The multifunctional APE1 protein is required for tumor progression and is associated with cancer resistance. It is shown that APE1 presents structural elements that function in distinct cellular roles, highlighting the molecular determinants of the multifunctional nature of this protein and providing the basis for a new role of the C65 residue.
Abstract
Apurinic/apyrimidinic endonuclease 1/redox factor-1 (APE1) protects cells from oxidative stress via the base excision repair pathway and as a redox transcriptional coactivator. It is required for tumor progression/metastasis, and its up-regulation is associated with cancer resistance. Loss of APE1 expression causes cell growth arrest, mitochondrial impairment, apoptosis, and alterations of the intracellular redox state and cytoskeletal structure. A detailed knowledge of the molecular mechanisms regulating its different activities is required to understand the APE1 function associated with cancer development and for targeting this protein in cancer therapy. To dissect these activities, we performed reconstitution experiments by using wild-type and various APE1 mutants. Our results suggest that the redox function is responsible for cell proliferation through the involvement of Cys-65 in mediating APE1 localization within mitochondria. C65S behaves as a loss-of-function mutation by affecting the in vivo folding of the protein and by causing a reduced accumulation in the intermembrane space of mitochondria, where the import protein Mia40 specifically interacts with APE1. Treatment of cells with (E)-3-(2-[5,6-dimethoxy-3-methyl-1,4-benzoquinonyl])-2-nonyl propenoic acid, a specific inhibitor of APE1 redox function through increased Cys-65 oxidation, confirm that Cys-65 controls APE1 subcellular trafficking and provides the basis for a new role for this residue.
INTRODUCTION
The apurinic/apyrimidinic endonuclease 1/redox factor-1 (APE1) has a crucial role in the maintenance of genome stability and redox signaling and has emerged as an excellent target for sensitizing tumor cells to chemotherapy (Bapat et al., 2009, 2010; Tell et al., 2010a; Tell and Wilson, 2010; Wilson and Simeonov, 2010). In fact, it acts as an essential enzyme in the base excision repair (BER) pathway of DNA damage caused by both endogenous and exogenous oxidizing/alkylating agents, including chemotherapeutic drugs. In combination with thioredoxin (Ueno et al., 1999; Seemann and Hainaut, 2005), it also acts as a regulatory redox agent to maintain cancer-related transcription factors (Egr-1, NF-κB, p53, HIF-1α, AP-1, and Pax proteins) in an active reduced state (Hirota et al., 1997; Wei et al., 2000; Ziel et al., 2004; Gray et al., 2005; Pines et al., 2005a, 2005b; Tell et al., 2005, 2010a). APE1 can also function as a transcriptional repressor through indirect binding to negative Ca2+-response elements, which is regulated by K6/K7 acetylation (Bhakat et al., 2003). Acetylation occurring on these Lys residues has been recently demonstrated to occur during genotoxic treatment conditions (Yamamori et al., 2010) and to inhibit the apoptotic extrinsic pathway triggered by Helicobacter pylori infection in gastric epithelial cells while the DNA repair function inhibits the intrinsic pathway (Chattopadhyay et al., 2010). Recently, APE1 was demonstrated to bind/cleave abasic RNA (Vascotto et al., 2009a; Fantini et al., 2010; Tell et al., 2010b) and to control c-Myc expression by nicking its mRNA (Barnes et al., 2009). These discoveries point to a new function of APE1 in regulating gene expression through posttranscriptional mechanisms and open new routes to consider this protein as a possible target for anticancer therapy, to be pursued only after elucidation of the molecular details responsible for its different biological functions.
In this context, we showed that the first 33 amino acids of APE1 are required for a stable interaction with RNA, nucleophosmin, and other proteins involved in ribosome biogenesis/RNA processing (Vascotto et al., 2009a). Conversely, the APE1 N-terminal domain (spanning the region 35–127) is principally devoted to the redox transcriptional activity, whereas the C-terminal domain (region 161–318) exerts enzymatic activity on DNA abasic sites (Xanthoudakis and Curran, 1992; Xanthoudakis et al., 1994). Redox and repair functions of APE1 are completely independent; in fact, mutation at C65 abolishes the redox function but does not affect the repair one (Luo et al., 2008), whereas replacements of a variety of amino acids required for DNA repair, such as H309 and others (McNeill and Wilson, 2007), do not affect protein redox activity. Although the molecular mechanism of APE1 repair activity has been delineated (Gorman et al., 1997), that of the redox function has not. In fact, C65, which is required for this function (Georgiadis et al., 2008), is not accessible and resides inside the APE1 structure (Gorman et al., 1997; Mol et al., 2000; Luo et al., 2008). The absence of a C-X-X-C motif, as in the majority of redox regulatory proteins, together with an improper position of other APE1 cysteines to act as redox exchangers (Luo et al., 2010), hinders the molecular assignment of such a regulatory function. Recent in vitro studies showed that APE1 adopts different unfolded conformations depending on the redox state of its Cys residues (Su et al., 2011); moreover, the APE1 redox inhibitor (E)-3-(2-[5,6-dimethoxy-3-methyl-1,4-benzoquinonyl)]-2-nonyl propenoic acid (E3330) was shown to decrease the amount of the redox-active protein by driving C65 into disulfide bonds (Kelley et al., 2011). Thus it is plausible that APE1 may adopt a partially unfolded state as the redox-active form of the enzyme, as recently suggested (Su et al., 2011). E3330 holds clinical potential as a specific inhibitor of APE1 redox function without interfering with its endonuclease activity (Fishel and Kelley, 2007). The importance of this function is highlighted by results demonstrating that NF-κB–mediated gene expression is regulated by APE1 redox activity without effects on IκBα degradation (Shimizu et al. 2000; Nishi et al. 2002). E3330 was also found to selectively inhibit growth/migration of human pancreatic cancer cells (Zou and Maitra, 2008), suggesting that the APE1 redox function could represent a good candidate for inhibition of tumor invasion and metastasis. However, knowledge is lacking on the detailed molecular mechanisms responsible for the C65-mediated APE1 redox function and on the effects of E3330 inhibition on APE1 in vivo.
APE1 subcellular distribution within different mammalian cell types is mainly nuclear and critically controls cellular proliferative rate (He et al., 2003; Fung and Demple, 2005; Izumi et al., 2005). Increased expression of APE1 is associated with different tumorigenic processes (Tell et al., 2005). In particular, a higher intracellular expression and a robust cytoplasmic localization of APE1 have been described in lung, ovarian, thyroid, and breast cancers (Tell et al., 2005), in which they correlate with higher tumor aggressiveness. The possible causal role played by this particular distribution in tumor progression is unknown. However, APE1's ability to activate transcription factors, such as p53 and Egr-1 (Huang and Adamson, 1993; Gaiddon et al., 1999; Seo et al., 2002; Hanson et al., 2005; Pines et al., 2005a, 2005b), which are deeply involved in controlling cell cycle arrest and apoptotic programs, leaves the debate open regarding the mechanisms responsible for modulating its different functions in several contexts.
In different cell types, APE1 was proved to localize also in mitochondria, where it is involved in BER activity (Tell et al., 2001; Chattopadhyay et al., 2006; Szczesny et al., 2008, 2010). Mechanisms regulating APE1 localization and subdistribution are unknown (Chattopadhyay et al., 2006; Mitra et al., 2007), even though the proteolytic removal of the first 31–33 amino acids at the protein N-terminus seems to be responsible, at least in part, for promoting nuclear exclusion of the protein (Chattopadhyay et al., 2006). Recent work suggested the occurrence of a mitochondrial targeting sequence within region 289–318, which is masked by the N-terminal domain (Li et al., 2010), thus suggesting the need for specific and regulated mechanisms of protein unfolding–refolding to ensure proper APE1 localization.
APE1 is essential for cell viability (Xanthoudakis et al., 1996; Ludwig et al., 1998; Fung and Demple, 2005), and attempts to isolate stable APE1-knockout cell lines have been unsuccessful. For these reasons, achieving a comprehensive understanding of the molecular mechanisms mediating APE1 different functions has been very difficult. In the recent years, however, conditional knockout and knockdown strategies (Fung and Demple, 2005; Izumi et al., 2005; Vascotto et al., 2009a, 2009b) have made it possible to establish cell models to inspect and characterize the major functions of APE1. Nonetheless, knowledge of molecular effectors regulated by APE1 in determining its biological roles is still scanty. The essentiality of APE1 for mammalian cells seems to be mainly due to its DNA-repair activity in the BER pathway. In fact, transcomplementation of APE1 deficiency by the yeast homologous Apn1 (Fung and Demple, 2005), which lacks the redox-transcriptional activation domain, were able to recover, although not completely, mammalian cells from apoptosis induced by APE1 knockdown. However, attempts to restore the repair activity in cells not expressing APE1 by using an APE1 mutant lacking the acetylation sites in Lys-6/7 but not the DNA-repair activity (Izumi et al., 2005) were unsuccessful. Moreover, by specifically blocking the APE1 redox activity with the drug E3330 but not its DNA repair activity, it was shown that cytokine-mediated hemangioblast development in vitro was significantly impaired (Zou et al., 2007). These observations led us to hypothesize that the essentiality of APE1 may be due to its pleiotropic biological effects rather than merely to its DNA-repair activity and underline the necessity to elucidate the molecular mechanisms responsible for the fine tuning of the different APE1 biological functions to fully consider this protein as an established target for anticancer therapy.
In this work, we used a reconstitution strategy to reintroduce mutant APE1 proteins in APE1-silenced cell clones to determine the role of crucial amino acids in protein activities. We generated triple stable cell clones expressing ectopic flagged forms of APE1 wild-type and specific mutant APE1 proteins in an inducible APE1 small interfering RNA (siRNA) background, checking their ability to rescue the biological effects due to endogenous APE1 loss of expression. Our data demonstrate that APE1C65S behaves as a loss-of-function mutation affecting the in vivo folding of the protein and affecting mitochondrial functionality. Experiments with E3330 confirmed that C65 is responsible for controlling APE1 subcellular trafficking. Overall, our data shed light on some new molecular aspects mediating the multifunctional nature of APE1 in regulating different biological mechanisms and provide the first in vivo evidence of the relevance of C65 in regulating the folding and subcellular distribution of APE1.
RESULTS
Reconstitution of APE1 wild type and mutants reveals a role for H309, C65, and N-terminal 33 amino acids in mediating protein functions responsible for cell viability and growth
Evidence provided by us and others on APE1 multifunctional roles in mammalian cells raise several important questions regarding the possibilities to 1) discriminate between direct and indirect effects due to APE1 loss of expression, 2) investigate how these pleiotropic actions are modulated to guarantee a functional specificity, and 3) examine what function of APE1 is mainly responsible for its vital role. To gain insights into these issues, we used a knock-in strategy applied to the inducible APE1-silenced clones described previously (Vascotto et al., 2009a, 2009b). We generated triple stable cell clones expressing ectopic flagged forms of wild-type (WT) and mutant APE1 proteins on an inducible APE1 siRNA background. In parallel to WT protein (APE1WT), we expressed the following mutant APE1 proteins: 1) the APE1H309N mutant lacking the DNA repair function (Masuda et al., 1998; Evans et al., 2000); 2) the APE1C65S mutant with no redox activity (Evans et al., 2000); 3) the nonacetylatable APE1K6K7/R6R7 mutant (Bhakat et al., 2003); 4) the APE1C310S mutant lacking a recently discovered S-nitrosilation site involved in cytoplasmic relocalization of the protein upon nitrosative stress (Qu et al., 2007); 5) the double mutant APE1H309N/C310S; and 6) the noncleavable APE131-34A mutant, which has been suggested to have protective effects on cell viability (Fan et al., 2003). We evaluated the rescuing ability of each of these APE1 forms on different biological parameters previously demonstrated to be affected by APE1 silencing (Vascotto et al., 2009b), that is, cell growth, apoptosis, mitochondrial functioning, and stress fiber formation. During cell clone isolation, although we were able to isolate stably expressing ectopic forms of APE1WT, APE1C65S, APE1C310S, APE1K6K7/R6R7, and APE131-34A proteins (with a 20–40% of success on 5–20 colonies; see Table 1), neither APE1H309N nor APE1C310S/H309N gave positive colonies despite the similar number screened for each mutant (n = 13–20). Our further unpublished data (also B. Demple, personal communication) clearly showed that this mutation, which abolishes the DNA-repair (Masuda et al., 1998) and RNA quality control (Vascotto et al., 2009a; unpublished data) activities of APE1 while retaining its ability to bind the substrate, results in a protein form toxic to cells. Therefore these data do not exclude the possibility that the APE1H309N mutant may act as a dominant-negative protein form, being responsible for counterselection mechanisms occurring during the colony selection process, such that no viable APE1H309N-expressing clones were isolated during cell screening.
TABLE 1:
Knock-in colony screening results obtained for wild-type and different APE1 mutants.
| Knock-in APE1 | Number of screened clones | Number of positive clones | Frequency (%) |
|---|---|---|---|
| Wild type | 17 | 4 | 23.5 |
| K6K7/R6R7 | 5 | 2 | 40.0 |
| 31-34A | 15 | 3 | 20.0 |
| C65S | 5 | 2 | 40.0 |
| C310S | 6 | 1 | 16.6 |
| H309N | 20 | 0 | 0 |
| H309N+C310S | 13 | 0 | 0 |
HeLa cell clones grown in triple-selective medium as described in Materials and Methods were replated at low density and incubated for additional 14 d for single colonies, which were isolated by using cloning cylinders and then expanded for 10 d in triple-selective medium. The number of positive cell clones expressing ectopic APE1 different forms is reported.
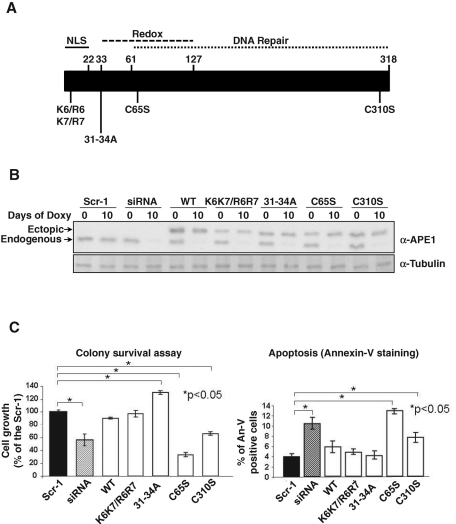
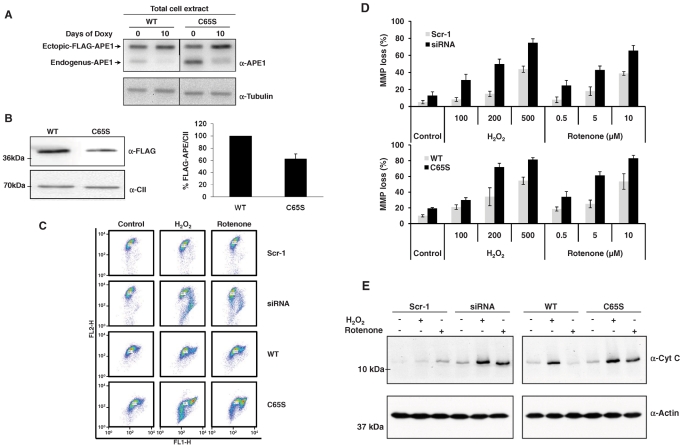
Expression of the other mutant APE1 proteins gave variable results. All positive clones displayed comparable expression levels with the ectopic APE1 protein upon endogenous APE1 silencing, as determined by Western blot analysis (Figure 1A), and all recombinant ectopic forms localized mainly within the nuclear compartment (Supplemental Figure S1). The ectopic expression was always comparable to that of the endogenous form before doxycycline treatment, thus showing that our cell model is much more similar to the physiological condition than that used in previous studies (Izumi et al., 2005). Comparison of cell growth and apoptosis upon endogenous APE1 silencing, as measured by clonogeneic assays (Figure 1B, left) and annexin V staining (Figure 1B, right), respectively, demonstrated a variable ability of the different ectopic proteins to rescue the biological effects due to the loss of endogenous APE1. In particular, whereas the APE1WT protein, as well as the nonacetylatable APE1K6K7/R6R7 mutant, resulted in almost complete rescue of cell growth, the noncleavable APE131-34A form resulted in a gain-of-function activity, having even higher viability than APE1WT-expressing cells. On the contrary, the APE1C65S mutant was completely unable to compensate the loss in cell viability due to endogenous APE1 silencing, thus demonstrating a dominant-negative functional activity. The APE1C310S mutant showed an intermediate behavior. Similar to cell growth results, the APE1C65S was completely unable to rescue from apoptosis induced by APE1 loss of expression. Further analysis to determine the mitochondrial function of the different cell clones showed that, with the exception of the APE1C65S mutant, all of the remainders were able to rescue the drop in mitochondrial membrane potential, Δψm, caused by endogenous loss of APE1 (see Table 2). This result suggests that the C65S mutation may play a substantial role in affecting the mitochondrial functionality of the protein, possibly affecting cell viability. Finally, we found that all APE1 mutants successfully prevented loss of stress fiber formation due to protein suppression (Table 2).
FIGURE 1:
Rescuing abilities of wild-type and different APE1 mutants. (A) Schematic representation of the human APE1 protein with indication of functional domains and mutants used for KI studies. (B) Inducible APE1 siRNA HeLa cell clones were stably transfected with the p3XFLAG-CMV-14 empty or encoding for APE1WT or APE1K6K7/R6R7, APE131-34A, APE1C65S, or APE1C310S mutant. After selection, different clones were assayed for the expression of the endogenous and the ectopic APE1 forms before and after expression of the specific APE1 siRNA sequence through doxycycline treatment. Scr-1 indicates the control-siRNA transfected cell line. Assays were performed by immunoblotting with the specific anti-APE1 antibody or anti-tubulin as loading control. (C) Left, rescuing abilities on cell growth of different ectopic APE1 forms after silencing of endogenous APE1 through inducible-expressed siRNA, as measured by colony survival assays. Five hundred cells of control and siRNA clones were seeded in Petri dishes and then treated with 1 μg/ml doxycycline for 10 d. Data are the mean ± SD of three independent experiments. Right, evaluation of apoptosis by annexin V staining on ectopic APE1-expressing cell lines. Details are as on the left. Data are the mean ± SD of three independent experiments.
TABLE 2:
Rescuing ability of wild-type and different APE1 mutants.
| Knock-in APE1 | Cell growth (colony survival assay) | Apoptosis | Δψm | Stress fibers |
|---|---|---|---|---|
| Wild type | +++ | ++ | +++ | +++ |
| K6K7/R6R7 | +++ | ++ | +++ | +++ |
| 31-34A | ++++ | +++ | +++ | +++ |
| C65S | - | - | + | ++ |
| C310S | + | + | +++ | +++ |
The rescue effect of the different APE1 ectopic forms was evaluated as described in Vascotto et al. (2009b).
C65S and 31-34A mutations affect APE1 functions through different mechanisms
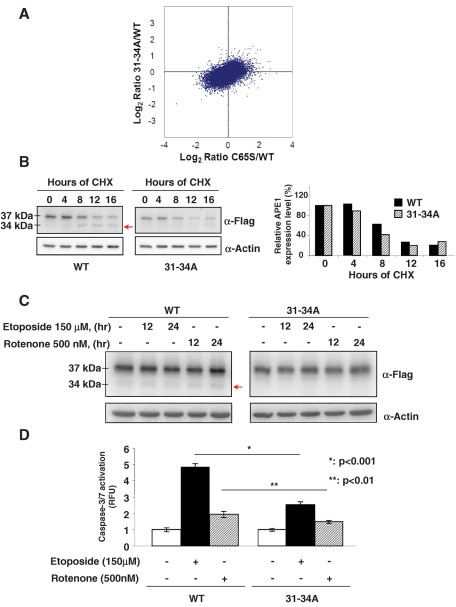
To gain more insight into the molecular effects caused by the knock-in experiments (Figure 1 and Table 2), we performed gene expression profiling through microarray analysis. We concentrated on two mutations because of their opposite effects: C65S, a loss-of-function mutation, and 31-34A, a gain-of-function mutation. The plot in Figure 2A shows that the APE1C65S mutant affected the global gene expression profile, in comparison with that of the wild type, more than the APE131-34A mutant. In fact, after statistical analysis of differential expression, no probe sets had a q value of <0.05 for APE131-34A versus APE1WT, whereas for APE1C65S versus APE1WT, 38 probe sets met this criterion, in line with the expectations suggested by the biological data shown in Figure 1. These 38 probe sets included genes involved in cell cycle (TGFA, DUSP4, and DUSP6) and growth (TGFA, AREG, NRG1, and GDF15; Supplemental Table S1). Owing to the pleiotropic action of APE1, it was impossible to drive any definitive conclusion on the main molecular mechanisms at the basis of the observed biological phenomena. These results suggest that the APE1C65S mutant exerts its effects in part through modulating, directly or indirectly, the APE1 target gene expression profile. However, the limited number of genes differentially expressed by the APE1C65S mutant with respect to APE1WT demonstrates that the biological effects of the APE1C65S mutation are not only associated with its impact on gene expression when compared with a previous observation showing that more than 1100 genes were found to be affected by APE1 silencing (Vascotto et al., 2009b).
FIGURE 2:
C65S and 31-34A mutations affect APE1 function by different mechanisms. C65S mutation, but not 31-34A, affects APE1 influence on gene expression. (A) Differential gene expression profiling in APE1C65S and APE131-34A vs. APE1WT-expressing clones. Log2 ratio scatter plots comparing expression profiles from the APE1 rescue experiments (mean expression from biological replicas made in triplicate). For APE131-34A vs. APE1WT, only 113 probe sets with a fold change greater than 2 were observed; for APE1C65S vs. APE1WT, 985 probe sets with a similar fold change were found. Only for the APE1C65S vs. APE1WT were statistically significant probe sets (38) found (Supplemental Table 1). (B) 31-34A mutation does not affect APE1 protein half-life. HeLa cells were transfected with APE1WT and APE131-34A constructs, as reported in Materials and Methods. The day after transfection, protein expression was inhibited by treatment with cycloheximide (100 μg/ml) up to 24 h to evaluate APE1 half-life. Thirty micrograms of total cellular extracts was separated by 12% SDS–PAGE and then transferred onto a nitrocellulose membrane. The membrane was immunoblotted with anti-FLAG or anti-actin antibodies. Left, CHX treatment causes a progressive disappearance of the 37-kDa protein band corresponding to the full-length APE1 protein and a concomitant accumulation of the truncated 34-kDa protein form (arrow) lacking the N-terminal part. This is evident only when expressing the APE1WT form but not the APE131-34A mutant. Right, normalized volume values of band intensities obtained after densitometric analysis of the ratio between the 37-kDa APE1 form and β-actin reported as histograms. Black and dashed boxes indicate APE1WT and APE131-34A expression levels, respectively. Data shown are the mean of two independent experimental sets. (C) Treatment with apoptosis-inducing agents causes proteolysis of APE1WT but not APE131-34A protein. HeLa cells were transfected with APE1WT- and APE131-34A-expressing plasmid, as reported in Materials and Methods, and then treated with etoposide and rotenone for the indicated times. Expression of ectopic APE1WT and APE131-34A proteins was assayed by immunoblotting. Forty micrograms of total cellular extracts was separated by 12% SDS–PAGE and then transferred onto a nitrocellulose membrane. The membrane was immunoblotted with anti-FLAG or anti–β-actin antibodies. Treatments induced the accumulation of the truncated APE1NΔ33 form of the protein only in the case of the APE1WT protein but not when expressing the APE131-34A mutant. (D) The APE131-34A mutant plays a protective role toward drug-induced apoptosis. After silencing of endogenous APE1 protein, APE1WT and APE131-34A clones were treated with etoposide and rotenone for 24 h and then caspase-3/7 activation was measured, as described in Materials and Methods. The histograms show the average ± SD of the fluorescence values obtained in three independent experiments. Cells expressing the APE131-34A mutant protein were significantly more resistant to apoptosis than the APE1WT-expressing cells. The p value was calculated by Student's t test.
Of interest, the APE131-34A mutant did not reveal a significantly different gene expression profile from that shown by APE1WT protein. To explore the molecular reasons for its gain-of-function phenotype, as shown in our cell growth data (Figure 1), additional experiments were performed. First, we evaluated whether this mutation increased APE1 stability. Half-lives of the APE1WT and the APE131-34A proteins were evaluated by treating reconstituted cells with the protein synthesis inhibitor cycloheximide (CHX) and by measuring the kinetics of APE1 disappearance through Western blot analysis. Figure 2B shows that CHX treatment promoted a time-dependent disappearance of the full-length APE1 band with a similar kinetics for both proteins, with an approximate half-life of 8 h. In the case of APE1WT, the disappearance of the full-length form was paralleled by a progressive increase of a truncated form (NΔ 33APE1) missing the first 33 amino acids at the protein N-terminus (Chattopadhyay et al., 2006), which was not visible in the case of the APE131-34A mutant. Thus the 31-34A mutation does not seem to directly affect protein half-life. We therefore evaluated the role of APE131-34A mutant in protecting cells from the apoptosis-inducing agents rotenone and etoposide. Similar to CHX treatment, we found that exposure of cells to these compounds induced the accumulation of the truncated form of APE1 in the case of APE1WT-expressing cells but not in the case of APE131-34A-expressing cells (Figure 2C). Indeed, the caspase-3/7 activation assay (Figure 2D) showed that the APE131-34A-expressing cells were significantly more resistant to apoptosis (induced by both rotenone and etoposide) than the APE1WT-expressing ones. On the basis of these data, we conclude that the proteolytic removal of the N-terminal part of APE1 is associated with the onset of an apoptotic phenotype but does not affect the overall protein half-life in vivo. This effect was not simply due to the loss of the NLS sequence leading to the inhibition of nuclear localization, since the APE1NΔ33 mutant retained the ability to reside within the nuclear compartment even if a significant cytoplasmic localization was evident (Vascotto et al., 2009a). Recent data from our laboratory demonstrated that the nonconserved protein N-terminus is essential to modulate APE1–protein interactions, APE1's role in the RNA-cleansing process (Vascotto et al., 2009a; Tell et al., 2010b), and APE1's DNA-repair function through modulation of its catalytic activity (Fantini et al., 2010). Thus this unstructured portion of the protein seems to act as an important molecular switch for regulating the different APE1 functions (Tell et al., 2010a).
C65 is critical for chaperone-mediated, redox-dependent folding of APE1 in vivo
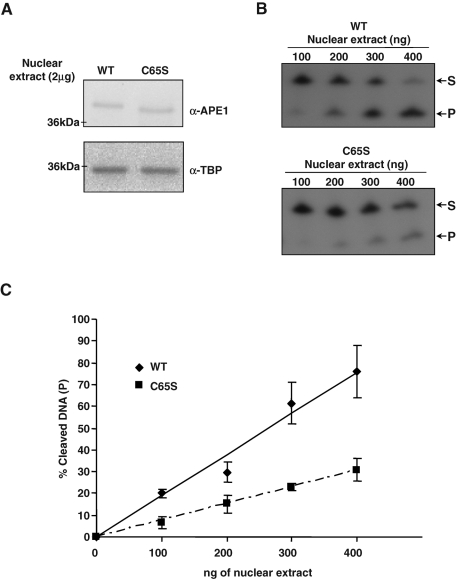
To better inspect the reasons for the loss-of-function behavior of the APE1C65S mutant, we analyzed the effect of this mutation on the protein endonuclease activity over abasic double-stranded DNA (dsDNA) by cleavage assay (Fantini et al., 2010). It was recently shown that the C65S mutation does not significantly affect the enzymatic activity of APE1 with respect to the WT protein when using recombinant purified proteins expressed in bacteria (Mantha et al., 2008). However, this finding cannot exclude the possibility that this mutation may affect the APE1 endonuclease activity in vivo. This is reinforced by the observation made by different laboratories that the endonuclease activity of purified bacterially expressed recombinant APE1 is significantly reduced (at least 100-fold lower) with respect to that obtained from mammalian cells (HeLa; data not shown). Thus we compared the APE1 enzymatic activity in nuclear extracts from APE1WT- and APE1C65S-expressing cells. Results shown in Figure 3C clearly demonstrate that the enzymatic activity of APE1C65S mutant is significantly lower than that of the APE1WT protein, accounting for a role in the biological effects seen in Figure 1.
FIGURE 3:
APE1 endonuclease activity on abasic dsDNA is reduced by the expression of the redox mutant C65S. (A) After silencing of endogenous APE1 protein, 2-μg nuclear extracts from APE1WT and APE1C65S mutant clones were separated on 12% SDS–PAGE and then blotted onto a nitrocellulose membrane, which was subsequently incubated with anti-FLAG or anti-TBP (TATA binding protein) antibodies as loading control. (B) Reported amounts of normalized nuclear extract from APE1WT and APE1C65S clones were used to evaluate APE1's endonuclease activity on abasic dsDNA. The conversion of the radiolabeled tetrahydrofuran-containing oligonucleotide substrate (S) to the shorter incised product (P) was evaluated on a denaturing 20% (wt/vol) polyacrylamide gel. In each reaction, 2.5 pmol of double-stranded tetrahydrofuran containing DNA oligonucleotide (dsFDNA) was used. A representative image from three independent experiments is shown. (C) Average values of incision percentage with standard deviations of three independent experiments.
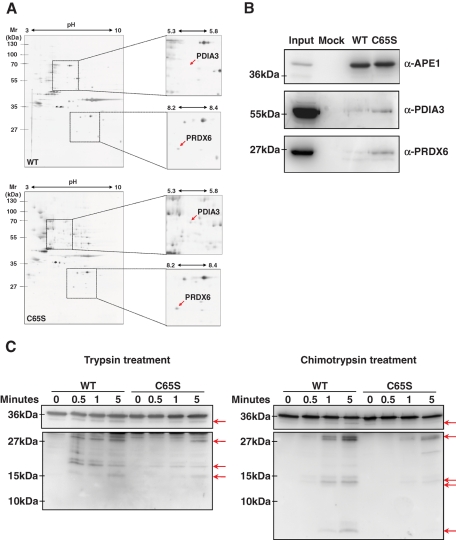
Because Cys residues can drive the folding process of a protein in vivo through sequential protein–protein interaction steps (Herrmann and Riemer, 2010b), we hypothesized that C65S mutation could affect the interactomic network of APE1. By using a proteomic approach described previously (Vascotto et al., 2009a), we analyzed the changes in the APE1 interactome as a consequence of C65S mutation. The interactomic map of APE1C65S clearly showed an increased presence of two protein species with respect to that of APE1WT (Figure 4, A and B). These were identified as peroxiredoxin-6 (PRDX6) and protein disulfide-isomerase A3 (PDIA3 or ERp57). Of interest, both are known to be involved in redox-assisted protein-folding processes in mammalian cells. Thus their increased interaction with the APE1C65S mutant may account for impairment in the kinetics of the redox-folding process for this mutant with respect to APE1WT.
FIGURE 4:
APE1C65S redox mutant interacts more strongly with PDIA3 and PRDX6 than APE1WT. (A) Differential two-dimensional electrophoresis analysis of the APE1-FLAG protein complex immunopurified under native conditions from APE1WT and APE1C65S mutants demonstrated the occurrence of a stronger interaction of the C65S mutant with PDIA3 and PRDX6 proteins. Results were obtained by silver staining and mass spectrometry identification of the proteins coimmunopurified with APE1-FLAG. (B) Western blotting analysis of a coimmunoprecipitated material from an independent experiment. Total cell extracts from HeLa cells (Mock) and APE1WT and APE1C65S clones were coimmunoprecipitated, normalized for immunoprecipitated APE1 protein, and then analyzed by 10% SDS–PAGE. Immunoblotting using specific antibodies (α-APE1, α-PDIA3, α-PRDX6) confirmed the differential interaction of APE1WT and APE1C65S clones with PDIA3 and PRDX6. Total cell extract was used as a positive control. Absence of signal in the mock line confirmed the specificity of these interactions. (C) Limited proteolysis analysis of the immunopurified APE1WT and APE1C65S proteins after trypsin (left) and chymotrypsin (right) treatment. After incubation with proteolytic enzymes for the reported times, proteins were separated on 12% SDS–PAGE and then blotted onto a nitrocellulose membrane, which was further immunoblotted with anti-APE1 antibody. Although the final pattern of proteolytic fragments generated upon digestion (highlighted by arrows) was similar for APE1WT and the APE1C65S mutant, their kinetics of proteolysis showed a significantly different behavior. These difference indicates a dissimilar folding state of the two proteins. A representative image from three independent experiments is shown.
The possible effect of C65 residue on the structural folding of APE1 in vivo was evaluated by limited proteolysis studies on immunopurified APE1WT and APE1C65S proteins. Time-course analyses by using trypsin and chymotrypsin as proteolytic enzymes are shown in Figure 4, C and D, respectively. Although the final pattern of proteolytic fragments generated upon digestion was similar for APE1WT and APE1C65S mutants, the kinetics of proteolysis showed a quite different behavior, thus suggesting a differential response of the proteins to proteolytic attack and confirming the role of C65 in controlling the proper folding of APE1 in vivo.
C65 is critical for mitochondrial localization of APE1 in cells, and C65S mutation causes mitochondrial impairment
Redox folding and interaction with PRDX6 and PDIA3 may constitute a mechanism affecting APE1 localization into the mitochondrial compartment. Moreover, based on the observations that APE1C65S-reconstituted cells showed a significant impairment of mitochondrial functionality (Table 2), we characterized the role of C65 residue by analyzing the effect of the C65S mutation on the amount of mitochondrial protein levels recoverable in the HeLa cells used in this work. Initially, we verified that the knock-in strategy (i.e., expression of an ectopic APE1 recombinant protein) did not affect the expression of the endogenous protein within mitochondria (unpublished data). As demonstrated in Figure 5A, the total amount of ectopic APE1 after 10 d of doxycycline treatment was comparable in APE1C65S and APE1WT clones. Of interest, the amount of APE1C65S protein in mitochondria was significantly lower (∼60%) than the WT protein (Figure 5B). These data demonstrated that this mutation affects proper accumulation of APE1 in mitochondria.
FIGURE 5:
APE1C65S mutation affects APE1 mitochondrial accumulation and causes mitochondrial impairment. (A) Western blotting analysis of total cell extracts from HeLa cells and APE1WT and APE1C65S clones confirmed that APE1WT and APE1C65S ectopic proteins were expressed at similar levels within the cells. (B) Western blotting analysis of mitochondrial extract from HeLa cells and APE1WT and APE1C65S clones by using anti-FLAG and anti–complex II (α-CII) antibodies showed that C65S mutation impairs APE1 translocation within mitochondria. Right, relative histogram plotting the average percentage values of ectopic recombinant flagged APE1WT and APE1C65S proteins normalized for α-CII and for total APE1 content into HeLa cells ± SD of three independent experiments. (C) JC-1 assay of mitochondrial membrane potential loss in APE1 knockdown (siRNA) and APE1C65S mutant clones after oxidative stress; Scr-1 and APE1WT clones were used as controls separately. To induce oxidative stress, different doses of H2O2 and rotenone were dissolved in the medium and incubated with cells for 3 h, then stained with JC-1 probe and immediately measured by using flow cytometry; only 200 μM H2O2 and 5 μM rotenone doses are displayed in the graph. (D) Histograms for JC-1 assays. The percentage of cell population undergoing MMP loss was scored in cells treated with different doses of both H2O2 and rotenone for 3 h. Data show the average values ± SD from three independent experiments. (E) The release of cytochrome c (α-CytC) after oxidative stress in APE1-knockdown (siRNA) and APE1C65S-mutant clones was assayed after 6 h of 200 μM H2O2 or 5 μM rotenone treatment. Scr-1 and APE1WT clones were also evaluated for comparison. Cytosolic fractionation was performed excluding the mitochondrial fraction by differential centrifugation. The content of cytochrome c in cytosolic fraction, representing the permeability of mitochondrial outer membrane and activation of mitochondrial pathways of apoptosis, was measured by Western blotting. β-Actin was also detected as a control for loading equality.
We then examined whether the biological effects of C65S mutation (Figure 1 and Table 2) are due to impairment of the mitochondrial function following oxidative stress; as oxidant stimuli we used H2O2 and rotenone. To measure the mitochondrial function alterations occurring after oxidative stress, we performed two different assays for evaluating changes in both Δψm and mitochondrial membrane permeability. As shown in the dot plots of JC-1 staining (Figure 5C), those cells with Δψm loss appeared to be associated with a decreased FL2 signal or/and an increased FL1 signal (right/lower shift). When treated with H2O2 and rotenone for 3 h, Δψm in all cell lines significantly dropped, and the amplitude of the Δψm decrease was dose dependent. In APE1-deficient cell lines, including knockdown (siRNA) and redox-mutant forms (APE1C65S), however, more cells underwent Δψm loss when compared with their counterparts (Scr-1 and APE1WT) separately (Figure 5D). Continuous Δψm loss usually causes mitochondrial impairment and membrane leakage, which consequently leads to the release of proapoptotic factors, such as cytochrome c, from mitochondria to cytoplasm, thus determining initiation of the apoptotic cascade. The results shown in Figure 5E indicate that cytochrome c was increased in cytosolic fraction after H2O2 or rotenone treatment; this occurred even more significantly in APE1-knockdown and APE1C65S mutant cell lines. These data are consistent with the observed Δψm alterations and clearly demonstrate that mitochondrial function of APE1-deficient and APE1C65S-reconstituted cells is impaired in response to oxidative stress and may induce apoptosis through a mitochondrial-dependent pathway.
APE1 interacts with the redox chaperone Mia40
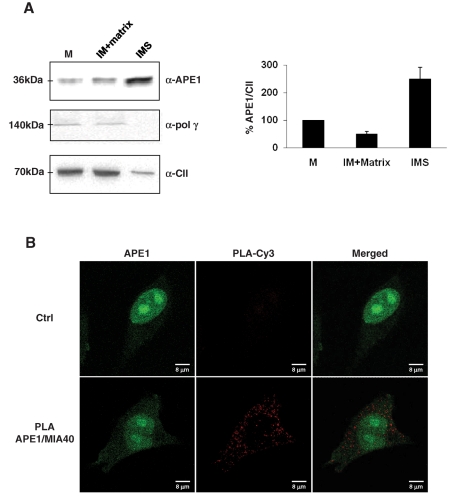
By using purified mitochondria from bovine heart as a reliable source for organelle subfractionation, we then investigated APE1 localization in the mitochondrial compartment. We treated mitochondria with digitonin to selectively disrupt the outer membrane and to separate the intermembrane space (IMS) and the matrix, two mitochondrial compartments enriched in soluble proteins. Figure 6A shows that APE1 in purified mitochondria is a full-length protein of ∼37 kDa and is mainly located within the IMS. The APE1 localization pattern was different from that of the complex II of the oxidative phosphorylation chain, used as a marker of the inner membrane in the different mitochondrial subfractions, and of polymerase γ, which was described as a non–free soluble matrix protein of the mitochondrial BER system (Stuart et al., 2005). Both were confirmed to be in the inner membrane/matrix fraction.
FIGURE 6:
Mitochondrial APE1 is accumulated within the IMS of mitochondria and interacts in vivo with the mitochondrial IMS import and assembly protein 40 (Mia40). (A) Mitochondria from bovine hearth were fractionated with digitonin and subsequent differential centrifugation to separate matrix and IMS, which are compartments rich in soluble proteins. Sixty micrograms of total mitochondrial extract (M), matrix plus inner membrane fraction (IM), and intermembrane space (IMS) fraction alone was separated onto 12% SDS–PAGE and then blotted onto a nitrocellulose membrane, which was further immunoblotted with anti-APE1, anti–Poly γ, and anti–complex II (α-CII) antibodies. Western blotting analysis showed that only the full-length protein was present within the mitochondrial fractions, and APE1 was predominantly found in the IMS fraction rich of soluble proteins. Polymerase γ, used as marker of non–freely soluble matrix protein of the mitochondrial BER system, was confirmed to be associated with the inner membrane–containing particulate fraction. Total mitochondria extracts were used as a positive control; complex II was used as a marker of the inner membrane. Average percentage values of APE1 normalized for CII ± SD of three independent experiments are plotted in the relative histogram (right) of Western blot analysis (left). (B) PLA technology was used to demonstrate the occurrence of the interaction between APE1 and Mia40. PLA reaction was performed following manufacturer's instructions. Confocal microscopy analysis highlighted the presence of distinct fluorescent red dots (PLA signals) indicating the occurrence of in vivo interaction between APE1 and Mia40; green fluorescence shows the APE1 localization within the cell. Negative control is represented by cells incubated only with α-APE1 antibody.
Due to its significant molecular mass, mitochondrial localization of APE1 should require active processes of transportation. The main chaperone responsible for redox-assisted folding processes within the IMS of mitochondria in mammals is Mia40 (Riemer et al., 2011). Therefore we determined whether Mia40 might interact with APE1 through proximity ligation assay (PLA) analysis, which allows in situ detection of two proteins that are at interacting distance range (<40 nm; Weibrecht et al., 2010). As shown in Figure 6B, we were able to demonstrate a specific signal of APE1 interaction with Mia40 within the cytoplasm of HeLa cells, accounting for the occurrence of a physical interaction between the two proteins, which may explain the localization of APE1 within the IMS.
E3330 treatment impairs APE1 subcellular trafficking, confirming that redox folding controls APE1 cellular distribution
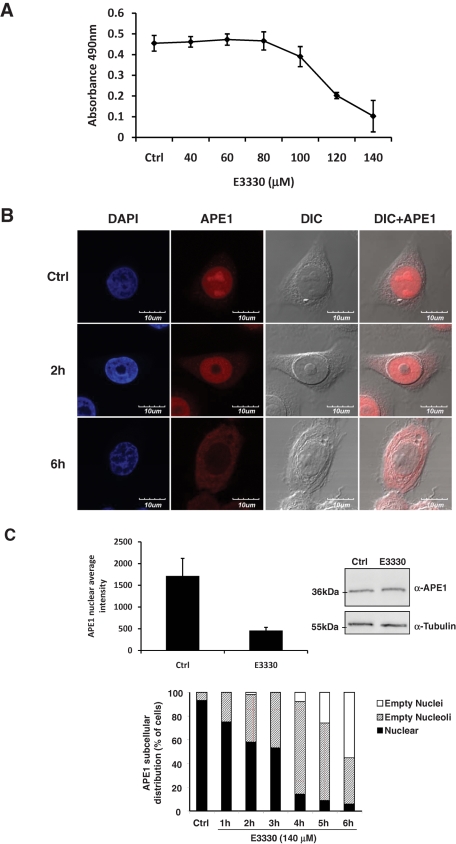
To elucidate the role of C65 in regulating the APE1 folding and thus its subcellular trafficking, we used the redox inhibitor E3330, which has been reported to directly interact with this protein and to inhibit its redox activity by increasing the formation of disulfide bonds by Cys-65 (Shimizu et al., 2000; Nishi et al., 2002) and decreasing the protein redox active population (Su et al., 2011). In this context, we used a tumor cell model represented by the human glioblastoma cell line SF767 (Hirose et al., 2003). Treatment of cells for 6 h with increasing doses of E3330 caused a significant reduction of mitochondrial functioning, as measured by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) cell viability assay (Figure 7A), without causing any apparent apoptotic process at early times of treatment (unpublished data). The maximal extent of this effect was obtained at 140 μM E3330 treatment. To confirm the effect of E3330 on APE1 subcellular trafficking, we used this dose of drug and performed kinetics experiments to evaluate APE1 subcellular distribution. Immunofluorescence experiments (Figure 7, B and C, left) clearly demonstrated that a significant relocalization of APE1 from the nuclear to the cytoplasmic compartment occurred with no significant alterations in total protein levels, as measured by Western blot analysis (Figure 7C, right). Moreover, kinetic experiments also showed that E3330 treatment caused a progressive emptying of the nucleoli, as confirmed by anti-nucleophosmin staining analysis (Figure 7C, bottom, and Supplemental Figure S2). Altogether, these data demonstrate that the redox state of C65 controls the subcellular trafficking of APE1 and that E3330 effects are associated with alterations of APE1 trafficking.
FIGURE 7:
APE1 redox inhibitor E3330 affects APE1 nuclear localization on human glioblastoma cell line SF767. (A) MTS cell viability assay on SF767 after 6 h of treatment with increased amounts of E3330 showed a progressive reduction of viable cells in proliferation as a consequence of the treatment. E3330 was resuspended in 100% dimethyl sulfoxide (DMSO); control is represented by cells treated with the same volume of DMSO used for the highest dose of E3330. (B) APE1 subcellular localization was detected through confocal analysis using a specific APE1 monoclonal antibody derivatized with Alexa Fluor 568. 4′,6-Diamidino-2-phenylindole was used for nuclear staining and differential interference contrast to observe cellular shape. In control cells, APE1 mainly localized within the nuclear compartment and accumulated into nucleoli. Treatment with 140 μM E3330 induced an early (1–4 h) and progressive reduction of APE1 nucleolar positivity, followed by a complete nuclear emptying and cytoplasm accumulation of APE1. (C) APE1 nuclear intensities of control and E3330-treated cells were measured with Metamorph analysis software, indicating a statistically significant (p < 0.001) reduction of APE1 nuclear positivity after 6 h of 140 μM E3330 treatment (upper left). On the contrary, this treatment did not affect APE1 protein expression, as demonstrated by Western blotting analysis on total cell extracts (upper right). As loading control, nitrocellulose membrane was incubated with anti-tubulin antibody. Histograms below represent the statistical analyses of APE1 subcellular distribution performed in up to 300 cells (nuclear positivity, nucleolar and nuclear emptying) for each time point of E3330 treatment (bottom, center).
DISCUSSION
Comprehension of the molecular signaling networks underlying the multifaceted action of APE1 is at its beginning. In particular, a pair of questions remain open in explaining why attempts to completely revert apoptosis due to APE1 suppression via sole targeting of the DNA-repair enzymatic activity of this protein were unsuccessful: 1) How are protein pleiotropic actions modulated to guarantee functional specificity? 2) What is the function of APE1 that is mainly responsible for its vital role?
To address these issues, we used an unbiased strategy to investigate the ability of different APE1 mutants, targeting specific known functions, to recover from the biological changes associated with APE1 loss of expression. Of interest, rescue experiments provided strong evidence for a determined and hierarchical role of specific APE1 amino acids in controlling the different biological protein functions. Our inability to isolate any cell clone expressing the APE1H309N mutant strongly indicated that complete abolition of DNA-repair and/or RNA quality control activities of this protein, although retaining its ability to binding substrate, is toxic for cells, as previously suggested (Fung and Demple, 2005). On the basis of gene expression profiling, the APE131-34A-mutant rescue was indistinguishable from that exerted by APE1WT, as also expected on the basis of the biological results obtained in this study; no genes with statistically significant differential expression were observed. Thus we inspected in more detail the mechanisms possibly explaining the gain-of-function effect of this mutant. On the basis of data reported in Figure 2, B–D, we can conclude that the proteolytic removal of the N-terminal part of APE1 leads to its functional inactivation in vivo without affecting protein half-life. We believe that the nonconserved protein N-terminus, which is responsible for modulating protein–protein interactions, is devoted to regulation of different APE1 functions in an indirect way, as suggested by our recent work (Vascotto et al., 2009a; Fantini et al., 2010; Tell et al., 2010a, 2010b). An additional intriguing hypothesis, which is under evaluation, is that the truncated APE1 may play an active role in apoptotic triggering. Further work is needed to clearly address this point.
The APE1K6K7/R6R7 mutant, in our cell model, was able to rescue from apoptosis induced by endogenous APE1 deprivation obtained through a specific siRNA silencing strategy in stable cell lines (Figure 1C). These data seem partially in contrast with those previously published (Izumi et al., 2005). However, it is remarkable to note the significantly different biological system used by these authors with respect to ours, in terms of: 1) cell type used (mouse normal fibroblasts vs. human tumor HeLa cells) and 2) generation of the model (Cre-knockout gene vs. siRNA knockdown in HeLa cells) and the procedure chosen for reconstitution with ectopic APE1 forms (microinjection vs. stable cell cloning). Thus, in our approach, alternative mechanisms able to compensate for some subtle APE1 functional deficiencies could have been selected during the cloning procedures. Thus these selection procedures could have hidden some differences particularly under basal conditions of cell growth, in which the majority of our experiments were performed while underscoring the most important functional deficiencies of the H309N and C65S mutants. In this regard, it is expected that using some genotoxic stimuli might highlight the functional relevance of further mutants, and work is in progress in our lab along these lines. Recent work showed increased caspase-3 activation occurring in APE1K6K7/R6R7 mutant with respect to the APE1WT protein. These results, which were obtained in gastric epithelial cells only upon H. pylori infection and not under basal conditions (Chattopadhyay et al. 2010), would confirm our hypothesis. On the other hand, our cell model has the advantage of having a strict and controlled amount of ectopic APE1 protein level, which is comparable in the different cell lines and, most important, is similar to the endogenous one, which has been replaced. Izumi et al. (2005) could not check for this important parameter in light of the relevance of a proper APE1 expression level in mammalian cells, and Chattopadhyay et al. (2010) used an overexpression cell model triggered with an oxidative stress–inducing stimulus (by H. pylori infection), two conditions quite different from ours. APE1 expression levels are homeostatically regulated in mammalian cells through control of protein turnover (Busso et al., 2009) and at the gene expression level through a negative feedback loop involving APE1 itself (Kuninger et al., 2002). Of interest, the absolute amount of APE1 protein is significantly variable, depending on cell type and growth condition (see later discussion). For these reasons, cell models based on overexpression strategies like those described by Chattopadhyay et al. (2010) and Izumi et al. (2005) might better resemble a condition of cell activation in terms of APE1 expression. Our reconstitution cell model, on the other hand, may well represent a more physiological condition in which cells are selected to survive by only expressing the ectopic recombinant protein under nonstressing conditions.
Mutation of C65 residue, which is responsible for controlling the redox activity of APE1, displayed a loss-of function-effect toward cell growth and viability. Of interest, the mild alteration of the gene expression profiling of APE1C65S-expressing cells with respect to APE1WT ones points to a role for the redox activity of APE1 in controlling expression of genes involved in cell cycle and cell growth. In any case, this alteration may not represent per se the major cause of the loss-of-function effect we observed, at least under basal cellular conditions. Rather, our data clearly highlight the central role of C65 in redox-assisted folding of APE1 in vivo, which affects mitochondrial localization of the protein. In fact, C65S mutation alters the folding process of APE1 and controls its interaction with PDIA3 (Figure 4), a protein containing thioredoxin-like domains and having chaperone activity (Grillo et al. 2006). Thus it could be hypothesized that 1) APE1C65S may induce a misshapen conformation associated with an increased affinity toward PDIA3 (Figure 4), and this could reduce the fraction of APE1 available to translocate into mitochondria; and 2) C65 may promote a redox-dependent conformational change toward a more translocatable protein form, thus behaving as an essential redox sensor. In this context, the absence of C65 would make APE1C65S unable to efficiently respond to oxidative stress and translocate into mitochondria. The reduced mitochondrial amount observed for the APE1C65S mutant should explain the biological effects associated with expression of this protein, which leads to a reduced cell viability as a consequence of mitochondrial impairment. The results reported in this study are then in agreement with recent evidence, obtained in vitro with purified recombinant proteins, showing a role for C65 in mediating the redox-assisted folding of APE1 (Kelley et al., 2011; Su et al., 2011). Our data extend these observations to the in vivo relevance for this unexpected mechanism and provide the molecular basis to explain apparently contradictory observations from different laboratories. Actually, the role of C65 in controlling APE1 redox function, as supported by published evidence (Walker et al., 1993; Su et al., 2011), contradicts the single work of Ordway et al. (2003), which reported the lack of any major phenotype alteration in APE1C64A (homologous to human APE1C65A) transgenic mice. We believe that our findings on APE1C65S mutant, on tumor cells, are not in contrast with previous work but rather reinforce current knowledge on the central role of C65 in redox-assisted folding of APE1 in vivo. Its impact on protein subcellular distribution should be interpreted on the basis of the cell context in which experimental observations were obtained and of the presence of compensatory mechanisms present in transgenic mice (Ordway et al., 2003). Our present data support a role for C65 in controlling subcellular trafficking of APE1 that may affect, in turn, the biological functions of the protein, including DNA-repair in cells. Owing to the differential essentiality of this activity for mammalian cells, an impairment of the redox-mediated folding process of APE1 may have different effects, depending on the cell type and cellular conditions. In addition, due to the relevance of APE1 protein for different cell lines, different biological mechanisms could be switched on to compensate for reduced/impaired APE1 function. It would be expected, in fact, that the biological impact of C65A mutation could be more relevant under pathological conditions associated with increased oxidative stress or tumorigenic process. Unpublished data confirm these hypotheses. Notably, the quite heterogeneous expression levels of APE1 protein in different cell lines, and thus its importance, should be taken into account. Whereas fibroblasts express ∼105 APE1 molecules/cell (Cappelli et al., 2001), tumor cells like the HeLa we used here express ∼107 molecules per cell (unpublished data). In the work by Ordway et al. (2003) all biochemical assays were performed with mouse embryo fibroblast cells under normal conditions, and thus it is possible that the discrepancies with our data could be ascribed to these profoundly different experimental conditions from a biological point of view. An alternative explanation about the difference from the results of Ordway et al. (2003) could in principle be ascribed to the choice of the mutation. In fact, the substitution of the Cys with a Ser could be different from Ala because Cys-65 is a buried residue in the three-dimensional structure and, as expected, the Cys-to-Ala mutation has no effect on the local folding of the molecule (Georgiadis et al., 2008). In this environment, Ser could be destabilizing for local structure due to its hydrophilic nature. In any case, since it has been recently demonstrated that Cys-65 (together with Cys-93) is the more oxidizable residue of APE1 (Su et al., 2011), thus supporting a model in which a conformational transition implying solvent exposure is required for exerting the redox activity of Cys-65, the Cys-to-Ser mutation better resembles this condition occurring during the redox function of APE1 and causing partial protein unfolding. Thus, Cys to Ser is a better mimic of an “open” conformation of this domain of APE1, whereas Cys to Ala better mimics a “closed” conformation. In this light, both mutations will give partial but not contrasting information about the real biological condition of the APE1 redox state.
We have also demonstrated here that APE1 accumulates within the IMS of mitochondria and that C65 integrity is essential for protein subcellular localization. Notably, despite the hundreds of proteins present within the matrix of mitochondria, only 50–100 have been described to occur within the IMS (Riemer et al., 2011). Of interest, those mitochondrial proteins present within the matrix are initially synthesized on cytosolic ribosomes as precursor proteins carrying N-terminal MTSs, which direct them into mitochondria, where they are proteolytically removed in the matrix by matrix processing peptidases (Chacinska et al., 2009). In contrast, mitochondrial proteins present within IMS mostly lack these classical MTS sequences and use alternative routes for mitochondrial import, generally relying on the Mia40-Erv1 disulfide relay system (Herrmann and Riemer, 2010a, 2010b). APE1 completely lacks a classic N-terminal MTS sequence (Tell et al., 2001; Chattopadhyay et al., 2006), and the molecular mechanisms regulating its mitochondrial trafficking are unknown. Recently, Li et al. (2010) suggested that residues spanning the region 289–318 are important for APE1 mitochondrial localization upon menadione-induced oxidative stress, being essential for interaction within the outer membrane Tom20cd translocase. They pointed that oxidative stress promotes APE1 mitochondrial accumulation, which is enhanced when testing K24/25/27/31>A APE1 mutants. They also discussed that their model could not explain the impact of an altered cellular redox state on modulating the amount of mtAPE1. The result reported in this study demonstrate that Mia40 specifically interacts with APE1, which provides further support to the hypothesis of the involvement of a redox-mediating folding process of APE1 affecting its subcellular distribution. Thus our data further extend the model proposed by Li et al. (2010) toward a plausible explanation causally linking the cellular redox state and APE1 mitochondrial trafficking. Further work is needed to characterize in detail the specific mechanisms regulating APE1 mitochondrial translocation and involving Mia40. However, this is the first time to our knowledge that such a model has been proposed. Notably, we recently demonstrated that some of the aforementioned K residues important for APE1 mitochondrial accumulation undergo acetylation in vivo (Fantini et al., 2010). It is plausible that both acetylation of critical K residues and C65 redox status may regulate the conditional translocation of APE1 within mitochondria, controlling protein accumulation there. This model would provide a molecular basis, based on quick and reversible posttranslational modifications (PTMs), for the rapid mechanism required for regulating APE1 subcellular trafficking on cellular vital sites, such as in mitochondria, where oxidative stress mainly exerts its damaging effects.
Our inability to fully rescue cells from APE1 silencing-induced apoptosis with the APE1C65S mutant strongly supports the redox activity of APE1 as an ideal candidate target for pharmacological intervention and cancer therapeutics. Data obtained with the E3330 inhibitor (Figure 7) strongly confirm the reliability of this approach and provide molecular evidence for a direct effect of this compound in vivo. Recent data clearly demonstrate that bifunctional therapy, including DNA-damaging agents (such as treatment with alkylating agents or ionizing radiation or radiomimetics treatment) and inhibition of DNA-repair enzymes activities, may represent a promising direction in cancer treatment (Helleday et al., 2008; Tell and Wilson, 2010). Therefore knowledge of the molecular mechanisms regulating key enzymes in DNA repair has a fundamental importance for developing new anticancer strategies.
In addition to basic research aimed at the understanding the molecular mechanisms responsible for the fine tuning of the different APE1 functions, development of small-molecule inhibitors of each function will be necessary to ultimately define which protein activity is preponderant in normal and cancer cells. Recent findings on redox inhibition of APE1 have potential clinical significance (Kelley and Fishel, 2008; Kelley et al., 2011; Reed et al., 2009; Fishel et al., 2010); they suggested that a redox inhibitor could be used as a single agent, in combination with current treatments, or as a potential antigrowth, cytostatic agent. Obviously, for proper modulation of different interconnected functions of multifunctional proteins like APE1, fine-tuned regulation of these activities is required. Better understanding of the processes controlling APE1 subcellular distribution, of its PTMs, and of its different interacting partners recruited as a function of the cellular status is required to fully address this complex issue, thus translating this unique protein from bench top to bedside.
In summary, our data represent the first attempt, to our knowledge, at gaining a global view of the molecular role of APE1 within the cell. By combining large-scale studies with classic biochemical approaches, we demonstrated that the wild-type APE1 protein has structural components functioning in several distinct cellular roles, providing a molecular basis for explaining its multifunctional biological activity. The data set coming from this work also offers several scientific leads for future studies aimed at shedding light on the subtle mechanisms of this control.
MATERIALS AND METHODS
Cell lines and materials
Both HeLa and SF767 (provided by the Brain Tumor Research Center, University of California at San Francisco, San Francisco, CA) cell lines were grown in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Euroclone, Milan, Italy), 100 U/ml penicillin, and 10 μg/ml streptomycin sulfate. All chemical reagents were supplied by Sigma (Milan, Italy) unless otherwise specified.
Inducible siRNA of APE1 and generation of APE1 knock-in cell lines
For inducible silencing of endogenous APE1 and reconstitution with different APE1 proteins, HeLa cell clones were developed as described (Vascotto et al., 2009a, 2009b) and as reported in the Supplementary Information. For inducible siRNA experiments, doxycycline (Sigma) was added to the cell culture medium at the final concentration of 1 μg/ml, and cells were grown for 10 d. All biological data were reproduced in at least two different cell clones for each model.
Preparation of cell extracts and anti-FLAG coimmunoprecipitation
Preparation of total cell lysates and coimmunoprecipitation analyses were performed as described (Vascotto et al., 2009a, 2009b) and as reported in the Supplemental Information.
Cell growth assays and apoptosis studies
All cell growth assays and apoptosis studies were performed as described (Vascotto et al., 2009a, 2009b) and detailed in the Supplemental Information.
Endonuclease assays
The determination of AP endonuclease activity of APE1 was performed as reported (Vascotto et al., 2009a) and detailed in the Supplemental Information.
Limited proteolysis studies
Approximately 4 μg of APE1WT and APE1C65S immunopurified proteins from reconstituted HeLa cells were subjected to limited proteolysis for the reported time by using trypsin (6 fmol) and chymotrypsin (24 fmol) in 0.1 M ammonium bicarbonate buffer (10 μl final volume). Proteolysis was blocked by adding 4 μl of Laemmli sample buffer 4× and incubating samples at 95°C for 5 min. Then samples were loaded onto 12% SDS–PAGE and transferred to nitrocellulose membranes for further Western blotting analysis with specific anti-APE1 antibody (Vascotto et al., 2009a).
Immunofluorescence analysis and proximity ligation assay
Immunofluorescence analyses on HeLa or SF767 cells were performed as described (Vascotto et al., 2009a) and detailed in the Supplemental Information. To study the interaction of APE1 with Mia40 in vivo, we used in situ PLA technology (Olink Bioscience, Uppsala, Sweden). After incubation with APE1 antibody (as previously described), cells were incubated with anti-Mia40 polyclonal antibody (Abcam, Cambridge, MA) diluted 1:50 in blocking solution, at 4°C, overnight. PLA was performed following manufacturer's instruction. Briefly, species-specific secondary antibodies, conjugated with oligonucleotides (PLA probes), were used, followed by hybridization with two oligonucleotides and ligation joining of the two hybridized oligonucleotides to a closed circle. During the amplification step, the oligonucleotide arm of one of the PLA probes acted as a primer for a rolling-circle amplification (RCA) reaction using the ligated circle as a template and generated a concatameric (repeated sequence) product extending from the oligonucleotide arm of PLA probe. In the detection step, fluorescently (Cy3) labeled oligonucleotides hybridized to the RCA product. The signal was easily visible as a distinct fluorescent red dot, which was detected through confocal microscopy. Technical controls, represented by the omission of Mia40 primary antibody, resulted in the complete lost of PLA signal.
Cells were visualized through a Leica TCS SP laser-scanning confocal microscope (Leica Microsystems, Wetzlar, Germany) and a 63× oil fluorescence objective (Figure 6B and Supplemental Figure S1) or an Olympus FluoView FV1000 MPE confocal/multiphoton system (Olympus, Center Valley, PA) and a Plan Apo N 60×/1.42 oil objective (Olympus; Figure 7B and Supplemental Figure S2). Determination of APE1 fluorescence and statistical analysis were performed with Metamorph analysis software, version 7.7.1 (Molecular Devices, Downingtown, PA).
Western blot analysis and antibodies
Reported amounts of nuclear and cytoplasmic extracts obtained from HeLa and SF767 cells were electrophoresed onto a 12% SDS–PAGE. Proteins were then transferred to nitrocellulose membranes (Schleicher & Schuell, Keene, NH). Membranes were saturated by incubation with 5% nonfat dry milk in PBS/0.1% Tween 20 for 1 h at 25°C and then incubated with the specific primary antibody [anti-APE1 monoclonal, 3 h at 25°C, dilution 1:2000; anti-FLAG (Sigma) monoclonal, 3 h at 25°C, dilution 1:1000; anti-PRDX6 (Abcam) polyclonal, overnight at 4°C, dilution 1:1000; anti-PDIA3 polyclonal (Abcam), overnight at 4°C, dilution 1:1000; anti-CytC monoclonal (Santa Cruz Biotechnology, Santa Cruz, CA), overnight at 4°C, dilution 1:500; anti–Pol γ polyclonal (Abcam) overnight at 4°C, dilution 1:500; anti-CII monoclonal (Invitrogen) overnight at 4°C, dilution 1:2000]. After three washes with PBS/0.1% Tween 20, membranes were incubated for 2 h at 25°C with a secondary antibody coupled to peroxidase (Sigma). Then membranes were washed three times with phosphate-buffered saline/0.1% Tween-20, and the blots were developed using the enhanced chemiluminescence procedure (GE Healthcare, Piscataway, NJ). Normalization was performed with the polyclonal anti-actin or anti–β-tubulin antibodies (Sigma). Blots were quantified by using a Gel Doc 2000 videodensitometer (Bio-Rad, Hercules, CA).
Interactomic analysis
Identification of differential protein-interacting partners of APE1C65S was performed as described (Vascotto et al., 2009a, 2009b); details are reported in the Supplemental Information.
Microarray analysis
Gene expression studies were performed as described previously (Vascotto et al., 2009b) and detailed in the Supplemental Information.
Transient transfection studies
HeLa cells (0.3 × 106 cells/60-mm dish) were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol with 200 ng of the pcDNA5.1-APE1-c-FLAG plasmid, encoding for the C-terminal FLAG-tagged WT APE1 or the 31-34A mutant; the pcDNA5.1-empty vector was used as control.
Preparation of crude mitochondria from cells and of mitochondrial subfractions from bovine heart
Mitochondria isolation was performed as previously reported (Abou-Khalil et al., 1985). Subconfluent HeLa cells were collected and suspended at a concentration of 2 × 107 cells/ml in mitochondrial isolation buffer (250 mM sucrose, 1 mg/ml BSA, 2 mM EDTA, pH 7.4) with 1:100 protease inhibitor cocktail (Sigma). Cell suspensions were sonicated on ice under mild controlled conditions to disrupt selectively plasma membranes (5-s sonication repeated three times, with 30-s intervals). The homogenates (∼95% of disrupted cells) were then fractionated by differential centrifugation at 800 × g and 16,000 × g at 4°C, to obtain the enriched mitochondria pellet. Purity of mitochondria was judged on the basis of low cross-contamination (6–16%) of marker proteins H3acK18, Hsp70, and calreticulin during semiquantitative analysis of Western blots for the nucleus, cytoplasm, and endoplasmic reticulum, respectively.
Bovine heart mitochondria (BHM) were isolated as previously described (Ferguson et al. 1977). Subfractions were prepared by incubation of BHM on ice with digitonin (0.18 mg/mg protein), followed by centrifugation at 35,000 × g for 15 min at 4°C to separate mitoplasts (matrix plus inner membrane) from intermembrane space soluble, protein-rich fractions (Bisetto et al., 2008). Membrane integrity of mitoplasts was judged on the basis of the malate dehydrogenase (matrix enzyme) activity, which was 83% of that in BHM (Gallet et al., 1999).
JC-1 assay for mitochondrial membrane potential
The mitochondrial membrane potential, Δψm, was assessed by using the JC-1 Mitochondria Staining Kit (Sigma-Aldrich, St. Louis, MO) for flow cytometry, following the manufacturer's recommendations. Cells were treated with various doses of different drugs for the indicated times, then incubated in medium containing JC-1 probe at desired working concentration for 30 min at 37°C. After double washing with JC-1 binding buffer, Δψm was measured immediately by using flow cytometry. JC-1 is a mitochondrial-selective sensor and aggregates in normal and highly polarized mitochondria, resulting in a red emission of 590 nm after excitation at 490 nm. On depolarization of the mitochondrial membrane, JC-1 forms green fluorescent monomers and determines an increase in the green/red (FL1/FL2) fluorescence intensity ratio. Thus the loss of JC-1 aggregates directly correlates with changes of Δψm. Valinomycin, a mitochondrial membrane potential disrupter, was used as a positive control.
Cytochrome c release assay
To measure the cytochrome c release from mitochondria to cytoplasm following oxidative stress, mitochondria-free cytosolic fractions were subjected to Western blot analysis by using an antibody against cytochrome c. Mitochondria-free cytosolic fraction was prepared by using a protocol described before (Li et al., 2008). Briefly, 5 × 107 cells were washed once with 10 ml of grinding buffer and collected by centrifugation at 800 × g for 5 min at 4°C. The pellet was resuspended in 200 μl of grinding buffer and sonicated on ice at 30 W for 15 s. Approximately 50–70% of cell lysis was ensured under microscope. The supernatant was immediately centrifuged at 8500 × g for 20 min at 4°C to pellet the mitochondria. The obtained supernatant contained the cytosolic fraction.
Statistical analysis
Statistical analysis was performed by using the Excel (Microsoft, Redmond, WA) data analysis program for Student's t test analysis. p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Malgorzata M. Kamocka from the Indiana Center for Biological Microscopy (Indiana University–Purdue University Indianapolis, Indianapolis, IN) and Milena Romanello for their helpful suggestions during immunofluorescence analysis, Marta Deganuto and Laura Cesaratto for cell cycle and apoptosis studies, and Damiano Fantini for help in immunoprecipitation analysis. We also thank Bruce Demple for critically reading the manuscript, making helpful suggestions, and sharing some unpublished data from his lab.
This work was supported by grants from Italian Ministry of Education, University and Research (FIRB_RBRN07BMCT and PRIN2008_CCPKRP_003) and the Italian Association for Cancer Research (IG10269) to G.T.; from the National Natural Science Foundation of China to M.L. (NSFC_30900553); from the Italian Ministry of Education, University and Research (PRIN2008_CCPKRP_002 and FIRB2008_RBNE08YFN3_003) to A.S.; and from the National Institute of Health and National Cancer Institute (R01 CA114571, CA94025, CA106298, and CA121168) and the Riley Children's Foundation to M.R.K. This work was also supported by a Union for International Cancer Control Yamagiwa-Yoshida Memorial International Cancer Study Grant to G.T.
Abbreviations used:
- APE1/Ref-1
apurinic/apyrimidinic endonuclease/redox effector factor-1
- BER
base excision repair
- E3330
(E)-3-(2-[5,6-dimethoxy-3-methyl-1,4-benzoquinonyl])-2-nonyl propenoic acid
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-05-0391) on August 24, 2011.
Gene expression data have been loaded into an ArrayExpress database and are accessible for review purposes at the following link: www.ebi.ac.uk/aerep/login. Username: Reviewer_E-MEXP-2941; password: 1287419348983.
The authors declare that they have no conflict of interest.
REFERENCES
- Abou-Khalil S, Abou-Khalil WH, Planas L, Tapiero H, Lampidis TJ. Interaction of rhodamine 123 with mitochondria isolated from drug-sensitive and -resistant Friend leukemia cells. Biochem Biophys Res Commun. 1985;127:1039–1044. doi: 10.1016/s0006-291x(85)80049-8. [DOI] [PubMed] [Google Scholar]
- Bapat A, Fishel ML, Kelley MR. Going ape as an approach to cancer therapeutics. Antioxid Redox Signal. 2009;11:651–668. doi: 10.1089/ars.2008.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapat A, Glass LS, Luo M, Fishel ML, Long EC, Georgiadis MM, Kelley MR. Novel small-molecule inhibitor of apurinic/apyrimidinic endonuclease 1 blocks proliferation and reduces viability of glioblastoma cells. J Pharmacol Exp Ther. 2010;334:988–998. doi: 10.1124/jpet.110.169128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes T, Kim WC, Mantha AK, Kim SE, Izumi T, Mitra S, Lee CH. Identification of apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res. 2009;37:3946–58. doi: 10.1093/nar/gkp275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakat KK, Izumi T, Yang SH, Hazra TK, Mitra S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J. 2003;1:6299–6309. doi: 10.1093/emboj/cdg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisetto E, Picotti P, Giorgio V, Alverdi V, Mavelli I, Lippe G. Functional and stoichiometric analysis of subunit e in bovine heart mitochondrial F(0)F(1)ATP synthase. J Bioenerg Biomembr. 2008;40:257–267. doi: 10.1007/s10863-008-9183-5. [DOI] [PubMed] [Google Scholar]
- Busso CS, Iwakuma T, Izumi T. Ubiquitination of mammalian AP endonuclease (APE1) regulated by the p53-MDM2 signaling pathway. Oncogene. 2009;28:1616–1625. doi: 10.1038/onc.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelli E, Hazra T, Hill JW, Slupphaug G, Bogliolo M, Frosina G. Rates of base excision repair are not solely dependent on levels of initiating enzymes. Carcinogenesis. 2001;22:387–393. doi: 10.1093/carcin/22.3.387. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay R, Bhattacharyya A, Crowe S. Dual regulation by apurinic/apyrimidinic endonuclease-1 inhibits gastric epithelial cell apoptosis during Helicobacter pylori infection. Cancer Res. 2010;70:2799–2808. doi: 10.1158/0008-5472.CAN-09-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay R, Wiederhold L, Szczesny B, Boldogh I, Hazra TK, Izumi T, Mitra S. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res. 2006;34:2067–2076. doi: 10.1093/nar/gkl177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutat Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- Fan Z, Beresford PJ, Zhang D, Xu Z, Novina CD, Yoshida A, Pommier Y, Lieberman J. Cleaving the oxidative repair protein APE1 enhances cell death mediated by granzyme A. Nat Immunol. 2003;4:145–153. doi: 10.1038/ni885. [DOI] [PubMed] [Google Scholar]
- Fantini D, et al. Critical lysine residues within the overlooked N-terminal domain of human APE1 regulate its biological functions. Nucleic Acids Res. 2010;38:8239–8256. doi: 10.1093/nar/gkq691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SJ, Harris DA, Radda GK. The adenosine triphosphatase-inhibitor content of bovine heart submitochondrial particles. Influence of the inhibitor on adenosine triphosphate-dependent reactions. Biochem J. 1977;162:351. doi: 10.1042/bj1620351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel ML, Colvin ES, Luo M, Kelley MR, Robertson KA. Inhibition of the redox function of APE1/Ref-1 in myeloid leukemia cell lines results in a hypersensitive response to retinoic acid-induced differentiation and apoptosis. Exp Hematol. 2010;38:1178–1188. doi: 10.1016/j.exphem.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol Aspects Med. 2007;28:375–395. doi: 10.1016/j.mam.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Fung H, Demple B. A vital role for APE1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol Cell. 2005;17:463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- Gaiddon C, Moorthy NC, Prives C. Ref-1 regulates the transactivation and pro-apoptotic functions of p53 in vivo. EMBO J. 1999;18:5609–5621. doi: 10.1093/emboj/18.20.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallet PF, Zachowski A, Julien R, Fellmann P, Devaux PF, Maftah A. Transbilayer movement and distribution of spin-labelled phospholipids in the inner mitochondrial membrane. Biochim Biophys Acta. 1999;1418:61–70. doi: 10.1016/s0005-2736(99)00022-x. [DOI] [PubMed] [Google Scholar]
- Georgiadis M, Luo M, Gaur R, Delaplane S, Li X, Kelley MR. Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease. Mutat Res. 2008;643:54–63. doi: 10.1016/j.mrfmmm.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman MA, Morera S, Rothwell DG, de La Fortelle E, Mol CD, Tainer JA, Hickson ID, Freemont PS. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J. 1997;16:6548–6558. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Zhang J, Ellis LM, Semenza GL, Evans DB, Watowich SS, Gallick GE. HIF-1alpha, STAT3, CBP/p300 and Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene. 2005;24:3110–3120. doi: 10.1038/sj.onc.1208513. [DOI] [PubMed] [Google Scholar]
- Grillo C, D'Ambrosio C, Scaloni A, Maceroni M, Merluzzi S, Turano C, Altieri F. Cooperative activity of Ref-1/APE and ERp57 in reductive activation of transcription factors. Free Radic Biol Med. 2006;41:1113–1123. doi: 10.1016/j.freeradbiomed.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Hanson S, Kim E, Deppert W. Redox factor 1 (Ref-1) enhances specific DNA binding of p53 by promoting p53 tetramerization. Oncogene. 2005;24:1641–1647. doi: 10.1038/sj.onc.1208351. [DOI] [PubMed] [Google Scholar]
- He T, Weintraub NL, Goswami PC, Chatterjee P, Flaherty DM, Domann FE, Oberley LW. Redox factor-1 contributes to the regulation of progression from G0/G1 to S by PDGF in vascular smooth muscle cells. Am J Physiol Heart Circ Physiol. 2003;285:H804–H812. doi: 10.1152/ajpheart.01080.2002. [DOI] [PubMed] [Google Scholar]
- Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Riemer J. The intermembrane space of mitochondria. Antioxid Redox Signal. 2010a;13:1341–1358. doi: 10.1089/ars.2009.3063. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Riemer J. Oxidation and reduction of cysteines in the intermembrane space of mitochondria: multiple facets of redox control. Antioxid Redox Signal. 2010b;13:1323–1326. doi: 10.1089/ars.2010.3270. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Kreklau EL, Erickson LC, Berger MS, Pieper RO. Delayed repletion of O6-methylguanine-DNA methyltransferase resulting in failure to protect the human glioblastoma cell line SF767 from temozolomide-induced cytotoxicity. J Neurosurg. 2003;98:591–598. doi: 10.3171/jns.2003.98.3.0591. [DOI] [PubMed] [Google Scholar]
- Hirota K, Matsui M, Iwata Z, Nishiyama A, Mori K, Yodoi J. AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc Natl Acad Sci USA. 1997;94:3633–3638. doi: 10.1073/pnas.94.8.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RP, Adamson ED. Characterization of the DNA-binding properties of the early growth response-1(Egr-1) transcription factor: evidence for modulation by a redox mechanism. DNA Cell Biol. 1993;12:265–273. doi: 10.1089/dna.1993.12.265. [DOI] [PubMed] [Google Scholar]
- Izumi T, Brown DB, Naidu CV, Bhakat KK, Macinnes MA, Saito H, Chen DJ, Mitra S. Two essential but distinct functions of the mammalian abasic endonuclease. Proc Natl Acad Sci USA. 2005;102:5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley MR, Fishel ML. DNA repair proteins as molecular targets for cancer therapeutics. Anticancer Agents Med Chem. 2008;8:417–425. doi: 10.2174/187152008784220294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley MR, Luo M, Reed A, Su D, Delaplane S, Borch RF, Nyland RL, Gross ML, Georgiadis MM. Functional analysis of novel analogues of E3330 that block the redox signaling activity of the multifunctional AP endonuclease/redox signaling enzyme APE1/Ref-1. Antioxid Redox Signal. 2011;14:1387–1401. doi: 10.1089/ars.2010.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuninger DT, Izumi T, Papaconstantinou J, Mitra S. Human AP-endonuclease 1 and hnRNP-L interact with a nCaRE-like repressor element in the AP-endonuclease 1 promoter. Nucleic Acids Res. 2002;30:823–829. doi: 10.1093/nar/30.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, et al. Identification and characterization of mitochondrial targeting sequence of human apurinic/apyrimidinic endonuclease 1. J Biol Chem. 2010;285:14871–1481. doi: 10.1074/jbc.M109.069591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MX, Wang D, Zhong ZY, Xiang DB, Li ZP, Xie JY, Yang ZZ, Jin F, Qing Y. Targeting truncated APE1 in mitochondria enhances cell survival after oxidative stress. Free Radic Biol Med. 2008;45:592–601. doi: 10.1016/j.freeradbiomed.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Ludwig DL, MacInnes MA, Takiguchi Y, Purtymun PE, Henrie M, Flannery M, Meneses J, Pedersen RA, Chen DJ. A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity. Mutat Res. 1998;409:17–29. doi: 10.1016/s0921-8777(98)00039-1. [DOI] [PubMed] [Google Scholar]
- Luo M, et al. Role of the multifunctional DNA repair and redox signaling protein Ape1/Ref-1 in cancer and endothelial cells: small molecule inhibition of Ape1's redox function. Antioxid Redox Signal. 2008;10:1853–1867. doi: 10.1089/ars.2008.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, He H, Kelley MR, Georgiadis MM. Redox regulation of DNA repair: implications for human health and cancer therapeutic development. Antioxid Redox Signal. 2010;12:1247–1269. doi: 10.1089/ars.2009.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantha AK, Oezguen N, Bhakat KK, Izumi T, Braun W, Mitra S. Unusual role of a cysteine residue in substrate binding and activity of human AP-endonuclease 1. J Mol Biol. 2008;379:28–37. doi: 10.1016/j.jmb.2008.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y, Bennett RA, Demple B. Dynamics of the interaction of human apurinic endonuclease (Ape1) with its substrate and product. J Biol Chem. 1998;273:30352–30359. doi: 10.1074/jbc.273.46.30352. [DOI] [PubMed] [Google Scholar]
- McNeill DR, Wilson DM., 3rd A dominant-negative form of the major human abasic endonuclease enhances cellular sensitivity to laboratory and clinical DNA-damaging agents. Mol Cancer Res. 2007;5:61–70. doi: 10.1158/1541-7786.MCR-06-0329. [DOI] [PubMed] [Google Scholar]
- Mitra S, Izumi T, Boldogh I, Bhakat KK, Chattopadhyay R, Szczesny B. Intracellular trafficking and regulation of mammalian AP-endonuclease 1 (APE1), an essential DNA repair protein. DNA Repair. 2007;6:461–469. doi: 10.1016/j.dnarep.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Mol CD, Izumi T, Mitra S, Tainer J A. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- Nishi T, et al. Spatial redox regulation of a critical cysteine residue of NF-kappa B in vivo. J Biol Chem. 2002;277:44548–56. doi: 10.1074/jbc.M202970200. [DOI] [PubMed] [Google Scholar]
- Ordway JM, Eberhart D, Curran T. Cysteine 64 of Ref-1 is not essential for redox regulation of AP-1 DNA binding. Mol Cell Biol. 2003;23:4257–4266. doi: 10.1128/MCB.23.12.4257-4266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines A, Bivi N, Romanello M, Damante G, Kelley MR, Adamson ED, D'Andrea P, Quadrifoglio F, Moro L, Tell G. Cross-regulation between Egr-1 and APE/Ref-1 during early response to oxidative stress in the human osteoblastic HOBIT cell line: evidence for an autoregulatory loop. Free Radic Res. 2005a;39:269–281. doi: 10.1080/10715760400028423. [DOI] [PubMed] [Google Scholar]
- Pines A, Perrone L, Bivi N, Romanello M, Damante G, Gulisano M, Kelley MR, Quadrifoglio F, Tell G. Activation of APE1/Ref-1 is dependent on reactive oxygen species generated after purinergic receptor stimulation by ATP. Nucleic Acids Res. 2005b;33:4379–4394. doi: 10.1093/nar/gki751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J, Liu GH, Huang B, Chen C. Nitric oxide controls nuclear export of APE1/Ref-1 through S-nitrosation of cysteines 93 and 310. Nucleic Acids Res. 2007;35:2522–2532. doi: 10.1093/nar/gkl1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed AM, Fishel ML, Kelley MR. Small-molecule inhibitors of proteins involved in base excision repair potentiate the anti-tumorigenic effect of existing chemotherapeutics and irradiation. Future Oncol. 2009;5:713–726. doi: 10.2217/FON.09.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemer J, Fischer M, Herrmann JM. Oxidation-driven protein import into mitochondria: Insights and blind spots. Biochim Biophys Acta. 2011;1808:981–989. doi: 10.1016/j.bbamem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Seemann S, Hainaut P. Roles of thioredoxin reductase 1 and APE/Ref-1 in the control of basal p53 stability and activity. Oncogene. 2005;24:3853–3863. doi: 10.1038/sj.onc.1208549. [DOI] [PubMed] [Google Scholar]
- Seo YR, Kelley MR, Smith ML. Selenomethionine regulation of p53 by a ref1-dependent redox mechanism. Proc Natl Acad Sci USA. 2002;99:14548–14553. doi: 10.1073/pnas.212319799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N, et al. High-performance affinity beads for identifying drug receptors. Nat Biotechnol. 2000;18:877–881. doi: 10.1038/78496. [DOI] [PubMed] [Google Scholar]
- Stuart JA, Mayard S, Hashiguchi K, Souza-Pinto NC, Bohr VA. Localization of mitochondrial DNA base excision repair to an inner membrane-associated particulate fraction. Nucleic Acids Res. 2005;33:3722–3732. doi: 10.1093/nar/gki683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D, Delaplane S, Luo M, Rempel DL, Vu B, Kelley MR, Gross ML, Georgiadis MM. Interactions of apurinic/apyrimidinic endonuclease with a redox inhibitor: evidence for an alternate conformation of the enzyme. Biochemistry. 2011;50:82–92. doi: 10.1021/bi101248s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J Biol Chem. 2008;283:26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny B, Tann AW, Mitra S. Age- and tissue-specific changes in mitochondrial and nuclear DNA base excision repair activity in mice: susceptibility of skeletal muscles to oxidative injury. Mech Ageing Dev. 2010;131:330–337. doi: 10.1016/j.mad.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tell G, Crivellato E, Pines A, Paron I, Pucillo C, Manzini G, Bandiera A, Kelley MR, Di Loreto C, Damante G. Mitochondrial localization of APE/Ref-1 in thyroid cells. Mutat Res. 2001;485:143–152. doi: 10.1016/s0921-8777(00)00068-9. [DOI] [PubMed] [Google Scholar]
- Tell G, Damante G, Caldwell D, Kelley MR. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid Redox Signal. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- Tell G, Fantini D, Quadrifoglio F. Understanding different functions of mammalian AP endonuclease (APE1) as a promising tool for cancer treatment. Cell Mol Life Sci. 2010a;67:3589–608. doi: 10.1007/s00018-010-0486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tell G, Wilson DM., 3rd Targeting DNA repair proteins for cancer treatment. Cell Mol Life Sci. 2010;67:3569–3572. doi: 10.1007/s00018-010-0484-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tell G, Wilson DM, 3rd, Lee CH. Intrusion of a DNA repair protein in the RNome world: is this the beginning of a new era. Mol Cell Biol. 2010b;30:366–371. doi: 10.1128/MCB.01174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M, Masutani H, Arai RJ, Yamauchi A, Hirota K, Sakai T, Inamoto T, Yamaoka Y, Yodoi J, Nikaido T. Thioredoxin-dependent redox regulation of p53-mediated p21 activation. J Biol Chem. 1999;274:35809–35815. doi: 10.1074/jbc.274.50.35809. [DOI] [PubMed] [Google Scholar]
- Vascotto C, et al. APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Mol Cell Biol. 2009a;29:1834–1854. doi: 10.1128/MCB.01337-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vascotto C, et al. Genome-wide analysis and proteomic studies reveal APE1/Ref-1 multifunctional role in mammalian cells. Proteomics. 2009b;9:1058–1074. doi: 10.1002/pmic.200800638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LJ, Robson CN, Black E, Gillespie D, Hickson ID. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol Cell Biol. 1993;13:5370–5376. doi: 10.1128/mcb.13.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SJ, Botero A, Hirota K, Bradbury CM, Markovina S, Laszlo A, Spitz DR, Goswami PC, Yodoi J, Gius D. Thioredoxin nuclear translocation and interaction with redox factor-1 activates the activator protein-1 transcription factor in response to ionizing radiation. Cancer Res. 2000;60:6688–6695. [PubMed] [Google Scholar]
- Weibrecht I, Leuchowius KJ, Clausson CM, Conze T, Jarvius M, Howell WM, Kamali-Moghaddam M, Söderberg O. Proximity ligation assays: a recent addition to the proteomics toolbox. Expert Rev Proteomics. 2010;7:401–409. doi: 10.1586/epr.10.10. [DOI] [PubMed] [Google Scholar]
- Wilson DM, 3rd, Simeonov A. Small molecule inhibitors of DNA repair nuclease activities of APE1. Cell Mol Life Sci. 2010;67:3589–608. doi: 10.1007/s00018-010-0488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthoudakis S, Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthoudakis S, Miao GG, Curran T. The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains. Proc Natl Acad Sci USA. 1994;91:23–27. doi: 10.1073/pnas.91.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamori T, DeRicco J, Naqvi A, Hoffman TA, Mattagajasingh I, Kasuno K, Jung SB, Kim CS, Irani K. SIRT1 deacetylates APE1 and regulates cellular base excision repair. Nucleic Acids Res. 2010;38:832–845. doi: 10.1093/nar/gkp1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziel KA, Campbell CC, Wilson GL, Gillespie MN. Ref-1/Ape is critical for formation of the hypoxia-inducible transcriptional complex on the hypoxic response element of the rat pulmonary artery endothelial cell VEGF gene. FASEB J. 2004;18:986–988. doi: 10.1096/fj.03-1160fje. [DOI] [PubMed] [Google Scholar]
- Zou GM, Luo MH, Reed A, Kelley MR, Yoder MC. APE1 regulates hematopoietic differentiation of embryonic stem cells through its redox functional domain. Blood. 2007;109:1917–1922. doi: 10.1182/blood-2006-08-044172. [DOI] [PubMed] [Google Scholar]
- Zou GM, Maitra A. Small-molecule inhibitor of the AP endonuclease 1/REF-1 E3330 inhibits pancreatic cancer cell growth and migration. Mol Cancer Ther. 2008;7:2012–2021. doi: 10.1158/1535-7163.MCT-08-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.