Abstract
Background
After positive selection, the newly generated single positive (SP) thymocytes migrate to the thymic medulla, where they undergo negative selection to eliminate autoreactive T cells and functional maturation to acquire immune competence and egress capability.
Methodology/Principal Findings
To elucidate the genetic program underlying this process, we analyzed changes in gene expression in four subsets of mouse TCRαβ+CD4+CD8- thymocytes (SP1 to SP4) representative of sequential stages in a previously defined differentiation program. A genetic signature of the migration of thymocytes was thus revealed. CCR7 and PlexinD1 are believed to be important for the medullary positioning of SP thymocytes. Intriguingly, their expression remains at low levels in the newly generated thymocytes, suggesting that the cortex-medulla migration may not occur until the SP2 stage. SP2 and SP3 cells gradually up-regulate transcripts involved in T cell functions and the Foxo1-KLF2-S1P1 axis, but a number of immune function-associated genes are not highly expressed until cells reach the SP4 stage. Consistent with their critical role in thymic emigration, the expression of S1P1 and CD62L are much enhanced in SP4 cells.
Conclusions
These results support at the molecular level that single positive thymocytes undergo a differentiation program and further demonstrate that SP4 is the stage at which thymocytes acquire the immunocompetence and the capability of emigration from the thymus.
Introduction
Acquisition of a complete and functional competent T cell repertoire requires that T cell progenitors undergo a tightly regulated developmental program. This eventful program includes T cell lineage commitment, V(D)J recombination at the γδ and αβ T cell receptor (TCR) loci, positive selection of T cells with appropriate MHC restriction, phenotypic proceeding of CD4-CD8- (double-negative, DN) cells through CD4+CD8+ (double-positive, DP) to CD4+CD8- or CD4-CD8+ (single positive, SP) cells, negative selection, and final maturation of SP cells [1], [2]. The latter two events occur in the thymic medulla whereas the rest take place in the thymic cortex.
For decades, SP thymocyte maturation has received the least attention and the cells are simply viewed as a nearly uniform population that has finished the positive selection and are waiting for the emigration from the thymus to the periphery. Recently, however, accumulating evidence suggested that SP thymocytes are heterogeneous and undergo an ordered differentiation program. The newly generated CD4+ and CD8+ SP cells gradually change their surface markers [3], [4], [5], acquire the resistance to apoptosis and capabilities to proliferate and produce cytokines when triggered with various stimuli [3], [6], [7], [8], [9]. Most importantly, T cells undergo negative selection to rid the repertoire of potentially autoreactive specificities [8], [10], [11]. After about 4–5 days of residence in the medulla, the proliferation competent “mature” CD4+ and CD8+ SP cells emigrate from the medulla and/or corticomedullary junction (CMJ) following homeostatic cues of the peripheral T cell pool [12], [13], [14], [15], [16]. In addition, some evidence suggests that the development of unique regulatory properties in subsets such as regulatory T (Treg) cells may take place in the medulla [17], [18].
Polarized migration from the cortex to the medulla, movement within the medulla, and normal medullary structure and components are essential for proper SP development, repertoire selection, and T cell function [19], [20], [21], [22]. Preventing SP cells from entering or accumulating in the medulla such as that in Ccr7 -/- and plt mice, or defects in the medulla such as those caused by Aire and Relb deficiency could result in autoimmunity, strongly supporting the important role of the medullary microenvironment in SP thymocyte development, in particular, negative selection against self-reactivity [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36]. However, the molecular mechanisms govern the SP migration and development remain elusive.
We have previously resolved CD4+ SP thymocytes into four subsets: SP1 (6C10+CD69+), SP2 (6C10-CD69+), SP3 (CD69-Qa-2-), and SP4 (CD69-Qa-2+), of which the proliferation, cytokine production, and survival capacities showed a trend of SP1<SP2<SP3<SP4. Multiple approaches were used to demonstrate that these four subsets define a linear, multiple-stage developmental program for the newly generated CD4+ SP thymocytes prior to their exportation to the periphery [37]. A severe blockage at the transition from SP3 and SP4 was found in mice deficient in Aire and Relb, indicating that this transition could be an important checkpoint [37]. Further studies of the mechanisms governing this developmental program will require a systemic analysis of molecular changes in these SP subsets. To this end, the array-based and/or high throughput sequencing-based analysis will be extremely useful to investigate gene expression signatures. However, all the data currently available treat the SP thymocytes as a homogenous population and are not ready to be applied for the analysis of the genes involved in various aspects of the differentiation within SP thymocytes [38], [39], [40], [41], [42], [43], [44], [45], [46]. Thus, we performed here a comparative microarray analysis of these four subsets of TCRαβ+CD4+CD8- thymocytes and found a clear signature of cell migration at the molecular level.
Results
Gene expression profile of single positive thymocytes
To identify genes involved in medullary thymocyte development, we compared genome-wide mRNA expression profiles of the four subsets of TCRαβ+CD4 SP thymocytes. We used antibodies specific for 6C10, CD69, and Qa-2 to separate these subsets. CD25+ and NK1.1+ cells were excluded to avoid the influence of mature and/or returning mature Treg and NKT cells (Figure 1a). All populations were purified to 95–99% (Figure 1b), and total RNA was used to generate biotinylated cRNA. Complementary RNA from three independent preparations of SP1 to SP4 subsets was hybridized to Affymetrix murine gene arrays, containing 45101 probe sets. The data have been deposited in NCBI's Gene Expression Omnibus [47] and are accessible through GEO Series accession number GSE30083 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30083).
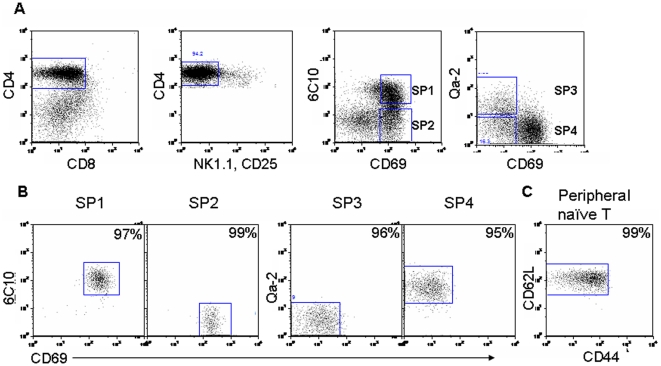
Figure 1. Isolation of CD4+ single positive thymocyte subsets and CD4+ naïve T cells from lymph nodes.
(A–B) Thymocytes pooled from 15–20 mice were treated with anti-CD8 (3.155) mAb and complement. Viable cells were then separated by density centrifugation and stained with mAbs against mouse CD4, CD8 (53-6.7), CD69, and 6C10 or Qa-2. To exclude the contamination of NKT and Treg cells, PE-conjugated anti-NK1.1 and anti-CD25 antibodies were included in the mixture of staining Abs. CD4+CD8-CD25-NK1.1- medullary thymocytes were gated (A) and sorted into the four subsets (B) defined in the text (i.e., SP1–SP4). The numbers in the top-right corners indicate the purity of the sorted cells. (C) Brachial, mesenteric, and inguinal lymph nodes were pooled from 5 to 8 mice and stained with mAbs against mouse CD4, CD8, CD44, CD62L, NK1.1, and CD25. CD4+CD8-CD44lo/intCD62LhiCD25-NK1.1- naïve cells were sorted. The number indicates the purity of the sorted cells.
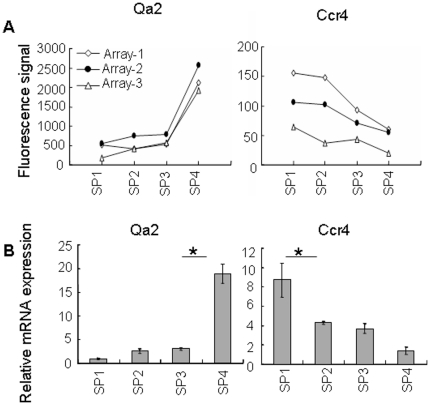
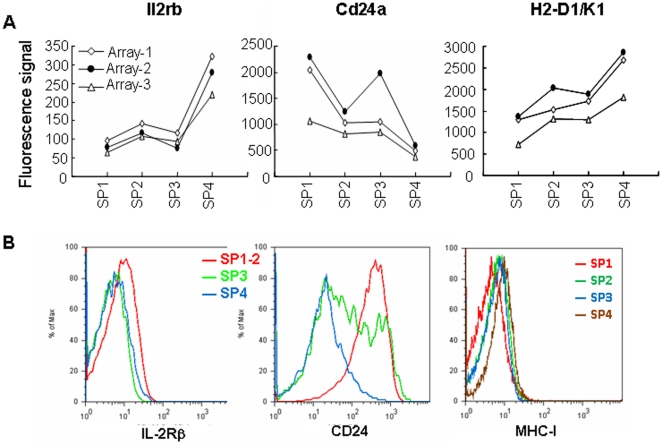
To test the general validity of the chip-based hybridization, quantitative RT-PCR (qRT-PCR) and flow cytometry were used to confirm the differential expression of several genes among the four subpopulations. In addition, our initial cell purification strategy for the microarray analysis did not exclude a mature conventional T cell population (Qa-2+) that migrate from the periphery back to the thymus [48], [49], [50], [51], [52], the acquired array data in the SP4 subset may thus be contaminated by the presence of less than 5% of mature T cells (data not shown). Since most of these thymus-returning T cells have an activated/memory phenotype (CD44hi, composed of 80% of GFP negative CD4+ SP thymocytes in RAG2p-GFP transgenic mice, data not shown), the RNAs of the four SP subsets used in the qRT-PCR validation were differently sorted to exclude the NK1.1+, CD25+, and CD44hi cells. Consistent with our cell purification strategy, CD25 and NK1.1 mRNAs were found to be expressed at extremely low levels both in arrays and in qRT-PCR (data not shown). Qa2 mRNA expression was largely restricted to SP4 cells in the array data and was confirmed by qRT-PCR (Figures 2a–b), agreeing well with the fact that Qa-2 was used as a marker to separate the SP4 cells. Other genes such as Ccr4, Cd24, Il2rb, and MHC class I also showed similar expression patterns in the array, qRT-PCR, and flow cytometry (Figures 2 and 3).
Figure 2. General validation of gene expression.
(A) Gene expressions of Qa2 and Ccr4 in the four thymocyte subsets are revealed by the microarray analysis. (B) Quantitative RT-PCR results of Qa2 and Ccr4 transcripts in the four thymocyte subsets. Asterisks indicate significant (P<0.05) changes in each two-subset comparison. The experiment was performed for three times and similar results were obtained.
Figure 3. Expressions of Il2rb, Cd24, and H2-D1/K1 among the four subsets of CD4+ SP thymocytes.
(A) Gene expression of Il2rb, Cd24a, and H2-D1/K1 by the microarray analysis. (B) Expression of IL-2Rβ, CD24, and MHC class I by flow cytometry.
Overview of the differentially expressed genes
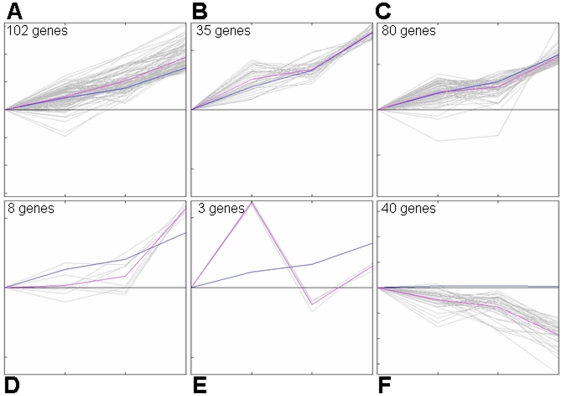
Sets of probes whose expression increased or decreased by two-fold or more in any comparison were identified (see Table 1 for the number of genes and Table S1 for details). It is interesting to note that the majority of differentially expressed genes were enhanced along the developmental pathway (from SP1 to SP4). Based on the Kohonen's self-organizing maps (SOM analysis), six patterns of expression kinetics of the differentially expressed genes in the four subsets were further established. Roughly, they could be grouped into three categories: gradual increase (Figures 4a–d), gradual decrease (Figure 4f), and others (Figure 4e). Gene ontology (GO) analysis indicated that the genes within the gradual-increase category were related to such functions as immune response, lymphocyte activation, metabolic process, transcription, migration, and regulation of apoptosis. The KEGG pathway analysis revealed that cytokine-cytokine receptor (including chemokine-chemokine receptor) interaction, cell adhesion molecules, Jak-STAT signaling pathway, and immune response-related pathways were among the most significant pathways (Table S2). These data suggest that acquiring appropriate capabilities of migration, cytokine production, and immune function may be the major events in SP thymocyte development.
Table 1. Numbers of genes that are differentially expressed among the four subsets.
| Numbers of genes increased | Numbers of genes decreased | |
| SP2 vs. SP1 | 4 | 0 |
| SP3 vs. SP1 | 22 | 0 |
| SP3 vs. SP2 | 20 | 20 |
| SP4 vs. SP1 | 253 | 28 |
| SP4 vs. SP2 | 160 | 14 |
| SP4 vs. SP3 | 93 | 3 |
The numbers of genes that had greater than a 2-fold change in each two-subset comparison were separately listed as numbers of genes increased and decreased.
Figure 4. Patterns of expression kinetics among the four subsets of CD4 single positive thymocytes based on SOM analysis.
(A–F) Six patterns of expression kinetics are shown.
Molecular signatures of functional maturation of SP thymocytes
By comparing various SP subsets directly purified from the thymus, we have shown that upon anti-CD3 and anti-CD28 stimulation in vitro, the SP1 and SP2 thymocytes undergo apoptosis and produce little or no cytokines whereas SP3 and SP4 cells have increased capabilities of proliferation and cytokine production [53]. This increase of functional competence along with the SP development was supported at the molecular level by the up-regulation of a variety of mRNAs including MHCs (Figures 3a–b), costimulatory molecules (Icos, Ctla4, Pdcd1, Tnfsf8), cytokines and their receptors (Il2rb, Il6ra, Tnfsf8, Tnfrsf25, Tgfbr2), and transcription factors and intracellular proteins that regulate immune responses (Pik3cd, Ppp3cc, Malt1, Ctss, Dtx1) (Figure 5a). Most of these significantly up-regulated molecules had highest expression in the cells at the SP4 stage, indicating the importance of this stage in the functional maturation of SP thymocytes. This gain of immune competence at the molecular level was further supported at the cellular level. When the same number of purified SP3, SP4 thymocytes and naïve CD4+ T cells from C57BL/6 Ly5.1 mice were adoptively transferred into congenic Ly5.2 mice, similar levels of donor T cell activation were observed seven days after Listeria monocytogenes infection of the host (Figure 5b). Surprisingly, unlike our previous finding that the SP3 cells proliferate less and produced less cytokines in vitro compared to the SP4 cells, we didn't see such significant differences in vivo. The functional maturation of the SP3 cells in the periphery, like that of the recent thymic emigrants (RTEs), may partially explain the differences between in vitro and in vivo [54], [55] (data not shown).
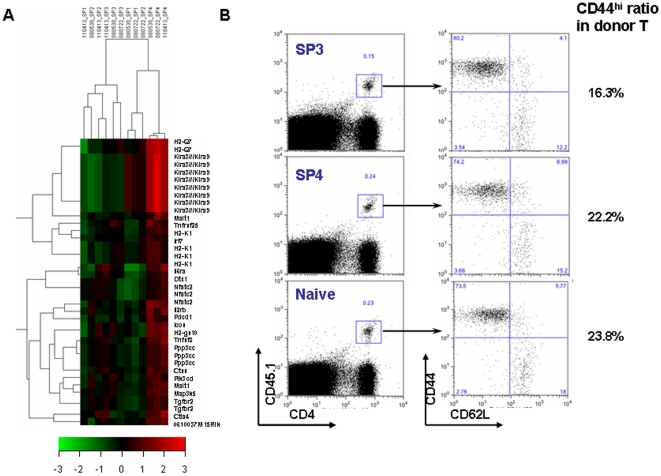
Figure 5. CD4 SP thymocytes gradually acquire immuno-competence.
(A) Expression profiles of the immune-related genes among the four subsets. The profile was generated by hierarchical clustering using GeneCluster 2.0 package. All data from three array repeats were shown (080530_SPx, 080722_SPx, and 110413_SPx, x represents 1 to 4). Red represents up-regulation of gene expression, and green represents down regulation of gene expression. The entire list of genes is available in Table S2. (B) Similar activation of SP3, SP4 thymocytes and naïve CD4+ T cells upon Listeria Monocytogenes (Lm) infection. Purified SP3, SP4 thymocytes and naïve CD4+ T cells from C57BL/6 Ly5.1 congenic mice were adoptively transferred into C57BL/6 Ly5.2 mice (9×106 per host mouse). One day later, the host mice were challenged with 2×104 live Lm via tail vein injection. On day 7 of the infection, the cells from the lymph nodes and spleens were harvested and stained with CD45.1, CD4, CD44, and CD62L. The donor cells with the expression of both CD45.1 and CD4 are gated (shown in left paneal). The expression of CD44 and CD62L of donor cells (CD45.1+CD4+) are shown on the right panel.
Regulation of migration and adhesion during SP maturation
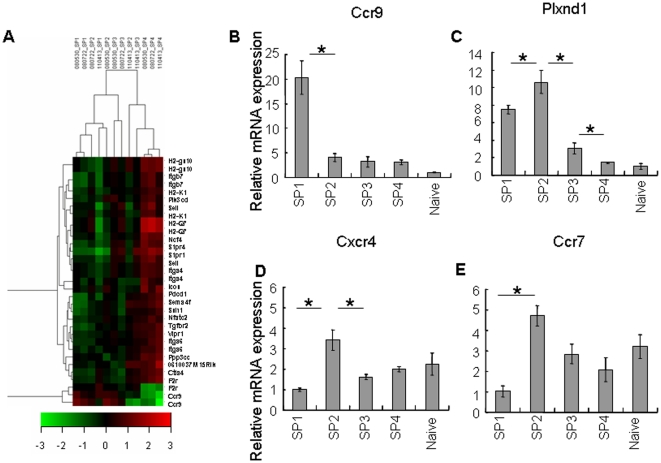
The migration of SP thymocytes from the cortex to the medulla and the migration of SP thymocytes on medullary stroma are important for proper SP maturation and negative selection. The egress of SP cells from the thymus to the periphery also requires the cells capable of responding to the guidance from the periphery. Adhesion molecules and directional cues that are either attractive or repulsive may all contribute to the cell migration. As can be seen in our array data, there are a variety of changes in gene expression of chemokine receptors, adhesion molecules, cytokeletal molecules, signaling molecules, and other factors known to regulate cell motility in other systems (Figure 6a). The kinetics of these changes can be grouped into gradual increase and gradual decrease patterns as described previously. Interestingly, only one gene, Ccr9, fell into the continuously decreased expression profile. The decrease of Ccr9 mRNA level was confirmed by qRT-PCR in which the amount of Ccr9 transcript was significantly down-regulated in the SP2 cells and was lowest in the SP4 (Figure 6b). CCR9 is expressed in DP thymocytes and activated DP respond to CCR9 ligand (CCL25) expressed by cortical epithelial cells [34], [56]. The down-regulation of Ccr9 in SP2 thymocytes may relieve this subpopulation from retaining in the thymic cortex. PlexinD1 has been shown to suppress CCR9/CCL25 signaling and facilitate the medullary entry of positively selected CD69+ thymocytes [41]. Unlike the results from the array data where no significant differences were found, those from qRT-PCR showed high levels of Plxnd1 transcripts in SP1 and SP2 cells but significantly lower levels in SP3 and SP4 cells (Figure 6c). Compared to SP1 cells, the higher level of Plxnd1 in SP2 seems to correlate with the lower level of Ccr9 in these cells, facilitating their cortex-to-medulla migration. CCR7 is also an essential molecule in this type of migration [33], [38], [57]. Its deficiency leads to the accumulation of SP thymocytes in the cortex and the development of autoimmunity [33], [57]. Interestingly, the transcripts of Ccr7 were rather low in the newly generated SP thymocytes (SP1 cells), but were quickly up-regulated and maintained at high levels afterwards with the highest expression at the SP2 stage (Figure 6e). These expression patterns between SP1 and SP2 cells indicate that the cells at the SP2 stage might be the main population capable of migrating from the cortex to the medulla. The down-regulation of Ccr9 and up-regulation of Ccr7 and Plxnd1 may be coordinated to direct the SP2 thymocytes to enter the medulla. Other molecules that may contribute to the cortex-to-medulla migration include CCR4 and CXCR4. The expression of CCR4 in the thymocytes was reported to increase after positive selection while its ligands were produced predominantly in the medullary CD80hi epithelial cells [58], [59], [60], [61]. CXCR4 also changes after positive selection but its role in the medullary entry is not very clear [62], [63], [64]. However, in our array and qRT-PCR results, the level of Ccr4 transcript was highest in the SP1 cells (a population that is likely derived from DPs after positive selection) and significantly down-regulated in the SP2 and other subgroups (Figures. 2a–b) whereas that of Cxcr4 showed a transient and mild (though significant) increase in the SP2 cells (Figure 6d). Thus, CCR4 and CXCR4 may not play a major role in directing the medullary entry.
Figure 6. Molecular basis of the adhesion and migration of CD4 SP thymocytes during their final maturation process.
(A) Expression profiles of the adhesion/migration-related genes among the four subsets. The profile was generated by hierarchical clustering using GeneCluster 2.0 package. All data from three array repeats were shown (080530_SPx, 080722_SPx, and 110413_SPx, x represents 1 to 4). Red represents up-regulation of gene expression, and green represents down regulation of gene expression. The entire list of genes is available in Table S2. (B–D) Gene expression of chemotactic molecules that have been reported to change after positive selection. Relative mRNA level of Plxnd1 (B), Ccr9 (C), Cxcr4 (D), and Ccr7 (E) in the four subsets of CD4 SP thymocytes as determined by qRT-PCR. Asterisks indicate significant (p<0.05) changes compared between the two subsets. Three experiments were performed on RNAs from three independently isolated cell populations for each thymocyte subset and similar results were seen.
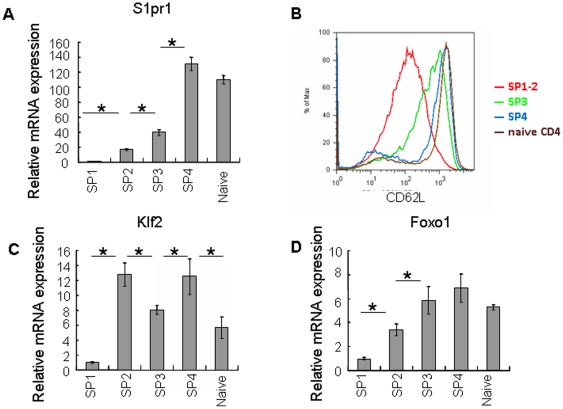
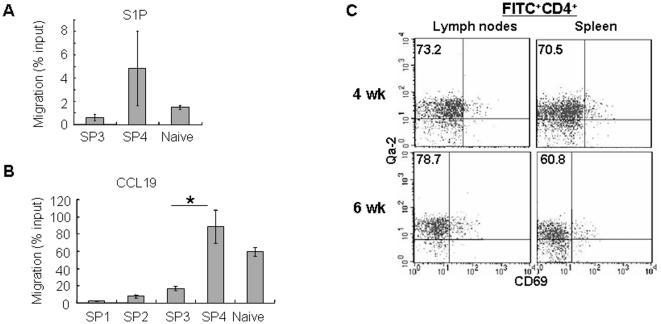
Eighteen genes demonstrated continuously increased kinetics in the microarray data. Among them, S1pr1 (S1P1) and Sell (CD62L) showed an up-regulation in the SP3 and SP4 subsets (data not shown). This trend of expression was confirmed by qRT-PCR of S1pr1 (Figure 7a) and FACS staining of CD62L (Figure 7b). It has been reported that both mature SP thymocytes and RTEs highly express S1P1 and CD62L [15], [21]. The surface receptor S1P1 allows SP thymocytes being exported from the thymus in response to S1P present in the circulation as well as produced by a thymic stromal cell type such as neural crest-derived perivascular cells [14], [15], [65], [66], [67]. CD62L enables the emigrants homing to the secondary lymphoid organs (SLO). We thus further examined the chemotaxis of SP thymocytes towards S1P. Consistent with the high expression of the receptor, S1pr1, in the SP4 subset, only cells from this group migrated towards S1P in a comparative way as naïve CD4+ T cells (Figure 8a). As a control of the migration assay, we chose CCL19 because the expression pattern of its receptor, CCR7, was different from that of S1pr1. The level of CCR7 transcripts was high in most of the CD4+ SPs except the cells at the SP1 stage (Figure 6e). Furthermore, CCR7 is essential in both cortex-to-medulla migration in the thymus and SLO homing in the periphery [68], [69]. Unexpectedly, however, the SP2 and SP3 cells did not respond to CCL19 as efficiently as the SP4 and naïve T cells (Figure 8b). Although the cause for this discrepancy is unclear, the results of both migration assays provide the functional evidence that the SP4 cells may be best equipped to leave the thymus. Indeed, when FITC was introduced into adult mice via intrathymic injection (a common approach to study RTEs), most of the FITC positive CD4+ T cells in the periphery (lymph nodes and spleen) had a Qa-2+CD69- phenotype (SP4 phenotype) (Figure 8c). When purified SP1 thymocytes were injected into the thymus, we also found that most of the cells being exported had a SP4 phenotype [53]. In addition, studies with RAG2p-GFP transgenic mice by Fink group and ours consistently found that most of the GFPhi T cells (representing RTEs) in the periphery of the adult mice have a Qa-2+ phenotype [54] (and data not shown). Together, these data suggest that the SP4 subset is the one in the thymus that acquires the capability to egress.
Figure 7. Expression of genes that are involved in the thymocyte migration and in Foxo1-Klf2 axis.
(A, C–D) Relative mRNA amount of S1pr1, Klf2, and Foxo1 in naïve CD4+ T cells and the four subsets of CD4 SP thymocytes as determined by real time RT-PCR. (B) Expression of CD62L as determined by flow cytometry in peripheral naïve CD4+ T cells, double positive thymocytes, and the four subsets of CD4 single positive thymocytes. Asterisks indicate significant (p<0.05) change compared between the two subsets. The experiment was repeated for two to three times and similar results were seen.
Figure 8. Cells with the SP4 phenotype are the main population in RTEs.
(A-B) Transwell migration of SP thymocytes and naïve CD4+ T cells to S1P (A) and CCL19 (B). Three experiments were performed and the mean and standard deviation were shown. Asterisks indicate significant (P<0.05) difference between SP3 and SP4 subsets. (C) Phenotype of RTEs. Twenty-four hours after intrathymic injection of FITC, the cells from the lymph nodes and spleen were harvested and stained with CD4, Qa-2 and CD69. The surface expression of Qa-2 and CD69 in FITC+CD4+ T cells are shown. The numbers represent the percentage of Qa-2+CD69- cells in FITC+CD4+ T cells. The experiment was repeated for two more times and similar results were obtained.
Kruppel Like Factor 2 (KLF2) is reported to directly regulate the expression of egress-enabling molecules S1P1 and CD62L [21], [70], [71]. Foxo1 has also been shown to be involved in regulating CD62L expression. Other molecules that are regulated by Foxo1 include KLF2 and CCR7. Mice deficient in Klf2 or Foxo1 were found to have similar failures in thymic egress as those lack S1P1 [68], [71], [72]. In our array data, both Klf2 and Foxo1 presented a gradually increased expression pattern from SP1 to SP4 cells whereas the qRT-PCR of Klf2 showed a significant (P<0.05) and sudden increase at SP2 (Figures 7c–d and data not shown). This significant (P<0.05) and abrupt up-regulation at the SP2 subpopulation was also seen in Ccr7 by both microarray and real time PCR (Figure 6e and data not shown). Interestingly, the transcripts of Sell and S1pr1 did not reach the highest levels until cells became SP4 (Figures 7a–b). Additionally, highest migratory capabilitiy towards CCL19, a CCR7 ligand, was not acquired until SP thymocytes reach the SP4 stage (Figure 8b). These unexpected late expressions in the SP4 subset suggest that additional events in the SP2 and/or SP3 stages may be required in the regulation of these molecules. In supporting this notion, SP3-to-SP4 transition was found to be blocked in Aire-/- mice and whether the blockage affects the egress is currently under investigation [37], [61]. Taken together, these expression data strongly suggest that SP4 cells are preferentially exported from the thymus. The molecular program that regulates this migration may start from the SP2 stage and continue in the SP3 stage.
Discussion
As many key events in T-lymphopoiesis including negative selection and Treg lineage commitment are found to take place in the thymic medulla, the importance of single positive thymocyte differentiation and medullary microenvironment is gradually appreciated. However, the molecular basis that regulates this developmental program lacks a systemic and detailed analysis, rendering a much slower progress in the research in the SP compared to that in the DN and DP cells. In the present study, mRNA expression profiling by microarray was performed in the four subsets of SP thymocytes which have been previously shown to represent sequential stages in a defined differentiation program in the medulla. A clear molecular signature of cells acquiring functional competence was found. Molecules such as MHCs, costimulatory molecules, cytokines and their receptors showed a gradual increase in their expressions starting from SP2 or SP3 subpopulation, in concert with our previous finding that the development of SP1 to SP4 follows a progressive phenotypic and functional maturation process [37], [53].
During this maturation process, a transition from SP3 to SP4 was found to be critical as a blockage of this transition in Aire -/- and Relb -/- mice is associated with high incidence of autoimmune diseases. Whether this blockage mainly affects the deletion of autoreactive T cells or affects the thymocyte maturation as well is not clear. In our array data, there is a sharp increase of multiple gene transcripts in the SP4 cells. Considering that the normality of thymic stromal cells is required for the SP3 to progress to SP4 [37], this type of abrupt and significant up-regulation of functional genes in the SP4 cells may be partially driven by the signals from medullary stromal cells. The expressional changes of which molecules require such a thymocyte-stromal cell interaction and/or facilitate the transition of SP3 to SP4 cells are currently under investigation. Another possibility of the significant expressional changes in the SP4 cells may be derived from our purification strategy and the impurity of SP4 cells. Although we made an effort to exclude Treg, NKT, and thymus-reentered activated T cells (CD25+, NK1.1+, and CD44hi) from the SP4 subset, SP4 thymocytes may still be heterogeneous, containing cells that are on their way to become mature Treg and NKT and trace amount of naïve T cells that migrate back to the thymus. The molecular signature but not necessarily the cellular function may be affected by this type of contamination. The use of RAG2p-GFP transgenic mice and/or MHC class II-restricted RAG knockout TCR transgenic mice may help to obtain more homogeneous cell population.
The orchestrated migration of developing thymocytes through various specialized microenvironments in the cortex is essential for proper T cell development. But less is known of the thymocyte trafficking from the cortex to the medulla and within the medullary environment. Similarly, although the widely accepted mechanism for thymocytes' exiting the thymus is S1P1-mediated migration, when do SP cells express S1P1 and how to prevent the premature exit of autoreactive cells at the cortico-medullary junction is still not clear [73]. Our array and qRT-PCR results on the four SP subsets suggest a route by which the newly generated SP thymocytes migrate within the thymus: the SP2 cells down-regulate CCR9, up-regulate CCR7 and plexinD1, and migrate from the cortex to the medulla; the SP2 and SP3 cells gradually up-regulate molecules involved in T cell functions and the Foxo1-KLF2-S1P1 axis; the high expressions of S1P1, CCR7, and CD62L in SP4 cells enable these functional competent cells to leave the medulla and enter the periphery. These results provide fine evidence to support the previous finding that the emigration of SP thymocytes is not random and only a subpopulation of SP cells that finish the series of maturational events are capable of exiting the thymus [16]. Development of self tolerant and fully matured SP cells rely heavily on this ordered trafficking as defects in SP thymocyte migration from the cortex to the medulla, such as Ccr7 deficiency, resulted in the ectopic development of SP in the cortex/cortex-medullary junction and autoimmunity in the periphery. Similarly, defects in the thymocytes-epithelial cells interaction in the medulla, such as Aire and Relb deficiency, result in the SP3-to-SP4 blockage and autoimmunity in the periphery [37], [53].
It remains unknown how this highly ordered process is regulated at the molecular level and whether positive selection in the thymic cortex alone or together with signals from the medullary microenvironment including negative selection drive these events to occur in a sequential order. A study of Egr1-deficient mice suggests that positive selection could have an impact on the survival of cells further down the thymocyte developmental pathway and even after entering the periphery [74]. Foxo1 and the Foxo1-KLF2-S1P1 axis could be affected in mice with defects in positive selection due to the fact that the expression and activity of Foxo1 requires TCR stimulation and PI3K [75]. Medullary stromal cells, however, also likely play an important role in SP development in addition to the elimination of autoreactive T cells. At least four pieces of evidence in our data support this, 1) the high expression of immune function-associated genes was mostly seen in the SP4 subset (Figure 5a); 2) although Foxo1 and Klf2 were up-regulated in the SP2 cells, their downstream genes, S1pr1 and Sell, were not fully expressed until cells reached the SP4 stage (Figures 7a-b); 3) only SP4 cells showed a comparative response to CCL19 as naïve T cells despite of an early upregulation of Ccr7 mRNA level in the SP2 subset; 4) the transition of cells from the SP3 to SP4 likely requires a normal medullary microenvironment as defects in the stromal cells (such as those in Aire -/- mice) could block SP3-to-SP4 transition [37], [53]. Thus, SP2 and especially SP3 may be key stages at which thymocytes receive signals from medullary environment including stromal cells.
Taken together, this is the first detailed analysis of the gene expression profiles of CD4 SP subsets. It reveals at a molecular level a clear map of how CD4 SP thymocytes gradually acquire the functionally matured, and ready-to-egress properties. It also provides transcriptomic evidence supporting the model of “maturation-dependent emigration” of SP thymocytes [16]. It established a molecular basis to further study the signals that initiate and govern these progressive changes and to understand what signals are obtained from the positive selection and what from the medullary microenvironment.
Materials and Methods
Mice and Ethics Statement
Congenic C57BL/6 Ly-5,1 (CD45.1) and C57BL/6 Ly-5.2 (CD45.2) mice between 6-8 weeks of age were used throughout this study. The animals were kept in a specific pathogen-free facility at Peking University Health Science Center (Beijing, China). The experimental procedures on use and care of animals had been approved by the ethics committee of Peking University Health Science Center with an approval number of bjmu20110301.
Antibodies
Anti-CD8 (3.155) was prepared from hybridomas obtained from the American Type Culture Collection. The 6C10 (SM6C10) was from Dr. Linna Ding (National Institutes of Health) and was subsequently labeled with FITC. Antibodies against CD4 (H129.19 or RM4-5), CD8 (53-6.7), TCRβ (H57–597), CD69 (H1.2.F3), Qa-2 (H1-1-2), NK1.1 (PK136), CD44 (IM7), CD62L (MEL-14), CD24 (M1/69), IL-2Rβ (5H4), streptavidin-PE/APC, and isotype control antibodies were purchased from BD Pharmingen. Anti-Qa-2 (H1-1-2) and CD25 (PC61) were from eBioscience.
Isolation of peripheral naïve CD4+ T cells and subsets of CD4 SP thymocytes
Single-cell suspensions of thymocytes derived from C57BL/6 mice at 6-8 weeks of age were treated with anti-CD8 (3.155) mAb and complement (guinea pig sera) to remove CD8+ cells. After two cycles of killing and removal of dead cells by density centrifugation, the viable cells were stained with CD4-PerCP-Cy7, CD8-APC-Cy7 (53-6.7), CD69-PerCP-Cy5.5, 6C10-FITC, Qa-2-Biotin-strepavidin-APC, CD25-PE, and NK1.1-PE and then subjected to cell sorting (FACSAria). For the RNAs used in qRT-PCR validation, the cells were further stained with CD44-PE to purify the CD44lo cells. Four subsets of CD4+CD8- thymocytes were thus obtained: SP1 (6C10+CD69+Qa2-), SP2 (6C10-CD69+Qa2-), SP3 (6C10-CD69-Qa2-), and SP4 (6C10-CD69-Qa2+). The naïve CD4+ T cells (CD4+CD44lo/intCD62LhiCD25-NK1.1-) were purified from the lymph node cells from the same mice. The purity of individually sorted groups was 95–99% as reanalyzed on FACS (Fig. 1a–b). The expression of TCRβ was checked among these purified cell subsets and 99.9% of the cells were TCRβ+ (data not shown).
Microarray analysis
Total RNA was isolated from each SP subset using Trizol (Invitrogen) according to the manufacturer's instructions. RNA concentrations were determined by spectrophotometry. Only those samples that showed no degradation (ratios approaching 2∶1 for the 28S and 18S bands) in formaldehyde agarose gel electrophoresis were used to generate labeled targets. The RNA samples were then sent to Bioassay Laboratory of CapitalBio Corporation (Beijing, China) for amplification, biotin labeling, hybridization to Affymetrix Mouse Genome 430 2.0 Arrays (Affymetrix). Three arrays were used for each SP subsets. The hybridization data were analyzed using Affymetrix® GeneChip® Operating Software Version 1.4 (GCOS 1.4). The scanned images were first assessed by visual inspection and analyzed to generate raw data files saved as CEL files using the default setting of GCOS 1.4. A Robust Multi-array Analysis normalization procedure was performed to normalize the different arrays using R bioconductor. The noise and 3′ to 5′ signal ratios of all arrays were listed in Table 2. The data have been deposited in NCBI's Gene Expression Omnibus [47] and are accessible through GEO Series accession number GSE30083 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30083).
Table 2. Quality control data from 16 arrays.
| Noise (Mean±STDEV) | Gapdh 3′/5′ signal ratios | |||||
| Repeat 1 | Repeat 2 | Repeat 3 | Repeat 1 | Repeat 2 | Repeat 3 | |
| SP1 | 1.21±0.04 | 0.9±0.03 | 1.16±0.03 | 1.01 | 1.00 | 1.10 |
| SP2 | 1.13±0.05 | 0.89±0.02 | 1.13±0.04 | 1.05 | 1.04 | 1.08 |
| SP3 | 1.16±0.05 | 1.00±0.04 | 1.07±0.02 | 1.03 | 0.99 | 1.17 |
| SP4 | 1.12±0.04 | 0.84±0.03 | 1.12±0.02 | 1.01 | 1.07 | 1.14 |
In the comparison analysis, a two-class unpaired method in the Significant Analysis of Microarray software (SAM) was applied to identify differentially expressed genes between test and control groups. The selection threshold of false discovery rate is FDR<5% and fold change>2.0 in the SAM output result.
Clustering of self-organizing maps (SOM) was performed by GeneCluster 2.0 package and 9 class (3×3) SOMs were constructed. To organize genes into hierarchical categories and uncover gene regulatory network on the basis of biological process and molecular function, the differentially expressed genes were mapped to Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using MAS (molecule annotation system, http://bioinfo.capitalbio.com/mas) platform. GO terms and KEGG pathways with false discovery rate (FDR)-corrected P values <0.05 were considered statistically significant.
Quantitative RT-PCR
RNA was purified from isolated thymocytes using TRIZOL (Invitrogen) and was used to make cDNA using random primers and the Reverse Transcription System (Promega). For quantitative Real-Time PCR, iQ™ SYBR® Green Supermix (Bio-Rad) was used according to the manufacturer's instructions. Quantitative PCR was performed on an iCycler (Bio-Rad Ltd, Hemel Hempstead, England, UK), with each sample in triplicate, at an RNA equivalence of 15 ng/well. Oligos for each gene are listed in Table 3. PCRs were performed for 40 cycles at 95°C for 30 s, 58°C for 30 s, and 72°C for 20 s. The quantification was based on ΔΔCT calculations and were normalized to GAPDH as loading controls and calibrated to SP1 levels in Figures 2b left, 6d–e, and 7a, c–d; to SP4 levels in Figure. 2b right; and to naïve T cell levels in Figures 6b–c. RT-PCR analysis was carried out on RNA from three independently isolated cell populations for each thymocyte subset and naïve T cells. Statistical analysis of the results obtained from quantitative RT-PCR was performed using GraphPad Prism 5 software (San Diego, CA, USA). Student's t-test was used to evaluate the significance of the differences between group means. Statistical significance was defined as P<0.05.
Table 3. Primers for selected genes analyzed using real-time PCR.
| Forward (5′ - 3′) | Reverse (5′ – 3′) | |
| GAPDH | CGGCCGCATCTTCTTGTGCA | GCAAATGGCAGCCCTGGTGAC |
| Qa2 | CAGGTCTTATGGTGCTGTCA | GCATGTGTAATTCTGCTCCTTC |
| Ccr4 | GGAAGGTATCAAGGCATTTGGG | GTACACGTCCGTCATGGACTT |
| S1pr1 | TCGCCGACAGCAGCAAGATG | AAAGCCAGGTCAGCGAGCAATC |
| Klf2 | CTCAGCGAGCCTATCTTGCC | CACGTTGTTTAGGTCCTCATCC |
| Foxo1 | AGGGCGACAGCAACAGCTCG | TCCATGGACGCAGCTCTTCTCC |
| Ccr7 | GCACCATGGACCCAGGGAAACC | TGAGAGGCAGGAACCAGGCCT |
| Plxnd1 | TGCAGGCTGCCATTCGTGCA | GGCCGCTCCGCAATCCAGTT |
| Ccr9 | CTTCAGCTATGACTCCACTGC | CAAGGTGCCCACAATGAACA |
| Cxcr4 | AGGCGTTTGGTGCTCCGGTA | TTCATCCCGGAAGCAGGGTTCCT |
Flow cytometry
Single-cell suspensions of thymocytes from C57BL/6 mice were removed of CD8+ cells and stained with CD4-PerCP-Cy7, CD8-APC-Cy7 (53-6.7), CD69-PerCP-Cy5.5, Qa-2-Biotin-strepavidin-APC, CD25-PE, CD44-PE, and NK1.1-PE. The expressions of CD62L, CD24, and IL-2Rβ among three subsets of CD4+CD8- cells, including SP1-2 (CD69+Qa2-), SP3 (6C10-CD69-Qa2-), and SP4 (6C10-CD69-Qa2+), were obtained. The lymph node cells from the same mice were stained with CD4-PerCP-Cy7, CD8-APC-Cy7 (53-6.7), CD44-APC, CD62L-FITC, CD25-PE, and NK1.1-PE and the CD62L level of the naïve CD4+ T cells (CD4+CD44lo/intCD25-NK1.1-) was obtained and compared with that of the thymocyte subsets.
RTE analysis
Mice were anesthetized with 1% napental via intraperitoneal injection. An incision was made in the sternum to reveal the thymus, and approximately 10–20 µl of FITC solution (1 mg/ml) was injected into each thymic lobe with a 30-gauge needle. Control mice were injected with PBS. The chest was closed with surgical clips in the overlying skin. FITC positive cells from the spleen and lymph nodes were analyzed 24 hours later.
Listeria infection
SP3, SP4 thymocytes and naïve CD4+ T cells from C57BL/6 Ly5.1 congenic mice were purified by flow cytometry and were adoptively transferred into C57BL/6 Ly5.2 mice at 6–8 weeks of age (9×106 per host mouse). Three mice in each group. One day later, the host mice were challenged with 2×104 live Listeria Monocytogenes (a kind gift from Prof. Qibin Leng, Laboratory of Immune Regulation, Institute Pasteur of Shanghai, Chinese Academy of Sciences) via tail vein injection. On day 7 of the infection, the mice were sacrificed and the cells from the lymph nodes and spleens were stained with CD45.1, CD4, CD44, and CD62L. The donor cells with the expression of both CD45.1 and CD4 were gated.
Transwell migration
Thymocyte subpopulations and naïve CD4+ T cells from C57BL/6 mice were purified and rested in migration medium at room temperature for one hour. The cells were then used for three-hour migration assays through Transwells (5 mm; Corning). S1P (Sigma) was used at 50 nM; CCL19 (R&D Systems) was used at 250 ng/ml.
Supporting Information
Differentially expressed genes among the four subsets of CD4 SP thymocytes. Sets of probes whose expression increased (red) or decreased (green) by two-fold or more in any comparison are listed.
(XLS)
KEGG pathway analysis of the differentially expressed genes using MAS platform. The false discovery rate (FDR)-corrected P values <0.05 are considered statistically significant.
(XLS)
Acknowledgments
We acknowledge the excellent technical assistance of Drs. Hounan Wu, Li Su, Qinghua Cui at Peking University Health Science Center, and Ya Xu at CapitalBio Corporation. We thank Prof. Qibin Leng, Laboratory of Immune Regulation, Institute Pasteur of Shanghai, Chinese Academy of Sciences, for providing Listeria Monocytogenes.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the Natural Basic Research Program of China (2010CB945300, Dr. Ge, 2011CB711000, Dr. Ge and 2011CB946100, Dr. Zhang), the National Natural Sciences Foundation of China (31070787, Dr. Ge, 30830091, Dr. Zhang), Program for New Century Excellent Talents in University (NCET-10-0175/BMU20100005), the Research Fund for the Doctoral Program of Higher Education (20100001110049), and Key Laboratory of Medical Immunology, Ministry of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rodewald HR. Thymus organogenesis. Annu Rev Immunol. 2008;26:355–388. doi: 10.1146/annurev.immunol.26.021607.090408. [DOI] [PubMed] [Google Scholar]
- 2.Takahama Y. Journey through the thymus: stromal guides for T-cell development and selection. Nat Rev Immunol. 2006;6:127–135. doi: 10.1038/nri1781. [DOI] [PubMed] [Google Scholar]
- 3.Ramsdell F, Jenkins M, Dinh Q, Fowlkes BJ. The majority of CD4+8- thymocytes are functionally immature. J Immunol. 1991;147:1779–1785. [PubMed] [Google Scholar]
- 4.Vicari A, Abehsira-Amar O, Papiernik M, Boyd RL, Tucek CL. MTS-32 monoclonal antibody defines CD4+8- thymocyte subsets that differ in their maturation level, lymphokine secretion, and selection patterns. J Immunol. 1994;152:2207–2213. [PubMed] [Google Scholar]
- 5.Lucas B, Vasseur F, Penit C. Production, selection, and maturation of thymocytes with high surface density of TCR. J Immunol. 1994;153:53–62. [PubMed] [Google Scholar]
- 6.Bendelac A, Schwartz RH. CD4+ and CD8+ T cells acquire specific lymphokine secretion potentials during thymic maturation. Nature. 1991;353:68–71. doi: 10.1038/353068a0. [DOI] [PubMed] [Google Scholar]
- 7.Dyall R, Nikolic-Zugic J. The majority of postselection CD4+ single-positive thymocytes requires the thymus to produce long-lived, functional T cells. J Exp Med. 1995;181:235–245. doi: 10.1084/jem.181.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kishimoto H, Sprent J. Negative selection in the thymus includes semimature T cells. J Exp Med. 1997;185:263–271. doi: 10.1084/jem.185.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCaughtry TM, Wilken MS, Hogquist KA. Thymic emigration revisited. J Exp Med. 2007;204:2513–2520. doi: 10.1084/jem.20070601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: learning self-control in the thymus. Nat Rev Immunol. 2005;5:772–782. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- 11.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 12.Yagi H, Kamba R, Chiba K, Soga H, Yaguchi K, et al. Immunosuppressant FTY720 inhibits thymocyte emigration. Eur J Immunol. 2000;30:1435–1444. doi: 10.1002/(SICI)1521-4141(200005)30:5<1435::AID-IMMU1435>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 13.Rosen H, Alfonso C, Surh CD, McHeyzer-Williams MG. Rapid induction of medullary thymocyte phenotypic maturation and egress inhibition by nanomolar sphingosine 1-phosphate receptor agonist. Proc Natl Acad Sci U S A. 2003;100:10907–10912. doi: 10.1073/pnas.1832725100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 15.Allende ML, Dreier JL, Mandala S, Proia RL. Expression of the sphingosine 1-phosphate receptor, S1P1, on T-cells controls thymic emigration. J Biol Chem. 2004;279:15396–15401. doi: 10.1074/jbc.M314291200. [DOI] [PubMed] [Google Scholar]
- 16.Gabor MJ, Godfrey DI, Scollay R. Recent thymic emigrants are distinct from most medullary thymocytes. Eur J Immunol. 1997;27:2010–2015. doi: 10.1002/eji.1830270827. [DOI] [PubMed] [Google Scholar]
- 17.Jordan MS, Boesteanu A, Reed AJ, Petrone AL, Holenbeck AE, et al. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol. 2001;2:301–306. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 18.Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, et al. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Koble C, Kyewski B. The thymic medulla: a unique microenvironment for intercellular self-antigen transfer. J Exp Med. 2009;206:1505–1513. doi: 10.1084/jem.20082449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ehrlich LI, Oh DY, Weissman IL, Lewis RS. Differential contribution of chemotaxis and substrate restriction to segregation of immature and mature thymocytes. Immunity. 2009;31:986–998. doi: 10.1016/j.immuni.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinreich MA, Hogquist KA. Thymic emigration: when and how T cells leave home. J Immunol. 2008;181:2265–2270. doi: 10.4049/jimmunol.181.4.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bunting MD, Comerford I, McColl SR. Finding their niche: chemokines directing cell migration in the thymus. Immunol Cell Biol. 2010 doi: 10.1038/icb.2010.142. [DOI] [PubMed] [Google Scholar]
- 23.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 24.Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC. Aire regulates negative selection of organ-specific T cells. Nat Immunol. 2003;4:350–354. doi: 10.1038/ni906. [DOI] [PubMed] [Google Scholar]
- 25.Weih F, Carrasco D, Durham SK, Barton DS, Rizzo CA, et al. Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-kappa B/Rel family. Cell. 1995;80:331–340. doi: 10.1016/0092-8674(95)90416-6. [DOI] [PubMed] [Google Scholar]
- 26.Burkly L, Hession C, Ogata L, Reilly C, Marconi LA, et al. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature. 1995;373:531–536. doi: 10.1038/373531a0. [DOI] [PubMed] [Google Scholar]
- 27.Heino M, Peterson P, Sillanpaa N, Guerin S, Wu L, et al. RNA and protein expression of the murine autoimmune regulator gene (Aire) in normal, RelB-deficient and in NOD mouse. Eur J Immunol. 2000;30:1884–1893. doi: 10.1002/1521-4141(200007)30:7<1884::AID-IMMU1884>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 28.Zhu M, Chin RK, Christiansen PA, Lo JC, Liu X, et al. NF-kappaB2 is required for the establishment of central tolerance through an Aire-dependent pathway. J Clin Invest. 2006;116:2964–2971. doi: 10.1172/JCI28326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boehm T, Scheu S, Pfeffer K, Bleul CC. Thymic medullary epithelial cell differentiation, thymocyte emigration, and the control of autoimmunity require lympho-epithelial cross talk via LTbetaR. J Exp Med. 2003;198:757–769. doi: 10.1084/jem.20030794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kajiura F, Sun S, Nomura T, Izumi K, Ueno T, et al. NF-kappa B-inducing kinase establishes self-tolerance in a thymic stroma-dependent manner. J Immunol. 2004;172:2067–2075. doi: 10.4049/jimmunol.172.4.2067. [DOI] [PubMed] [Google Scholar]
- 31.Chin RK, Lo JC, Kim O, Blink SE, Christiansen PA, et al. Lymphotoxin pathway directs thymic Aire expression. Nat Immunol. 2003;4:1121–1127. doi: 10.1038/ni982. [DOI] [PubMed] [Google Scholar]
- 32.Akiyama T, Maeda S, Yamane S, Ogino K, Kasai M, et al. Dependence of self-tolerance on TRAF6-directed development of thymic stroma. Science. 2005;308:248–251. doi: 10.1126/science.1105677. [DOI] [PubMed] [Google Scholar]
- 33.Kurobe H, Liu C, Ueno T, Saito F, Ohigashi I, et al. CCR7-dependent cortex-to-medulla migration of positively selected thymocytes is essential for establishing central tolerance. Immunity. 2006;24:165–177. doi: 10.1016/j.immuni.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 34.Ueno T, Saito F, Gray DH, Kuse S, Hieshima K, et al. CCR7 signals are essential for cortex-medulla migration of developing thymocytes. J Exp Med. 2004;200:493–505. doi: 10.1084/jem.20040643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davalos-Misslitz AC, Rieckenberg J, Willenzon S, Worbs T, Kremmer E, et al. Generalized multi-organ autoimmunity in CCR7-deficient mice. Eur J Immunol. 2007;37:613–622. doi: 10.1002/eji.200636656. [DOI] [PubMed] [Google Scholar]
- 36.Davalos-Misslitz AC, Worbs T, Willenzon S, Bernhardt G, Forster R. Impaired responsiveness to T-cell receptor stimulation and defective negative selection of thymocytes in CCR7-deficient mice. Blood. 2007;110:4351–4359. doi: 10.1182/blood-2007-01-070284. [DOI] [PubMed] [Google Scholar]
- 37.Li J, Li Y, Yao JY, Jin R, Zhu MZ, et al. Developmental pathway of CD4+CD8- medullary thymocytes during mouse ontogeny and its defect in Aire-/- mice. Proc Natl Acad Sci U S A. 2007;104:18175–18180. doi: 10.1073/pnas.0708884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mick VE, Starr TK, McCaughtry TM, McNeil LK, Hogquist KA. The regulated expression of a diverse set of genes during thymocyte positive selection in vivo. J Immunol. 2004;173:5434–5444. doi: 10.4049/jimmunol.173.9.5434. [DOI] [PubMed] [Google Scholar]
- 39.DeRyckere D, Mann DL, DeGregori J. Characterization of transcriptional regulation during negative selection in vivo. J Immunol. 2003;171:802–811. doi: 10.4049/jimmunol.171.2.802. [DOI] [PubMed] [Google Scholar]
- 40.Schmitz I, Clayton LK, Reinherz EL. Gene expression analysis of thymocyte selection in vivo. Int Immunol. 2003;15:1237–1248. doi: 10.1093/intimm/dxg125. [DOI] [PubMed] [Google Scholar]
- 41.Choi YI, Duke-Cohan JS, Ahmed WB, Handley MA, Mann F, et al. PlexinD1 glycoprotein controls migration of positively selected thymocytes into the medulla. Immunity. 2008;29:888–898. doi: 10.1016/j.immuni.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puthier D, Joly F, Irla M, Saade M, Victorero G, et al. A general survey of thymocyte differentiation by transcriptional analysis of knockout mouse models. J Immunol. 2004;173:6109–6118. doi: 10.4049/jimmunol.173.10.6109. [DOI] [PubMed] [Google Scholar]
- 43.Hoffmann R, Bruno L, Seidl T, Rolink A, Melchers F. Rules for gene usage inferred from a comparison of large-scale gene expression profiles of T and B lymphocyte development. J Immunol. 2003;170:1339–1353. doi: 10.4049/jimmunol.170.3.1339. [DOI] [PubMed] [Google Scholar]
- 44.Tabrizifard S, Olaru A, Plotkin J, Fallahi-Sichani M, Livak F, et al. Analysis of transcription factor expression during discrete stages of postnatal thymocyte differentiation. J Immunol. 2004;173:1094–1102. doi: 10.4049/jimmunol.173.2.1094. [DOI] [PubMed] [Google Scholar]
- 45.Tanzola MB, Kersh GJ. The dual specificity phosphatase transcriptome of the murine thymus. Mol Immunol. 2006;43:754–762. doi: 10.1016/j.molimm.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 46.Huang YH, Li D, Winoto A, Robey EA. Distinct transcriptional programs in thymocytes responding to T cell receptor, Notch, and positive selection signals. Proc Natl Acad Sci U S A. 2004;101:4936–4941. doi: 10.1073/pnas.0401133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bosco N, Kirberg J, Ceredig R, Agenes F. Peripheral T cells in the thymus: have they just lost their way or do they do something? Immunol Cell Biol. 2009;87:50–57. doi: 10.1038/icb.2008.83. [DOI] [PubMed] [Google Scholar]
- 49.Agus DB, Surh CD, Sprent J. Reentry of T cells to the adult thymus is restricted to activated T cells. J Exp Med. 1991;173:1039–1046. doi: 10.1084/jem.173.5.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hale JS, Boursalian TE, Turk GL, Fink PJ. Thymic output in aged mice. Proc Natl Acad Sci U S A. 2006;103:8447–8452. doi: 10.1073/pnas.0601040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gopinathan R, DePaz HA, Oluwole OO, Ali AO, Garrovillo M, et al. Role of reentry of in vivo alloMHC peptide-activated T cells into the adult thymus in acquired systemic tolerance. Transplantation. 2001;72:1533–1541. doi: 10.1097/00007890-200111150-00011. [DOI] [PubMed] [Google Scholar]
- 52.Tian C, Bagley J, Forman D, Iacomini J. Induction of central tolerance by mature T cells. J Immunol. 2004;173:7217–7222. doi: 10.4049/jimmunol.173.12.7217. [DOI] [PubMed] [Google Scholar]
- 53.Jin R, Wang W, Yao JY, Zhou YB, Qian XP, et al. Characterization of the in vivo dynamics of medullary CD4+CD8- thymocyte development. J Immunol. 2008;180:2256–2263. doi: 10.4049/jimmunol.180.4.2256. [DOI] [PubMed] [Google Scholar]
- 54.Boursalian TE, Golob J, Soper DM, Cooper CJ, Fink PJ. Continued maturation of thymic emigrants in the periphery. Nat Immunol. 2004;5:418–425. doi: 10.1038/ni1049. [DOI] [PubMed] [Google Scholar]
- 55.Makaroff LE, Hendricks DW, Niec RE, Fink PJ. Postthymic maturation influences the CD8 T cell response to antigen. Proc Natl Acad Sci U S A. 2009;106:4799–4804. doi: 10.1073/pnas.0812354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wurbel MA, Philippe JM, Nguyen C, Victorero G, Freeman T, et al. The chemokine TECK is expressed by thymic and intestinal epithelial cells and attracts double- and single-positive thymocytes expressing the TECK receptor CCR9. Eur J Immunol. 2000;30:262–271. doi: 10.1002/1521-4141(200001)30:1<262::AID-IMMU262>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 57.Nitta T, Nitta S, Lei Y, Lipp M, Takahama Y. CCR7-mediated migration of developing thymocytes to the medulla is essential for negative selection to tissue-restricted antigens. Proc Natl Acad Sci U S A. 2009;106:17129–17133. doi: 10.1073/pnas.0906956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campbell JJ, Pan J, Butcher EC. Cutting edge: developmental switches in chemokine responses during T cell maturation. J Immunol. 1999;163:2353–2357. [PubMed] [Google Scholar]
- 59.Chantry D, Romagnani P, Raport CJ, Wood CL, Epp A, et al. Macrophage-derived chemokine is localized to thymic medullary epithelial cells and is a chemoattractant for CD3(+), CD4(+), CD8(low) thymocytes. Blood. 1999;94:1890–1898. [PubMed] [Google Scholar]
- 60.Annunziato F, Romagnani P, Cosmi L, Beltrame C, Steiner BH, et al. Macrophage-derived chemokine and EBI1-ligand chemokine attract human thymocytes in different stage of development and are produced by distinct subsets of medullary epithelial cells: possible implications for negative selection. J Immunol. 2000;165:238–246. doi: 10.4049/jimmunol.165.1.238. [DOI] [PubMed] [Google Scholar]
- 61.Laan M, Kisand K, Kont V, Moll K, Tserel L, et al. Autoimmune regulator deficiency results in decreased expression of CCR4 and CCR7 ligands and in delayed migration of CD4+ thymocytes. J Immunol. 2009;183:7682–7691. doi: 10.4049/jimmunol.0804133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kwan J, Killeen N. CCR7 directs the migration of thymocytes into the thymic medulla. J Immunol. 2004;172:3999–4007. doi: 10.4049/jimmunol.172.7.3999. [DOI] [PubMed] [Google Scholar]
- 63.Uehara S, Song K, Farber JM, Love PE. Characterization of CCR9 expression and CCL25/thymus-expressed chemokine responsiveness during T cell development: CD3(high)CD69+ thymocytes and gammadeltaTCR+ thymocytes preferentially respond to CCL25. J Immunol. 2002;168:134–142. doi: 10.4049/jimmunol.168.1.134. [DOI] [PubMed] [Google Scholar]
- 64.Suzuki G, Sawa H, Kobayashi Y, Nakata Y, Nakagawa K, et al. Pertussis toxin-sensitive signal controls the trafficking of thymocytes across the corticomedullary junction in the thymus. J Immunol. 1999;162:5981–5985. [PubMed] [Google Scholar]
- 65.Zachariah MA, Cyster JG. Thymic egress: S1P of 1000. F1000 Biol Rep. 2009;1:60. doi: 10.3410/B1-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pappu R, Schwab SR, Cornelissen I, Pereira JP, Regard JB, et al. Promotion of lymphocyte egress into blood and lymph by distinct sources of sphingosine-1-phosphate. Science. 2007;316:295–298. doi: 10.1126/science.1139221. [DOI] [PubMed] [Google Scholar]
- 67.Zachariah MA, Cyster JG. Neural crest-derived pericytes promote egress of mature thymocytes at the corticomedullary junction. Science. 2010;328:1129–1135. doi: 10.1126/science.1188222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kerdiles YM, Beisner DR, Tinoco R, Dejean AS, Castrillon DH, et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10:176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carpenter AC, Bosselut R. Decision checkpoints in the thymus. Nat Immunol. 2010;11:666–673. doi: 10.1038/ni.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bai A, Hu H, Yeung M, Chen J. Kruppel-like factor 2 controls T cell trafficking by activating L-selectin (CD62L) and sphingosine-1-phosphate receptor 1 transcription. J Immunol. 2007;178:7632–7639. doi: 10.4049/jimmunol.178.12.7632. [DOI] [PubMed] [Google Scholar]
- 71.Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 72.Gubbels Bupp MR, Edwards B, Guo C, Wei D, Chen G, et al. T cells require Foxo1 to populate the peripheral lymphoid organs. Eur J Immunol. 2009;39:2991–2999. doi: 10.1002/eji.200939427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scollay R, Godfrey DI. Thymic emigration: conveyor belts or lucky dips? Immunol Today. 1995:273. doi: 10.1016/0167-5699(95)80179-0. discussion 273–264. [DOI] [PubMed] [Google Scholar]
- 74.Schnell FJ, Kersh GJ. Control of recent thymic emigrant survival by positive selection signals and early growth response gene 1. J Immunol. 2005;175:2270–2277. doi: 10.4049/jimmunol.175.4.2270. [DOI] [PubMed] [Google Scholar]
- 75.Fabre S, Carrette F, Chen J, Lang V, Semichon M, et al. FOXO1 regulates L-Selectin and a network of human T cell homing molecules downstream of phosphatidylinositol 3-kinase. J Immunol. 2008;181:2980–2989. doi: 10.4049/jimmunol.181.5.2980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially expressed genes among the four subsets of CD4 SP thymocytes. Sets of probes whose expression increased (red) or decreased (green) by two-fold or more in any comparison are listed.
(XLS)
KEGG pathway analysis of the differentially expressed genes using MAS platform. The false discovery rate (FDR)-corrected P values <0.05 are considered statistically significant.
(XLS)