Summary
Naturally occurring mutants and genetically manipulated strains of mice are widely used to model a variety of human diseases. Atlases are an invaluable aid in understanding the impact of such manipulations by providing a standard for comparison and to facilitate the integration of anatomic, genetic, and physiologic observations from multiple subjects and experiments. We have developed digital atlases of the C57BL/6J mouse brain (adult and neonate) as comprehensive frameworks for storing and accessing the myriad types of information about the mouse brain. Along with raw and annotated images, these contain database management systems and a set of tools for comparing information from different techniques and different animals. Each atlas establishes a canonical representation of the mouse brain and provides the tools for the manipulation and analysis of new data. We describe both these atlases and discuss how they may be put to use in organizing and analyzing data from mouse models of epilepsy.
Keywords: Anatomy, Atlas, Neonatal, Probabilistic
Historically, atlas construction comprised sectioning, staining, and recording of photomicrographs along with their descriptions within a Cartesian coordinate system. Most every brain atlas, at the very least, incorporates three critical elements: (1) graphical reconstructions highlighting important anatomical detail; (2) nomenclature and description of anatomical structures; and (3) a 3D coordinate system.
Traditional mouse brain atlases are published in book form and are very widely used (Hof et al., 2000; Paxinos and Franklin, 2001). However, in book form, the intrinsically three-dimensional brain must be viewed as a series of two-dimensional sections, making it difficult to follow three-dimensional structures or to compare one’s own invariably oblique sections with the orthogonal planes of the atlas. Data acquired from strains or ages that differ from the atlas, with subtle variations in technique, or in different planes of section, make it nearly impossible to compare results accurately. Additionally, there is no way to combine a variety of anatomic and/or physiologic observations or statistically measure the similarity or differences among them.
Digital atlases provide solutions to these confounding problems (Toga et al., 1995; Gibaud et al., 1998; Swanson, 2001). There are several commercially available rat atlases (Swanson, 1992, 1998; Franklin and Paxinos, 1997) some containing CD-ROMs, at least two such mouse atlases (Hof et al., 2000; Paxinos and Franklin, 2001), and other noncommercial CD-ROM undertakings (Ghosh et al., 1994; Smith et al., 1994). A variety of 2D data is presented by a number of world wide web sites: the High Resolution Mouse Brain Atlas (http://www.hms.harvard.edu/research/brain), the Edinburgh Mouse Atlas Project (http://genex.hgu.mrc.ac.uk), and the Mouse Brain Library (Rosen et al., 2003) (http://www.mbl.org). Software packages are available for visualization of different data modalities and brain alignment (the Neuroterrain project, URL: http://www.neuroterrain.org) and some with an aim toward being three-dimensional atlases (Toga et al., 1995). The work of the Informatics Center of the Mouse Neurogenetics (http://www.nervenet.org) includes a comprehensive library of Nissl-stained images of brains of over 100 strains of mice and a set of software tools for two-and three-dimensional visualization and reconstruction of different brain regions of interest. Additionally, the Edinburgh Mouse Atlas Project has made a significant effort to create a gene expression database (Ringwald et al., 1994) based upon the Atlas of Mouse Development (Kaufman, 1992).
We ourselves have produced electronic atlases of the rat (Toga et al., 1995), neonatal and adult mouse (MacKenzie-Graham et al., 2004; Lee et al., 2005), and Nemestrina monkey (Cannestra et al., 1997). The effort of the International Consortium for Brain Mapping (ICBM) is based on digital 3D representations of a population’s anatomy (Mazziotta et al., 1995). We have also created a variety of human atlases that describe anatomic detail from multiple modalities and disease states (Toga et al., 1994; Sowell et al., 2000; Thompson et al., 2000; Narr et al., 2001).
Digital atlases can easily be extended to include an entire animal. Such a data set could potentially embody all quantitative information known about the organism in a digital framework.
In this article, we describe the structure and function of multimodal, multidimensional atlases of the mouse. These systems provide mechanisms to facilitate the examination of anatomy and physiology of disease models in a quantitatively and visually integrated format.
METHODS
Mice
Hundred-day-old male C57BL/6J mice (Jackson Laboratories Bar Harbor, ME, U.S.A.) were used for the adult atlas, though a volume database contains data from mice a various ages. Newborn C57BL/6J mice (Jackson Laboratories) were used for the neonatal atlas. Data was collected from 165 adult mice using magnetic resonance imaging, blockface imaging, classical histology, and immunohistochemistry. All animals were housed and treated in accordance with the UCLA Animal Research Committee guidelines.
Adult MR
Mice were anesthetized initially with ketamine/xylzaine and then maintained on isofluorane for the duration of the imaging experiment. Magnetic resonance imaging was done at 37°C using an 89 mm vertical bore 11.7 T Bruker Avance imaging spectrometer with a microimaging gradient insert and 30 mm birdcage RF coil (Bruker Instruments). Typical imaging parameters were as follows: T2-weighted RARE 3D imaging protocol (eight echoes), matrix dimensions = 256 × 256 × 256; FOV = 3 cm × 1.5 cm × 1.5 cm; repetition time (TR) = 1500 ms; effective time (TE) = 10 ms; number of averages = 4. The images were padded with zeros to double the number of time domain points in each dimension and the Fourier transformed to yield a matrix of 512 × 256 × 256. This procedure is commonly called “zero-filling” and is a well known interpolation method (Farrar and Becker, 1971; Fukushima and Roeder, 1981). Typical spatial resolution was approximately 60 × 60 × 60 μm3 per voxel.
Neonatal MR
Neonatal mice were sacrificed hypothermally within 24 h after birth. After intracardial perfusion with 10 ml of phosphate-buffered saline (PBS) and then 10 ml of 2% paraformaldehyde (PFA), the animals were decapitated and the heads postfixed with 2% PFA for 24 h prior to the MR scans. Heads were soaked in 5% ProHance (paramagnetic MR contrast agent) for 5 days, then immersed in Fomblin (an embedding medium to limit tissue dehydration) for the MR scans. T2-weighted 3D spin-echo MRI images were acquired using an 11.7 T Bruker Avance imaging spectrometer with a microimaging gradient insert and 20 mm birdcage RF coil (Bruker Instruments). The following data acquisition parameters were used: TR/TE = 300/6.8 ms, two averages, FOV = 12.8 × 9 × 9 mm3, matrix size = 256 × 128 × 128, T = 288.1 K. Spatial resolution of the obtained images was 70 × 50 × 70 μm3 per voxel or 40 × 40 × 40 μm3 per voxel.
Blockface and histology
Mice were sacrificed and brains cut serially in 50μm thick transverse (coronal) sections on a modified CM3050S cryostat (Leica Microsystems, Wetzlar, Germany). A DMCIe digital camera (Polaroid Corporation, Waltham, MA, U.S.A.) captured images of the blockface prior to each section at a resolution of 1600 × 1200 (approximately 6.7 μm/pixel) in 24-bit color. Sections were Nissl-stained (thionin) as described (Simmons and Swanson, 1993) or myelin-stained using a modified myelin impregnation stain (Gallyas, 1979). Immunohistochemistry was done on free floating sections with anti-bovine GFAP (1:500) (Dako, Glostrup, Denmark). Stained preparations were digitized using a 1.25× objective on an AX70 microscope (Olympus Corporation, Tokyo, Japan) with a microfire digital camera (Optronics, Goleta, CA, U.S.A.) and a motorized stage (Ludl Electronic Products Ltd., Hawthorne, NY, U.S.A. ) driven by Neurolucida software (Microbrightfield, Inc. Williston, VT, U.S.A.) at a resolution of approximately 1 μm/pixel in 24-bit color.
Image processing
The sequence of two-dimensional digital images were brought into linear register with Alignlinear (Automated Image Registration 5.2.5) (Woods et al., 1998a, 1998b) using a rigid-body transformation. The registered images were reconstructed into 3D volumes using Reunite (AIR). 3D digital volumes were subsequently brought into register with a diffusion-weighted MRM in a common coordinate system (defined by the mid-sagittal plane and the interaural line (Paxinos and Franklin, 2001)), once again using Alignlinear. All image processing was done in the LONI Pipeline Processing Environment (Rex et al., 2003).
Nomenclature and delineations
In the development of a comprehensive, standardized, and mutually exclusive nomenclature (Bowden and Martin, 1995; Bard et al., 1998) and anatomic delineation, our primary references were the mouse brain atlases of Hof (2000) and Franklin and Paxinos (1997) and inconsistencies were resolved by Swanson (1998). Neural structures (including cell groups, fiber tracts, and gross anatomical features such as the ventricles) were determined under the microscope from the histologically stained sections. 3D label volumes were “painted” onto coregistered MR, Nissl-, and myelin-stained volumes using BrainSuite 2 (Shattuck and Leahy, 2002). Anatomic delineations were prepared by tracing digital images from these serially stained sections. Three-dimensional surfaces were produced by tessellation in BrainSuite 2.
The MAP atlas viewer
The MAP Atlas Viewer is a self-contained, platform independent software tool capable of visualizing multiple two- or three-dimensional image datasets simultaneously at several levels of magnification (Fig. 1). It provides the user with powerful, intuitive tools for the manipulation, labeling, and analysis of that image data. Images may be reoriented manually, with immediate visual feedback of the manipulation. Images may be manually edited to remove confounding structures and for manual correction of image volumes that have been inaccurately edited. Labels may be drawn upon image files to provide direct estimates of structure volumes, areas, and lengths, or can be exported for more sophisticated analyses.
FIG. 1.
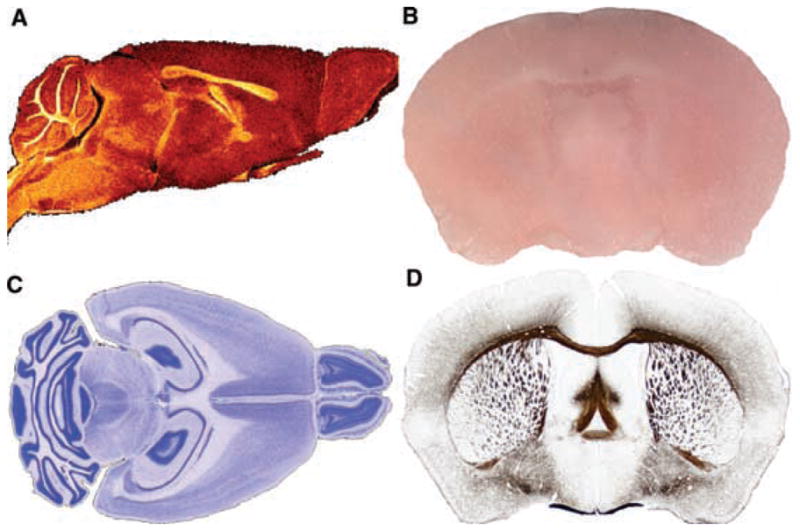
A digital atlas: orthogonal sections and high-resolution display. (A) Horizontal, (B) transverse (coronal), and (C) sagittal sections through two volumes, an MRM volume and a Nissl-stained volume from a d100 mouse, shown overlaid. Small, low-resolution thumbnails are for navigation, whereas, (D) a high-resolution view of the same data allows one to visualize both nuclei and white matter tracts.
The MAP Atlas Viewer is also capable of volume and surface rendering, greatly facilitating the visualization of data acquired at arbitrary angles. The user can select an arbitrary plane of section to view a volume-rendered representation of the data. The anatomic delineations, stored as surfaces overlaid upon the data volumes, can then be viewed as two-dimensional curves upon that plane or as three-dimensional surfaces in space (Fig. 2).
FIG. 2.
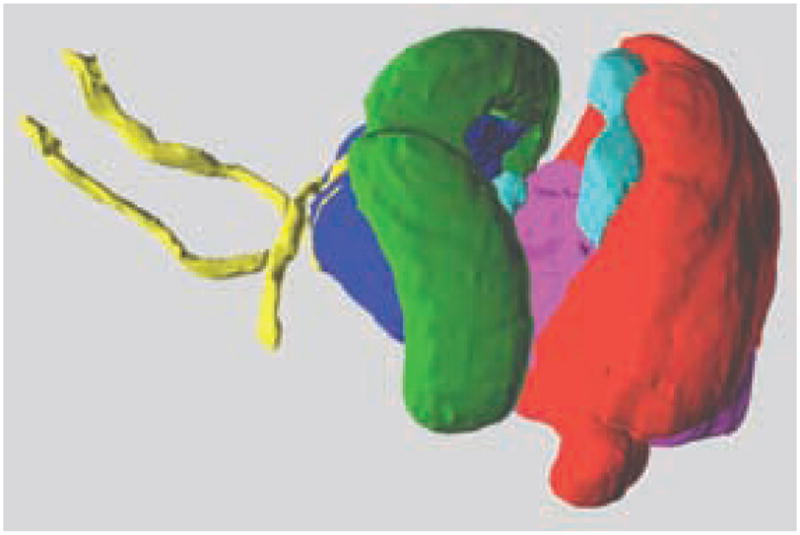
3D surfaces displayed in space. 3D structures were created by tessellating volumetric anatomical delineations. The outer surface of the brain and some deep structures have been rendered transparent to visualize the anterior commissure, the hippocampus, and the thalamus in the anterior part of the brain, and the cerebellum, inferior colliculi, and hindbrain in the posterior part.
The MAP Atlas Viewer contains workspaces for manipulating image data, such as image reorientation, masking, and labeling. The reorientation workspace allows the user to reorient their image data so that it oriented to other images or an atlas. The image masking workspace can be used to generate masks by thresholding the image and then manually editing the mask. The labeling workspace allows the user to delineate regions of interest for measurements of area or volume.
RESULTS
The atlases incorporate magnetic resonance imagery, blockface images, classical histology, and immunohistochemistry. Complete volumes range from a spatial resolution of approximately 100 μm3 (128 × 128 × 256 voxels, 4.2 Mb uncompressed) for a low-resolution grayscale MR volume to 1 × 1 × 50 μm3 (14400 × 13200 × 330 voxels, 178 Gb uncompressed) for a high-resolution full-color Nissl-stained volume. These volumes are available for download at http://www.loni.ucla.edu/MAP/Atlas/Databases.html. The data were reconstructed into three-dimensional volumes, transformed quantitatively into a defined and common coordinate system, and described anatomically. A set of visualization, database, and mapping tools complete the atlases.
Modeling, comparative neuroanatomy, measurements of tissue and cell dimensions, and analyses of genes influences on strain differences all require original, undistorted representations of tissue. However, all histological processing methods introduce distortions. To minimize histological distortions, our data was registered to a MR image volume of the same animal obtained prior to sectioning (Fig. 3A). This allowed us to represent the brain in a form closer to its in vivo morphology.
FIG. 3.
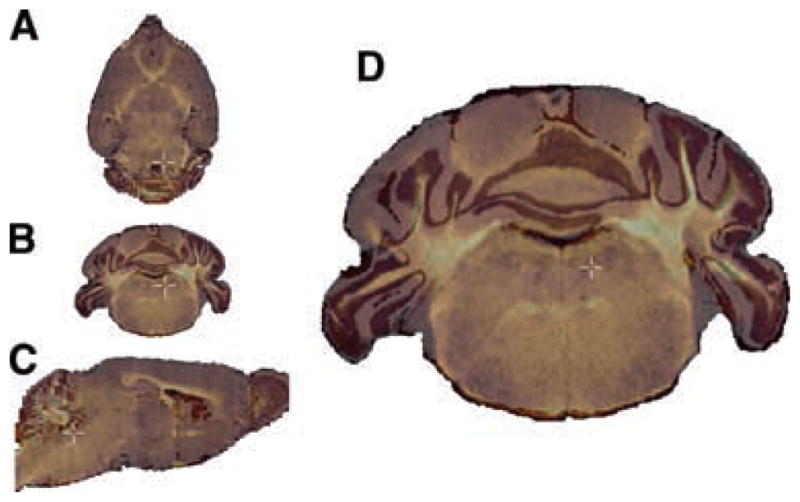
Multiple modalities and planes of section. Data are shown in several planes of section to demonstrate the inherently 3D nature of the atlas. (A) An MRM scan of a d100 mouse brain using a z-direction diffusion-weighted imaging protocol. (B) A transverse (coronal) section from a blockface imaging volume of a d100 mouse brain. (C) A horizontal section from a Nissl-stained volume of a d123 mouse brain. (D) A transverse (coronal) section from a myelin-stained volume of a d100 mouse brain.
Blockface imaging is a imaging modality that does not suffer from many of the spatial artifacts that affect serially-stained sections mounted on glass slides: shatter, tears, bubbles, and other mechanical distortions. High-resolution color images of the frozen blockface are acquired as it is sectioned, relying on the contrast inherent between white and gray matter to discriminate anatomical boundaries. A number of gross structural subdivisions of the brain are visible, even without the benefit of histological staining. For example, the internal capsule and the hippocampus are clearly discernable (Fig. 3B).
Traditionally atlases have comprised of collections of histologically stained sections produced to visualize anatomy. Cortical lamination and the topography of subcortical nuclei can be visualized in Nissl-stained sections (Fig. 3C). Myelin-stained sections complement the Nissl stain’s cytoarchitectural data with myeloarchitecture, delineating fiber tracts and helping in turn to define nuclei (Fig. 3D). Two dimensional images of serial sections were imported into the atlas using the methods described above.
Visualizing and measuring gene expression patterns may also aid in discerning the relationship between genotype and phenotype. The atlases were originally developed with the intent of allowing researchers to import their own gene expression patterns into the atlas. Immunohistochemistry for various neuronal and glial markers (such as GFAP) was carried out on serial sections (Fig. 4), demonstrating that immunohistochemistry can also be readily imported into the atlases.
FIG. 4.
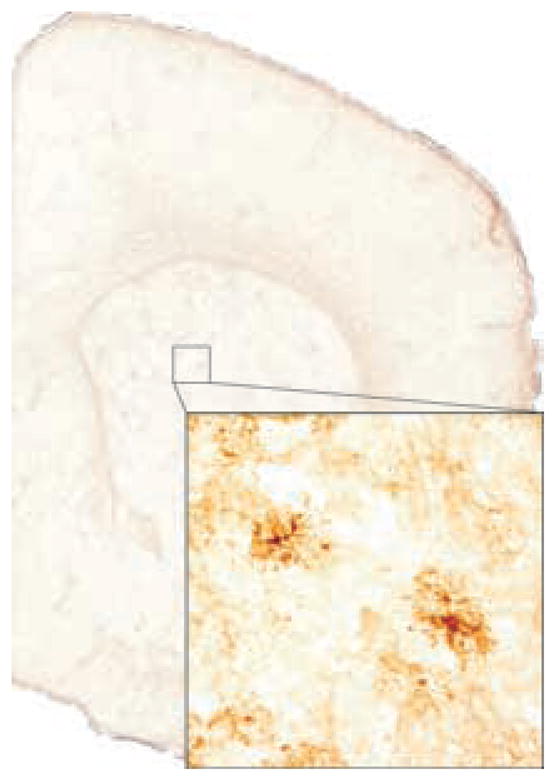
Gene expression maps: Immunohistochemistry. A low (0.5×) magnification image of a transverse (coronal) section from a glial fibrillary acidic protein stained volume of a d100 mouse. The inset is a magnified (10×) image demonstrating individual staining astrocytes visible in the dorsal striatum.
Anatomic delineations are fundamental to the atlases. They serve to help orient the user with graphical representations highlighting important anatomical detail and providing a standard description and nomenclature of the region of interest. Manual input was necessary for even approximate parcellation of brain in its fine details. Accurate delineation of brain nuclei requires an expert neuroanatomist to draw on high-level knowledge, accumulated over a lifetime of careful study of disparate materials (Swanson, 1998).
Two sets of delineations are bundled with the adult atlas, a set of volumetric labels and a set of three-dimensional surfaces. The label volume provides the basis for the interaction between image-based data volumes and text-based information networks. They allow us to reference the name of a given structure and synchronize the location of the cursor with the appropriate structure in any system so indexed. Three-dimensional surfaces may be viewed in relation to each other or sectioned at arbitrary angles, permitting the user a more intuitive grasp of the neuroanatomy of the C57BL/6J mouse.
The Probability maps were generated for the neonatal atlas by superimposing the anatomical labeled volumes of the coregistered brains used to define the atlas. The incidence that any of the delineated anatomical structures occupied a given spatial coordinate was used to calculate the probabilities. Each voxel in the probability map for a given anatomical structure therefore describes the probability that the voxel belongs to this anatomical structure and indicated the confidence levels of the anatomical space for each structure. The label volume retrieved from a given threshold is the collection of voxels with probabilities greater than a set threshold (Lee et al., 2005).
DISCUSSION
While traditional atlases have used histological observations almost exclusively to describe the anatomy of the brain, we can now augment those descriptions with a nondestructive, comprehensive survey collected in vivo and in three-dimensions. Magnetic resonance (MR) imaging has revolutionized our ability to investigate brain structure and function. MR techniques are now available to capture features of anatomy and function at both the molecular and whole-brain scales. Thus, neuronal dynamics and gene expression patterns can be mapped at microscopic resolutions in normal adult and developing animals, as well as in pathological or genetically modified states (Ahrens et al., 1998; Benveniste et al., 1998; Jacobs et al., 1999; Benveniste et al., 2000; Dhenain et al., 2001).
In a multimodal digital atlas many different kinds of data can be encompassed, allowing the investigator to visualize covarying patterns simultaneously. Maps can be generated that amalgamate data from various experimental techniques, and quantitative measures of anatomy can be determined (e.g., structure volume, cross-sectional area, orientation, or complexity). Temporal and spatial gene and protein expression patterns, axonal trajectories, patterns of vasculature, and specific functional responses all can be combined to obtain a standard or canonical representation. In a digital atlas the anatomical space can be described with a common numerical system, the coordinate representation can then serve as an index for database entries facilitating integration of multimodality data based on their spatial relationships. These data are not limited to image data, but easily extend to electrophysiological data, text-based information such as nomenclature, descriptions of gene expression, or even literature citations. All are easily accessible from electronic databases and links to sources on the web.
One of the primary uses of atlases is the mapping of structural or functional attributes in an individual or population. Establishing the relationship between multiple observations from different data collections can be realized using modern computational atlases and appropriately integrated tools. For example, establishing the localization of gene or protein expression data to an anatomical structure would be a difficult task with a paper atlas due to the variability of cutting planes and tissue processing. Using the computational multimodal, multidimensional atlas described here, gene or protein expression data can be aligned to anatomical delineations within the atlas, allowing for unambiguous localization. The atlas will facilitate collaboration by providing researchers a common framework for comparing results. Examples of such studies would be the comparison of gene or protein expression data collected in different laboratories, or finding the histological correlates of an MR signal.
Most brain atlases facilitate the structural mapping by providing coordinates that define brain orientations and positions with grid systems. However, bone calcification is incomplete in early development, making it impossible to utilize skull based bony landmarks to establish a coordinate system. In addition, neonatal brains are undergoing massive volume expansion at this stage and minor differences in growth rates could result in significant variations in brain size, generally much larger than in fully developed brains. Therefore, the conventional definition of adult brain coordinate systems which utilize landmarks on the skull to orient and scale the brain are too simple to represent the anatomy in the early developmental stages, even for the animals from the same genetic background and the same gestational stage. Thus, to date, no published atlases for the developing mouse brain include a coordinate system (Schambra et al., 1992; Jacobowitz and Abbott, 1998; Baldock et al., 2001). As genes involved in morphogenesis, functional development, and susceptibility to disease may have specific spatial expression patterns, a coordinate system for each development stage is required to correlate gene expression to brain anatomy.
In addition to providing a stable anatomical framework for the task of cross-subject comparison, a protocol for registration to this template framework was also defined. Atlas-based registration limited the geometrical variation problem for cross-subject comparison. The associated anatomical probability maps, which were derived from the coregistered brains used in the atlas construction, were used to evaluate the registration accuracy and variation factors other than geometrical variation. We have demonstrated the validation of the atlas-based segmentation when applying the atlas to other subject as well as data acquired using different modalities (Lee et al., 2005).
There is no simple way of creating an “average” anatomy or representing 3D anatomic variations, let alone variations across strains, genetically manipulated animals, and disease states. To address the wide variation in the anatomies of experimental animals and human populations, population-based atlases have been developed to better represent the anatomies of distinct subgroups (Mazziotta et al., 1995; Toga and Thompson, 1998). These approaches can be applied to generate a disease-specific atlas (Thompson et al., 2000). An “average” or canonical brain atlas can be produced using probabilistic atlasing approaches, allowing observations and comparisons to be made across a large number of subjects which are age, strain, and disease matched.
Atlases can be generated from disease states as a way of better understanding the process and effects of disease. Additionally, atlases can be made from genetically-modified or mutant animals. These atlases could produce great insight into the effect of genetic manipulation or mutation on the phenotype of the mouse.
It should also be clear that comparisons across different atlases, each representing a particular subpopulation (i.e., disease versus matched control), are also made possible within this digital atlas framework. Five spontaneously occurring mutants produce a phenotype that resembles epileptic seizures with features similar to those occurring in human idiopathic generalized epilepsy (IGE). They exhibit bilaterally synchronous spike-wave discharges (SWDs) on cortical electroencephalogram (EEG) recordings that are suppressed by the antiabsence drug etho-suximide (Noebels et al., 1998). Mouse models such as tottering (Noebels and Sidman, 1979), lethargic (Hosford et al., 1992), ducky (Barclay et al., 2001), and stargazer (and the related mutant, waggler) (Noebels et al., 1990), have demonstrated the importance of voltage-dependent calcium channels in maintaining normal electrical activity in the brain. The slow-wave epilepsy mouse suffers from a mutation in the gene encoding the sodium hydrogen exchanger, NHEI (Cox et al., 1997). Several of these models have a highly restricted anatomical expression of the affected gene. All of these models lend themselves to analysis with a digital atlas.
The slow-wave epilepsy mouse demonstrates progressive neuronal degeneration in the deep cerebellar nuclei at 3 weeks of age, coincident with the onset of ataxia. Most DCN large neurons are gone at four months of age (Cox et al., 1997). Even though the sodium hydrogen exchanger NHEI is expressed ubiquitously, the effects of the swe mutation are anatomically localized. Voltage-dependent calcium channel subunits have very specific patterns of expression. In the mouse models of epilepsy described above, many are expressed in the cerebellum. Purkinje cell loss has been observed in the tottering mutant and severe dendritic abnormalities have been documented in the ducky mutant (Barclay et al., 2001).
The ability to visually localize not only the genes of interest, but the anatomical changes that are occasioned by these genes is an immensely powerful tool for understanding mechanisms that underlie normal physiology and disease. This perhaps will lead us to a greater understanding of the connection between genotype and phenotype.
Acknowledgments
This work was generously supported by research grants from the National Institute of Mental Health (5 RO1 MH61223) and the National Institutes of Health/National Center for Research Resources (P41 RR13642). The authors also wish to acknowledge their deep appreciation to the members of the Laboratory of Neuro Imaging.
We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Footnotes
Disclosure Statement: None of the authors have any conflicts of interest in the publication of this work.
References
- Ahrens ET, Laidlaw DH, Readhead C, Brosnan CF, Fraser SE, Jacobs RE. MR microscopy of transgenic mice that spontaneously acquire experimental allergic encephalomyelitis. Magn Reson Med. 1998;40:119–132. doi: 10.1002/mrm.1910400117. [DOI] [PubMed] [Google Scholar]
- Baldock R, Bard J, Brune R, Hill B, Kaufman M, Opstad K, Smith D, Stark M, Waterhouse A, Yang Y, Davidson D. The Edinburgh Mouse Atlas: using the CD. Brief Bioinform. 2001;2:159–169. doi: 10.1093/bib/2.2.159. [DOI] [PubMed] [Google Scholar]
- Barclay J, Balaguero N, Mione M, Ackerman SL, Letts VA, Brodbeck J, Canti C, Meir A, Page KM, Kusumi K, et al. Ducky mouse phenotype of epilepsy and ataxia is associated with mutations in the Cacna2d2 gene and decreased calcium channel current in cerebellar Purkinje cells. J Neurosci. 2001;21:6095–6104. doi: 10.1523/JNEUROSCI.21-16-06095.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard JL, Kaufman MH, Dubreuil C, Brune RM, Burger A, Baldock RA, Davidson DR. An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature. Mech Dev. 1998;74:111–120. doi: 10.1016/s0925-4773(98)00069-0. [DOI] [PubMed] [Google Scholar]
- Benveniste H, Qui H, Hedlund LW, D’Ercole F, Johnson GA. Spinal cord neural anatomy in rats examined by in vivo magnetic resonance microscopy. Reg Anesth Pain Med. 1998;23:589–599. doi: 10.1016/s1098-7339(98)90087-6. [DOI] [PubMed] [Google Scholar]
- Benveniste H, Kim K, Zhang L, Johnson GA. Magnetic resonance microscopy of the C57BL mouse brain. Neuroimage. 2000;11:601–611. doi: 10.1006/nimg.2000.0567. [DOI] [PubMed] [Google Scholar]
- Bowden DM, Martin RF. NeuroNames brain hierarchy. Neuroimage. 1995;2:63–83. doi: 10.1006/nimg.1995.1009. [DOI] [PubMed] [Google Scholar]
- Cannestra AF, Santori EM, Holmes CJ, Toga AW. A three-dimensional multimodality brain map of the nemestrina monkey. Brain Res Bull. 1997;43:141–148. doi: 10.1016/s0361-9230(96)00429-7. [DOI] [PubMed] [Google Scholar]
- Cox GA, Lutz CM, Yang CL, Biemesderfer D, Bronson RT, Fu A, Aronson PS, Noebels JL, Frankel WN. Sodium/hydrogen exchanger gene defect in slow-wave epilepsy mutant mice. Cell. 1997;91:139–148. doi: 10.1016/s0092-8674(01)80016-7. [DOI] [PubMed] [Google Scholar]
- Dhenain M, Ruffins SW, Jacobs RE. Three-dimensional digital mouse atlas using high-resolution MRI. Developmental Biology. 2001;232:458–470. doi: 10.1006/dbio.2001.0189. [DOI] [PubMed] [Google Scholar]
- Farrar TC, Becker ED. Pulse and Fourier Transform NMR. Academic Press; New York: 1971. [Google Scholar]
- Franklin KBJ, Paxinos G. The mouse brain in stereotaxic coordinates. Academic Press; San Diego: 1997. [Google Scholar]
- Fukushima E, Roeder SBW. Experimental Pulse NMR. Addison-Wesley; Reading, MA: 1981. [Google Scholar]
- Gallyas F. Silver staining of myelin by means of physical development. Neurol Res. 1979;1:203–209. doi: 10.1080/01616412.1979.11739553. [DOI] [PubMed] [Google Scholar]
- Ghosh P, O’Dell M, Narasimhan PT, Fraser SE, Jacobs RE. Mouse lemur microscopic MRI brain atlas. Neuroimage. 1994;1:345–349. doi: 10.1006/nimg.1994.1019. [DOI] [PubMed] [Google Scholar]
- Gibaud B, Garlatti S, Barillot C, Faure E. Computerized brain atlases as decision support systems: a methodological approach. Artif Intell Med. 1998;V14:83–100. doi: 10.1016/s0933-3657(98)00017-7. [DOI] [PubMed] [Google Scholar]
- Hof PR, Young WG, Bloom FE, Belichenko PV, Celio MR. Comparative cytoarchitectonic atlas of the C57BL 6 and 129 Sv mouse brains. Elsevier; Amsterdam; New York: 2000. [Google Scholar]
- Hosford DA, Clark S, Cao Z, Wilson WA, Jr, Lin FH, Morrisett RA, Huin A. The role of GABAB receptor activation in absence seizures of lethargic (lh/lh) mice. Science. 1992;257:398–401. doi: 10.1126/science.1321503. [DOI] [PubMed] [Google Scholar]
- Jacobowitz DM, Abbott LC. Chemoarchitectonic atlas of the developing mouse brain. CRC Press; Boca Raton: 1998. [Google Scholar]
- Jacobs RE, Ahrens ET, Dickinson ME, Laidlaw D. Towards a microMRI atlas of mouse development. Comput Med Imaging Graph. 1999;23:15–24. doi: 10.1016/s0895-6111(98)00059-7. [DOI] [PubMed] [Google Scholar]
- Kaufman MH. The atlas of mouse development. Academic Press; London; San Diego: 1992. [Google Scholar]
- Lee EF, Jacobs RE, Dinov I, Leow A, Toga AW. Standard atlas space for C57BL/6J neonatal mouse brain. Anat Embryol (Berl) 2005;210:245–263. doi: 10.1007/s00429-005-0048-y. [DOI] [PubMed] [Google Scholar]
- MacKenzie-Graham A, Lee EF, Dinov ID, Bota M, Shattuck DW, Ruffins S, Yuan H, Konstantinidis F, Pitiot A, Ding Y, Hu G, Jacobs RE, Toga AW. A multimodal, multidimensional atlas of the C57BL/6J mouse brain. J Anat. 2004;204:93–102. doi: 10.1111/j.1469-7580.2004.00264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta JC, Toga AW, Evans A, Fox P, Lancaster J. A probabilistic atlas of the human brain: theory and rationale for its development. The International Consortium for Brain Mapping (ICBM) Neuroimage. 1995;2:89–101. doi: 10.1006/nimg.1995.1012. [DOI] [PubMed] [Google Scholar]
- Noebels JL, Sidman RL. Inherited epilepsy: spike-wave and focal motor seizures in the mutant mouse tottering. Science. 1979;204:1334–1336. doi: 10.1126/science.572084. [DOI] [PubMed] [Google Scholar]
- Narr K, Thompson P, Sharma T, Moussai J, Zoumalan C, Rayman J, Toga A. Three-dimensional mapping of gyral shape and cortical surface asymmetries in schizophrenia: gender effects. Am J Psychiatry. 2001;158:244–255. doi: 10.1176/appi.ajp.158.2.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noebels JL, Qiao X, Bronson RT, Spencer C, Davisson MT. Stargazer: a new neurological mutant on chromosome 15 in the mouse with prolonged cortical seizures. Epilepsy Res. 1990;7:129–135. doi: 10.1016/0920-1211(90)90098-g. [DOI] [PubMed] [Google Scholar]
- Noebels JL, Fariello RG, Jobe PC, Lasley SM, Marescaux C. Genetic models of generalized epilepsy. In: Engel J, Pedley TA, editors. Epilepsy: a comprehensive textbook. Lippincott-Raven; Philadelphia: 1998. pp. 457–465. [Google Scholar]
- Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. 2. Academic; San Diego: 2001. [Google Scholar]
- Rex DE, Ma JQ, Toga AW. The LONI pipeline processing environment. Neuroimage. 2003;19:1033–1048. doi: 10.1016/s1053-8119(03)00185-x. [DOI] [PubMed] [Google Scholar]
- Ringwald M, Baldock R, Bard J, Kaufman M, Eppig JT, Richardson JE, Nadeau JH, Davidson D. A database for mouse development. Science. 1994;265:2033–2034. doi: 10.1126/science.8091224. [DOI] [PubMed] [Google Scholar]
- Rosen GD, La Porte NT, Diechtiareff B, Pung CJ, Nissanov J, Gustafson C, Bertrand L, Gefen S, Fan Y, Tretiak OJ, et al. Informatics center for mouse genomics: the dissection of complex traits of the nervous system. Neuroinformatics. 2003;1:327–342. doi: 10.1385/NI:1:4:327. [DOI] [PubMed] [Google Scholar]
- Schambra UB, Lauder JM, Silver J. Atlas of the prenatal mouse brain. Academic Press; San Diego: 1992. [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Leahy RM. BrainSuite: an automated cortical surface identification tool. Med Image Anal. 2002;6:129–142. doi: 10.1016/s1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- Simmons DM, Swanson LW. The Nissl Stain. In: Wouterlood FG, editor. Neuroscience protocols. Elsevier; Amsterdam: 1993. pp. 93-050-012-001–093-050-012-007. [Google Scholar]
- Smith BR, Johnson GA, Groman EV, Linney E. Magnetic Resonance Microscopy of Mouse Embryos. Proc Natl Acad Sci USA. 1994;V91:3530–3533. doi: 10.1073/pnas.91.9.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell ER, Levitt J, Thompson PM, Holmes CJ, Blanton RE, Kornsand DS, Caplan R, McCracken J, Asarnow R, Toga AW. Brain abnormalities in early-onset schizophrenia spectrum disorder observed with statistical parametric mapping of structural magnetic resonance images. Amer J Psychiatry. 2000;157:1475–1484. doi: 10.1176/appi.ajp.157.9.1475. [DOI] [PubMed] [Google Scholar]
- Swanson LW. Brain maps: structure of the rat brain. Elsevier; Amsterdam; New York: 1992. [Google Scholar]
- Swanson LW. Brain maps: structure of the rat brain: a laboratory guide with printed and electronic templates for data, models, and schematics. 2. Elsevier; Amsterdam: 1998. [Google Scholar]
- Swanson LW. Interactive brain maps and atlases. In: Arbib MA, Grethe JS, editors. Computing the brain: a guide to neuroinformatics. Academic Press; San Diego: 2001. pp. 167–177. [Google Scholar]
- Thompson PM, Giedd JN, Woods RP, MacDonald D, Evans AC, Toga AW. Growth patterns in the developing brain detected by using continuum mechanical tensor maps. Nature. 2000;404:190–193. doi: 10.1038/35004593. [DOI] [PubMed] [Google Scholar]
- Toga AW, Ambach K, Quinn B, Hutchin M, Burton JS. Postmortem anatomy from cryosectioned whole human brain. J Neurosci Methods. 1994;54:239–252. doi: 10.1016/0165-0270(94)90196-1. [DOI] [PubMed] [Google Scholar]
- Toga AW, Santori EM, Hazani R, Ambach K. A 3D digital map of rat brain. Brain Res Bull. 1995;38:77–85. doi: 10.1016/0361-9230(95)00074-o. [DOI] [PubMed] [Google Scholar]
- Toga AW, Thompson PM. Multimodal Brain Atlases. In: Wong S, editor. Advances in biomedical image databases. Kluwer Academic Press; Dordrecht, The Netherlands: 1998. pp. 53–88. [Google Scholar]
- Woods RP, Grafton ST, Holmes CJ, Cherry SR, Mazziotta JC. Automated image registration: I. General methods and intrasubject, intramodality validation. J Comput Assist Tomogr. 1998a;V22:139–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Watson JDG, Sicotte NL, Mazziotta JC. Automated image registration: II. Intersubject validation of linear and nonlinear models. J Comput Assist Tomogr. 1998b;V22:153–165. doi: 10.1097/00004728-199801000-00028. [DOI] [PubMed] [Google Scholar]


