Abstract
Gβ5 is a divergent member of the signal-transducing G protein β subunit family encoded by GNB5 and expressed principally in brain and neuronal tissue. Among heterotrimeric Gβ isoforms, Gβ5 is unique in its ability to heterodimerize with members of the R7 subfamily of the regulator of G protein signaling (RGS) proteins that contain G protein-γ like domains. Previous studies employing Gnb5 knockout (KO) mice have shown that Gβ5 is an essential stabilizer of such RGS proteins and regulates the deactivation of retinal phototransduction and the proper functioning of retinal bipolar cells. However, little is known of the function of Gβ5 in the brain outside the visual system. We show here that mice lacking Gβ5 have a markedly abnormal neurologic phenotype that includes impaired development, tiptoe-walking, motor learning and coordination deficiencies, and hyperactivity. We further show that Gβ5-deficient mice have abnormalities of neuronal development in cerebellum and hippocampus. We find that the expression of both mRNA and protein from multiple neuronal genes is dysregulated in Gnb5 KO mice. Taken together with previous observations from Gnb5 KO mice, our findings suggest a model in which Gβ5 regulates dendritic arborization and/or synapse formation during development, in part by effects on gene expression.
Keywords: guanine nucleotide-binding regulatory protein, regulator of G protein signaling, R7BP, Purkinje cell, dendrite, dentate gyrus, limbic cortex
Introduction
Heterotrimeric guanine nucleotide-binding regulatory proteins (G proteins), composed of Gα, Gβ, and Gγ subunits, relay signals from activated G protein-coupled cell surface receptors to various signaling pathways (Cabrera-Vera et al. 2003, Gutkind 2000). The mammalian genome contains genes encoding a dozen Gγ subunits and five Gβ isoforms suggesting a large potential combinatorial assortment of distinct Gβγ signaling complexes. While the Gβ1–4 isoforms are highly homologous (80–90%) and widely expressed (Gautam et al. 1998), Gβ5 exhibits much less homology with other isoforms (~50%) and is preferentially expressed in the brain and nervous system (Watson et al. 1994, Witherow & Slepak 2003). The primary transcript of Gβ5 can be alternatively spliced to produce two isoforms: a short isoform (referred to hereafter simply as Gβ5), expressed in brain and neurons, and a long isoform expressed in the retinal photoreceptor outer segments. Besides its restricted pattern of expression and its divergent primary structure, Gβ5 is unique in its ability to heterodimerize with regulator of G protein signaling (RGS) proteins of the R7 subfamily (R7-RGS) (Cabrera et al. 1998, Snow et al. 1998, Zhang & Simonds 2000, Makino et al. 1999). R7-RGS proteins, in common with other members of the larger RGS protein family, contain a core RGS homology domain that acts as a GTPase activating protein (GAP) for heterotrimeric G protein α subunits to rapidly terminate G protein signaling (Ross & Wilkie 2000). As a consequence of its binding to R7-RGS proteins, Gβ5 can be found in a heterotrimeric complex with one of two SNARE-like membrane-anchoring proteins, R7-binding protein (R7BP) or R9-anchoring proteins (R9AP) [see (Jayaraman et al. 2009, Anderson et al. 2009) for recent reviews].
Knockout experiments often provide strong clues as to gene function. Yet while study of Gnb5 knockout mice has demonstrated destabilization of R7-RGS proteins in brain (Chen et al. 2003), most investigations have focused on the visual system (Krispel et al. 2003, Rao et al. 2007) and as a result the physiological role of Gβ5 in brain outside the visual system is still not well understood.
To address this problem we have examined the phenotype of Gnb5 KO mice in greater depth. We report here that mice lacking Gβ5 have a markedly abnormal neurologic behavioral phenotype that includes developmental impairment, wide-based gait, gross motor coordination and learning deficiencies and hyperactivity. We further show the absence of Gβ5 causes improper neuronal maturation and brain development. Based on our results and previous findings, we propose a model in which Gβ5 regulates dendritic arborization and/or synapse formation during development, in part by gene regulation.
Materials and methods
Mouse husbandry and genotyping
Generation of Gβ5 KO mice with heterozygous deletion of exon 3 of in the germline was previously described (Chen et al. 2003). Mice heterozygous for Gnb5 with mixed 129SvEv and C57BL/6 background were back-crossed with wild-type C57BL/6 for three to four generations, with selection for the mutant Gnb5 allele by genotyping, and the resulting Gnb5 heterozygotes mated to generate litters of wild-type and Gnb5 heterozygous and homozygous pups. Age-matched littermate cohorts were used for all the experiments to account for potential strain differences. During approximately 36 months of animal handling, testing and experimentation on littermate cohorts, no obvious sex differences were observed in the developmental, behavioral or microscopic anatomical phenotypes reported here. Mice were housed and treated in strict accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals and maintained in a pathogen-free facility, with 4–5 animals per cage in a temperature-controlled room with a 12-h light/dark cycle and access to food and water ad libitum. At most we identified 1 or 2 Gnb5 KO pups per litter. To reduce the high mortality of Gnb5 KO pups prior to weaning (Chen et al. 2003), cages with litters that included Gnb5 KO pups were supplemented for three weeks with a high fructose corn syrup-sweetened nutritional gel (Transgel, Charles River Laboratories, Wilmington, MA). In order to generate the large number of Gnb5 KO mice needed for this study a large-scale mating plan was employed, using up to 30 mating cages at any given time. Details of the mouse genotyping methods are included in the supplementary online data.
Testing of developmental milestones in mouse pups
Age-matched littermates were tested for the development of neurobehavioral milestones including the surface righting reflex and the placing response according to published protocols (Heyser 2003). For categorical assessment of the surface righting reflex by Fisher’s exact test, the success in righting on a solid surface was determined in one or two week-old mouse pups 5 seconds after placement on their backs. Fisher’s exact test of the placing response was applied by assessing placing activity in three week-old mouse pups 90 seconds after gentle suspension upside down.
Gait analysis, motor coordination and activity testing
Footprint analysis of one to three-month old age-matched littermate mice was performed primarily as described with results expressed as the ratio of stride length to hind-base width (Klapdor et al. 1997). Briefly, forelimbs and hind limbs of mice were dipped in different non-toxic watercolors and the mice were made to walk on an inclined gangway lined by a white paper (100 × 7.5 × 7.5 cm) leading to a darkened enclosure. Footprints were recorded in duplicate for each mouse after which the limbs were washed and dried before placing the mouse back to its original cage. Footprint patterns recorded on paper were measured and analyzed. Details of motor coordination and activity testing methodology are included in the supplementary online data.
Histology, immunofluorescence, and laser confocal scanning
Mice were anesthetized with avertin and transcardially perfused by PBS followed by 4% paraformaldehyde in PBS. Brains were carefully removed from the cranium and fixed overnight in the same fixative at 4° C. Mouse brains were transferred sequentially in 10%, 20% and 30% sucrose dissolved in PBS in a 50 ml centrifuge tube until they settled to the bottom. Brains were then sectioned sagittally on a freezing cryotome (Leica CM3050S, Leica Microsystems Inc., Bannockburn, IL) at a thickness of 20 μm and sections were collected either on sialanated slides or in a 6-well plate free floating in PBS. Details of immunofluorescence methodology, cresyl violet staining, hippocampal histomorphometry, and laser confocal scanning and intensity analysis are included in the supplementary online data.
mRNA quantification
Gene expression levels were estimated based on transcript abundance in total RNA isolated from the cerebella of 2-week old mice as measured by quantitative RT-PCR and oligo microarray analysis. Quantitative RT-PCR was performed with one step quantitative RT-PCR master mix (Agilent Technologies) using a Stratagene MX 3000P real time PCR machine and analyzed using the accompanying software. For each experiment, the transcript levels of tested genes were normalized to the corresponding β-actin mRNA levels and compared using the comparative quantitation algorithm of the software. The sequences of primers employed were as follows: Synpo: Fwd: 5′-AGG AAG AGG AAG TGC CAT TGG T-3′, Rev: 5′-TGT GGT GAG TGT GGC ATT AGG T-3′; Tmod3: Fwd: 5′-TGT GCG ACC TTG CAG CTA TTC T-3′, Rev: 5′-ACG CTC TTG GTT GAC ACC GTT A-3′; Guca1b: Fwd: 5′-ACA AGG ACC GAA ATG GCT GCA T-3′, Rev: 5′-GGC AGG CTT TCT TCA GCT TGT A-3′; Grid2: Fwd: 5′-TGA CAC TTT GCC AAC ACG CCA A-3′; Rev: 5′-TTT GCT GAG AGT GTG CGG CTA-3′; Trim37: Fwd 5′-ATG GCA CCC CTT CTC TCA GTT ATG-3′, Rev: 5′-CAT AAC TGA GAG AAG GGG TGC CAT-3′. Each reaction was conducted in triplicate and 3–9 biological samples prepared independently were used in data analysis. The Prism software version 5.0c (GraphPad Software, Inc., La Jolla, CA, USA) was used for graphing of the analyzed data set.
Microarray analysis
Microarray analysis was performed with mRNA isolated from the cerebella and non-cerebellar regions of the 2 week old Gnb5 KO mice and their wild type littermates using RNeasy® mini kit (QIAGEN). The RNA quality was checked by an Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Additional details of microarray analysis are included in the supplementary online data.
Immunoblotting, chemiluminescence and infrared imaging
Cell lysates were boiled with equal volume of Laemmli’s 2 X gel loading buffer and the hot solution was loaded onto 4–20% Tris-Glycine SDS-PAGE gels (EC6025BOX; Invitrogen, Carlsbad, CA, USA) to separate the proteins, followed by transfer of the proteins on to 0.2-micron nitrocellulose membrane using iBlot Gel Transfer Stacks and iBlot Gel Transfer System (IB3010–01, IB1001; Invitrogen). Membranes were blocked with TBS (pH 7.4) containing 0.1% Tween 20 and 5% nonfat dry milk (blocking buffer) and incubated overnight with primary antibodies in the same buffer. The membranes were then washed seven times for 5 minutes each with the above buffer without milk, followed by a 1-hour incubation in blocking buffer including appropriate secondary IR antibodies (dilution 1: 20,000) (details regarding primary and secondary antibodies employed are included in the supplementary online methodology data). Membranes were then washed three times as above, and the protein signals were detected by the Odyssey near-infrared fluorescence imaging system (LI-COR Bioscience, Lincoln, NE, USA). For the quantification of the intensity of the protein bands membranes were dually probed, with the b-actin used as a loading control.
Statistical analysis
Data were analyzed using Prism software version 5.0c (GraphPad Software, Inc.). Data are expressed as means ± SEM, where n equals the number of animals tested with the indicated genotype. The significance of differences was assessed using two-tailed unpaired Student’s t-tests for continuous data or Fisher’s exact test for categorical data, as appropriate. The confidence level for significance was 95%.
Results
Mice lacking Gβ5 exhibit impaired neurobehavioral development
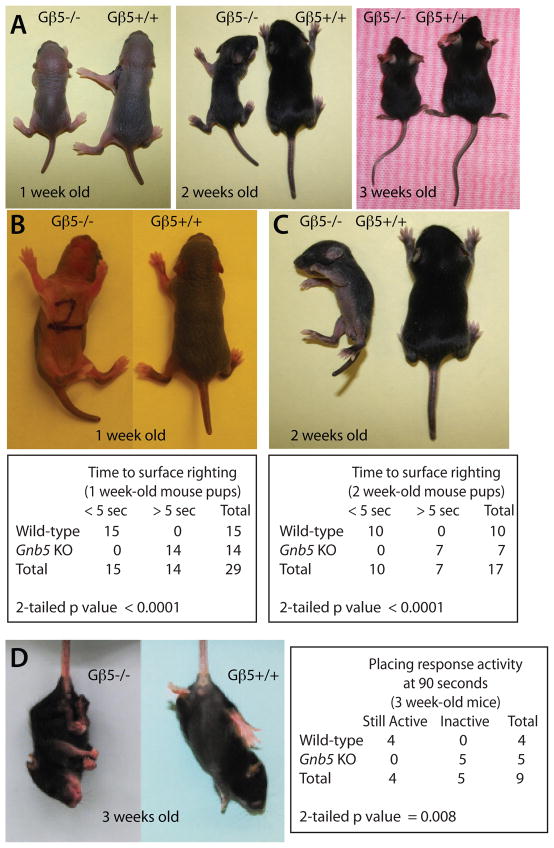
Homozygous Gβ5 knockout mice are runty at birth and exhibit a persistent smaller body size than their wild-type litter mates as previously described (Fig. 1A) (Chen et al. 2003). By five days of age (with a normal range of one to 10 days), a typical mouse pup rights itself promptly when placed on its back on a level surface (Heyser 2003). When tested at one and two weeks of age, Gβ5 homozygous knockout pups showed a significantly delayed or absent surface righting reflex when placed on their backs compared to their wild-type littermates (Fig. 1B, C). Gβ5 heterozygotes showed surface righting reflexes indistinguishable from the wild-type. While it is suspended gently upside down by its tail, a normal mouse pup raises its head and extend its limbs in anticipation of correctly placing itself on a nearby solid surface, in a so-called placing response. The placing response developmental milestone is typically achieved at 15 days of age, with a normal range of 11–18 days (Heyser 2003). When tested at three weeks of age, homozygous Gβ5 knockout pups had markedly abnormal placing responses compared to wild-type littermates and, after a brief attempt to anticipate surface placement, became passive and retracted instead of extending their limbs (Fig. 1D, left). When assessed at 90 seconds, 4/4 wild-type but 0/5 Gnb5 KO pups were still actively anticipating surface placement (Fig. 1D, right). Gnb5 heterozygous mouse pups behaved like wild-type mice and achieved the placing response developmental milestone within the normal age range.
Fig 1. Developmental impairment in Gβ5-deficient mice.
(A) Gβ5 homozygous knockout mice, shown here next to their wild-type littermates at the indicated ages, have a runty phenotype as previously described (Chen et al. 2003). Impaired surface righting reflex is evident in Gβ5-deficient, but not wild-type, mice at one week (B) or two weeks (C) of age in photographs taken approximately 30 seconds after placing mice on their backs on a solid surface. (Below) 2 × 2 contingency table showing the progress toward surface righting of wild-type and Gnb5 KO mice assessed after 5 seconds with the number of mice tested of each genotype shown (one-week old mice, n=15 wild-type and n=14 Gnb5 KO, Fisher’s exact test, 2-tailed p value <0.0001; two-week old mice, n=10 wild-type and n=7 Gnb5 KO, Fisher’s exact test, 2-tailed p value <0.0001). (D) Abnormal placing reflex in 3-week old Gβ5-deficient, but not wild-type, mice suspended by their tails near a solid surface. (Right) 2 × 2 contingency table showing placing response activity of wild-type and Gnb5 KO mice assessed at 90 seconds with the number of mice tested of each genotype shown (n=4 wild-type and n=5 Gnb5 KO, Fisher’s exact test, 2-tailed p value = 0.008).
Motor coordination, balance and learning abnormalities in mice lacking Gβ5
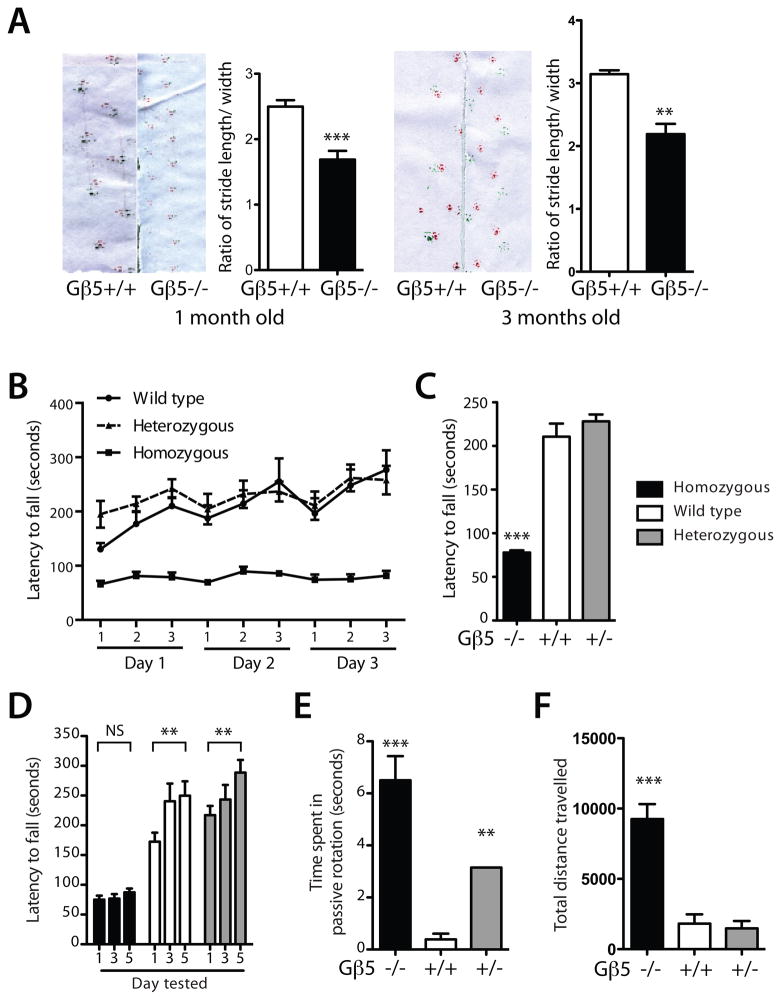
Compared to the wild-type, Gβ5 homozygous knockout mice lacked motor coordination and exhibited an abnormal wide-based gait on footprint analysis, a defect that persisted as the mice grew older and was demonstrable in mice tested at one and three months of age (Fig. 2A). An abnormal style of tiptoe-walking was also observed in homozygous Gβ5 KO mice (supplemental video No. 1). The gait of Gβ5 heterozygous mice was not distinct from the wild-type. Testing of mice at 3 to 4 months of age on an accelerating rotarod demonstrated balance and motor learning abnormalities in Gβ5 homozygous knockout mice (Fig. 2B-E). Mean latency to fall was significantly shorter in Gβ5 homozygous knockout mice compared to wild-type or Gβ5 heterozygotes (Fig. 2B, C). When repeatedly tested on the rotarod over a five-day period, wild-type and Gβ5 heterozygous mice, but not their Gβ5 homozygous KO littermates, showed significant motor learning between days one and five (Fig. 2D). It is noteworthy that R7BP (Anderson et al. 2010) and RGS9 (Blundell et al. 2008) KO mice also show deficiencies in gross motor coordination, since Gβ5 and R7BP govern the stability of RGS9 (Chen et al. 2003, Anderson et al. 2007).
Figure 2. Gait and motor abnormalities in Gβ5-homozygous knockout mice.
(A) Footprint analysis in one- and three-month old mice shows abnormal stride length to hind limb width ratio in Gβ5-deficient mice compared to wild-type. (one-month old mice, n=6 wild-type and n=6 Gnb5 KO, **P = 0.0016, two-tailed unpaired t-test; three-month old mice, n=4 wild-type and n=4 Gnb5 KO, ***P = 0.0006, two-tailed unpaired t-test) (B) Accelerating rotarod testing of littermates with the indicated genotypes for three successive trials repeated over a three-day period showing latency to fall. (C) Rotarod latency to fall results averaged over entire three-day trial (n=8 wild-type, n=9 Gnb5 heterozygotes and n=12 Gnb5 KO, ***P < 0.0001; versus wt, two-tailed unpaired t-test). (D) Five day testing on the accelerating rotarod for mice with the indicated genotypes (n=8 wild-type, n=10 Gnb5 heterozygotes and n=20 Gnb5 KO, **P < 0.02; versus day one results, two-tailed unpaired t-test). (E) Average time spent in passive rotation per trial over the five-day rotarod testing period, by Gβ5 genotype (n=8 wild-type, n=10 Gnb5 heterozygotes and n=20 Gnb5 KO, **P < 0.02; ***P < 0.0001; versus wt, two-tailed unpaired t-test). (F), Total distance travelled in open field-testing, by Gβ5 genotype. For open field-testing, mice were tested at three months of age (n=6 wild-type, n=4 Gnb5 heterozygotes and n=11 Gnb5 KO, ***P < 0.0001; versus wt, two-tailed unpaired t-test). After rotarod testing, mice from the same cohort were tested for locomotor activity by the open field test and then for footprint/gait analysis when three months old. The one month-old mice used for footprint/gait analysis represented a separate cohort.
Mice unable to maintain their balance on the rotarod sometimes adopt a passive strategy, by flattening themselves, clinging onto the rotating drum and riding around until they fall off, a behavior favored by some mice with known neurologic deficits (Le Marec & Lalonde 1997). During the five-day rotarod testing period, the time spent in such passive rotation by Gβ5 homozygous mice was markedly greater than that of wild-type mice (Fig. 2E). Interestingly the Gβ5 heterozygous mice also spent more time in passive rotation than their wild-type littermates, albeit less time than the KO mice (Fig. 2E).
Mice lacking Gβ5 are hyperactive
In open field-testing, Gβ5 homozygous KO mice were markedly hyperactive compared to wild-type mice (Fig. 2F, supplemental Fig. S1, and supplemental video No. 2). This hyperactivity was evident by measurement of several parameters: increased total travel distance (Fig. 2F, supplemental video No. 2), horizontal and ambulatory activity and decreased rest time (Fig. S1). In contrast, Gβ5 heterozygotes showed no differences from their wild-type littermates in open field testing (Figs. 2F, S1).
Abnormal cerebellar development in mice lacking Gβ5
Gβ5 is expressed in multiple regions throughout the rodent brain (Liang et al. 1998, Betty et al. 1998, Zhang et al. 2000). Gβ5 KO mice in this study exhibited abnormal gait, balance, and motor learning, functions governed in part by the cerebellum. Since Gβ5 is found in relative abundance in Purkinje cells and other cells in the cerebellar cortex (Liang et al. 1998, Zhang et al. 2000), it seemed plausible that the developmental impairment and constellation of neurologic abnormalities in Gβ5 KO mice might reflect in part abnormalities of the cerebellum. We therefore decided to focus on cerebellar development in the Gβ5 mutant mice, and included experiments employing a mouse transgene expressing enhanced green fluorescent protein under the direction of the mouse Purkinje cell protein 2 promoter (Pcp2-GFP) as a marker of Purkinje cell maturation and identity (Tomomura et al. 2001).
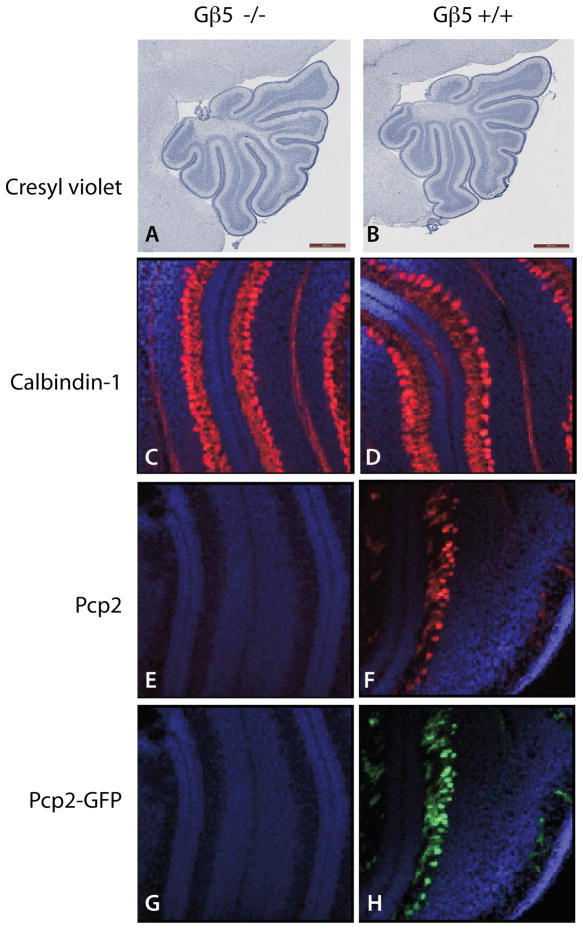
Initial studies employed one week-old mice. Cerebella of wild-type and Gβ5 homozygous KO mouse pup littermates, both hemizygous for Pcp2-GFP, were compared by light microscopy, immunohistochemistry, and histofluorescence (Figs. 3, S2). Cresyl violet staining did not reveal any obvious differences in the cerebellar lobar architecture or patterning of the sulci and gyri at the light microscopic level (Fig. 3A, B). Immunoreactivity of calbindin-1, a protein normally expressed in the precursors of Purkinje cells beginning at day E12 (Hatten et al. 1997), was comparable in wild-type and Gβ5 homozygous KO pups (Fig. 3C, D). In contrast, both Pcp2 immunoreactivity and expression of the Pcp2-GFP transgene were barely detectable in one-week old Gnb5 KO mouse pups (Fig. 3E, G), while readily demonstrable in wild-type littermates (Fig. 3F, H). Since Pcp2 (also known as L7) is normally expressed in immature Purkinje precursor cells beginning at birth (day P0) (Hatten et al. 1997), these findings indicate markedly delayed Purkinje cell development in the Gβ5 KO pups (Hatten & Heintz 1995).
Figure 3. Delayed Purkinje cell maturation in the cerebellar cortex of seven-day old Gβ5-homozygous knockout mice.
(A, B) Cresyl violet staining of sagittal sections of cerebella of seven-day old Pcp2-GFP hemizygous Gβ5 KO and wild-type mouse littermates (scale bars = 500 μm). (C, D) Merged DAPI (blue) and calbindin-1-immunostaining (red) images of frozen sections of cerebella of Gβ5 KO and wild-type mouse littermates (50X). (E, F) Merged DAPI (blue) and Pcp2-immunostaining (red) images of frozen sections of cerebella of Gβ5 KO and wild-type mouse littermates (50X). (G, H) Merged DAPI (blue) and Pcp2-GFP reporter fluorescence (cyan) images of frozen sections of cerebella of Gβ5 KO and wild-type mouse littermates (50X). Results shown in A-H representative of three littermate pairs with similar findings.
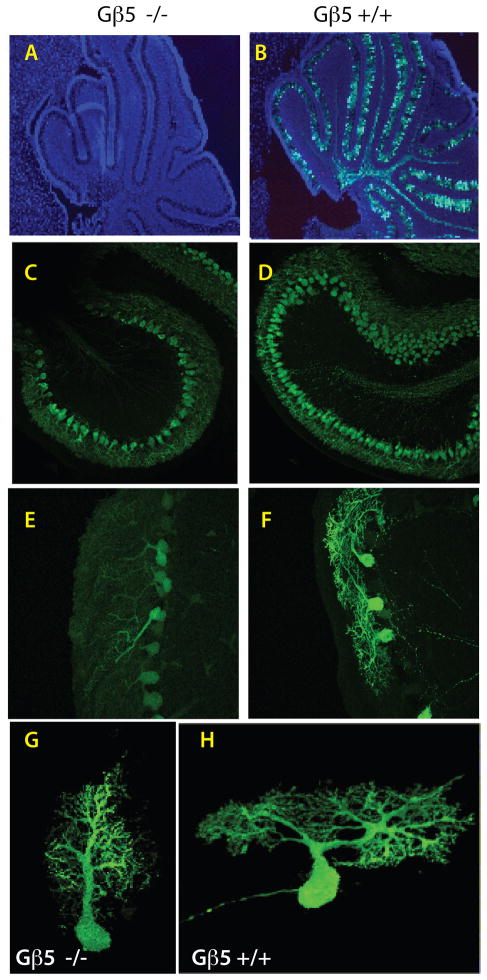
Additional longitudinal studies of cerebellar development and Purkinje cell maturation employing wild-type and Gβ5 homozygous KO mice hemizygous for Pcp2- GFP were performed on mice between four and 15 days old (Figs. 4A, B; Fig. S3). Discernable fluorescence of the Pcp2-GFP reporter was evident in the periphery of vibratome sections taken from the cerebella of 4 to 6 day old wild-type pups but not in their Gβ5 homozygous KO littermates (Fig. S3A, B). In whole brains dissected from seven to 10 day old mice, the extent and intensity of gross cerebellar fluorescence paled in Gβ5 homozygous KO mice compared to their wild-type littermates (Fig. S3C, D). By 14 to 15 days of age, however, such differences in gross cerebellar fluorescence were no longer apparent (Fig. S3E, F). Analysis of cerebellar sections from seven-day old mice again confirmed the absence of the Pcp2-GFP reporter signal in Gβ5 homozygous KO mice (Fig. 4A, cf. 4B) noted above (Fig. 3G, H). Laser confocal histofluorescence analysis of cerebella from 7 to 10-day old mice demonstrated that the maturity of Purkinje cells, evidenced by the degree of dendritic arborization, was substantially diminished in Gβ5 homozygous KO mice compared to their wild-type littermates (Fig. 4C-H; supplemental videos 3 and 4). Quantitative immunoblotting confirmed that Pcp2 protein expression was diminished compared to wild-type littermates in 10-day old Gβ5 KO mice (not shown).
Figure 4. Delayed cerebellar development and deficient Purkinje cell maturation in Gβ5-homozygous knockout mice.
Analysis of Pcp2-GFP hemizygous Gβ5 KO (A, C, E, G) and Pcp2-GFP hemizygous Gβ5 wild-type (B, D, F, H) littermate mice. (A, B) Merged DAPI (blue) and Pcp2-GFP reporter histofluorescence (green) images from cerebellar sections harvested from seven-day old Gβ5 homozygous KO and wild-type littermates (50X). (C-H) Laser confocal analysis of Pcp2-GFP reporter histofluorescence in cerebellar sections from 10-day old mice comparing the degree of dendritic arborization in Gβ5 homozygous KO mice and their wild-type littermates. Magnification in C, D, 20 X; in E, F, 40 X. See also supplemental videos 3 and 4, corresponding to images G and H, showing 3-D images of the corresponding neurons reconstructed from a stack of confocal images acquired at 0.17 μm step size, as described in supplementary online materials and methods. Results in A-F are representative of results from three wild-type and Gnb5 KO sibling pairs.
Abnormal hippocampal development in mice lacking Gβ5
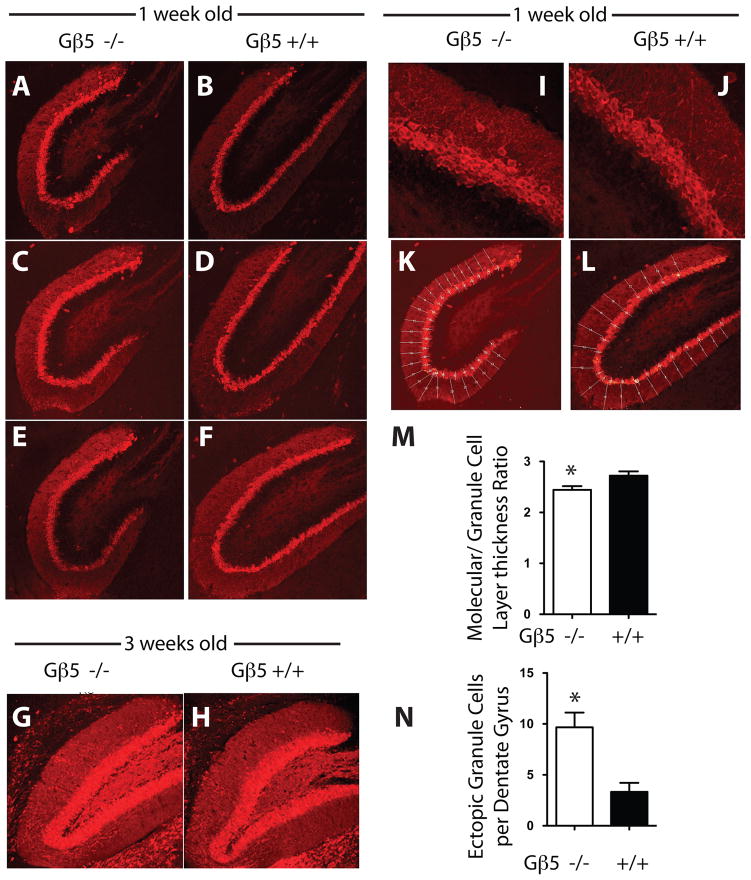
The impaired motor learning demonstrated in Gβ5 KO mice prompted us to also examine the hippocampus in wild-type and Gβ5 mutant mice, given the established importance of this brain region in learning and memory. In wild-type mice the angular zone (AZ) of the dentate gyrus (DG), joining its supra- and infrapyramidal blades, undergoes a shape change during early postnatal development, from a more rounded contour on days P1 to P4 to a more acute contour at 2 and 4 weeks, as seen in Nissl stained brain sections of developing wild-type mice (Fig. S4). Using calbindin-1 immunoreactivity to label granule cells of the DG of the hippocampus (Baimbridge & Miller 1982), the development of AZ of the DG in Gnb5 KO mice lagged behind the wild-type (Fig. 5A-H). In 1-week old mice the contour of the AZ was rounder and less acute in Gnb5 KO mice than in their wild-type littermates (Fig. 5A-F). By 3-weeks of age the differences in AZ contour between Gnb5 KO and wild-type mice had largely resolved (Fig. 5G, H).
Figure 5. Morphology and immunohistochemistry of the hippocampal dentate gyrus in wild-type and Gnb5-knockout mice.
The shape of the dentate gyrus in Gnb5 KO mice, in sections of hippocampus stained for calbindin-1 immunoreactivity and examined at 20X magnification, was found to be more circular and immature (A, C and E; cf. Fig. S3) than in wild-type littermates in which its shape was fuller and more elongated (B, D and F). Data shown are from three independent age-matched littermates. At 3 weeks there was no discernible visual difference in the dentate gyrus morphology between the Gnb5 KO mice (G) and their wild type littermates (H). Sections shown are representative of results from three age-matched littermates. At higher magnification (63X) in the Gnb5 KO mice, calbindin-1 positive neurons were found to be more disarranged and a significant number of ectopic cells were found outside the dentate gyrus layer (I) as compared to the wild type which were more organized (J). Hippocampal sections (20X) from Gnb5 KO mice (K) and their wild type littermates (L) were subdivided into multiple regions as shown to allow for quantitative histomorphometry using ImageJ software as described in the supplementary online methods. M. The thickness of the granule cell layer relative to the molecular layer was determined as described in the supplementary online methods. N. The number of ectopic calbindin-1 positive cells present in the molecular layer of the hippocampus in 1-week old wild-type or Gβ5-deficient mice was determined as described in the supplementary online methods. In M, N, n = 3 littermate pairs of wild-type and Gnb5 KO mice.
The organization of the granule cell layer was also abnormal in the DG of young Gnb5 KO mice (Fig. 5I-N). The granule cell layer was more loosely arranged, less compact, and slightly thicker (relative to the adjacent molecular layer) in 1-week old Gnb5 KO mice than in wild-type littermates (Fig. 5I, J, M). Furthermore ectopic calbindin 1-positive granule cells were found more frequently in the molecular layer in KO mice (Fig. 5I, J, N).
Multiple genes are dysregulated in the brains of Gnb5 knockout mice
The cerebellar and hippocampal developmental delay illustrated by the immunohistochemical and Pcp2-GFP reporter studies suggests dysregulation of critical gene expression in Gnb5 KO mice. To address this possibility we (a) studied the expression of several candidate genes previously implicated in neuronal development or function, and (b) employed microarray analysis to compare gene transcripts harvested from wild-type and Gnb5 KO mouse brain.
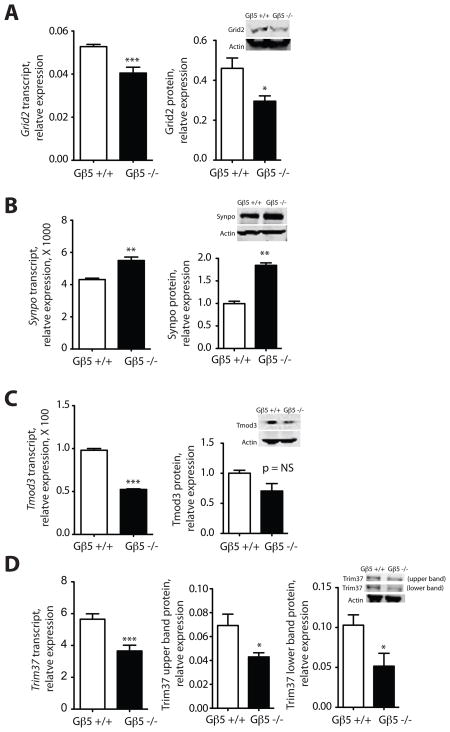
Quantitative RT-PCR of transcripts encoding candidate proteins implicated in neuronal development or function and harvested from the brains of two week-old Gnb5 KO mice and their wild-type littermates provided examples of such dysregulation (Fig. 6A, B). Compared to the wild-type, transcript and protein levels for glutamate receptor, ionotropic, delta 2 (Grid2), a gene implicated in motor coordination and Purkinje cell synapse formation (Kashiwabuchi et al. 1995), were down-regulated in the Gnb5 KO mice (Fig. 6A). In contrast, Gnb5 KO mice demonstrated higher transcript and protein expression than controls of synaptopodin (Synpo), a gene expressed in the hippocampus and implicated in synaptic plasticity (Deller et al. 2003) (Fig. 6B).
Figure 6. Quantitative analysis of neuronal gene expression in two-week old Gnb5 wild-type and homozygous knockout mice.
Total RNA and protein were harvested from the non-cerebellar brain regions of two week-old Gnb5 KO mice and their wild-type littermates and the expression of transcripts, estimated by quantitative RT-PCR, and protein, estimated by immunoblotting with near-infrared fluorescence quantification, were determined as described in the main and supplementary online Materials and Methods, and shown in the indicated histograms. Inset images (right) show immunoblotting results from a representative experiment. A. Glutamate receptor, ionotropic, delta 2 (Grid2); B. Synaptopodin (Synpo); C. Tropomodulin 3 (Tmod3); D. Tripartite motif containing 37 (Trim37). The upper and lower immunoreactive Trim37 bands were analyzed and are shown separately. (*P < 0.05; **P < 0.01; ***P < 0.0002; versus wt, two-tailed unpaired t-test; NS= not significant; n = 3 to 5 biological samples of total RNA or protein, with mRNA quantification from each biological sample by quantitative RT-PCR conducted in triplicate.)
Microarray analysis was also used to compare the expression of transcripts harvested from the brains of Gβ5 homozygous KO mice and their wild-type littermates. Expression of transcripts from the cerebellum and non-cerebellar regions of brain were analyzed separately. The analysis showed that 150 genes expressed in the cerebellum and 228 genes from non-cerebellar brain regions were significantly affected (either positively or negatively) in the Gnb5 mutant mice (Supp. Fig. 5, Supp. Tables 1, 2). An overlapping set of 39 genes common to both brain regions was identified (Supp. Fig. 5, Supp. Table 3). This microarray data has been deposited in the NCBI Gene Expression Omnibus (GEO) repository, with GEO Reference Series accession number GSE29083.
Changes in the expression of several transcripts from the pool of 69 genes identified in the microarray experiments were tested by quantitative RT-PCR and correlated where possible with protein expression (Fig. 6C, D). Transcripts for both tropomodulin 3 (Tmod3) (Fig. 6C) and tripartite motif containing 37 (Trim37) (Fig. 6D) were down-regulated in Gnb5 KO mice. Both the upper and lower anti-Trim37 immunoreactive protein bands were reduced in the Gnb5 KO mice, correlating with the reduced Trim37 transcript expression (Fig. 6D), although the reduction in Tmod3 protein levels in the Gnb5 KO mice did not reach statistical significance (Fig. 6C). Transcript for guanylate cyclase activator 1B (Guca1b) was significantly up-regulated in the Gnb5 KO mice (P < 0.0002), consistent with the microarray results, although anti-Guca1b antibody suitable to assess protein expression was not available (not shown).
Discussion
This study further analyzes the phenotype of mice deficient for Gβ5 and finds marked neurobehavioral developmental delay, impaired gait and motor learning, and hyperactivity, associated with widespread abnormalities of neuronal development. The present findings complement earlier studies of Gβ5 KO mice documenting destabilization of R7-RGS proteins (Chen et al. 2003), defective visual adaptation (Krispel et al. 2003), and abnormal development and functioning of retinal bipolar cells (Rao et al. 2007). The striking behavioral abnormalities documented here in the Gnb5 KO mice appear more severe than those previously reported in R7BP (Anderson et al. 2010) and RGS9 (Blundell et al. 2008) KO mice. Since Gβ5 stabilizes not only RGS9, but also the three other R7-RGS subfamily proteins found in brain, viz. RGS6, RGS7 and RGS11, the more severe behavioral phenotype in the Gnb5 KO mice likely reflects additional signaling deficiencies in pathways involving these latter RGS proteins (Chen et al. 2003).
There are shortcomings in the methods used here to compare gene expression in wild-type and Gnb5 KO mice. Comparison of microarray and quantitative RT-PCR methodologies to assess gene expression has shown that, while there is generally a strong correlation between the two methods, a minority of genes can yield disparate results (Dallas et al. 2005). Such difficulty is illustrated in our own assessment of Trim37 expression for which the microarray results (Supp. Tables 1, 2) and quantitative RT-PCR results (Fig. 6D) were of opposite sign. Quantitative RT-PCR is still considered the “gold” standard of mRNA quantification despite its shortcomings however (Nolan et al. 2006), and such results that we present here generally correlate with companion protein expression analysis (Fig. 6).
Some part of the neurologic abnormalities in Gβ5-deficient mice can be ascribed to impaired development of the cerebellum and hippocampus, documented in the present work by immunohistochemical and Pcp2-GFP reporter analysis. Indeed, hyperactivity has been reported in association with cerebellar (Boy et al. 2009) and hippocampal (Goddyn et al. 2006) neuropathology in mouse models. The migration and chemotaxis of neuronal precursors is critical for the development of both the cerebellum and hippocampal formation. Within the cerebellar cortex, the proper migration and maturation of Purkinje cells depends on local signals and cell-cell interactions that, in the postnatal period, include critical interactions with cerebellar granular cells (Hatten & Heintz 1995) where Gβ5 is also expressed (Liang et al. 1998, Betty et al. 1998, Zhang et al. 2000).
Because Gβ5 is normally expressed in multiple brain regions (Liang et al. 1998, Betty et al. 1998, Zhang et al. 2000), we suspect that the cerebellar and hippocampal abnormalities in the Gβ5 KO mice demonstrated here are indicative of diffuse neuronal defects in the brains of Gβ5-deficient mice. Given the high mortality among newborn Gnb5 KO mice (Chen et al. 2003), a disruption in the regulation of multiple genes required for neuronal development and/or function, so severe as to be frequently fatal, likely accounts for the marked developmental impairment in the surviving Gnb5 KO mice studied here. At the cell biological level, the defect in Purkinje cell dendritic arborization described in the present work is reminiscent of a previously described defect in the retinas of Gnb5 KO mice (Rao et al. 2007). In the retinas of such mice, the dendrites of the rod bipolar cells were shorter and more disorganized than in controls, a finding associated with a reduced number of synaptic triads and gross morphological abnormalities of the retinal outer plexiform layer (Rao et al. 2007). Thus it is conceivable that dendritic and/or synaptic development in multiple brain regions normally depends on the function of the Gβ5 complex, a role consistent with the documented localization of R7-RGS/Gβ5 complexes to dendritic tips and postsynaptic densities (Cao et al. 2008, Song et al. 2006).
Based on the marked neurobehavioral developmental delay described here in Gnb5 KO mice, we speculate that loss of Gβ5 might also be associated in humans with developmental delay. Deletions involving human chromosomal region 15q21.2, a locus that includes GNB5, have been associated in at least four cases with mental retardation and moderate to severe developmental delay (Liehr et al. 2003, Pramparo et al. 2005, Lalani et al. 2006, Tempesta et al. 2008). Whether GNB5 loss-of-function contributes to the developmental delay in patients with such interstitial chromosomal deletions, or in a subset of patients with autosomal recessive mental retardation, are questions that must await further clinical and molecular genetic investigation.
Supplementary Material
Acknowledgments
The authors thank Kimya Davani and William Jou of the NIDDK Mouse Metabolism Core for their expert assistance. This research was supported by the Intramural Research Programs of the National Institute of Diabetes and Digestive and Kidney Diseases and National Institute of Allergy and Infectious Diseases. C-K.C. is supported in part by an NIH grant EY013811.
Abbreviations used
- RGS
regulator of G protein signaling
- R7-RGS
regulators of G protein signaling of R7 subfamily
- R7BP
R7-RGS binding protein
- R9AP
RGS9/11 anchoring protein
- KO
knockout
- GFP
green fluorescent protein
- EGFP
enhanced green fluorescent protein
- PCR
polymerase chain reaction
- RT-PCR
real time-PCR
- PBS
phosphate-buffered saline
- BSA
bovine serum albumin
- Pcp2
Purkinje cell protein 2
- PAGE
polyacrylamide gel electrophoresis
- AZ
angular zone of dentate gyrus
- DG
dentate gyrus
- GEO
Gene Expression Omnibus
Footnotes
Conflict of Interest: None of the authors has a financial, advisory, or consultative conflict of interest with the data or its interpretation as presented in this manuscript.
References
- Anderson GR, Cao Y, Davidson S, Truong HV, Pravetoni M, Thomas MJ, Wickman K, Giesler GJ, Jr, Martemyanov KA. R7BP complexes with RGS9-2 and RGS7 in the striatum differentially control motor learning and locomotor responses to cocaine. Neuropsychopharmacol. 2010;35:1040–1050. doi: 10.1038/npp.2009.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GR, Posokhova E, Martemyanov KA. The R7 RGS protein family: multi-subunit regulators of neuronal G protein signaling. Cell Biochem Biophys. 2009;54:33–46. doi: 10.1007/s12013-009-9052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GR, Semenov A, Song JH, Martemyanov KA. The membrane anchor R7BP controls the proteolytic stability of the striatal specific RGS protein, RGS9-2. J Biol Chem. 2007;282:4772–4781. doi: 10.1074/jbc.M610518200. [DOI] [PubMed] [Google Scholar]
- Baimbridge KG, Miller JJ. Immunohistochemical localization of calcium-binding protein in the cerebellum, hippocampal formation and olfactory bulb of the rat. Brain Res. 1982;245:223–229. doi: 10.1016/0006-8993(82)90804-6. [DOI] [PubMed] [Google Scholar]
- Betty M, Harnish SW, Rhodes KJ, Cockett MI. Distribution of heterotrimeric G-protein beta and gamma subunits in the rat brain. Neuroscience. 1998;85:475–486. doi: 10.1016/s0306-4522(97)00623-4. [DOI] [PubMed] [Google Scholar]
- Blundell J, Hoang CV, Potts B, Gold SJ, Powell CM. Motor coordination deficits in mice lacking RGS9. Brain Res. 2008;1190:78–85. doi: 10.1016/j.brainres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy J, Schmidt T, Wolburg H, et al. Reversibility of symptoms in a conditional mouse model of spinocerebellar ataxia type 3. Hum Mol Genet. 2009;18:4282–4295. doi: 10.1093/hmg/ddp381. [DOI] [PubMed] [Google Scholar]
- Cabrera JL, De Freitas F, Satpaev DK, Slepak VZ. Identification of the G®5-RGS7 protein complex in the retina. Biochem Biophys Res Commun. 1998;249:898–902. doi: 10.1006/bbrc.1998.9218. [DOI] [PubMed] [Google Scholar]
- Cabrera-Vera TM, Vanhauwe J, Thomas TO, Medkova M, Preininger A, Mazzoni MR, Hamm HE. Insights into G protein structure, function, and regulation. Endocr Rev. 2003;24:765–781. doi: 10.1210/er.2000-0026. [DOI] [PubMed] [Google Scholar]
- Cao Y, Song H, Okawa H, Sampath AP, Sokolov M, Martemyanov KA. Targeting of RGS7/Gbeta5 to the dendritic tips of ON-bipolar cells is independent of its association with membrane anchor R7BP. J Neurosci. 2008;28:10443–10449. doi: 10.1523/JNEUROSCI.3282-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Eversole-Cire P, Zhang H, Mancino V, Chen YJ, He W, Wensel TG, Simon MI. Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc Natl Acad Sci U S A. 2003;100:6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas PB, Gottardo NG, Firth MJ, et al. Gene expression levels assessed by oligonucleotide microarray analysis and quantitative real-time RT-PCR -- how well do they correlate? BMC Genomics. 2005;6:59. doi: 10.1186/1471-2164-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deller T, Korte M, Chabanis S, et al. Synaptopodin-deficient mice lack a spine apparatus and show deficits in synaptic plasticity. Proc Natl Acad Sci U S A. 2003;100:10494–10499. doi: 10.1073/pnas.1832384100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam N, Downes GB, Yan K, Kisselev O. The G-protein ®© complex. Cell Signal. 1998;10:447–455. doi: 10.1016/s0898-6568(98)00006-0. [DOI] [PubMed] [Google Scholar]
- Goddyn H, Leo S, Meert T, D’Hooge R. Differences in behavioural test battery performance between mice with hippocampal and cerebellar lesions. Behav Brain Res. 2006;173:138–147. doi: 10.1016/j.bbr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Gutkind JS. Regulation of mitogen-activated protein kinase signaling networks by G protein-coupled receptors. Sci STKE. 2000;2000:RE1. doi: 10.1126/stke.2000.40.re1. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Alder J, Zimmerman K, Heintz N. Genes involved in cerebellar cell specification and differentiation. Curr Opin Neurobiol. 1997;7:40–47. doi: 10.1016/s0959-4388(97)80118-3. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Heintz N. Mechanisms of neural patterning and specification in the developing cerebellum. Annu Rev Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- Heyser CJ. Assessment of developmental milestones in rodents. Current Protocols in Neuroscience. 2003;25:8.18.11–18.18.15. doi: 10.1002/0471142301.ns0818s25. [DOI] [PubMed] [Google Scholar]
- Jayaraman M, Zhou H, Jia L, Cain MD, Blumer KJ. R9AP and R7BP: traffic cops for the RGS7 family in phototransduction and neuronal GPCR signaling. Trends Pharmacol Sci. 2009;30:17–24. doi: 10.1016/j.tips.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwabuchi N, Ikeda K, Araki K, et al. Impairment of motor coordination, Purkinje cell synapse formation, and cerebellar long-term depression in GluR delta 2 mutant mice. Cell. 1995;81:245–252. doi: 10.1016/0092-8674(95)90334-8. [DOI] [PubMed] [Google Scholar]
- Klapdor K, Dulfer BG, Hammann A, Van der Staay FJ. A low-cost method to analyse footprint patterns. J Neurosci Methods. 1997;75:49–54. doi: 10.1016/s0165-0270(97)00042-3. [DOI] [PubMed] [Google Scholar]
- Krispel CM, Chen CK, Simon MI, Burns ME. Prolonged photoresponses and defective adaptation in rods of Gbeta5−/− mice. J Neurosci. 2003;23:6965–6971. doi: 10.1523/JNEUROSCI.23-18-06965.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani SR, Sahoo T, Sanders ME, Peters SU, Bejjani BA. Coarctation of the aorta and mild to moderate developmental delay in a child with a de novo deletion of chromosome 15(q21.1q22.2) BMC Med Genet. 2006;7:8. doi: 10.1186/1471-2350-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Marec N, Lalonde R. Sensorimotor learning and retention during equilibrium tests in Purkinje cell degeneration mutant mice. Brain Res. 1997;768:310–316. doi: 10.1016/s0006-8993(97)00666-5. [DOI] [PubMed] [Google Scholar]
- Liang JJ, Cockett M, Khawaja XZ. Immunohistochemical localization of G protein beta1, beta2, beta3, beta4, beta5, and gamma3 subunits in the adult rat brain. J Neurochem. 1998;71:345–355. [PubMed] [Google Scholar]
- Liehr T, Starke H, Heller A, et al. Evidence for a new microdeletion syndrome in 15q21. Int J Mol Med. 2003;11:575–577. doi: 10.3892/ijmm.11.5.575. [DOI] [PubMed] [Google Scholar]
- Makino ER, Handy JW, Li TS, Arshavsky VY. The GTPase activating factor for transducin in rod photoreceptors is the complex between RGS9 and type 5 G protein ® subunit. Proc Nat Acad Sci USA. 1999;96:1947–1952. doi: 10.1073/pnas.96.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- Pramparo T, Mattina T, Gimelli S, Liehr T, Zuffardi O. Narrowing the deleted region associated with the 15q21 syndrome. Eur J Med Genet. 2005;48:346–352. doi: 10.1016/j.ejmg.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Rao A, Dallman R, Henderson S, Chen CK. Gbeta5 is required for normal light responses and morphology of retinal ON-bipolar cells. J Neurosci. 2007;27:14199–14204. doi: 10.1523/JNEUROSCI.4934-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: Regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- Snow BE, Krumins AM, Brothers GM, et al. A G protein gamma subunit-like domain shared between RGS11 and other RGS proteins specifies binding to G®5 subunits. Proc Nat Acad Sci USA. 1998;95:13307–13312. doi: 10.1073/pnas.95.22.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JH, Waataja JJ, Martemyanov KA. Subcellular targeting of RGS9-2 is controlled by multiple molecular determinants on its membrane anchor, R7BP. J Biol Chem. 2006;281:15361–15369. doi: 10.1074/jbc.M600749200. [DOI] [PubMed] [Google Scholar]
- Tempesta S, Sollima D, Ghezzo S, et al. Mild mental retardation in a child with a de novo interstitial deletion of 15q21.2q22.1: a comparison with previously described cases. Eur J Med Genet. 2008;51:639–645. doi: 10.1016/j.ejmg.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Tomomura M, Rice DS, Morgan JI, Yuzaki M. Purification of Purkinje cells by fluorescence-activated cell sorting from transgenic mice that express green fluorescent protein. Eur J Neurosci. 2001;14:57–63. doi: 10.1046/j.0953-816x.2001.01624.x. [DOI] [PubMed] [Google Scholar]
- Watson AJ, Katz A, Simon MI. A fifth member of the mammalian G-protein ®-subunit family. Expression in brain and activation of the ®2 isotype of phospholipase C. J Biol Chem. 1994;269:22150–22156. [PubMed] [Google Scholar]
- Witherow DS, Slepak VZ. A novel kind of G protein heterodimer: the G®5-RGS complex. Receptors Channels. 2003;9:205–212. [PubMed] [Google Scholar]
- Zhang JH, Lai ZN, Simonds WF. Differential expression of the G protein ®5 gene: Analysis of mouse brain, peripheral tissues, and cultured cell lines. J Neurochem. 2000;75:393–403. doi: 10.1046/j.1471-4159.2000.0750393.x. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Simonds WF. Copurification of brain G-protein ®5 with RGS6 and RGS7. J Neurosci. 2000;20:RC59, 1–5. doi: 10.1523/JNEUROSCI.20-03-j0004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.