Summary
Many replication initiators form higher-order oligomers that process host origins to promote replisome formation. In addition to dedicated duplex DNA-binding domains, cellular initiators possess AAA+ (ATPases Associated with various cellular Activities) elements that drive functions ranging from protein assembly to origin recognition. In bacteria, the AAA+ domain of the initiator DnaA has been suggested to bind single-stranded DNA formed during origin melting. Here we show crystallographically and in solution that the ATP-dependent assembly of DnaA into a spiral oligomer creates a continuous surface that allows successive AAA+ domains to bind and extend single-stranded DNA segments. The mechanism of binding is unexpectedly similar to that of RecA, a homologous recombination factor, but it differs in that DnaA promotes a nucleic acid conformation that prevents pairing of a complementary strand. These findings, combined with strand-displacement assays, indicate that DnaA melts replication origins by a direct ATP-dependent stretching mechanism. Comparative studies reveal remarkable commonalities between the approach used by DnaA to engage DNA substrates and other, nucleic acid-dependent AAA+ systems.
Introduction
All organisms depend on ring- and spiral-shaped ATPase assemblies to carry out essential processes ranging from proteolysis and membrane trafficking, to signaling events and nucleic acid transactions. DNA replication onset in cells reflects one such process, employing ATP-dependent initiation factors to coordinate replisome assembly1, 2. Replication initiators of eukaryotes and prokaryotes contain AAA+-family ATPase domains, whose activity is augmented by duplex DNA-binding domains (DBDs) and specialized protein-protein interaction elements that assist with origin recognition and recruit specific replication factors3, 4. AAA+ enzymes share a common structural core with RecA-type ATPases, together forming the Additional Strand Catalytic glutamatE (ASCE) supergroup of P-loop NTPases5; the molecular logic that allows a common nucleotidyl-hydrolase module to control the disparate activities of replication initiators, and ASCE proteins in general, is not understood.
In bacteria, replication initiation relies on the DnaA protein6–8. In Escherichia coli, multiple DnaA molecules bind to the replication origin, oriC, through several duplex DNA-binding sites, forming a large nucleoprotein complex in the presence of ATP9–11. With the aid of of appropriate architectural proteins (such as Integration Host Factor) and negatively-supercoiled DNA, this complex subsequently melts an AT-rich, DNA-unwinding element (DUE) located adjacent to the duplex DnaA binding sites12, 13. ATP also activates a secondary DNA-binding site within DnaA, postulated to reside within the AAA+ domain, which engages single-stranded regions of the DUE to form a stable open complex12, 14–16. DnaA then collaborates with the bacterial helicase loader (DnaC in E. coli), to recruit two hexamers of the DnaB helicase to the origin and promote replisome assembly17–19.
Although most AAA+ enzymes form closed-ring assemblies20, 21, structural studies have indicated that initiators and polymerase clamp-loaders form openring structures14, 22–24. Among initiator/loader systems, DnaA is particularly unusual in that it has been seen to oligomerize into a right-handed, spiral filament14. Two models have been proposed to explain how this structure might aid origin melting (Fig S1). In one, the wrapping of duplex DNA about a DnaA super-helix would constrain a positive supercoil, generating compensatory negative writhe that could aid opening of the neighboring DUE. In the other, the wrapped DnaA/DNA complex would serve as a nucleation center, allowing DnaA protomers to directly engage and melt the DUE, possibly through the initiator's ATPase elements. Thus far, experimental evidence has supported both models9, 14–16, 25, leaving open the question as to how DnaA catalyzes origin melting. The relationship of this mechanism to other initiation systems, or to AAA+/ASCE proteins overall, is also unclear.
A DnaA-ssDNA crystal structure
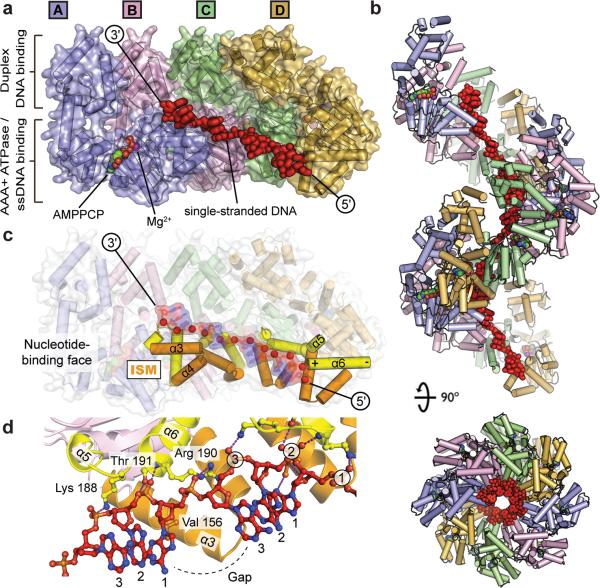
To examine these issues, we set out to determine the structure of DnaA bound to single-stranded DNA. Employing a truncation of Aquifex aeolicus DnaA consisting of the AAA+ and duplex-DNA-binding domains (which, like its E. coli counterpart16, 17, is active for both ATP-stimulated assembly and single-stranded-DNA (ssDNA) binding25), we first grew DNA-free crystals in the presence of Mg2+ and the non-hydrolyzable ATP mimic AMPPCP14. DNA substrates were then soaked into these crystals under low-salt conditions (Methods). Data collection and phasing by molecular replacement revealed four DnaA protomers per asymmetric unit, arranged in a spiral configuration that propagates into a continuous protein helix by the action of crystal-symmetry elements (Fig 1a, 1b), along with bound single-stranded DNA. Of the multiple substrates screened (Methods), dA12 yielded the highest-quality density (Fig S2a, S2b), and served as the best target for model building and refinement. The final structure, containing a DnaA:AMPPCP:Mg2+:dA12 stoichiometry of 4:4:4:1, was refined to an Rwork/Rfree of 24.9/26.8% at 3.35 Å resolution (Table S1).
Figure 1. The ATPase pore of assembled DnaA binds ssDNA.
a, Side view of the asymmetric unit, with DnaA subunits differentially colored. Single-stranded DNA is displayed as red sticks. AMPPCP and Mg2+, bound to chain A, are shown as spheres colored by element and in magenta, respectively; AMPPCP•Mg2+ bound to chains B-D are occluded in this view. b, Side and top views of oligomerized DnaA, reconstructed through crystal packing, showing twelve DnaA subunits and three strands of ssDNA. Coloring as per panel A. c, Side view of the DnaA tetramer with helices α3/α4 and α5/α6 highlighted in orange and yellow, respectively (“ISM” – initiator specific motif). Single-stranded DNA is shown as a transparent stick-and-surface representation colored by element; phosphates are further highlighted as red spheres. d, Protein-DNA contacts. Protein chains B (left) and C (right) are displayed with the same coloring as in c. Single-stranded DNA is colored by element.
DnaA-ssDNA interactions
The overall arrangement of DnaA subunits in the helical assembly is highly similar to a DNA-free form reported previously (0.7 Å r.m.s.d. between all Cα positions)14. AMPPCP•Mg2+ binds at the interface between neighboring subunits, with the γ-phosphate of AMPPCP coordinated by catalytic amino acids from pairs of adjoining AAA+ domains. Single-stranded DNA associates exclusively with the AAA+ elements of the initiator, with each protomer binding three nucleotides of the dA12 strand (Fig 1a). Almost all contacts are made through the phosphodiester backbone, exposing the DNA bases to solvent. Each trinucleotide segment adopts a B-form DNA conformation (Fig S3) with the bases between consecutive segments separated by large (~10 Å) gaps, extending the substrate by ~50% (Table S2; Supplemental Material).
DnaA binds single-stranded DNA using just two pairs of helices, α3/α4 and α5/α6, both of which line the central channel of the protein assembly (Fig 1c). The geometry of these two elements creates a single conduit along the length of the DnaA superhelix that allows substrate to traverse consecutive DnaA protomers. Interestingly, helices α3/α4 also comprise the Initiator Specific Motif (ISM), which both promotes filament formation14, 19 and distinguishes DnaA as a member of initiator clade of the AAA+ superfamily26, 27.
DnaA uses a simple network of interactions to coordinate ssDNA. The ISM forms a shelf for each trinucleotide, in which a conserved hydrophobic residue, Val156, forms van der Waal contacts with the sugar and base of the first nucleotide in the triplet (Fig 1d). The central phosphate of each trinucleotide is bound by the electropositive, N-terminal helix dipole of α6 and hydrogen bonded by Thr191 (Fig 1c, 1d). These contacts are flanked by two positively charged residues, Arg190 and Lys188, which make salt-bridge interactions with the phosphates of nucleotides 1 and 3, respectively. Importantly, mutant initiators containing substitutions in these observed DNA-binding residues show reduced affinity for ssDNA in solution (Fig S4) confirming that the crystals captured a physiologically-meaningful initiator state. Moreover, mutations of the same positions in E. coli DnaA (amino acids Arg245, Lys243 and Val211) also disrupt ssDNA binding and origin melting15. Thus, the single-stranded DNA engagement strategy seen here appears conserved across bacterial species.
Structural similarities between DnaA and RecA
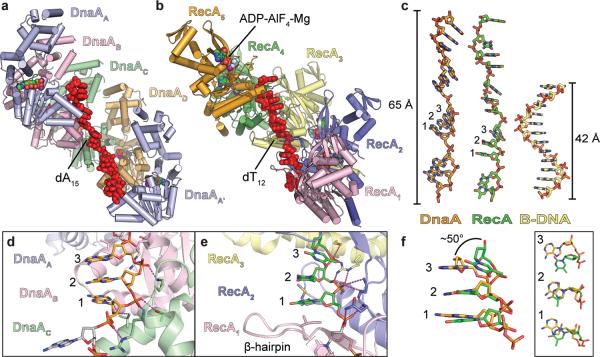
In considering the assembly patterns of oligomeric ATPases, we were struck by the similarity of DnaA to one system in particular: the homologous recombination protein, RecA. Although the cellular functions of these two proteins are fundamentally different (catalysis of DNA strand-exchange reactions versus replication origin melting and coordination of replisome assembly), both RecA and DnaA are predicated upon an ASCE ATPase fold27, 28. Like DnaA, RecA (and its Rad51/RadA orthologs) forms a helical assembly that engages DNA with its pore regions28–32. These shared physical properties led us to undertake a more detailed comparison of RecA and DnaA. Of the multiple models available, the structure of a RecA oligomer bound to single-stranded DNA33, representing the presynaptic complex formed during the initial stages of homologous recombination, is globally most similar to the DnaA state we observe (Fig 2a, 2b). As with DnaA, RecA contacts DNA almost exclusively through the phosphodiester backbone, which sits in the interior of a positively-charged filament pore. Each RecA protomer binds three nucleotides in a B-DNA conformation, with the base stacking between each triplet interrupted such that single-stranded DNA is extended ~1.5 fold compared to a B-form duplex (Fig 2c).
Figure 2. DnaA engages ssDNA in a manner similar to RecA.
a, View of a DnaA-AMPPCP-ssDNA pentamer (consisting of one full tetramer, as well as chain A (DnaAA') and its associated triplet from the adjacent asymmetric unit). AMPPCP•Mg2+ is shown as spheres colored by atom, ssDNA as red sticks. b, View of a RecA-ADP-AlF4-ssDNA pentamer (PDB ID 3CMW)33. ADP•AlF4•Mg2+ is shown as spheres colored by atom, ssDNA as red sticks. c, Comparison of ssDNA bound to DnaA (orange), RecA (green) and a strand of B-DNA (yellow). d, Close-up view of triplet bound to DnaA (chain C) with magenta dashed lines indicating key contacts. e, Close-up view of triplet bound to RecA (protomer 2) with magenta dashed lines indicating key contacts. f, Side (left) and top (right) views of the triplets displayed in d and e aligned with each other.
RecA and DnaA also exhibit some interesting and significant differences. A visual examination of each triplet shows that RecA uses a more extensive network of contacts for engaging single-stranded DNA than does DnaA (Fig 2d, 2e), burying twice as much surface area per triplet (318 Å2 and 639 Å2 for DnaA and RecA, respectively). This difference derives largely from an additional β-hairpin in RecA that fills the gap between each triplet and reinforces each three-base stack33. Moreover, while two of the three nucleotides within each RecA triplet (positions 1 and 2) align well with those seen in DnaA, position 3 of the DnaA trinucleotide rotates away from the pore axis by ~50° (Fig 2f). This difference skews consecutive DnaA triplets away from one another, disrupting the formation of a smoothly spiral arrangement as seen in RecA (Fig 2c).
DNA extension is ATP- and assembly-dependent
The ability of RecA to stretch DNA to the extent observed crystallographically has been amply substantiated by a wealth of methodologies34–36. Using these efforts as a guide, we set out to determine whether the DNA conformation we observe bound to DnaA accurately represents the state of the substrate in solution. To accomplish this, we employed a bulk-phase Fluorescence Resonance Energy Transfer (FRET)-based ssDNA extension assay analogous to single-molecule approaches applied to RecA37. Using a poly-thymine DNA labeled with Cy3 and Cy5 (FR-dT21) (Table S3), we monitored changes in the length of single-stranded DNA resulting from DnaA binding (Fig 3a). Analogous studies were performed with RecA as a control. As both RecA and DnaA require ATP for formation of the oligomers observed in the structural models, we expected ATP-dependent extension to lead to a loss of FRET signal. We tested for extension both in the presence of ADP•BeF3, to avoid complications that might arise from nucleotide hydrolysis, and in the presence of ADP, which is known to promote DnaA disassembly. Pronounced extension was observed only in the presence of the ATP analog (Fig 3b and 3c), and not in the presence of ADP. The lengths of single-stranded DNA in the ATP-assembled states of both proteins, as calculated from the FRET data, were in close agreement with those observed in the crystal structures (Table S6). Likewise, mutations in ssDNA-binding amino acids and residues required for DnaA assembly all significantly reduced ssDNA extension (Fig S6), demonstrating that this activity depends on substrate binding to the pore of an initiator oligomer that forms only when activated by ATP.
Figure 3. DnaA extends ssDNA in solution.
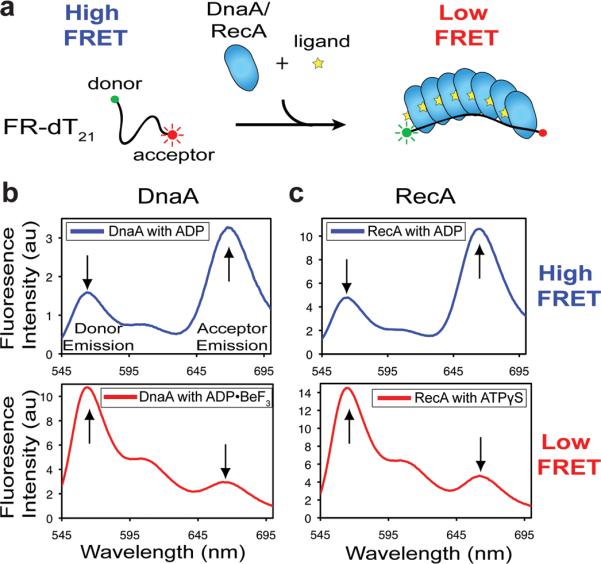
a, Cartoon of ssDNA extension assay. b, Emission scan (donor excitation) of FR-dT21 in the presence of 10 μM DnaA with either ADP•BeF3 (top) or ADP (bottom). c, Emission scan (donor excitation) of FR-dT21 in the presence of 10 μM RecA with either ATPγS (top) or ADP (bottom). Reported transfer efficiencies and distances were calculated using donor emission as described in the Methods.
DnaA directly catalyzes duplex melting
How replication origins are opened for replisome assembly is an important, unanswered question. Given the similarities between the ssDNA binding and extension activities of DnaA and RecA, we reasoned that the initiator might directly destabilize and disrupt DNA duplexes. This activity is a known property of RecA38, albeit one that permits the recombinase to actively exchange DNA strands between target substrates30, 33.
To test this idea, we developed a DNA strand-displacement assay for DnaA. First, the initiator was incubated with a short duplex containing one fluorescently-labeled strand. Unlabeled competitor strand was then added to capture any unwound species (Fig 4a). Both ADP and ADP•BeF3 were tested to determine whether initiator assembly affected the outcome of the experiment, as were DNAs of different lengths and stabilities. Analysis of the resultant products by gel electrophoresis shows that DnaA readily unwinds a 15mer duplex DNA of moderate stability (Tm=43°C) in the presence of the ATP mimic (Fig 4b). By contrast, increasing the stability of the DNA substrate by ~30% (using a 20mer, Tm=55°C) weakens the unwinding activity of DnaA (Fig 4b), whereas increasing DNA stability even further (30mer, Tm=62°C) abrogates melting completely (Fig S7a). Importantly, ADP did not support strand displacement, nor did ssDNA binding and DnaA assembly mutants (Fig S7b, S7c). These controls indicate that dsDNA melting is dependent not only upon formation of an assembled DnaA oligomer, but that the initiator is fine-tuned to specifically disrupt DNAs of modest stability.
Figure 4. DnaA directly melts duplex DNA.
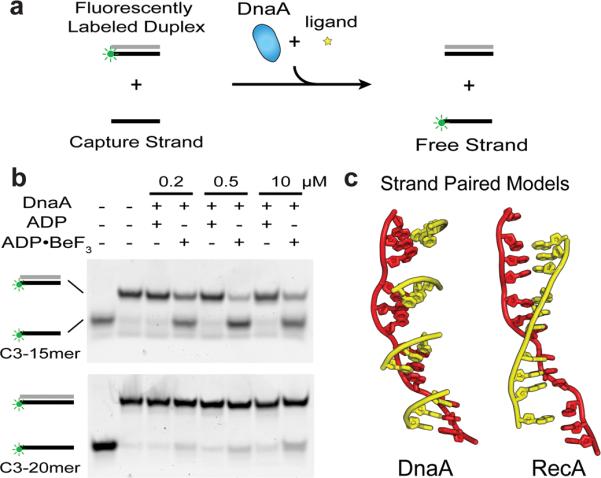
a, Schematic of strand displacement assay. The green circle represents the Cy3 fluorescent end-label used to follow the status of one DNA strand. Complementary strands of duplex substrates are colored grey and black. b, Strand displacement assay conducted with 15 and 20mer duplex substrates (C3–15mer and C3–20mer) in the presence and absence of different nucleotides. DnaA concentrations used are indicated above each lane. c, (left) Cartoon model showing how complementary base triplets (yellow) would pair (in a B-DNA manner) with ssDNA bound to DnaA (red). The orientation of successive DnaA-bound triplets is such that it prevents the formation of a continuous base-paired strand favoring duplex separation. (right) Same DNA view, but as seen in RecA, where triplets are oriented to allow pairing of an extended complementary strand to promote duplex formation and strand exchange (PDB ID 3CMX)33.
One significant functional difference between RecA and DnaA is that the recombination protein can drive a true strand-exchange reaction; that is, in addition to displacing one strand of a duplex, RecA can also pair homologous single-stranded DNA segments into a double-stranded molecule. By contrast, DnaA's function is to separate double-stranded origin regions. Inspection of the RecA and DnaA complexes reveals a physical basis for these differing properties: in DnaA, successive trinucleotide elements are arranged in a state incompatible with the formation of a continuous duplex, whereas ssDNA bound to RecA adopts a smoothly spiraled arrangement permitting the contiguous pairing of a complementary strand (Fig 4c). This distinction arises primarily from the 50° rotation between the nucleotides at the third position of each triplet seen in the RecA and DnaA models (Fig 2f). In DnaA, the orientation of this nucleotide appears to be stabilized by base stacking, whereas in RecA the β-hairpin insertion helps sculpt the configuration of the DNA to create a contiguous base-pairing surface.
Implications for origin melting
Together, our findings present the strongest evidence yet that DnaA melts replication origins by directly assisting with the separation and sequestration of duplex DNA strands (Fig S1c). Notably, this activity does not contradict the demonstrated need for other factors capable of reshaping and/or destabilizing DNA (e.g., IHF and negative supercoiling) during initiation12, 13. Rather, these elements likely help promote DnaA assembly and prime the origin for melting by what otherwise would be an inefficient unwindase. In this view, the AAA+ domains of DnaA may first engage only one of the two strands of duplex DNA with their ssDNA binding elements (possibly at reported ssDNA or ATP-DnaA binding sites15, 16). In the presence of ATP, which triggers initiator assembly, subunit/subunit interactions would help restructure the DNA backbone, stretching the contacted strand to facilitate melting. Reannealing would be disfavored by the non-contiguous arrangement of base triplets in the extended state (Fig 2d). Future studies will be needed to define the specific order and effect of these events further.
We envision that the propensity of DnaA to open DNA could be adjusted in other bacterial species by strengthening or weakening the association of its ATPase domains with DNA and/or each other. An attractive feature of such a mechanism is that it is amenable to additional layers of control by changes to DUE sequence, superhelical density, and co-resident architectural factors to ensure that a replication origin fires only when DnaA is both present and assembled properly. Such flexibility may have played a role in allowing DnaA to persist as the primary initiator in bacteria that have adapted to markedly different environmental niches.
Thematic patterns of substrate recognition in ASCE ATPases
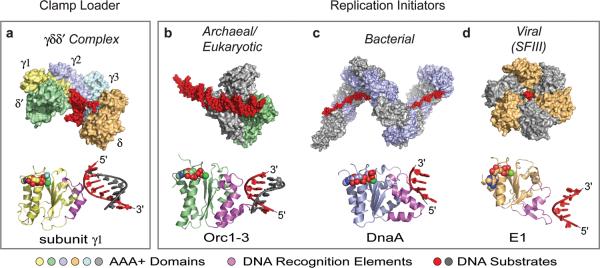
The mechanism by which DnaA coordinates ssDNA also comports well with findings in other replication initiation systems and with ASCE ATPases in general. For instance, many oligomeric RecA and AAA+ enzymes bind substrate in the interior pore of a closed- or cracked-ring particle33, 39–41. DnaA follows this pattern. A comparison of DnaA to other, disparate nucleic acid-dependent AAA+ systems – e.g., polymerase clamp loaders and processive helicases – further shows that these factors also associate with client substrates in a remarkably analogous manner, using the same face of the core αβα ATP-binding fold to engage a short backbone stretch of their target DNAs (Fig 5). For AAA+ proteins involved in initiation, these similar contact mechanisms have been differentially co-opted to assist with specific protein functions, ranging from the control of origin recognition (as seen in archaeal Orc1 proteins26, 42), to mediating processive DNA unwinding (viral superfamily 3 helicases40, 43). DnaA, with its ability to melt (but not translocate along) DNA, appears to employ an intriguing mix of some of the activities exhibited by related initiation systems. Future efforts will be needed to determine how subtle differences in the position and nature of substrate-binding surfaces, combined with specific alterations in the assembly patterns of central AAA+ domains, endow such molecular motors and switches with their distinct biochemical properties.
Figure 5. Common DNA recognition strategies of AAA+ proteins.
Structures of DNA-bound assemblies (top) and individual domains (bottom) for AAA+ proteins involved in replication. All recognize DNA using the same face of the AAA+ fold (violet) (bottom). a, Bacterial clamp-loader (γδδ′) complex (AAA+ domains – differentially colored) bound to primer-template DNA (PDB ID 3GLF)41. b, Archaeal initiators Orc1–1 (gray) and Orc1–3 (AAA+ domain - green) bound to origin DNA (PDB ID 2QBY)26. c, Bacterial initiator DnaA (AAA+ domains – gray/blue) bound to ssDNA. d, Viral initiator/helicase E1 (AAA+ domains – orange/gray) bound to ssDNA (PDB ID 2GXA)40. For all panels, DNA is shown as either red spheres (top), or as a red/grey cartoon (bottom).
Nucleotide co-factors bound to AAA+ domains (bottom) are represented as spheres colored by atom.
Methods Summary
Detailed information regarding experimental methods, substrate sequences, binding constants, and FRET efficiencies and distances can be found in the Online Methods section and in the Supplementary Material.
Supplementary Material
Acknowledgements
We would like to thank Karl Drlica, James Keck, Thomas Murray and the Berger laboratory for helpful comments, and Michael M. Cox for his generous contribution of RecA protein. This work was supported by the NIGMS (GM071747) and the National Institute of Health Molecular Biophysics Training Grant T32 GM008295.
Footnotes
Author Information Coordinates have been deposited in the RSCB PDB under the accession number 3R8F.
Author Contributions K.E.D. and J.M.B. designed the experiments, analyzed the data and wrote the paper. Protein purification, crystallization and ssDNA binding assays were performed by K. C. and K.E.D. K.E.D. performed the other experiments.
References
- 1.Kornberg A, Baker TA. DNA Replication. W.H. Freeman and Company; New York: 1992. [Google Scholar]
- 2.Stillman B. Origin recognition and the chromosome cycle. FEBS Lett. 2005;579:877–884. doi: 10.1016/j.febslet.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Duderstadt KE, Berger JM. AAA+ ATPases in the initiation of DNA replication. Crit. Rev. Biochem. Mol. Biol. 2008;43:163–187. doi: 10.1080/10409230802058296. [DOI] [PubMed] [Google Scholar]
- 4.Lee DG, Bell SP. ATPase switches controlling DNA replication initiation. Curr. Opin. Cell Biol. 2000;12:280–285. doi: 10.1016/s0955-0674(00)00089-2. [DOI] [PubMed] [Google Scholar]
- 5.Leipe DD, Koonin EV, Aravind L. Evolution and classification of P-loop kinases and related proteins. J. Mol. Biol. 2003;333:781–815. doi: 10.1016/j.jmb.2003.08.040. [DOI] [PubMed] [Google Scholar]
- 6.Katayama T, Ozaki S, Keyamura K, Fujimitsu K. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 2010;8:163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- 7.Kaguni JM. DnaA: controlling the initiation of bacterial DNA replication and more. Annu. Rev. Microbiol. 2006;60:351–375. doi: 10.1146/annurev.micro.60.080805.142111. [DOI] [PubMed] [Google Scholar]
- 8.Leonard AC, Grimwade JE. Regulating DnaA complex assembly: it is time to fill the gaps. Curr. Opin. Microbiol. 2010 doi: 10.1016/j.mib.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuller RS, Funnell BE, Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984;38:889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- 10.Funnell BE, Baker TA, Kornberg A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J. Biol. Chem. 1987;262:10327–10334. [PubMed] [Google Scholar]
- 11.Crooke E, Thresher R, Hwang DS, Griffith J, Kornberg A. Replicatively active complexes of DnaA protein and the Escherichia coli chromosomal origin observed in the electron microscope. J. Mol. Biol. 1993;233:16–24. doi: 10.1006/jmbi.1993.1481. [DOI] [PubMed] [Google Scholar]
- 12.Bramhill D, Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988;52:743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- 13.Kowalski D, Eddy MJ. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 1989;8:4335–4344. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- 15.Ozaki S, et al. A Common Mechanism for the ATP-DnaA-dependent Formation of Open Complexes at the Replication Origin. J. Biol. Chem. 2008;283:8351–8362. doi: 10.1074/jbc.M708684200. [DOI] [PubMed] [Google Scholar]
- 16.Speck C, Messer W. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 2001;20:1469–1476. doi: 10.1093/emboj/20.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sutton MD, Carr KM, Vicente M, Kaguni JM. Escherichia coli DnaA protein. The N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J. Biol. Chem. 1998;273:34255–34262. doi: 10.1074/jbc.273.51.34255. [DOI] [PubMed] [Google Scholar]
- 18.Fang L, Davey MJ, O'Donnell M. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol. Cell. 1999;4:541–553. doi: 10.1016/s1097-2765(00)80205-1. [DOI] [PubMed] [Google Scholar]
- 19.Mott ML, Erzberger JP, Coons MM, Berger JM. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell. 2008;135:623–634. doi: 10.1016/j.cell.2008.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 21.Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure--diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 22.Jeruzalmi D, O'Donnell M, Kuriyan J. Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III. Cell. 2001;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- 23.Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat. Struct. Mol. Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarey MG, et al. Nucleotide-dependent conformational changes in the DnaA-like core of the origin recognition complex. Nat. Struct. Mol. Biol. 2006;13:684–690. doi: 10.1038/nsmb1121. [DOI] [PubMed] [Google Scholar]
- 25.Duderstadt KE, et al. Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator. J. Biol. Chem. 2010;285:28229–28239. doi: 10.1074/jbc.M110.147975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dueber EL, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317:1210–1213. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- 27.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 28.Story RM, Weber IT, Steitz TA. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 29.Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 30.Cox MM. Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell Biol. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- 31.Egelman E. A common structural core in proteins active in DNA recombination and replication. Trends Biochem. Sci. 2000;25:179–182. doi: 10.1016/s0968-0004(00)01555-8. [DOI] [PubMed] [Google Scholar]
- 32.Conway AB, et al. Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 33.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–484. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 34.Stasiak A, Di Capua E. The helicity of DNA in complexes with recA protein. Nature. 1982;299:185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- 35.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 36.Nishinaka T, Ito Y, Yokoyama S, Shibata T. An extended DNA structure through deoxyribose-base stacking induced by RecA protein. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6623–6628. doi: 10.1073/pnas.94.13.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joo C, et al. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 38.Bianchi M, Riboli B, Magni G. E. coli recA protein possesses a strand separating activity on short duplex DNAs. EMBO J. 1985;4:3025–3030. doi: 10.1002/j.1460-2075.1985.tb04039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 41.Simonetta KR, et al. The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell. 2009;137:659–671. doi: 10.1016/j.cell.2009.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural basis of DNA replication origin recognition by an ORC protein. Science. 2007;317:1213–1216. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- 43.Liu X, Schuck S, Stenlund A. Adjacent residues in the E1 initiator beta-hairpin define different roles of the beta-hairpin in Ori melting, helicase loading, and helicase activity. Mol. Cell. 2007;25:825–837. doi: 10.1016/j.molcel.2007.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.