Abstract
Background:
To date, few administrative diabetes mellitus (DM) registries have distinguished type 1 diabetes mellitus (T1DM) from type 2 diabetes mellitus (T2DM).
Objective:
Using a classification tree model, a prediction rule was developed to distinguish T1DM from T2DM in a large administrative database.
Methods:
The Medical Archival Retrieval System at the University of Pittsburgh Medical Center included administrative and clinical data from January 1, 2000, through September 30, 2009, for 209,647 DM patients aged ≥18 years. Probable cases (8,173 T1DM and 125,111 T2DM) were identified by applying clinical criteria to administrative data. Nonparametric classification tree models were fit using TIBCO Spotfire S+ 8.1 (TIBCO Software), with model size based on 10-fold cross validation. Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of T1DM were estimated.
Results:
The main predictors that distinguished T1DM from T2DM are age <40 years; International Classification of Disease, 9th revision, codes of T1DM or T2DM diagnosis; inpatient oral hypoglycemic agent use; inpatient insulin use; and episode(s) of diabetic ketoacidosis diagnosis. Compared with a complex clinical algorithm, the tree-structured model to predict T1DM had 92.8% sensitivity, 99.3% specificity, 89.5% PPV, and 99.5% NPV.
Conclusion:
The preliminary predictive rule appears to be promising. Being able to distinguish between DM subtypes in administrative databases will allow large-scale subtype-specific analyses of medical care costs, morbidity, and mortality.
Keywords: administrative database, classification and regression tree, diabetes, Medical Archival Retrieval System, negative predictive value, positive predictive value, sensitivity, specificity
Introduction
Diabetes mellitus (DM) and its complications pose a major challenge to health and to health care systems. According to a World Health Organization report, more than 220 million people worldwide suffer from DM.1 As of 2007, in the United States, 23.6 million people (7.8% of the population) had DM,2 and total costs associated with DM exceed $218 billion nationally.3 Type 1 diabetes mellitus (T1DM) and type 2 diabetes mellitus (T2DM) differ with respect to temporal evolution, complications, and prognosis, as well as the planning, implementation, and monitoring of appropriate interventions. Administrative records have limited ability to distinguish between T1DM and T2DM due to the lack of required information and other limitations inherent to administrative data [e.g., missing values and possibly nonspecific International Classification of Disease codes].4,5 Accurate and valid methods to distinguish T1DM from T2DM must be developed before administrative data can be used to study T1DM and T2DM.
Decision tree methods, also called recursive partitioning methods, were developed to classify a target population for the purpose of clinical diagnosis and/or prognosis. Classification and regression trees (CART) are a non-para-metric approach to estimate a decision tree.6 Classification and regression tree identify easily defined, mutually exclusive population subgroups whose members share characteristics that are important predictors of the outcome of interest. For analyzing large administrative data sets, advantages of CART include the ability to use surrogate splits (to account for missing data), to handle nonlinear relationships and complex interactions, and to provide intuitive information regarding the predictive tree.
The purpose of the present study is to build a tree-structured model to distinguish between T1DM and T2DM among probable cases of DM, using administrative data from the Medical Archival Retrieval System (MARS) database at the University of Pittsburgh Medical Center (UPMC). Specifically, the authors (i) identify probable DM patients using published criteria,5 (ii) distinguish probable cases of T1DM from T2DM using detailed clinical criteria and medical record review, and (iii) assess whether a relatively simple classification tree can reproduce clinical classification.
Methods
Setting
The UPMC is one of the largest nonprofit integrated health care systems in the United States. Each year, the UPMC has more than 187,000 inpatient admissions, 4.5 million outpatient visits, and 480,000 emergency visits.7 The UPMC provides services for a large majority of people with DM throughout Western Pennsylvania.8
Diabetes Registry from Medical Archival Retrieval System Data
The UPMC has used the MARS electronic medical record system since 1987. The MARS is a repository of information forwarded from the health system's electronic clinical, administrative, and financial databases, including laboratory results, patient demographics, visits, and charges. The MARS is indexed on every word in the medical record and can recover all encounters for a given patient between specified dates. Medical Archival Retrieval System-based data sources used to establish the DM registry include (1) medical records discharge abstract file, consisting of all visits coded by the medical records department, with up to 25 ICD-9 diagnosis codes assigned per patient; (2) the Hospital Laboratory Information System that includes inpatient, emergency room, hospital-based clinics, outpatient surgery, and mail-in specimens (laboratory data are sent to the MARS using the Misys® laboratory information system, which supports all UPMC hospitals and clinics); and (3) the Hospital Pharmacy Information System, which includes inpatient information on medication dispensed in the emergency room, hospital-based clinics, and outpatient surgery settings.5 The MARS database is audited continually by internal audit at the UPMC as part of the capability maturity model process that is required of all UPMC databases and as part of the UPMC policies and procedures for maintaining data integrity.5
For the period January 1, 2000, through September 30, 2009, 140,781,751 laboratory reports, 5,720,470 visits, and 139,750,158 charge records representing approximately 290,552 patients were searched. First, the initial source population was identified by the presence of any one of the following six criteria: ICD-9 code 250 (DM) for either inpatient, emergency room (ER), or outpatient visits (treated as three separate indicators); any hemoglobin A1c (HbA1c) result (regardless of value); a blood glucose >200 mg/dl; or use of any DM medication (i.e., acarbose, acetohexamide, chlorpropamide, exenatide, glimepiride, glipizide, glucagon, glyburide, insulin, metformin, miglitol, nateglinide, pioglitazone, repaglinide, rosiglitazone, sitagliptin, tolazamide, tolbutamide, troglitazone, or pramlintide).5 Using the previously validated criteria of Zgibor and colleagues,5 two or more of these indicators or an outpatient diagnosis identified DM patients. Zgibor and colleagues5 showed that using any HbA1c result (regardless of values) plus another indicator performed better than choosing a specific HbA1c cut point. Patients were excluded who did not meet these criteria (n = 80,145) or were younger than 18 years of age (n = 818). The sample included 209,647 DM patients aged ≥18 years. Data management was done using SAS 9.2 (SAS Institute, Inc., Cary, NC).
Identifying Probable Type 1 and Type 2 Diabetes Mellitus Cases
Based on a literature review and clinical input, indicators for T1DM9–14 or T2DM15 were defined according to the strength of associations with T1DM or T2DM. Available variables obtained from the MARS database included ICD-9 codes for T1DM (250.×1 or 250.×3) and T2DM (250.×0 or 250.×2); diabetic ketoacidosis (DKA); hypoglycemic coma; comorbidities (i.e., Addison's, thyroid, or celiac disease); complications (i.e., myocardial infarction, coronary artery bypass graft, dialysis, amputation, retinopathy, or neuropathy); inpatient use of insulin, pramlintide, or other oral hypoglycemic agent (OHA); age at time of first entry in the MARS database; and ages at the time of the first confirmed diagnosis of diabetic complications in the MARS database during the study period and at the times of DM diagnoses at inpatient, outpatient, or ER visits. These variables were defined mainly by ICD-9 codes or parsed notes in the electronic medical records.
The outcome variable was the type of DM (i.e., T1DM or T2DM). Indicators for T1DM included (1) inpatient insulin use with no records of OHA use; (2) specific ICD-9 code for T1DM (250.×1 or 250.×3); (3) parsed notes for T1DM-related diagnoses, including childhood-onset DM, juvenile DM, and insulin-dependent DM; (4) DKA diagnosis; (5) hypoglycemia diagnosis; (6) other autoimmune-related comorbidities; (7) any complication diagnosis (especially before the age of 40 years); (8) younger age; and (9) DM diagnosis at ER or inpatient visits.
Indicators for T2DM included (1) inpatient use of OHA with or without insulin; (2) ICD-9 code for T2DM or unspecified type DM (250.×0 or 250.×2); (3) parsed notes for T2DM-related diagnoses, including adult DM and non-insulin-dependent DM; (4) no DKA diagnosis; (5) no hypoglycemic coma diagnosis; (6) no other autoimmune-related comorbidities; (7) presence or absence of other complications at certain ages (e.g., none after age 40 or diagnoses after age 65); and (8) older age. We considered ICD-9 codes for DM diagnosis, medication use, and absence of DKA as relatively strong indicators for T2DM.
To reduce misclassification between T1DM and T2DM cases, we identified “probable” T1DM and T2DM cases using sequential clinical rules (Appendix 1) developed by two pharmacists who are also epidemiologists (Zgibor and Lo-Ciganic). These authors classified patients sequentially by applying the first criterion to all DM patients, then the second criterion to the subset of remaining of patients, and so on. For example, using the first rule, patients with ICD-9 diagnosis of T1DM only, inpatient insulin use, and no inpatient OHA use were considered to be T1DM cases (n = 1,680). Then using rule 2, the remaining patients from the first rule who had an ICD-9 diagnosis of T2DM only, no inpatient insulin use, and inpatient OHA use were considered to be T2DM cases (n = 8,918). The authors randomly selected 30~50% of patients who met each criterion and verified that their electronic medical records were consistent with the assigned category. We excluded 79,963 patients (38.1%) who had no strong indicators for either T1DM or T2DM and a substantial amount of missing data for the weaker indicators. A cohort of 129,684 identified probable cases (8,173 with T1DM and 121,511 with T2DM) was used to construct the tree models. The first 19 clinical rules in Appendix 1 accounted for 92.9% of these probable cases.
Statistical Analysis: Developing a Tree-Structured Model
The “recursive partitioning and regression tree” routines in Spotfire S+® 8.1 (TIBCO Software Inc., Palo Alto, CA) were used to estimate classification tree models in two stages. Starting at the tree root, the most discriminating variable is selected first, to partition data into two nodes. This process is applied recursively to each node until either no improvement is possible or the nodes reach a minimum size. Patients with missing values for any splitting variable are classified using surrogate splits.16–19 The initial tree may be large and complex. In the second stage, 10-fold cross validation is used to prune back the initial tree.17,18 During the 10-fold cross-validation process, trees are fit sequentially to 9/10 of the data, with each remaining 10th withheld in turn; for each tree, a discrepancy measure (the deviance) is computed based on the independent predictions for the subsets not included in the estimation. The resulting cross-validation error rates can be used to compare trees of different sizes (i.e., different numbers of terminal nodes) and to identify an optimal tree that does not overfit the data. The 1-SE rule chooses the smallest sized tree whose cross-validation error rate is within one standard error of the error rate for the tree with the minimum error rate.16 The 1-SE rule and a complexity parameter of 0.10 were used to choose an optimally sized tree. The predictive accuracy of the cross-validated tree to identify T1DM cases was quantified in terms of sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV).
Results
Table 1 shows the distributions of demographic and clinical factors that distinguish T1DM from T2DM. Compared with probable T2DM cases, probable T1DM cases were more likely to be younger at first entry in the MARS database, to be African American, and to have inpatient insulin use, ICD-9 codes for T1DM diagnosis, complications at younger ages, autoimmune-related comorbidities, DKA or hypoglycemic coma diagnoses, and outpatient or ER diagnoses of DM.
Table 1.
Distribution of Each Predictor Variable among Probable Type 1 and Type 2 Diabetes Mellitus Cases in the MARS Data Seta
| Variable | Probable T1DM (n = 8,173) | Probable T2DM (n = 121,511) |
|---|---|---|
| Age at first entry into the MARS database, | ||
| years (mean, SD) | 44.2 (18.2) | 64.9 (14.8) |
| Missing, % (n) | 6.0 (487) | 10.7 (12,966) |
| Gender, % (n) | ||
| Male | 50.4 (4,118) | 47.3 (57,470) |
| Female | 48.5 (3,963) | 51.8 (63,000) |
| Missing | 1.1 (92) | 0.9 (1,041) |
| Race, % (n) | ||
| White | 76.0 (6,208) | 80.0 (97,288) |
| African American | 15.5 (1,268) | 12.8 (15,574) |
| Asian | 0.1 (10) | 0.2 (177) |
| Hispanic | 0.3 (22) | 0.2 (276) |
| Other | 0.9 (74) | 0.9 (1,079) |
| Missing | 17.2 (59) | 5.9 (7,117) |
| Inpatient insulin use, % (n) | 71.2 (5,819) | 51.2 (62,238) |
| Inpatient OHA use, % (n) | 2.4 (193) | 41.9 (50,911) |
| ICD-9 code of DM, % (n)b | ||
| T1DM only | 47.2 (3,858) | 0.2 (241) |
| T2DM or unspecified DM only | 0.5 (38) | 78.6 (95,525) |
| Both T1DM and T2DM | 24.5 (2,010) | 7.0 (8,493) |
| Missing | 27.7 (2,267) | 14.2 (17,252) |
| Age at first complicationc in the MARS database, years (mean, SD) | 48.2 (15.0) | 70.4 (12.1) |
| History of CABG or MI, % (n) | 6.8 (558) | 9.6 (11,686) |
| CABG | 3.8 (311) | 5.9 (7,215) |
| MI | 4.7 (380) | 6.0 (7,236 ) |
| Age at first CABG or MI in the MARS database, years (mean, SD) | 53.7 (14.4) | 71.6 (11.5) |
| CABG | 56.7 (13.9) | 72.1 (10.6) |
| MI | 51.8 (14.2) | 71.0 (12.1) |
| History of dialysis, % (n) | 2.9 (233) | 0.6 (679) |
| Age at first dialysis in the MARS database, years (mean, SD) | 41.1 (13.8) | 66.6 (13.2) |
| History of amputation, % (n) | 1.1 (90) | 0.4 (518) |
| Age at first amputation in the MARS database, years (mean, SD) | 48.2 (16.6) | 69.2 (12.8) |
| History of retinopathy, % (n) | 4.2 (345) | 0.7 (903) |
| Age at first retinopathy in the MARS database, years (mean, SD) | 50.2 (13.1) | 65.9 (12.0) |
| History of neuropathy, % (n) | 5.7 (466) | 2.6 (3,103) |
| Age at first neuropathy in the MARS database, years (mean, SD) | 51.2 (12.8) | 66.8 (12.6) |
| History of DKA, % (n) | 17.6 (1,435) | 0.2 (284) |
| History of hypoglycemic coma, % (n) | 9.2 (750) | 0.1 (170) |
| History of thyroid disease, % (n) | 0.9 (73) | 0.6 (680) |
| History of celiac disease, % (n) | 0.3 (25) | 0.06 (76) |
| History of Addison's disease, % (n) | 1.4 (110) | 0.3 (376) |
| Inpatient DM diagnosis, % (n) | 67.8 (5,547) | 65.8 (79,896) |
| Outpatient DM diagnosis, % (n) | 20.4 (1,669) | 11.23 (13,647) |
| ER DM diagnosis,% (n) | 41.3 (3,371) | 34.1 (41,435) |
SD, standard deviation; CABG, coronary artery bypass graft; MI, myocardial infarction.
ICD-9 code of DM: T1DM only = only with ICD-9 code for T1DM-specific diagnosis (250.×1 or 250.×3); T2DM or unspecified DM only = only with ICD-9 code for T2DM or unspecified type DM diagnosis (250.×0 or 250.×2); both T1DM and T2DM = with both ICD-9 codes for T1DM and T2DM or unspecified diagnosis [(250.×1 or 250.×3) and (250.×0 or 250.×2)]; missing values = without any ICD-9 code diagnosis for DM.
Complications include coronary artery bypass graft, myocardial infarction, dialysis, retinopathy, neuropathy, and amputations.
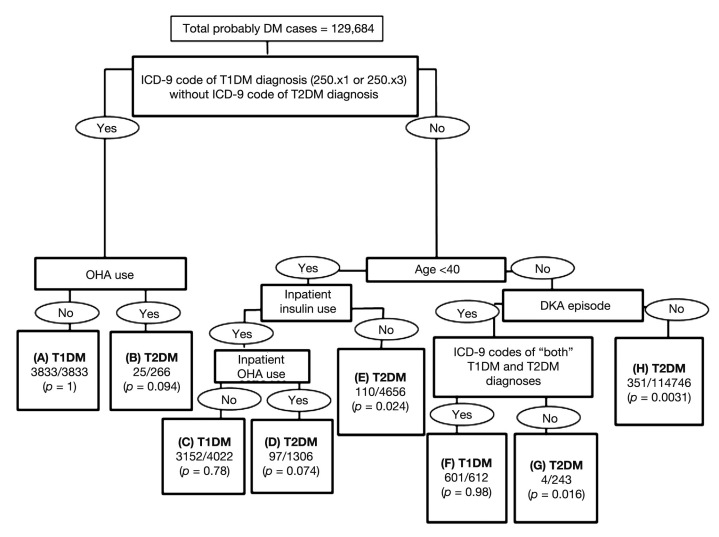
Figure 1 summarizes the tree-based model; successive partitions of data into relatively homogenous nodes are shown with the rule labeling each split. The predictors in this model were ICD-9 codes for DM diagnoses, inpatient OHA use, history of DKA diagnosis, inpatient insulin use, and age <40 years. For example, in terminal node H in Figure 1, 351 patients were probable T1DM cases and 114,395 patients were probable T2DM cases and the estimated probability of T1DM is 0.0031. Node H is defined by having ICD-9 codes of T2DM diagnosis only, both T1DM and T2DM diagnoses (or missing ICD-9 codes of DM diagnosis), age ≥40 (or missing), and no DKA episode (or missing information about history of DKA) in the MARS data during the study period.
Figure 1.
A tree-based model for predicting T1DM and T2DM cases. In each terminal node, the first line represents the predicted category (T1DM or T2DM) and the second line represents the empirical probability of being a T1DM case (i.e., the number of probable T1DM cases divided by total number in that terminal node). The distance between the splits represents the relative importance of the splits. For example, the most important indicator defines the first split.
Table 2 shows node-specific distributions of clinically classified probable T1DM and T2DM cases. Terminal nodes A, F, and C had highest predicted probabilities of T1DM and correctly classified 92.9% of clinically defined T1DM cases. Terminal node C had a higher misclassification rate (22%) than did nodes A and F; this node defines a subgroup more likely to be misclassified because insulin use data were available only on inpatients, and T2DM patients could have used insulin while they were hospitalized. Terminal node H had the highest predicted probability of T2DM and correctly classified 94.1% of probable T2DM cases.
Table 2.
Node-Specific Distributions of the Clinically Classified Probable Type 1 and Type 2 Diabetes Mellitus Cases
| Terminal node | Predicted classification from the tree | Total number of patients | Predicted probability of T1DM | Clinically classified T1DM (n) | Percentage of total T1DM (n = 8173) | Predicted probability of T2DM | Clinically classified T2DM (n) | Percentage of total T2DM (n = 121,511) |
|---|---|---|---|---|---|---|---|---|
| A | T1DM | 3,833 | 1.0 | 3,833 | 46.9% | 0 | 0 | 0% |
| F | T1DM | 612 | 0.98 | 601 | 7.4% | 0.02 | 11 | 0.009% |
| C | T1DM | 4,022 | 0.78 | 3,152 | 38.6% | 0.22 | 870 | 0.7% |
| B | T2DM | 266 | 0.094 | 25 | 0.3% | 0.91 | 241 | 0.2% |
| D | T2DM | 1,306 | 0.074 | 97 | 1.2% | 0.93 | 1,209 | 0.99% |
| E | T2DM | 4,656 | 0.024 | 110 | 1.3% | 0.98 | 4,546 | 3.8% |
| G | T2DM | 243 | 0.016 | 4 | 0.05% | 0.98 | 239 | 0.2% |
| H | T2DM | 114,746 | 0.0031 | 351 | 4.3% | 0.99 | 114,395 | 94.1% |
Considering T1DM as the positive category, sensitivity, specificity, PPV, and NPV of T1DM cases for the tree-structured model were 92.8%, 99.3%, 89.5%, and 99.5%, respectively (Table 3). Approximately 7.2% of T1DM cases were misclassified as T2DM, and 0.73% of T2DM cases were misclassified as T1DM, with an overall misclassification rate of 1.1%.
Table 3.
Summary of the Eight-Node Predictive Tree Model of Probable Type 1 and Type 2 Diabetes Mellitus Cases
| Tree model | ||||
|---|---|---|---|---|
| T1DM | T2DM | Total | ||
| T1DM | 7,586 sensitivity = 92.8% | 587 | 8,173 | |
| Clinical rules | T2DM | 881 | 120,630 specificity = 99.3% | 121,511 |
| Total | 8,467 PPV = 89.5% | 121,217 NPV = 99.5% | 129,684 | |
Discussion
Predictors of T1DM in this tree-structured model were ICD-9 codes of T1DM or T2DM, history of DKA, age <40, inpatient insulin use, and inpatient OHA use. This tree model performed well in identifying T2DM cases in a combined cohort of patients with DM, with 99.3% specificity and 99.5% NPV. To the best of our knowledge, no other studies have distinguished between T1DM and T2DM using a large administrative/clinical database. Although both the clinical prediction rule and the tree model depend on variables coded in the MARS database, the strategy would generalize to other settings. In addition, the tree model developed for this large diverse cohort of UPMC patients may generalize to other populations of DM patients.
Some limitations in this study were inherent in the MARS administrative database. First, patient age was available at the time of first seeking UPMC services between 2001 and 2009, but not at the time of DM diagnosis. Also, age is missing for 10.4% of probable DM patients. Second, some informative factors of T1DM status (e.g., body mass index, presence of islet antibodies, low C-peptide, and other genetic and environmental factors) rarely are described in the charts of patients with DM. Third, medication data on insulin and/or OHA use were available only for inpatients. Inpatient insulin use alone is not a reliable indicator of T1DM status, because DM patients are more likely to use insulin while they are hospitalized, e.g., after surgery. The performance of these prediction rules could be improved if outpatient insulin and OHA data were available. Fourth, results from this adult cohort (age ≥18 years) would not apply to children. Fifth, laboratory data were missing for samples sent to non-UPMC laboratories that do not supply data to the MARS.
Another limitation is the incomplete ascertainment of DM cases in the initial sample. It is likely some patients with only a single indicator who did not meet Zgibor and colleagues'5 criteria may truly have DM. The number of actual cases is likely to be underestimated, because most people with T2DM remain asymptomatic for years after onset of the disease. Furthermore, misclassification between T1DM and T2DM may occur. For example, patients with T2DM may present with DKA, or patients with late onset T1DM and a slow but virulent progression of disease may show features of autoimmune disease. No independent source or “gold standard,” such as anti-body titers, was used to distinguish T1DM and T2DM cases. Our reference standard is expert clinical judgment applied to administrative records. In addition, the sequential clinical rules for identifying probable cases in this study are limited by a substantial amount of missing clinical information; missing data among patients without clear indicators for either T1DM or T2DM diagnosis precluded classification of 38.1% of the cohort. Finally, the accuracy of our predictive rule has not yet been ascertained through any external validation.
While inherent limitations of this administrative database affect both the clinical classification rule and the tree model, the tree efficiently distinguishes almost all identified T2DM cases from a cohort of adult DM patients quickly and efficiently using multiple criteria. However, the tree model less reliably and accurately identifies T1DM cases.
Accurate information about magnitude, distribution, and types of DM can inform policy and support health care evaluation and clinical prognosis. Being able to distinguish between these DM subtypes will facilitate subtype-specific analyses of medical care costs, morbidity, and mortality. This predictive model allows researchers to easily identify subtypes of a population of patients with DM who are more or less likely to exhibit outcomes of interest; to track different processes of care, costs of DM care delivery, and progression of clinical outcomes from large databases; and to identify characteristics that are important barriers to or facilitators of relevant health-related behaviors among T1DM or T2DM cases.
Conclusion
The preliminary predictive rule to distinguish between T1DM and T2DM cases in a large administrative database appears to be promising and needs further validation. However, the clinical rules to distinguish T1DM and T2DM are limited by a substantial amount of missing clinical information. Furthermore, the clinical rules more accurately identify T2DM than T1DM cases. Future work will focus on ascertaining the validity of our predictive rule in a database of patients with definitive diagnoses of T1DM or T2DM and assessing the generalizability of the rule to other administrative databases. Accurate classification will facilitate subtype-specific analysis for patients with T1DM and T2DM.
Acknowledgments
We especially thank John McMichael for his help with the MARS database.
Abbreviations
- (CART)
classification and regression tree
- (DKA)
diabetic ketoacidosis
- (DM)
diabetes mellitus
- (ER)
emergency room
- (HbA1c)
hemoglobin A1c
- (ICD-9)
International Classification of Disease, 9th revision
- (MARS)
Medical Archival Retrieval System
- (NPV)
negative predictive value
- (OHA)
oral hypoglycemic agents
- (PPV)
positive predictive value
- (T1DM)
type 1 diabetes mellitus
- (T2DM)
type 2 diabetes mellitus
- (UPMC)
University of Pittsburgh Medical Center
References:
- 1.World Health Organization. Diabetes fact sheet N°312. 2011. http://www.who.int/mediacentre/factsheets/fs312/en/. Accessed December 7, 2010.
- 2.Centers for Disease Control and Prevention. Atlanta: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention; 2008. National diabetes fact sheet: general information and national estimates on diabetes in the United States, 2007. [Google Scholar]
- 3.Dall TM, Zhang Y, Chen YJ, Quick WW, Yang WG, Fogli J. The economic burden of diabetes. Health Aff (Millwood) 2010;29(2):297–303. doi: 10.1377/hlthaff.2009.0155. [DOI] [PubMed] [Google Scholar]
- 4.Asghari S, Courteau J, Carpentier AC, Vanasse A. Optimal strategy to identify incidence of diagnostic of diabetes using administrative data. BMC Med Res Methodol. 2009;9:62. doi: 10.1186/1471-2288-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zgibor JC, Orchard TJ, Saul M, Piatt G, Ruppert K, Stewart A, Siminerio LM. Developing and validating a diabetes database in a large health system. Diabetes Res Clin Pract. 2007;75(3):313–319. doi: 10.1016/j.diabres.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Breiman L, Friedman JH, Olshen RA, Stone CJ. Belmont: Wadsworth International Group; 1984. Classification and regression trees. [Google Scholar]
- 7.University of Pittsburgh Medical Center. UPMC Fast facts. http://www.upmc.com/AboutUPMC/Pages/default.aspx. Accessed December 13, 2010. [Google Scholar]
- 8.AADE. Siminerio LM, Drab SR, Gabbay RA, Gold K, McLaughlin S, Piatt GA, Solowiejczyk J, Weil R. Diabetes educators: implementing the chronic care model. Diabetes Educ. 2008;34(3):451–456. doi: 10.1177/0145721708316627. [DOI] [PubMed] [Google Scholar]
- 9.Sperling MA, Weinzimer SA, Tamborlane WV. Diabetes mellitus. In: Sperling MA, editor. Pediatric endocrinology. 3rd ed. Philadelphia: Saunders; 2008. [Google Scholar]
- 10.American Diabetes Association. Standards of medical care in diabetes–2009. Diabetes Care. 2009;32(Suppl 1):S13–S61. doi: 10.2337/dc09-S013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daneman D. Type 1 diabetes. Lancet. 2006;367(9513):847–858. doi: 10.1016/S0140-6736(06)68341-4. [DOI] [PubMed] [Google Scholar]
- 12.Lipton RB. Is now the time for an intervention to prevent autoimmune type 1 diabetes? Pediatr Diabetes. 2001;2(1):12–16. doi: 10.1046/j.1399-543x.2001.020103.x. [DOI] [PubMed] [Google Scholar]
- 13.Polonsky KS, Licinio-Paixao J, Given BD, Pugh W, Rue P, Galloway J, Karrison T, Frank B. Use of biosynthetic human C-peptide in the measurement of insulin secretion rates in normal volunteers and type I diabetic patients. J Clin Invest. 1986;77(1):98–105. doi: 10.1172/JCI112308. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson C, Susan L, Lynch A, Saria R, Peterson D. Patients with diagnosed diabetes mellitus can be accurately identified in an Indian Health Service patient registration database. Publice Health Rep. 2001;116(1):45–50. doi: 10.1016/S0033-3549(04)50021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCall AL, Saunders JT. Diabetes mellitus in adults. In: Rakel RE, Bope ET, editors. Conn's current therapy. Philadelphia: Saunders; 2009. [Google Scholar]
- 16.Venables WN, Ripley BD. tree-based methods. 4th ed. New York: Springer; 2002. Modern applied statistics with S. [Google Scholar]
- 17.TIBCO Spotfire S+® 8.1 for Windows®. User's guide. TIBCO Software, Inc. tn.spotfire.com/pdfud/TIBCO_Spotfire_SPlus_Users_Guide.pdf. Accessed April 26, 2011. [Google Scholar]
- 18.Therneau TM, Atkinson EJ. Rochester: Mayo Foundation; 1997. An introduction to recursive partitioning using the RPART routines. [Google Scholar]
- 19.Clark LA, Pregibon D. Tree-based models. In: Chambers JM, Hastie TJ, editors. Statistical models in S. Pacific Grove. Wadsworth & Brooks; 1992. [Google Scholar]