Applied Biosystems) and are listed in Supplemental Table S4. Primers were designed in a gene-specific manner unless stated otherwise (Supplemental Table S1). Cycling conditions were as follows: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. Gene expression was normalized to that of five control genes (replicative DNA helicase [AYWB_007], DNA gyrase subunit A [AYWB_254], pyruvate kinase [AYWB_434], 6-phosphofructokinase [AYWB_440], and docking protein FtsY [AYWB_064]). No amplification similar to that of infected samples was detected from healthy controls. Amplification of dilution series was performed for the calculation of amplification efficiencies of each gene assay, and the results obtained were incorporated into the data analysis to correct for differences in PCR efficiency. The ΔΔCt method (Applied Biosystems, User Bulletin 2, 1997) and REST 2009 software (Qiagen and Pfaffl) were used for data analysis and statistical tests.
Generation and Analysis of Transgenic Arabidopsis Lines
All intermediate DNA constructs were maintained in Escherichia coli DH5α cells. To obtain AY-WB DNA, two leaves of AY-WB-infected Arabidopsis leaves were ground in 400 μL of buffer (200 mm Tris-HCl, pH 7.5, 250 mm NaCl, 25 mm EDTA, and 0.5% SDS) and centrifuged to precipitate the plant debris. The liquid layer was then transferred to a new tube, and the DNA was precipitated with isopropanol. The DNA pellet was resuspended in 100 μL of distilled water, and 1 μL of the solution was used per 20 μL of PCR reaction. Each effector sequence without the signal peptide was amplified using the primers listed in Supplemental Table S2 by Phusion Taq polymerase (Thermo Fisher Scientific) following the manufacturer’s instructions. The DNA fragment was amplified with attB1 and attB2 adapter primers (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGT-3′) and was cloned into pDONR207 (Invitrogen) using BP Clonase (Invitrogen) following the manufacturer’s instructions. Clones were sequenced to verify the origin and sequence of the insert. The effector fragment in pDONR207 was then Clonase LR cloned into the Gateway destination vector pB7WG2, which has a 35S promoter at the 5′ end of the Clonase-compatible insertion site (Karimi et al., 2002), or pHW59, which has a phloem-specific AtSuc2 promoter at the 5′ end (Mathieu et al., 2007). The resulting plasmids were transformed into Agrobacterium tumefaciens strain GV3101. Arabidopsis Col-0 was transformed by floral dip (Clough and Bent, 1998). T1 seeds were germinated in soil, and the transformants were selected for resistance to the herbicide BASTA (glufosinate) in a glasshouse. We obtained a minimum of 10 individual transformants per construct for the majority of effectors to ensure that any alteration in phenotype was due to the expression of the effector (Supplemental Table S2). T2 seeds of transgenic BASTA-resistant lines were plated on Murashige and Skoog medium containing 20 μg mL−1 phosphinothricin. The surviving plants were used to generate T3 lines for further analyses.
Scanning Electron Microscopy
Transgenic plants were sown on soil and grown in a chamber set to a SD photoperiod (10/14 h of light/dark at 20°C). Infected plants were generated as described above. Samples were harvested approximately 1 to 2 weeks after bolting and were mounted on an aluminum stub using Optimal Cutting Temperature compound (BDH Laboratory Supplies). The stub was then immediately plunged into liquid nitrogen slush at approximately −210°C to cryopreserve the material. The sample was transferred onto the cryostage of an ALTO 2500 cryotransfer system (Gatan) attached to a Zeiss Supra 55 VP FEG scanning electron microscope. Sublimation of surface frost was performed at −95°C for 3 min before sputter coating the sample with platinum for 2 min at 10 mA at less than −110°C. After sputter coating, the sample was moved onto the cryostage in the main chamber of the microscope and held at approximately −130°C. The sample was imaged at 3 kV.
RT-PCR of SAP54
The two T3 heterozygous SAP54 lines were sown in soil and grown in the LD photoperiod. A cluster of the flowers was harvested from each line when plants were 5 weeks old, and the sample was subjected to RNA isolation. The RNA samples were treated with DNase (Qiagen) and cleaned up with the RNeasy mini kit (Qiagen). Five hundred nanograms of total RNA was used for cDNA synthesis by Moloney murine leukemia virus reverse transcriptase (Invitrogen) following the manufacturer’s instructions. The cDNA samples were diluted 10-fold by distilled water, and 0.5 μL of the diluted cDNA samples was used per 20 μL of PCR reaction using Go-Taq polymerase (Promega) and the gene-specific primers listed in Supplemental Table S3. Cycling conditions were as follows: 94°C for 2 min, 40 cycles of 95°C for 30 s, 45°C for 30 s, and 72°C for 30 s, followed by 72°C for 5 min. Fifteen microliters of the PCR samples was subjected to electrophoresis on a 2% agarose gel containing ethidium bromide.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. AY-WB infection stunts leaf growth and leads to anthocyanin accumulation in Arabidopsis.
Supplemental Figure S2. AY-WB-infected plants produce narrower and paler leaves than healthy plants.
Supplemental Figure S3. Rosettes of AY-WB-infected Arabidopsis plants have a reduced diameter.
Supplemental Figure S4. Plants infected with AY-WB have a decreased height.
Supplemental Figure S5. AY-WB-infected Arabidopsis plants produce an increased number of axillary stems (witches’ broom phenotype) at an early stage of infection.
Supplemental Figure S6. AY-WB-infected plants produce leaf-like flowers.
Supplemental Figure S7. AY-WB genes encoding effector proteins are expressed in infected Arabidopsis.
Supplemental Figure S8. SAP54 expression in Arabidopsis transgenic lines.
Supplemental Figure S9. Expression of SAP54 induces leaf-like stamens.
Supplemental Figure S10. Carpels of 35S::SAP54 Arabidopsis line 1 are bulbous and possess trichomes.
Supplemental Figure S11. Alignment of an AY-WB phytoplasma effector that induces morphological changes in flowers.
Supplemental Table S1. Fold increase of gene expression levels and P values of AY-WB candidate effector (SAP) genes.
Supplemental Table S2. Primers and other information for transgenic Arabidopsis lines that express AY-WB candidate effectors.
Supplemental Table S3. Primers used for RT-PCR to examine SAP54 expression of transgenic lines.
Supplemental Table S4. Primers used in the expression analysis of AY-WB genes.
Acknowledgments
We gratefully acknowledge Christian Ruzanski, Megan Brewer, Shirley Aris, and Heather N. Kingdom for helping to generate transgenic Arabidopsis lines, Gitte C. Christiansen for helping to establish the AY-WB plant-insect system at Aarhus University, and members of the John Innes Centre insectary and horticultural staff for their excellent services. We thank Andrew Davis for providing high-quality photographs.
References
- An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, et al. (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Bai X, Ammar ED, Hogenhout SA. (2007) A secreted effector protein of AY-WB phytoplasma accumulates in nuclei and alters gene expression of host plant cells, and is detected in various tissues of the leafhopper Macrosteles quadrilineatus. Bull Insectol 60: 217–218 [Google Scholar]
- Bai X, Correa VR, Toruño TY, Ammar D, Kamoun S, Hogenhout SA. (2009) AY-WB phytoplasma secretes a protein that targets plant cell nuclei. Mol Plant Microbe Interact 22: 18–30 [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795 [DOI] [PubMed] [Google Scholar]
- Bonnot F, de Franqueville H, Lourenço E. (2010) Spatial and spatiotemporal pattern analysis of coconut lethal yellowing in Mozambique. Phytopathology 100: 300–312 [DOI] [PubMed] [Google Scholar]
- Busch W, Benfey PN. (2010) Information processing without brains: the power of intercellular regulators in plants. Development 137: 1215–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Schwarz-Sommer Z, Davies B. (2010) Floral organ identity: 20 years of ABCs. Semin Cell Dev Biol 21: 73–79 [DOI] [PubMed] [Google Scholar]
- Cettul E, Firrao G. (2010) Effects of phytoplasma infection on Arabidopsis thaliana development. Brown DR, Bertaccini A, , 18th Congress of The International Organization for Mycoplasmology. The International Organization for Mycoplasmology, Chianciano Terme, Italy, p 82 [Google Scholar]
- Christensen NM, Nicolaisen M, Hansen M, Schulz A. (2004) Distribution of phytoplasmas in infected plants as revealed by real-time PCR and bioimaging. Mol Plant Microbe Interact 17: 1175–1184 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM. (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Constable FE. (2009) Phytoplasma epidemiology: grapevines as a model. Weintraub PGJ, , Phytoplasmas: Genomes, Plant Hosts and Vectors; CABI, Wallingford, UK, pp 188–212 [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Depuydt S, Dolezal K, Van Lijsebettens M, Moritz T, Holsters M, Vereecke D. (2008) Modulation of the hormone setting by Rhodococcus fascians results in ectopic KNOX activation in Arabidopsis. Plant Physiol 146: 1267–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14: 1935–1940 [DOI] [PubMed] [Google Scholar]
- Firrao G, Carraro L, Gobbi E, Locci R. (1996) Molecular characterization of a phytoplasma causing phyllody in clover and other herbaceous hosts in northern Italy. Eur J Plant Pathol 102: 817–822 [Google Scholar]
- Ghosh P. (2004) Process of protein transport by the type III secretion system. Microbiol Mol Biol Rev 68: 771–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno M, Neriya Y, Minato N, Miura C, Sugawara K, Ishii Y, Yamaji Y, Kakizawa S, Oshima K, Namba S. (2011) Unique morphological changes in plant pathogenic phytoplasma-infected petunia flowers are related to transcriptional regulation of floral homeotic genes in an organ-specific manner. Plant J 67: 971–979 [DOI] [PubMed] [Google Scholar]
- Hogenhout SA, Loria R. (2008) Virulence mechanisms of gram-positive plant pathogenic bacteria. Curr Opin Plant Biol 11: 449–456 [DOI] [PubMed] [Google Scholar]
- Hogenhout SA, Oshima K, Ammar D, Kakizawa S, Kingdom HN, Namba S. (2008) Phytoplasmas: bacteria that manipulate plants and insects. Mol Plant Pathol 9: 403–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenhout SA, Van der Hoorn RA, Terauchi R, Kamoun S. (2009) Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact 22: 115–122 [DOI] [PubMed] [Google Scholar]
- Hoshi A, Oshima K, Kakizawa S, Ishii Y, Ozeki J, Hashimoto M, Komatsu K, Kagiwada S, Yamaji Y, Namba S. (2009) A unique virulence factor for proliferation and dwarfism in plants identified from a phytopathogenic bacterium. Proc Natl Acad Sci USA 106: 6416–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RG, Kaufmann K, Angenent GC. (2010) The ‘ABC’ of MADS domain protein behaviour and interactions. Semin Cell Dev Biol 21: 87–93 [DOI] [PubMed] [Google Scholar]
- Jović J, Cvrković T, Mitrović M, Krnjajić S, Petrović A, Redinbaugh MG, Pratt RC, Hogenhout SA, Tosevski I. (2009) Stolbur phytoplasma transmission to maize by Reptalus panzeri and the disease cycle of maize redness in Serbia. Phytopathology 99: 1053–1061 [DOI] [PubMed] [Google Scholar]
- Kakizawa S, Oshima K, Nishigawa H, Jung HY, Wei W, Suzuki S, Tanaka M, Miyata S, Ugaki M, Namba S. (2004) Secretion of immunodominant membrane protein from onion yellows phytoplasma through the Sec protein-translocation system in Escherichia coli. Microbiology 150: 135–142 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Lee IM, Gundersen-Rindal DE, Davis RE, Bottner KD, Marcone C, Seemüller E. (2004) ‘Candidatus Phytoplasma asteris’, a novel phytoplasma taxon associated with aster yellows and related diseases. Int J Syst Evol Microbiol 54: 1037–1048 [DOI] [PubMed] [Google Scholar]
- Lee IM, Martini M, Bottner KD, Dane RA, Black MC, Troxclair N. (2003) Ecological implications from a molecular analysis of phytoplasmas involved in an aster yellows epidemic in various crops in Texas. Phytopathology 93: 1368–1377 [DOI] [PubMed] [Google Scholar]
- Marois E, Van den Ackerveken G, Bonas U. (2002) The Xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol Plant Microbe Interact 15: 637–646 [DOI] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M. (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Naito T, Tanaka M, Taba S, Toyosato T, Oshiro A, Takaesu K, Hokama K, Usugi T, Kawano S. (2007) Occurrence of chrysanthemum virescence caused by “Candidatus Phytoplasma aurantifolia” in Okinawa. J Gen Plant Pathol 73: 139–141 [Google Scholar]
- Pracros P, Renaudin J, Eveillard S, Mouras A, Hernould M. (2006) Tomato flower abnormalities induced by stolbur phytoplasma infection are associated with changes of expression of floral development genes. Mol Plant Microbe Interact 19: 62–68 [DOI] [PubMed] [Google Scholar]
- Replogle A, Wang J, Bleckmann A, Hussey RS, Baum TJ, Sawa S, Davis EL, Wang X, Simon R, Mitchum MG. (2011) Nematode CLE signaling in Arabidopsis requires CLAVATA2 and CORYNE. Plant J 65: 430–440 [DOI] [PubMed] [Google Scholar]
- Sablowski R. (2007) Flowering and determinacy in Arabidopsis. J Exp Bot 58: 899–907 [DOI] [PubMed] [Google Scholar]
- Seemüller E, Kiss E, Sule S, Schneider B. (2010) Multiple infection of apple trees by distinct strains of ‘Candidatus Phytoplasma mali’ and its pathological relevance. Phytopathology 100: 863–870 [DOI] [PubMed] [Google Scholar]
- Sugio A, Maclean AM, Kingdom HN, Grieve VM, Manimekalai R, Hogenhout SA. (2011) Diverse targets of phytoplasma effectors: from plant development to defense against insects. Annu Rev Phytopathol 49: 175–195 [DOI] [PubMed] [Google Scholar]
- Toruño TY, Musić MS, Simi S, Nicolaisen M, Hogenhout SA. (2010) Phytoplasma PMU1 exists as linear chromosomal and circular extrachromosomal elements and has enhanced expression in insect vectors compared with plant hosts. Mol Microbiol 77: 1406–1415 [DOI] [PubMed] [Google Scholar]
- Wang K, Hiruki C. (2001) Molecular characterization and classification of phytoplasmas associated with canola yellows and a new phytoplasma strain associated with dandelions. Plant Dis 85: 76–79 [DOI] [PubMed] [Google Scholar]
- Zamharir MG, Mirabolfathi M. (2011) Association of a phytoplasma with pistachio witches’ broom disease in Iran. J Phytopathol 159: 60–62 [Google Scholar]
- Zhang J, Hogenhout SA, Nault LR, Hoy CW, Miller SA. (2004) Molecular and symptom analyses of phytoplasma strains from lettuce reveal a diverse population. Phytopathology 94: 842–849 [DOI] [PubMed] [Google Scholar]