Summary
The instruction of the immune system to be tolerant of self, thereby preventing autoimmunity, is facilitated by the education of T cells in a specialized organ, the thymus, where self-reactive cells are either eliminated or differentiated into tolerogenic Foxp3+ regulatory T(Treg) cells1. However, it is unknown whether T cells are also educated to be tolerant of foreign antigens, such as those from commensal bacteria, in order to prevent immunopathology such as inflammatory bowel disease2–4. Here, we show that encounter with commensal microbiota results in the peripheral generation of Treg cells, rather than pathogenic effectors. We observed that colonic Treg cells utilized T cell antigen receptors (TCRs)different from those used by Treg cells in other locations, implying an important role for local antigens in shaping the colonic Treg cell population. Many of the local antigens appeared to be derived from commensal bacteria based on the in vitro reactivity of common colon Treg TCRs. Interestingly, these TCRs did not facilitate thymic Treg cell development, implying that manycolonic Treg cells arise instead via antigen-driven peripheral Treg cell development. Further analysis of two of these TCRs by the creation of retroviral bone marrow chimeras and a TCR transgenic linerevealed that microbiota indigenous to our mouse colony was required for the generation of colonic Treg cells from otherwise naive T cells. If T cells expressing these TCRs fail to undergo Treg cell development and instead become effector cells, they have the potential to induce colitis, as evidenced by adoptive transfer studies. These results suggest that the efficient peripheral generation of antigen-specific populations of Treg cells in response to an individual’s microbiota provides important post-thymic education of the immune system to foreign antigens, thereby providing tolerance to commensal microbiota.
Although Treg cells are required for maintaining gut tolerance3, commensal bacteria are not necessary for colonic Treg cell generation5, 6 (Supplementary Fig. 1). Moreover, Treg cells from germ-free mice can protect against colitis7. On the other hand, extra-thymic generation of Treg cells to foreign antigens has been demonstrated using TCR transgenic models of oral tolerance8, 9. Peripheral Treg cell development is also increased in the gut9, 10, potentially related to the presence of specialized antigen presenting cells9, 11–13. Finally, Clostridiales species14, and B. fragilis6 via a protease-resistant capsular polysaccharide, can increase the frequency or function of colonic Treg cells, but may do so via innate immune receptors15. Thus, it remains unclear whether the protective colonic Treg cell population is generated against self-antigens or foreign antigens derived from the commensal bacteria found in each individual.
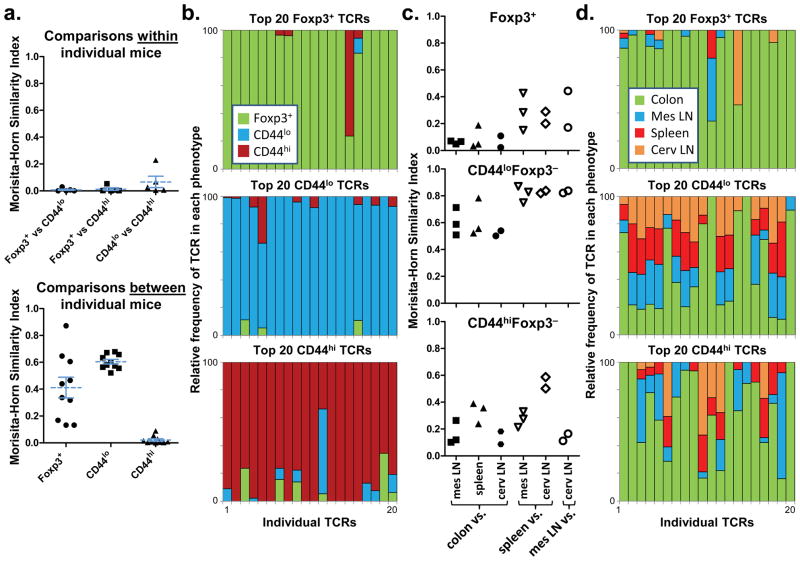
While TCR transgenic lines to antigens derived from commensal bacteria have been described16, the normal in vivo frequency of those TCRs in the Treg versus effector T cell subsets is unknown. To study the TCRs normally found in the colonic Treg cell population, we analyzed the colonic TCR repertoire. Due to the great diversity of the fully polyclonal TCR repertoire, we and others have utilized genetically engineered mice with limited polyclonal repertoires17–19. The analysis of TCRα chain repertoires of colonic lamina propria CD4+ T cells from mice expressing a fixed transgenic TCRβ chain revealed that Foxp3+ Treg cells utilize TCRs that are quite distinct from those of effector/memory (CD44hi) and naive (CD44lo) Foxp3− cells(Fig. 1a-b; Supplementary Fig. 2–3). This is illustrated using the Morisita-Horn similarity index (Fig. 1a), in which values from 0 to 1 represent low to high similarity between two data sets, as well as by the analysis of the relative frequencies of individual common TCRs in each T cell subset (Fig. 1b). Moreover, the analysis of TCRα chains of cells pooled from the secondary lymphoid organs and colons of additional mice showed that Treg TCR usage in the colon differed greatly from that in the other organs(Fig. 1c-d; Supplementary Fig. 3). Like the Treg cell population, the effector/memory T cell population expressed TCRs largely unique to the colon; however, these two subsets showed very little overlap. Thus, consistent with our previous observations in other peripheral locations20, these TCR repertoire data suggest that the colonic Treg cell population is strongly shaped by the local antigenic milieu.
Figure 1. The colonic Treg TCR repertoire is unique.
(a) TCR usage between colonic T cell subsets. A total of 2,892 TRAV14 TCRα sequences from colonic naive, memory, and Treg cells of five individual mice were compared (Supplementary Fig. 2). Each symbol represents a Morisita-Horn similarity comparison between two different T cell subsets within each mouse (top), or a comparison of the same T cell subset between different mice (bottom). Bars represent mean ± S.E.M. (b) Analysis of individual TCRs. The relative distribution within all T cell subsets is shown for the 20 most common individual TCRs in each colonic T cell subset. For example, a TCR with equal percentage in the Foxp3+ and CD44hi subset would be shown as a half-green/half-orange bar. This analysis uses the pooled data set, which includes sequences from individual mice as well as 9,680 sequences from Expt. 1–3, each consisting of cells from 3–5 mice (Supplementary Fig. 2). Note that 1 TCR is found in both Foxp3+ (#15) and CD44hi (#3) plots, and 1 in both CD44lo (#5) and CD44hi (#12) plots; all others appear in only one plot. (c) Anatomic distribution of colonic TCRs. Morisita-Horn indices comparing the colon data to other locations(closed symbols), or between each of the other locations (open symbols), are shown. Abbreviations: mes, mesenteric; cerv, cervical. (d) Analysis of individual TCRs. The 20 most prevalent colon TCRs for each subset in the pooled data set are shown, and their presence at other locations represented in a manner analogous to (b).
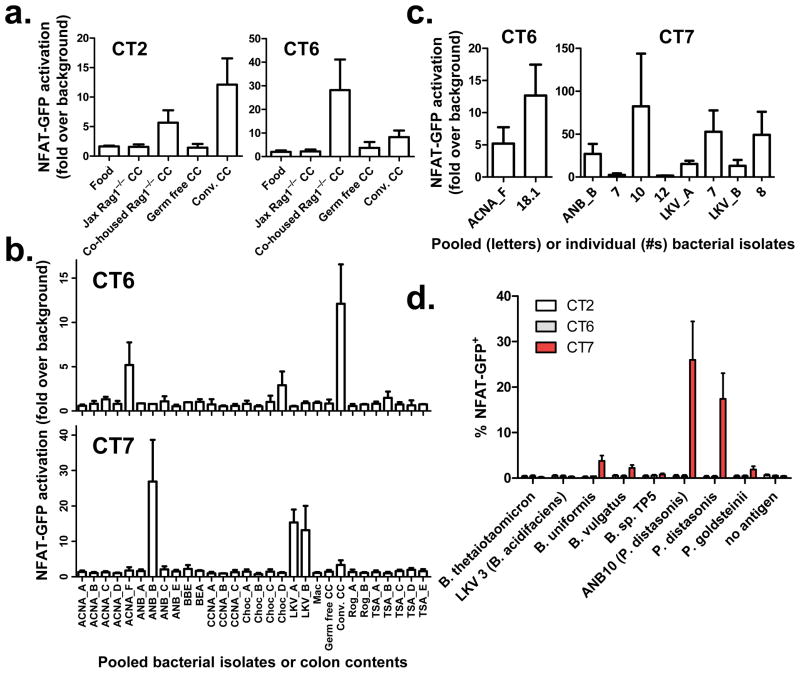
To assess whether the local antigens were bacterial in origin, we expressed colon Treg TCRs (Supplementary Fig. 3) in a hybridoma cell line that contains a GFP reporter for NFAT activation as a readout for TCR engagement21. We initially screened these hybridomas against autoclaved colonic contents(CC), and were surprised to find that many (5 of 8) showed some degree of reactivity to preparations from conventionally-housed (conv.), but not germ-free, mice (Fig. 2a, Supplementary Fig. 4a-b, Table 1). Importantly, colonic contents from Jackson Labs-sourced (Jax) Rag1−/− mice were not recognized by 4 of these colonic Treg TCRs unless they were first co-housed with mice from our colony. In contrast to the colonic Treg TCRs, none of the eight abundant colonic activated/memory (CD44hi) TCRs (Supplementary Fig. 5), nor four other TCRs tested (including B8 and TR520) showed any reactivity (not shown). Also, TCR recognition did not occur in the absence of dendritic cells, and was blocked by antibodies against MHC class II (not shown). Thus, these data suggest that the antigens responsible for TCR activation are derived from microbes that can be passed between co-housed mice.
Figure 2. In vitro reactivity of colonic Treg TCRs to colonic contents and bacterial isolates.
(a) Reactivity to colonic contents. Colonic Treg TCR -expressing NFAT-GFP hybridoma cells were cultured with Flt3L-induced dendritic cells in the presence of autoclaved food homogenate, or autoclaved colonic contents (CC) isolated from Rag1−/− mice from Jackson Labs (Jax Rag1−/−), Jax Rag1−/− mice co-housed with mice from our colony(Co-housed Rag1−/−), germ-free mice, and conventionally housed (Conv.) mice in our colony.(b) Reactivity to bacterial pools. Cultures of heat-killed commensal bacteria isolated from our colony (Supplementary Fig. 6) were pooled (denoted by culture conditions and a letter) and screened for their ability to stimulate colonic Treg TCR expressing hybridomas. For (a,b), see Supplementary Fig. 4 for additional TCRs. (c)Reactivity to individual isolates. Hybridomas showing reactivity against a pool of bacterial isolates were re-screened against the individual constituents (numbered). Data shown in (a-c) are the mean fold change in %GFP+ over the no antigen control ± S.E.M. from 2–4 experiments. (d) Specificity of colonic Treg TCRs. A panel of heat-killed Parabacteroides and Bacteroides spp. (Supplementary Fig. 7) was tested against CT2, CT6, and CT7-expressing hybridomas. Data shown are the mean ± S.E.M. from 3 experiments.
Table 1.
Summary of in vitro screening of colonic Treg TCRs
| Colon Treg TCR | Reactivity | ||
|---|---|---|---|
|
| |||
| Name and CDR3 a.a. seq. | Conv. CC | transferred by co-housing | Bacterial Isolate |
|
| |||
| CT1 AASWASGYNKLT | Yes | Yes | |
| CT2 AASAIWNTGYQNFY | Yes | Yes | |
| CT4 AASEYSALGRLH | |||
| CT6 AASGYSALGRLH | Yes | Yes | Clostridiales sp. ACNA18.1 |
| CT7 AASATGDNRIF | Parabacteroides distasonis | ||
| CT8 AASLTGGYKVV | |||
| CT9 AASADNRAGNKLT | Yes | Yes | |
| G57 AASELYQGGRALI | Yes | ||
We therefore attempted to identify bacteria recognized by these colonic Treg TCRs by screening small pools of 2–3 heat-killed bacteria isolated in pure culture from the colonic contents of mice in our colony (Fig. 2b, Supplementary Fig. 4c,6). Two colonic Treg TCRs (CT6 and CT7) reacted to one or more pools. Testing of individual isolates from these pools revealed that CT6 reacted with isolate ANCA18.1, identified by 16S ribosomal RNA gene sequencing as a previously uncharacterized Clostridiales species. Notably, all three isolates recognized by CT7, but none of 34 other sequenced isolates, were identified as Parabacteroides distasonis (Fig. 2c, Supplementary Fig. 6). To assess whether CT7 broadly recognize commensal species within the Bacteroideceae family, we screened it against an additional panel of closely related mouse-derived commensal Parabacteroides and Bacteroides species22. CT7, but not CT2 or CT6, recognized only a subset of these bacterial species, including a second isolate of P. distasonis (Fig. 2d). Importantly, isolates that did not stimulate CT7 were recognized by another TCR hybridoma, DP1 (Supplementary Fig. 7a), indicating that these preparations contained antigens capable of TCR stimulation. The almost mutually-exclusive specificity of CT7 and DP1 within the Bacteroideceae family makes it unlikely that these TCRs recognize host self-antigens that are differentially induced by these closely related bacteria22 (Supplementary Fig. 7b). The TCR-specific reactivity patterns further suggest that TCR activation is not due to non-specific stimulation by generic immunostimulatory bacterial components or superantigens. Rather, these TCRs likely recognize distinct bacterial protein antigens, as predigestion of heat-killed P. distasonis with proteinase K abrogated recognition by the CT7 hybridoma (Supplementary Fig. 7c), unlike what has been reported for Treg cell induction by protease-insensitive capsular polysaccharide from B. fragilis6. While proof of direct bacterial recognition will require the identification of specific epitopes, these data strongly argue for recognition of a bacterial-derived peptide by colonic Treg TCR CT7.
More than half of the tested colonic Treg TCRs recognized colonic contents and/or bacterial isolates (Table 1). However, this may underestimate the true frequency of colonic Treg TCRs which respond to bacterial antigens. A lack of reactivity in our screen cannot be interpreted to mean that the TCR does not recognize bacteria, as the antigens may be rare in unfractionated colonic contents, lost upon autoclaving, or derived from an organism that was not isolated in our screen. Thus, the specificity of common colonic Treg TCRs appears to be skewed towards recognition of bacterial antigens.
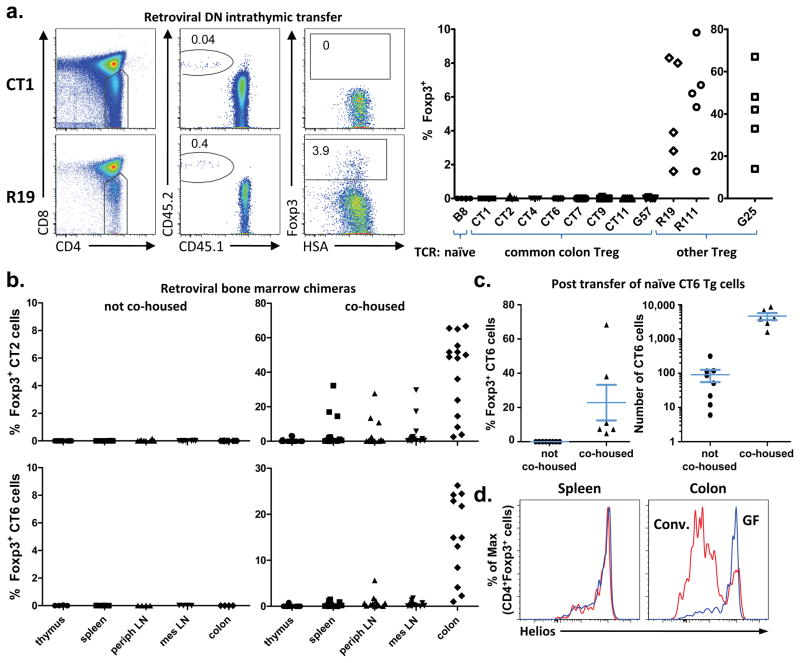
Although Treg cells may develop extra-thymically due to encounter with bacterial antigens, it is also possible that these Treg cells are selected via self-antigen recognition in the thymus, followed by expansion in the periphery due to cross-reactivity. To assess the ability of these colonic Treg TCRs to facilitate thymic Treg cell selection, we tracked the development of immature Foxp3gfp Rag1−/− thymocytes that were retrovirally transduced with a colonic Treg TCR. Remarkably, none of the colonic Treg TCRs generated an appreciable frequency of Foxp3+ thymocytes (Fig. 3a, Supplementary Fig. 8–9a), in contrast with Treg TCRs normally found at other peripheral locations(R19, G25, and R111; Supplementary Fig. 3). Note that G25 and R111 can be found at low frequency in the colonic Treg cell subset, suggesting that the colon does contain some thymically-derived Treg cells. The lack of Treg development in cells expressing colonic Treg TCRs cannot be attributed to an overwhelmed thymic niche23, 24 (Supplementary Fig. 9b). Thus, these data demonstrate that many common colonic Treg TCRs facilitate thymic Treg cell selection poorly, if at all, implying that these TCRs instead mediate peripheral Treg cell development.
Figure 3. Colonic Treg TCRs facilitate thymic Treg cell development poorly, if at all.
(a) Assessment of thymic Treg cell development from TCRαβ transduced Rag1−/− thymocytes. The gating strategy (left), and summary of 2–4 experiments per TCR (right) is shown. See Supplementary Fig. 3 for additional TCR information, and Supplementary Fig. 8–9 for plots and analysis of clonal frequencies. Comparison of colon versus other Treg TCRs revealed p-values < 0.01. (b) Mixed retroviral bone marrow chimeras. The percentage of Foxp3+ cells in the CT2 or CT6 expressing CD45.2+ CD4+ population is shown in hosts with (right) or without (left) co-housing with mice from our colony. See Supplementary Figs. 10–11 for additional analyses. (c) Peripheral conversion of CT6 TCR transgenic cells. Naive CD45.2 CT6 cells and CD45.1 CD4+ filler cells were adoptively transferred into Tcrb−/− hosts for 5 weeks. The percentage (left) of Foxp3+ CT6 cells (Vα2+Vβ6+CD45.2+CD45.1−CD4+) are shown. The number of CT6 cells (right) was determined by flow cytometry of the entire colonic lamina propria. Data are from 3 experiments; bars represent mean ± S.E.M. See Supplementary Fig. 13 for flow cytometric plots. (d) Helios expression in Treg cells. Representative intracellular Helios staining in CD4+ Foxp3+ cells from conventionally-housed and germ-free Foxp3gfp mice is shown, and summarized in Supplementary Fig. 14. For all plots in this Figure, each symbol represents data from an individual host.
The retroviral transduction of thymocytes does not result in the emergence of sufficient numbers of transduced T cells from the thymus to allow for their reliable detection in the periphery. We therefore retrovirally transduced self-renewing bone marrow progenitors and used them to create stable chimeras, selecting colonic Treg TCRs CT2 and CT6 based on their in vitro reactivity to colonic contents (Fig. 2a). In these chimeras, we were surprised to observe virtually no development of Foxp3 expression in CT2-or CT6-expressing cells (Fig. 3b, left; Supplementary Fig. 10). We reasoned that CT2 and CT6 may not recognize the microbiota in these commercially-sourced host mice(Fig. 2a), and performed experiments in which the chimeras were co-housed with mice from our colony. This resulted in the induction of CT2-or CT6-expressing Treg cells preferentially localized in the colon(Fig. 3b, right, Supplementary Fig. 11). In an observation paralleling that previously made in the thymus23, 24, there also appears to be a saturable, antigen-specific Treg cell niche in the periphery25 (Supplementary Fig. 11c).
Although these data strongly suggest that many colonic Treg cells are generated extra-thymically upon bacterial antigen encounter, it remained possible that a rare population of thymically-generated Treg cells below the limit of our detection expanded upon peripheral antigen encounter25. We therefore generated CT6 TCR transgenic mice (Supplementary Fig. 12), and adoptively transferred CD44lo Foxp3−CT6 T cells mixed with congenic polyclonal CD4+ “filler” T cells into T cell deficient Tcrb−/− hosts. Consistent with the bone marrow chimera data, the transferred CT6 T cells expanded and induced expression of Foxp3 only if the recipients were co-housed with mice from our colony (Fig. 3c, Supplementary Fig. 13). Together with the observed lack of Treg cell development by thymocytes expressing colonic Treg TCRs (Fig. 3a), these data suggest that a substantial proportion of the colonic Treg population arises extra-thymically from antigen-specific interactions with the colonic microbiota.
The notion that most colonic Treg cells are generated due to microbial interactions is at odds with the observation that germ-free mice have normal Treg cell frequencies6 (Supplementary Fig. 1). However, we and others14 have observed that most colonic Treg cells in conventionally housed, but not germ-free, mice are likely of peripheral origin, as these cells express low levels of the transcription factor Helios (Fig. 3d, Supplementary Fig. 14), a putative marker for thymically-derived Treg cells26. Thus, we hypothesize that germ-free conditions skew the colonic Treg TCR repertoire towards thymically-derived Treg TCRs.
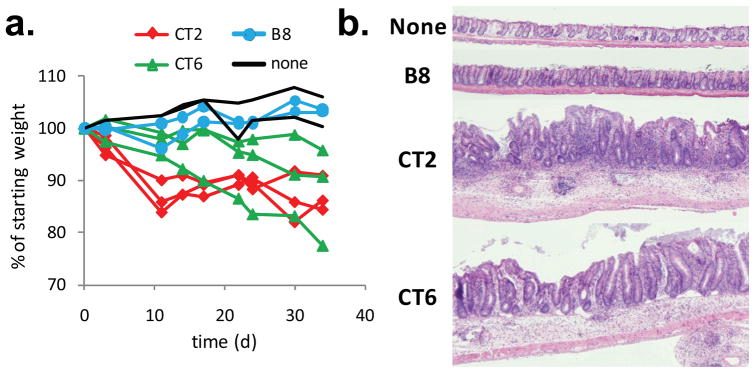
The efficient differentiation of naive T cells into Treg, rather than effector, cells may be important for generating colonic tolerance, as it has been observed that TCRs which facilitate thymic Treg cell development can be pathogenic when expressed on effector T cells17, 27. To address this possibility, we performed an initial analysis of colonic TCR repertoires in mice expressing the fixed TCRβ chain and undergoing spontaneous colitis due to genetic deficiencies in IL-2, IL-10, or TGFβ receptor signaling (Supplementary Fig. 2a). We observed that a number of colonic TCRs almost exclusively found in the Foxp3+ datasets in normal mice were found in the effector/memory data sets in the diseased animals (Supplementary Fig. 15a). While these genetic manipulations may affect Treg cell development or survival, the relatively high abundance of some of these TCRs in the CD44hi subset suggests that effector cells expressing these TCRs are expanding in the colitic environment. To test for pathogenic potential of colonic Treg TCRs, we retrovirally expressed CT2 and CT6 TCRs on peripheral, monospecific TCRαβ transgenic cells with known specificity for a foreign antigen (human CLIP peptide). Adoptive transfer of these cells, which were virtually all Foxp3−, into co-housed Rag1−/− hosts induced weight loss and colitis (Fig. 4, Supplementary Fig. 15b-c). In contrast, cells expressing only the transgenic TCR or a TCR from the naive T cell subset (B8) did not. The failure of these retrovirally-transduced T cells to upregulate Foxp3 and become regulatory in this situation is likely due to expansion in a lymphopenic environment, as well as in vitro T cell activation–a requirement for retroviral transduction. Thus, these data illustrate the potential pathologic consequences of T cell recognition of commensal bacterial antigens under conditions that disfavor Treg development.
Figure 4. Pathogenic potential of colonic Treg TCRs.
(a) Adoptive transfer of peripheral T cells transduced with CT2 or CT6 into co-housed Rag1−/− hosts. Non-transduced (none) or naive TCR (B8) transduced T cells were used as controls. Each line represents an individual recipient. One representative experiment is shown (summary in Supplementary Fig. 15b). (b) A representative H&E section of the descending colon is shown at 4x magnification 7–10 weeks after T cell transfer.
In summary, this analysis of common colonic Treg TCRs in a fixed TCRβ repertoire suggests a model (Supplementary Fig. 16) in which T cells expressing these TCRs exist as naive T cells in the absence of antigen (Fig. 3b left; Supplementary Figs. 10–11). Encounter with bacterial-derived foreign antigens in the colon appears to efficiently drive the generation of Foxp3+ Treg cells(Fig. 3b), as it typically does not result in substantial co-development of CD44hi cells of the same specificity(Fig. 1a-b). This diversion of naive T cells with bacterial TCR specificity into the Treg cell lineage may be crucial for preventing the generation of colitogenic effector cells(Fig. 4). Thus, these data support a model in which an individual’s T cell population is not only instructed by classic self/non-self discrimination mechanisms during thymic development, but is also educated in the periphery to accommodate the variety of non-self antigens derived from the commensal microbiota at mucosal sites.
Methods Summary
Mice
TCli TCRβ Foxp3gfp Tcra+/−; Foxp3IRES-GFP;IL-2−/−;IL-10−/−; and dnTGFβRII strains have been described (see Online Methods). C57BL/6 Rag1−/− and CD45.1 mice were obtained from Jackson Labs and NCI, respectively. Germ-free mice were generated in collaboration with Dr. Jeff Gordon (Wash. U.). CT6 transgenic mice were generated as described23.
TCR repertoire analysis
Analyses of TRAV14 (Vα2) TCR sequences from TCliβ transgenic mice were performed as described20. Lamina propria cell suspensions from the entire colon were prepared as described9 and CD4+ subsets sorted using a FACSAria(Becton Dickenson).
Hybridoma assays
Hybridoma cells expressing GFP under an NFAT promoter21 were retrovirally transduced with TCli TCRβ-IRES-mCD4 and an individual TCRα chain. Hybridomas were cultured with flt3-ligand elicited dendritic cells with the indicated antigen preparations and analyzed by flow cytometry after 1.5 days.
Antigen preparations
Whole colonic contents and food pellets were diluted with PBS, vortexed, homogenized, filtered, and autoclaved for 15 minutes. Colonic bacterial isolation was performed as described22 (see Supplementary Fig. 6).
In vivo developmental assays
Retroviral transduction and intrathymic transfer of Rag1−/− thymocytes was performed as described in online Methods. Analysis of CD4 SP thymocytes was done ~2.5 weeks later. Retroviral bone marrow chimeras were created as described23. Some recipients were co-housed with mice from our colony 2 weeks after bone marrow reconstitution, for a period of 1 week.
In vivo peripheral T cell assays
Retroviral transduction of peripheral TCli-αβ Foxp3gfp Rag1−/− T cells was performed as described27 and cells were intravenously transferred into co-housed Rag1−/− hosts. 5×104 sorted CD4+CD44lo Foxp3− cells from CT6 transgenic mice were co-transferred with 5×105 CD45.1+CD4+ “filler” cells into Tcrb−/− mice. Recovered cells were analyzed by flow cytometry 5 weeks later.
Statistics
The Wilcoxon rank sum test is used unless otherwise indicated.
Supplementary Material
Acknowledgments
We thank Ken Murphy, Takeshi Egawa (Wash. U.), Ye Zheng (Salk), and James Scott-Browne (LIAI), Jason Fontenot (Biogen Idec)and Scott Wetzel (U. of Montana)for discussion and reading of the manuscript; Andrew Kau and Jeff Gordon (Wash. U) for discussions and generation of germ-free animals; Nicole P. Malvin for assistance with bacteriology; and Jeremy Hunn for technical assistance. C.S.H. and co-workers are funded by the NIAID and the Burroughs-Wellcome Fund. S.M.B. was supported by NIH training grant 5T32AI0071632.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions S.K.L., S.R., K.N., and N.S. performed most of the experiments; S.M.B. designed and performed the bacteriology; C.W.L developed and assisted with the intrathymic transfer experiments; D.P. and T.S. were involved in study design; S.K.L. and C.S.H. designed the experiments and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
The authors declare no competing financial interests.
References
- 1.Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance. Immunity. 2009;30:616–25. doi: 10.1016/j.immuni.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belkaid Y, Tarbell K. Regulatory T cells in the control of host-microorganism interactions (*) Annu Rev Immunol. 2009;27:551–89. doi: 10.1146/annurev.immunol.021908.132723. [DOI] [PubMed] [Google Scholar]
- 3.Barnes MJ, Powrie F. Regulatory T cells reinforce intestinal homeostasis. Immunity. 2009;31:401–11. doi: 10.1016/j.immuni.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 5.Min B, et al. Gut flora antigens are not important in the maintenance of regulatory T cell heterogeneity and homeostasis. Eur J Immunol. 2007;37:1916–23. doi: 10.1002/eji.200737236. [DOI] [PubMed] [Google Scholar]
- 6.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–9. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh B, et al. Control of intestinal inflammation by regulatory T cells. Immunological Reviews. 2001;182:190–200. doi: 10.1034/j.1600-065x.2001.1820115.x. [DOI] [PubMed] [Google Scholar]
- 8.Curotto de Lafaille MA, et al. Adaptive Foxp3+ regulatory T cell-dependent and -independent control of allergic inflammation. Immunity. 2008;29:114–26. doi: 10.1016/j.immuni.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Sun CM, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–85. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng Y, et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010 doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coombes JL, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–64. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mucida D, et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–60. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 13.Benson MJ, Pino-Lagos K, Rosemblatt M, Noelle RJ. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J Exp Med. 2007;204:1765–74. doi: 10.1084/jem.20070719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–7. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci USA. 2009;106:19256–61. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsieh CS, et al. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 2004;21:267–277. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Pacholczyk R, Ignatowicz H, Kraj P, Ignatowicz L. Origin and T cell receptor diversity of Foxp3+CD4+CD25+ T cells. Immunity. 2006;25:249–59. doi: 10.1016/j.immuni.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 19.Wong J, et al. Adaptation of TCR repertoires to self-peptides in regulatory and nonregulatory CD4+ T cells. J Immunol. 2007;178:7032–41. doi: 10.4049/jimmunol.178.11.7032. [DOI] [PubMed] [Google Scholar]
- 20.Lathrop SK, Santacruz NA, Pham D, Luo J, Hsieh CS. Antigen-specific peripheral shaping of the natural regulatory T cell population. J Exp Med. 2008;205:3105–3117. doi: 10.1084/jem.20081359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ise W, et al. CTLA-4 suppresses the pathogenicity of self antigen-specific T cells by cell-intrinsic and cell-extrinsic mechanisms. Nat Immunol. 2010;11:129–35. doi: 10.1038/ni.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bloom SM, et al. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell host & microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bautista JL, et al. Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat Immunol. 2009;10:610–7. doi: 10.1038/ni.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung MW, Shen S, Lafaille JJ. TCR-dependent differentiation of thymic Foxp3+ cells is limited to small clonal sizes. J Exp Med. 2009;206:2121–30. doi: 10.1084/jem.20091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishio J, Feuerer M, Wong J, Mathis D, Benoist C. Anti-CD3 therapy permits regulatory T cells to surmount T cell receptor-specified peripheral niche constraints. J Exp Med. 2010;207:1879–89. doi: 10.1084/jem.20100205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thornton AM, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184:3433–41. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol. 2006;7:401–10. doi: 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.