Abstract
Originally identified as cytokine inhibitors, protein inhibitors of activated STAT (PIAS) are shown to regulate activities of a plethora of proteins and influence diverse processes such as immune response, cancer formation, and cell cycle progression. However, the roles of PIAS during vertebrate embryogenesis are less understood. In this study, we report isolation and initial characterization of all four PIAS genes from Xenopus laevis. The Xenopus PIAS genes are expressed throughout early development and have overlapping and distinct expression patterns, with for example high levels of PIAS2 in the notochord and strong expression of PIAS4 in the neural and neural crest derivatives. Overexpression of PIAS disrupts mesoderm induction and impairs body axis formation. PIAS proteins have differential ability to regulate signals from the growth factors activin, bone morphogenetic protein 4 (BMP4), and Wnt8. Our data suggest that Xenopus PIAS play important roles in mesodermal induction and patterning during early frog development.
Keywords: PIAS, mesoderm, neural, activin/nodal, BMP, Wnt, Xenopus laevis
INTRODUCTION
As their names imply, the protein inhibitors of activated STAT (PIAS) proteins were initially identified via their ability to bind directly to phosphorylated STATs (Signal Transducer and Activator of Transcription) and inhibit DNA-recognition and binding by these transcription factors (Chung et al., 1997; Liu et al., 1998). Four PIAS proteins have since been characterized: PIAS1, PIAS2 (originally named PIASx, with two splicing variants α and β), PIAS3, and PIAS4 (a.k.a. PIASy). Though they share a high degree of sequence homology with each other, they display specificity in regulating particular STAT proteins (Shuai, 2006). For example, PIAS1, but not the other PIAS members, blocks DNA binding by STAT1; and PIAS2, but not PIAS3, inhibits STAT4 (Liu et al., 1998; Arora et al., 2003). Subsequently, PIASs are shown to interact with a variety of proteins, most of which are transcription factors, such as NF-κB, nuclear hormone receptors, p53 family members, Msx1, GATA1/2, and LEF1. PIAS proteins can act both as activators and repressors of transcription, depending on particular genes they regulate. They do so by modulation of DNA binding activities, recruitment of other co-regulators (e.g. histone deacetylases HDACs), post-translational protein modification (see below), or alteration of subnuclear localization of the transcription factors (Jackson, 2001; Schmidt and Muller, 2003; Shuai, 2006; Palvimo, 2007; Rytinki et al., 2009).
One prominent activity of the PIAS proteins is that they facilitate modification of their target proteins by adding small ubiquitin-related modifier (SUMO) molecule. Similar to ubiquitination, sumoylation normally involves E1 activating enzyme, E2 conjugating enzyme and E3 protein ligases (Geiss-Friedlander and Melchior, 2007). PIAS proteins are shown to act as SUMO E3 ligases. All PIAS members contain in their central sequences a highly conserved cysteine-rich SP-RING (Siz/PIAS RING) motif. This motif is related to the classical RING-type zinc finger that is present in many ubiquitin E3 ligases and may mediate protein-protein interaction. Indeed, the integrity of the SP-RING motif, defined as CX12–42CX1–3HX2–3C/HX7–51CX2C, seems to be important for mammalian PIAS to facilitate covalent attachment of SUMO to their substrates. However, unlike in the ubiquitin pathway, PIAS E3 ligases do not seem to be absolutely required for sumoylation in certain cases, especially when the E2-conjugation enzyme Ubc9 is over-expressed (Schmidt and Muller, 2003). In addition to the SP-RING motif, PIAS proteins also possess several other conserved domains (Fig.1A; reviewed in (Palvimo, 2007; Rytinki et al., 2009). The N-terminal SAP (Scaffold Attachment Factor/Acinus/PIAS) domain contains 35 amino acid that includes a nuclear receptor box “LXXLL” sequence shown to play a part in the assembly of complexes between nuclear receptors and their co-activators. The SAP domain is found in a number of proteins that associate with chromatin and may thus confer PIAS proteins the ability to bind chromatin and regulate chromatin architecture. Following the SAP domain is a PINIT motif, which in the case of PIAS3 plays a role in nuclear retention of the protein (Duval et al., 2003). C-terminal to the SP-RING is a SUMO interacting motif, SIM. SIM normally consists of an essential hydrophobic core, a serine triplet and a C-terminal cluster of acidic amino acids and serves as a SUMO recognition motif to bind directly to SUMO (Palvimo, 2007; Rytinki et al., 2009). Sumoylation of a target protein by PIAS thus further enhances the interaction of PIAS with its target.
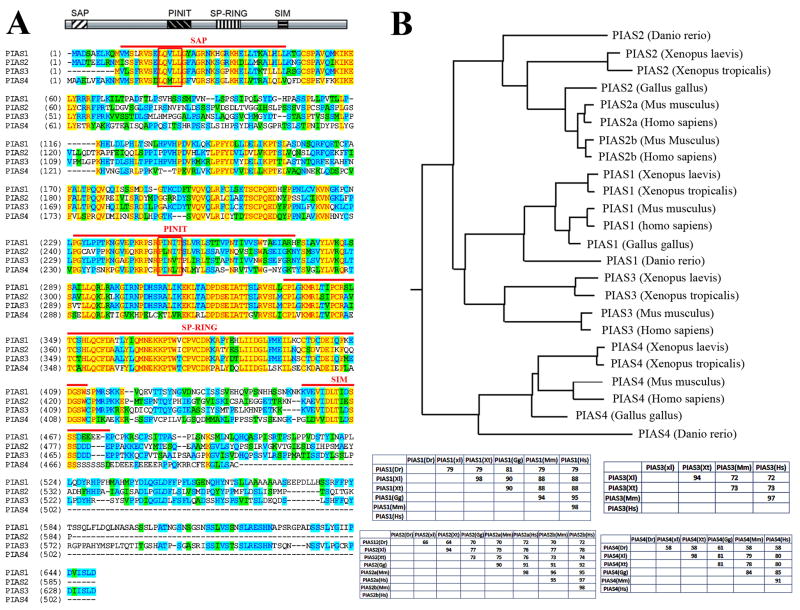
Figure 1. PIAS proteins are conserved across the vertebrate species.
A) Alignment of the four Xenopus laevis PIAS proteins reveals that they share a high degree of homology within the known functional domains, including the SAP, PINIT, SP-RING and SIM domains (marked by the red lines above the sequences), but are more divergent in the carboxyl termini. The LXXLL motif in the SAP domain and the PINIT motif in the PINIT domain are marked by the closed boxes. B) Alignment of vertebrate PIAS proteins show that PIAS are highly conserved in vertebrate. The phylogenic tree constructed using the neighbor-joining algorithm is shown here, and the identity tables for each vertebrate PIAS protein are shown at the bottom.
Despite the demonstration that PIAS proteins regulate many transcriptional events in mammalian cells, in vivo functions of PIAS members are less understood. Deletion of Pias1, Pias2 and Pias4 genes individually does not impair early development of mouse embryos, though Pias1−/−Pias4−/− double knockout mice die before embryonic day 11.5. No detailed analyses on tissue defects are reported for these mice (Tahk et al., 2007), and no overexpression study has been performed to examine its effects on early mouse development. In the frog Xenopus laevis, PIAS4 (PIASy) has been shown to inhibit Smad2 activities, though sumoylation may not be absolutely required for this function (Daniels et al., 2004). A single PIAS ortholog is also identified in both the fruit fly Drosophila and the worm C. elegans. Deletion of dPIAS in Drosophila results in severe abnormalities in structure, condensation and segregation of chromosomes and causes embryonic lethality, while silencing the PIAS ortholog in C. elegans by RNAi leads to defects in pharyngeal development and retardation of embryonic growth (Rytinki et al., 2009). These data suggest that PIAS regulate crucial processes during animal development.
To further comprehend the activities of PIAS genes during early vertebrate embryogenesis, we isolated the full length clones for all four PIAS genes from Xenopus laevis and examined their expression. We showed that all PIAS genes were expressed from maternal stage onward during early frog embryogenesis, and their distribution in mesodermal and neural derivatives implied that they play a role in the genesis of these tissues. Functional studies indicated that PIAS proteins modulated growth factor signals to influence induction and patterning of the mesoderm during early frog development.
RESULTS AND DISCUSSION
The PIAS genes are conserved among vertebrate species
Database searches revealed that similar to other vertebrate animals, Xenopus laevis has four PIAS genes. Using RT-PCR, we isolated full length clones for all four Xenopus PIAS genes. Sequence alignment of the PIAS proteins showed that they share between 46% to 60% identity with each other. The most conserved region in the proteins is the SP-RING domain, which is required for E3 sumoylation activity of PIAS. In addition, the other functional domains identified through studies in cultured mammalian cells, including SAP, PINIT and SIM domains, also show high degree of homology. In contrast, the carboxyl termini of the proteins encode more divergent sequences (Fig. 1A). Alignment of PIAS proteins from Xenopus laevis with those from other vertebrate animals revealed that PIAS proteins are highly conserved across vertebrate species (Fig. 1B). Xenopus PIAS proteins are more than 70% identical to their mammalian homologues. Among the four PIAS proteins, mammalian PIAS2 has two isoforms, PIAS2a and PIAS2b (also named as PIASx-α and PIASx-β), which are generated by differential splicing of the messenger RNA. However, only one PIAS2 cDNA is found in the database for zebrafish (Danio rerio), Xenopus (including both laevis and tropicalis) and chick (Gallus gallus), and the protein encoded by the gene shows equal distance to the two isoforms of mammalian PIAS2 (Fig. 1B). Phylogenetic tree constructed using neighbor-joining algorithm suggests that PIAS1 and PIAS3 are more closely related than the other two PIAS proteins (Fig. 1B).
Expression of the PIAS genes during early Xenopus development
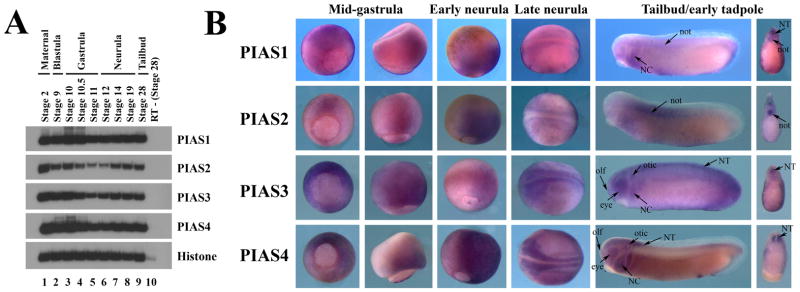
To understand the function of the PIAS genes during early Xenopus development, we first examined their expression patterns. Analyses of temporal expression by RT-PCR demonstrated that all four Xenopus PIAS genes were expressed maternally. Their expressions persisted until at least tailbud stages. Though the levels of PIAS1 and PIAS4 seemed to remain constant throughout early frog embryogenesis, PIAS2 and PIAS3 levels were lower at mid- to late gastrula stages before they were up-regulated again during neurulation (Fig. 2A).
Figure 2. Expression of the PIAS genes during early Xenopus development.
A) Temporal expression analyses by RT-PCR showed that all four PIAS genes were maternally expressed, and their expression persisted until at least tailbud stages. B) Spatial distribution of PIAS transcripts assayed by in situ hybridization revealed that the PIAS genes had overlapping and distinct expression patterns. The embryos were shown at mid-gastrula stages in columns 1 (vegetal view) and 2 (lateral view), late gastrula/early neurula stages in column 3 (lateral view), neurula stages in column 4 (dorsal view), and tailbud/early tadpole stages in column 5 (lateral view). The bisected embryos from tailbud stage embryos were shown in column 6. The labeled structures were: NT, neural tube; not, notochord; NC, neural crest; otic, otic placode; olf, olfactory placode; and eye.
To inspect spatial distribution of PIAS transcripts, we next performed in situ hybridization. At cleavage, blastula and early gastrula stages, all PIAS genes were widely expressed in both animal and marginal zone regions (not shown). As gastrulation progressed, PIAS RNAs were more restricted to the dorsal tissues. At the mid-gastrula stages (e.g. stage 11), though PIAS transcripts were still distributed all around the blastopore, ventral expression was reduced while dorsal expression was retained (Fig. 2B). By the end of gastrulation and at the early neurula stages (e.g. stage 12), PIAS RNAs were seen in the dorsal mesoderm and the neural tissues. At the neurula stages, all the PIAS genes were expressed in the neural and neural crest cells. At the tailbud and tadpole stages, PIAS transcripts were detected in overlapping and distinct regions. PIAS1 was seen in the neural tube (NT), the notochord (not), and the migrating neural crest (NC) cells, while PIAS2 was expressed strongly in the notochord as well as above the dorsal midline of the neural tube that was indicative of the delineating neural crest. Both PIAS3 and PIAS4 transcripts were highly enriched in the neural and neural crest derivatives, including the brain, the spinal cord, eyes, olfactory (olf) and otic placodes, and migrating cranial neural crest cells (Fig. 2B). The spatial patterns of PIAS expression indicate that they may have redundant activities early in development when they are co-expressed, but they also have specific functions in tissues where their expressions diverge, such as in the notochord.
Disruption of early Xenopus embryogenesis by ectopic expression of the PIAS genes
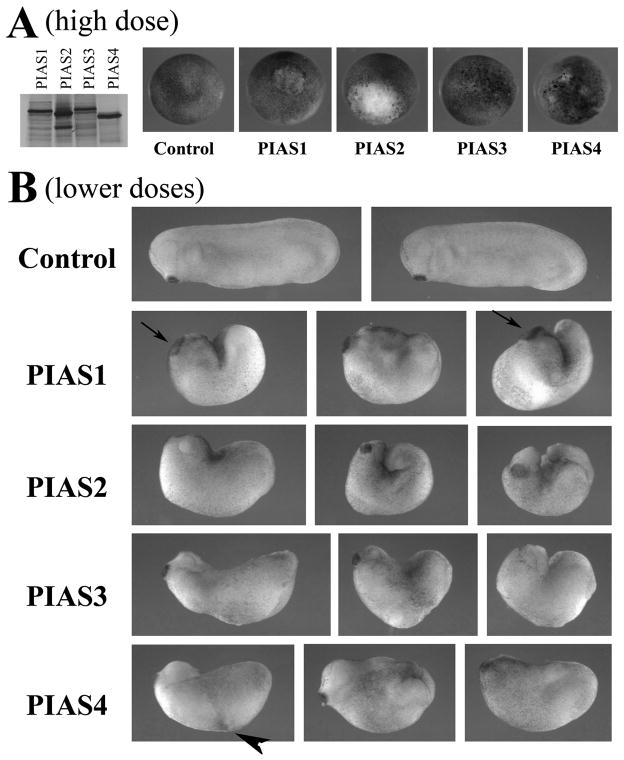
To investigate the roles of the PIAS genes in early Xenopus development, we synthesized PIAS RNAs and examined their protein production by in vitro translation. As shown in Fig. 3A, PIAS proteins were translated efficiently, though the levels of PIAS1 and PIAS3 were slightly lower. When injected into early frog embryos at high doses (e.g. 2ng), PIAS1, 2 and 4 induced lesions in early gastrula embryos, so that blastomeres displayed high tendency to dissociate and form white patches (Fig. 3A). These embryos normally did not survive beyond neurula to tailbud stages. Embryos injected with PIAS3 were more tolerant and showed only minor alteration in blastomere morphology at the same dose (Fig.3A); increasing the amount of PIAS3 RNA (e.g. 4ng) caused embryonic lesions similar to those induced by the other PIAS (not shown). When the doses of the RNAs were adjusted to allow embryo survival, severe defects were observed in developing tailbud to tadpole embryos. The anterior-posterior body axis was shortened in all injected embryos, and many failed to close the blastopore completely, leaving the endoderm and mesoderm exposed at the back. This phenotype reflects gastrulation defects and is often associated with failed convergent extension movements. It is interesting to note that one of the PIAS substrates, STAT3, is involved in gastrulation movements in zebrafish (Yamashita et al., 2002). Most embryos also displayed reduced head structures with malformation of the eyes. Interestingly, despite the reduction of the head, PIAS1 often induced expansion of the cement gland, a tissue located at the anterior end of the embryo that marks the position of the future mouth (arrows). Moreover, overexpression of PIAS4 sometimes led to formation of pigmented aggregate at the flank (arrowhead), with occasional induction of a secondary axis (Fig. 3B and data not shown). The data indicate that though the four PIAS genes have overlapping activities in reducing embryonic body length and disrupting the gastrulation process, they also have unique functions in other developmental processes.
Figure 3. Ectopic expression of PIAS genes in early frog embryos disrupts Xenopus development.
A) In vitro translation showed that all PIAS proteins were synthesized efficiently from their cognate RNAs. When RNAs were injected at high doses (2ng), PIAS1, 2 and 4 induced severe lesions in embryos at early gastrula stages and embryonic lethality before tailbud stages. PIAS3 was less effective in inducing the lesions. B) When the doses of the RNAs were adjusted to allow embryo survival (shown here were embryos injected with RNAs of 1ng PIAS1, 0.5ng PIAS2, 2ng PIAS3 and 1ng PIAS4), the resulting tailbud embryos showed failure in blastopore closure, shortened body axis, and defective head formation. The embryos with ectopic PIAS1 often displayed enlarged cement gland (arrows), and the embryos with elevated PIAS4 sometimes formed pigmented aggregates at the flank (arrowhead).
Overexpression of the PIAS genes impairs mesodermal induction
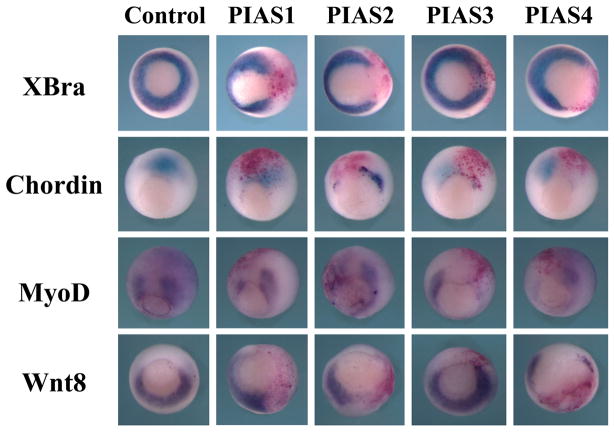
The gastrulation defects induced by the elevated levels of PIAS imply that either mesodermal induction is impaired or mesodermal cell movements are interrupted even though mesodermal cell fates are specified correctly. To distinguish these two possibilities, we examined the expression of several mesodermal markers at the gastrula stages by in situ hybridization. In this experiment, PIAS RNAs were coinjected with a lineage tracer, the RNA encoding nuclear β-galactosidase, into one blastomere of two-cell stage embryos. The embryos were collected at the gastrula stages and stained with the Red-Gal substrate to label the injected region. Expression of the pan-mesodermal marker Brachyury (XBra, (Smith et al., 1991), the dorsal mesodermal marker Chordin (Sasai et al., 1994), the dorsal-lateral marker MyoD (Hopwood et al., 1989) and the ventral lateral marker Wnt8 (Christian et al., 1991; Smith and Harland, 1991) was assayed by in situ hybridization. As shown in Fig. 4, all PIAS genes inhibited XBra, Chordin and MyoD expression, though PIAS3 was less efficient in blocking XBra. PIAS1, 2 and 4 also interfered with transcription of Wnt8, but PIAS3 did not reduce the Wnt8 level. The results demonstrate that ectopic expression of PIAS genes disrupts early Xenopus embryogenesis by inhibition of mesodermal marker induction, which then affected gastrulation secondarily.
Figure 4. Overexpression of PIAS disrupts early mesodermal formation.
In situ hybridization was performed to assess mesodermal formation in embryos injected with PIAS RNAs. While all four PIAS genes inhibited expression of Chordin, XBra and MyoD, they displayed different efficiency in doing so, with PIAS3 having weak XBra inhibitory activities. Unlike PIAS1, 2 and 4, PIAS3 did not block expression of Wnt8 efficiently. The doses of the RNAs used in this experiment were: 1ng PIAS1, 0.5ng PIAS2, 2ng PIAS3, and 1ng PIAS4.
PIAS genes differentially modulate signals from different growth factors
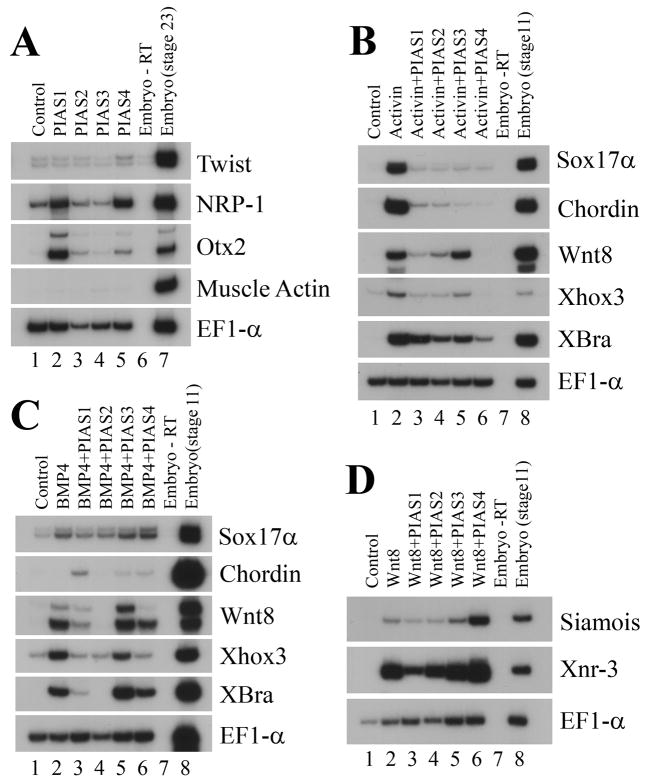
Induction and patterning of mesodermal tissues in early frog embryos depend on signals from several growth factor families. The signal that plays the most prominent role in inducing mesoderm is that of activin/nodal. In addition, bone morphogenetic protein (BMP) and Wnt signals have been shown to pattern the mesoderm to form the ventral or the dorsal types (Heasman, 2006). To address whether PIAS modulate any of these signals during development, we performed the animal cap assays. PIAS RNAs were coinjected with the signaling molecules encoding activin, BMP4 or Wnt8 into the animal region of two-cell stage embryos. Ectodermal explants (animal caps) were excised from the injected embryos at the late blastula stages and incubated to gastrula or neurula stages before they were collected for RT-PCR analyses of marker expression. When expressed alone, PIAS genes did not induce mesodermal markers. Instead, PIAS1 and PIAS4 directly induced the neural markers NRP-1 and Otx2 (Fig. 5A). Co-expression of PIAS with activin suppressed mesodermal and endodermal induction by this growth factor. The endodermal gene Sox17α and an array of the mesodermal markers, including chordin, Wnt8, Xhox3 and Xbra, were down-regulated (Fig. 5B). Interestingly, though PIAS3 reduced the expression of other markers, it was inefficient in blocking Wnt8, a result consistent with our in situ hybridization data. This implied that PIAS family members may interact with and modify distinct subsets of downstream targets to influence activin signal. Co-expression of PIAS with BMP4 also showed different effects of PIAS genes on marker induction by this growth factor (Fig. 5C). While none of the PIAS genes had significant effect on induction of the endoderm marker Sox17α by BMP4, the induction of the pan-mesodermal gene Xbra and the ventral and ventral-lateral markers Xhox3 and Wnt8 was reduced by PIAS1, PIAS2 and PIAS4. PIAS3 was weak in blocking BMP signaling, resulting in minor or no changes in marker induction. Among the four PIAS members, PIAS1 alone consistently switched on chordin expression in this assay (Fig. 5C). Unlike activin or BMP4, Wnt8 was not efficiently inhibited by the PIAS genes. PIAS1 reduced the induction of the direct targets of Wnt signaling Xnr3 and Siamois, but the other PIAS members were generally inefficient. In fact, PIAS4 enhanced, rather than suppressed, the expression of Xnr3 and Siamois (Fig. 5D). Taken together, our data reveal that different members of the PIAS family have differential ability to regulate distinct growth factor signals.
Figure 5. PIAS genes differentially regulate signals from various growth factors.
A) PIAS induced the neural markers with different efficiency, but none induced the expression of the mesodermal marker muscle actin. B) PIAS inhibited mesodermal induction by activin. C) PIAS blocked mesoderm induction by BMP4 with different effectiveness. D) PIAS1 reduced marker induction by Wnt8, but the other PIAS genes were ineffective; and PIAS4 enhanced, rather than inhibited, induction of the direct targets of Wnt signaling. The doses of the RNAs used in the experiments were: 1ng PIAS1, 0.5ng PIAS2, 2ng PIAS3, 1ng PIAS4; 2pg activin, 20pg BMP4, and 100pg Wnt8.
Conclusion
In this study, we isolated all four PIAS genes from Xenopus laevis and showed their temporal and spatial expression patterns. The wide distribution of PIAS transcripts during early stages of development indicated that they might play important roles in regulation of Xenopus embryogenesis. Consistent with this idea, our functional studies demonstrated that PIAS genes differentially regulated signals from several growth factors and interfered with early embryonic development when overexpressed. Since PIAS proteins are conserved in several identified functional domains, the differences in their activities likely lie within the less conserved carboxyl termini. Future studies will be conducted to pinpoint the regions that are responsible for specific activities of each PIAS member. In addition, loss-of-function approaches should be used to dissect the endogenous function of PIAS in different developmental processes, and the requirement for protein sumoylation or protein-protein interaction in PIAS-regulated development will be investigated.
EXPERIMENTAL PROCEDURES
Cloning of Xenopus PIAS genes
RT-PCR-based approach was used to clone the full length PIAS genes from cDNA obtained at the gastrula stages. The N- and the C-terminal primers used were: PIAS1-N(RI): 5′-GGAATTCACCATGGCGGACAGTGCGGAACTGAA-3′, PIAS1-C(XhoI): 5′-CCGCTCGAGTCAGTCCAAGGAGATAACATCTGG-3′; PIAS2-N(RI): 5′-GGAATTCACCATGGCGGACACCGAAGAGCTGCG-3′, PIAS2-C(XhoI): 5′-CCGCTCGA GTCAGGG TTTGCCAGTTATTTGG-3′; PIAS3-N(RI): 5′-GGAATTCACCATGGTTCTGAG CTTTAGA GTGTC-3′, PIAS3-C(XhoI): 5′-CCGCTCGAGTCAGTCCAAGGATATGATG TCTG-3′; PIAS4-N(RI): 5′-GGAATTCACCATGGCGGCGGAGTTAGTGGAGG-3′, and PIAS4-C(XhoI): 5′-CCGCTCGAGTCAACAAGCTGATAAGAGTCCTTTC-3′. The PCR products were cloned between the EcoRI and XhoI sites of the pCS105 or the pBSKS vectors. For mRNA synthesis for injection, AscI-linearized pCS105-based constructs were transcribed with the SP6 RNA polymerase; for making antisense probes for in situ hybridization, EcoRI-linearized pBSKS-based vectors were transcribed with the T3 RNA polymerase using digoxygenin-UTP and other NTPs.
Embryonic manipulations
Embryos were obtained and maintained as described previously (Nie and Chang, 2007). The doses of RNAs used for injections are indicated in the figure legends. For phenotypic observation, the RNAs were injected into the two-cell stage embryos and incubated to the gastrula or tailbud stages. For marker examination by in situ hybridization, the RNAs were injected into one blastomere of two-cell stage embryos together with a lineage tracer that encoded nuclear β-galactosidase (0.2ng RNA was used). Embryos were collected at gastrula stages, stained with the Red-Gal substrate, and proceed with in situ hybridization. For animal cap assays, the RNAs were injected into the animal regions of two-cell stage embryos. Ectodermal explants from the injected embryos were excised at the blastula stage 9 and incubated to gastrula or neurula stages before RNA was extracted and RT-PCR was performed.
Acknowledgments
This study is supported by the NIH grant GM083029.
References
- Arora T, Liu B, He H, Kim J, Murphy TL, Murphy KM, Modlin RL, Shuai K. PIASx is a transcriptional co-repressor of signal transducer and activator of transcription 4. J Biol Chem. 2003;278:21327–21330. doi: 10.1074/jbc.C300119200. [DOI] [PubMed] [Google Scholar]
- Christian JL, McMahon JA, McMahon AP, Moon RT. Xwnt-8, a Xenopus Wnt-1/int-1-related gene responsive to mesoderm-inducing growth factors, may play a role in ventral mesodermal patterning during embryogenesis. Development. 1991;111:1045–1055. doi: 10.1242/dev.111.4.1045. [DOI] [PubMed] [Google Scholar]
- Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, Shuai K. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- Daniels M, Shimizu K, Zorn AM, Ohnuma S. Negative regulation of Smad2 by PIASy is required for proper Xenopus mesoderm formation. Development. 2004;131:5613–5626. doi: 10.1242/dev.01449. [DOI] [PubMed] [Google Scholar]
- Duval D, Duval G, Kedinger C, Poch O, Boeuf H. The ‘PINIT’ motif, of a newly identified conserved domain of the PIAS protein family, is essential for nuclear retention of PIAS3L. FEBS Lett. 2003;554:111–118. doi: 10.1016/s0014-5793(03)01116-5. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Heasman J. Patterning the early Xenopus embryo. Development. 2006;133:1205–1217. doi: 10.1242/dev.02304. [DOI] [PubMed] [Google Scholar]
- Hopwood ND, Pluck A, Gurdon JB. MyoD expression in the forming somites is an early response to mesoderm induction in Xenopus embryos. EMBO J. 1989;8:3409–3417. doi: 10.1002/j.1460-2075.1989.tb08505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PK. A new RING for SUMO: wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes Dev. 2001;15:3053–3058. doi: 10.1101/gad.955501. [DOI] [PubMed] [Google Scholar]
- Liu B, Liao J, Rao X, Kushner SA, Chung CD, Chang DD, Shuai K. Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci U S A. 1998;95:10626–10631. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie S, Chang C. Regulation of Xenopus gastrulation by ErbB signaling. Dev Biol. 2007;303:93–107. doi: 10.1016/j.ydbio.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palvimo JJ. PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochem Soc Trans. 2007;35:1405–1408. doi: 10.1042/BST0351405. [DOI] [PubMed] [Google Scholar]
- Rytinki MM, Kaikkonen S, Pehkonen P, Jaaskelainen T, Palvimo JJ. PIAS proteins: pleiotropic interactors associated with SUMO. Cell Mol Life Sci. 2009;66:3029–3041. doi: 10.1007/s00018-009-0061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont LK, De Robertis EM. Xenopus chordin: a novel dorsalizing factor activated by organizer-specific homeobox genes. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Muller S. PIAS/SUMO: new partners in transcriptional regulation. Cell Mol Life Sci. 2003;60:2561–2574. doi: 10.1007/s00018-003-3129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai K. Regulation of cytokine signaling pathways by PIAS proteins. Cell Res. 2006;16:196–202. doi: 10.1038/sj.cr.7310027. [DOI] [PubMed] [Google Scholar]
- Smith JC, Price BM, Green JB, Weigel D, Herrmann BG. Expression of a Xenopus homolog of Brachyury (T) is an immediate-early response to mesoderm induction. Cell. 1991;67:79–87. doi: 10.1016/0092-8674(91)90573-h. [DOI] [PubMed] [Google Scholar]
- Smith WC, Harland RM. Injected Xwnt-8 RNA acts early in Xenopus embryos to promote formation of a vegetal dorsalizing center. Cell. 1991;67:753–765. doi: 10.1016/0092-8674(91)90070-f. [DOI] [PubMed] [Google Scholar]
- Tahk S, Liu B, Chernishof V, Wong KA, Wu H, Shuai K. Control of specificity and magnitude of NF-kappa B and STAT1-mediated gene activation through PIASy and PIAS1 cooperation. Proc Natl Acad Sci U S A. 2007;104:11643–11648. doi: 10.1073/pnas.0701877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S, Miyagi C, Carmany-Rampey A, Shimizu T, Fujii R, Schier AF, Hirano T. Stat3 Controls Cell Movements during Zebrafish Gastrulation. Dev Cell. 2002;2:363–375. doi: 10.1016/s1534-5807(02)00126-0. [DOI] [PubMed] [Google Scholar]