Abstract
Conditional mutations and transcription-based reporters are important new tools for exploring the dynamic functions of biological pathways in vivo. While studying the role of the Wnt signaling pathway in cartilage we observed that the β-catenin dependent reporter TOPGAL was expressed in chondrocytes in which β-catenin was conditionally inactivated using a Col2a1::cre driver. Here we show that in these embryos recombination is complete and full-length β-catenin protein is absent in chondrocytes. Although a null allele in this context, the recombined β-catenin locus produces a stable transcript that encodes a truncated protein. The truncated protein alone fails to activate TOPFLASH, but strongly potentiates reporter activity in the presence of expressed β-catenin or Tcf4. Together, these data show that each mouse model exhibits specific undesirable properties, findings that strongly suggest the need for specific standards to ensure proper validation of this new generation of genetic tools.
Keywords: TOPGAL, β-catenin, Wnt signaling, mouse, conditional mutant
Introduction
Wnt signaling plays an integral role in many developmental processes, including tissue specification, cell polarity, morphology, and proliferation (as reviewed in (Moon et al., 2002)). In the canonical pathway, Wnt ligands bind to Frizzled receptors that activate the intracellular signaling mediator Dishevelled (Dvl). Dvl activity inhibits the Axin/GSK3/APC complex, which normally phosphorylates, and thereby targets, β-catenin for degradation. In the absence of GSK3β activity, β-catenin is stabilized and translocates to the nucleus where it regulates transcription of target genes in combination with the cofactors of the T-cell factor/lymphoid enhancer-binding factor1 (TCF/LEF1) gene families.
Analysis of cell signaling in vivo has benefited from the development of tools that permit the visualization or manipulation of signaling pathways in specific cell or tissue types. One common tool is the TOPFLASH luciferase reporter system for measuring β-catenin dependent, canonical Wnt signaling. TOPFLASH is composed of three multimerized consensus TCF/LEF binding motifs upstream of the firefly luciferase gene (Korinek et al., 1997). Induction of luciferase activity from this reporter cassette is strongly dependent on the presence of both β-catenin and Tcf (Korinek et al., 1997). Thus, TOPFLASH is a faithful reporter of β-catenin activity in vitro.
The transgenic reporter mouse line TOPGAL, which was modeled after the TOPFLASH reporter system, is composed of three TCF/LEF binding sites that regulate a minimal c-fos promoter upstream of lacZ, which encodes β-galactosidase (DasGupta and Fuchs, 1999). Tests of the reporter cassette in cell culture showed a dependence on β-catenin and Lef1 similar to TOPFLASH. TOPGAL transgenic mice expressed β-galactosidase activity in a subset of skin cells and in de novo hair germs in response to constitutively active β-catenin. By contrast, β-galactosidase was not detected in the epidermis of non-transgenic mice (DasGupta and Fuchs, 1999). Altogether, these results suggested that the TOPGAL accurately reports β-catenin activity in vivo.
A second important tool for studies of cell signaling is the conditional allele that provides the spatial and temporal control of genetic manipulations necessary to study molecules with pleiotropic activities. A conditional allele of Ctnnb1, which encodes β-catenin, has been extensively employed to study canonical Wnt signaling (Brault et al., 2001). This was an important addition to the Wnt signaling toolkit because β-catenin knockout mice die during gastrulation (Haegel et al., 1995). In this mouse line, Cre mediated recombination removes exons 2–6, eliminating the initiator methionine and thereby generates a presumed null allele.
In our analyses of growth plate cartilage development using TOPGAL transgenic mice and Ctnnb1 conditional mutant mice, several issues arose that led us to question the functions of these alleles (Ahrens et al., 2009; Li and Dudley, 2009). In particular, TOPGAL shows patchy activation in the growth plate cartilage and the level of signal does not vary between regions that express distinct subsets of Wnt ligands (Yang et al., 2003). Moreover, TOPGAL activity is strong in the Piga mouse model, which lacks glycosylphosphatidylinositol (GPI)-anchored cell surface proteins (Ahrens et al., 2009). This is important because in Drosophila, loss of the GPI-anchored glypicans Dally and Dally-like downregulated canonical Wnt signaling and phenocopied a null mutation in wingless, which encodes a ligand of Frizzled receptors (Han et al., 2005). Proper interpretation of our results required confidence in the function of these mouse genetic tools.
Here we show that TOPGAL signal is not only present in Ctnnb1 null growth plate cartilage, but that the signal strength is comparable to the transgene in a wild type background. Extensive tests using PCR, Western blot analysis, and immunofluorescence methods demonstrate that conditional inactivation is efficient and that the growth plate cartilage is null for Ctnnb1. However, the recombined Ctnnb1flox/flox locus generates a stable truncated transcript that corresponds to the predicted Cre-mediated excision event. In cell culture, expression of the truncated sequence generates stable proteins that alone fail to promote transcriptional activation from TCF/LEF1 binding sites. In contrast, these mutant proteins potentiate TOPFLASH reporter activation in combination with full-length β-catenin or Tcf-4 and therefore could produce a gain of function in certain contexts. Together, these results suggest the need to more thoroughly validate new genetic tools to reduce the possibility of producing off-target effects.
Results
TOPGAL in a β-catenin loss of function background
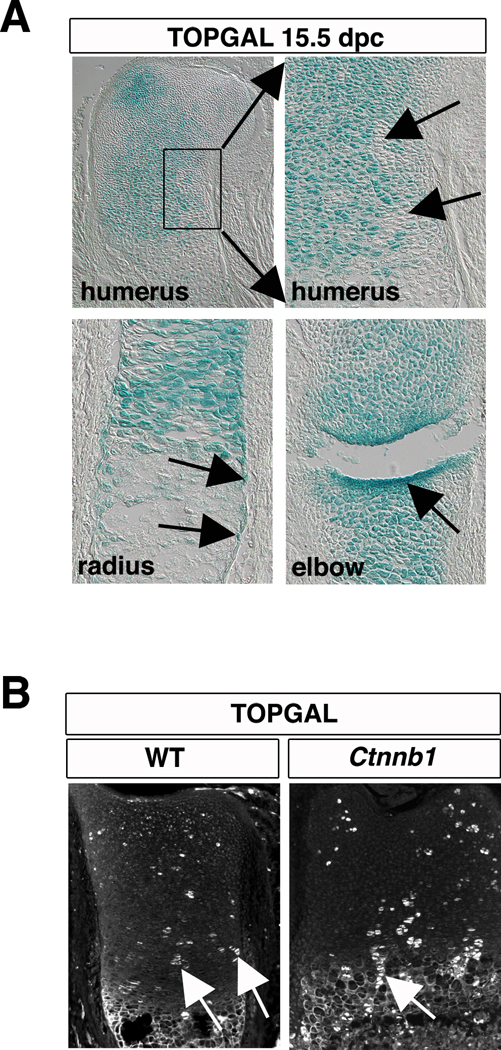
TOPGAL is a commonly used in vivo transgenic reporter for canonical Wnt signaling. In the developing skeleton, TOPGAL activity is readily detected in chondrocytes, in cells of the perichondrium (the presumptive osteoblast progenitors), and in the forming joints (Fig 1A) (Day et al., 2005). The higher level of signal in the developing joints is consistent with genetic experiments showing a requirement for Wnt signaling in the formation and maintenance of joints (Spater et al., 2006). However, the apparent uniformity of signal, suggestive of a fixed level of β-catenin activity across the cartilage, was surprising given the known expression domains of canonical and noncanonical Wnt ligands in the growth plate (Hartmann and Tabin, 2001; Yang et al., 2003). Moreover, the patchy expression of TOPGAL in each zone of the growth plate cartilage suggests that Wnt signaling is not the only mechanism that regulates transgene function (Ahrens et al., 2009).
Figure 1. TOPGAL is expressed in β-catenin mutant chondrocytes.
(A) Sections of cartilage from TOPGAL embryos at 15.5 dpc. display broad β-galactosidase activity in growth plate chondrocytes (top, right). A magnified image of the boxed region shows clusters of chondrocytes do not express Topgal. Topgal is also expressed in the perichondrium and in the joints. (B) In sections of growth plate cartilage from TOPGAL transgenics (P0) that are additionally either wild type or null for Ctnnb1 (TOPGAL; Ctnnb1flox/flox; Col2a1cre), immunofluorescence methods detect β-galactosidase in patches of chondrocytes in each maturation zone. Examples of columnar clones are highlighted with a white arrow. RZ, resting zone; PZ, proliferative zone; PHZ, prehypertrophic zone; HZ, hypertrophic zone. Scale bar equals 200 µm in all panels.
To ask whether the TOPGAL reporter allele is sensitive to endogenous levels of β-catenin activity, we generated embryos mutant for Ctnnb1 that carry the TOPGAL transgene by crossing females heterozygous for TOPGAL and homozygous for the conditional floxed β-catenin (TOPGAL; Ctnnb1flox/Ctnnb1flox) to males carrying Ctnnb1flox, TOPGAL, and the cartilage specific cre recombinase line Col2a1::cre (TOPGAL; cre; Ctnnb1flox/+). All analyses were performed on 18.5 days post coitum (dpc) embryos because mutants die upon birth. To our surprise, we found that β-galactosidase is robustly expressed in both wild type and mutant cartilage (Fig 1B); consistent with the possibility that activation of TOPGAL does not require β-catenin activity.
Conditional knockout mice lack full-length β-catenin
The above conclusion is dependent on demonstrating that Cre-mediated recombination is efficient in cartilage. We tested this prediction using five experimental approaches. First, we isolated DNA from growth plate cartilage to determine the level of genomic recombination. Analysis of genomic DNA isolated from growth plate chondrocytes revealed the presence of a robust band for the recombined (flox delete) allele in mutants (Fig 2A, 2B). Second, RT-PCR analysis of growth plate derived cDNA detected the presence of a truncated transcript, but not the full-length wild type transcript, in mutant mice (Fig 2C). Sequencing of RT-PCR generated amplicons demonstrated that the truncated transcript lacks exons 2 through 6, as predicted from the locations of the loxP sites in the engineered Ctnnb1 locus (Fig 2A, (Brault et al., 2001)). Third, Western blot analysis with an anti-β-catenin antibody did not detect either full-length or truncated β-catenin protein in mutant cartilage (Fig 2D and data not shown). Fourth, we mated the Col2a1::cre driver males used to generate Ctnnb1 mutants to homozygous Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J females (tdTomato), in which Cre recombinase converts chondrocytes from constitutive expression of tdTomato (a red fluorescent protein) to constitutive expression of green fluorescent protein (GFP). Recombination is observed in essentially 100% of chondrocytes by 15.5 dpc, a full three days before the mutant analysis is performed (Fig 3A). Fifth, we performed double immunofluorescence detection of β-catenin and β-galactosidase in wild type and mutant cartilage. In chondrocytes, as in epithelial cells, β-catenin is predominately localized to the cell membrane (Fig 3B, C). Diffuse cytoplasmic signal is equivalent to background and therefore is not indicative of β-catenin activity (data not shown). β-catenin signal was not detected at the plasma membrane of Ctnnb1 mutant chondrocytes, although these cells display robust expression of β-galactosidase, and β-catenin was strongly expressed in the unaffected surrounding epithelium (Fig 3B,C). Together, these data demonstrate that, within the limits of detection of the various analytical tools used, Col2a1::cre mediated recombination generates Ctnnb1 null mutant cartilage and that β-catenin activity is not essential for TOPGAL expression in chondrocytes.
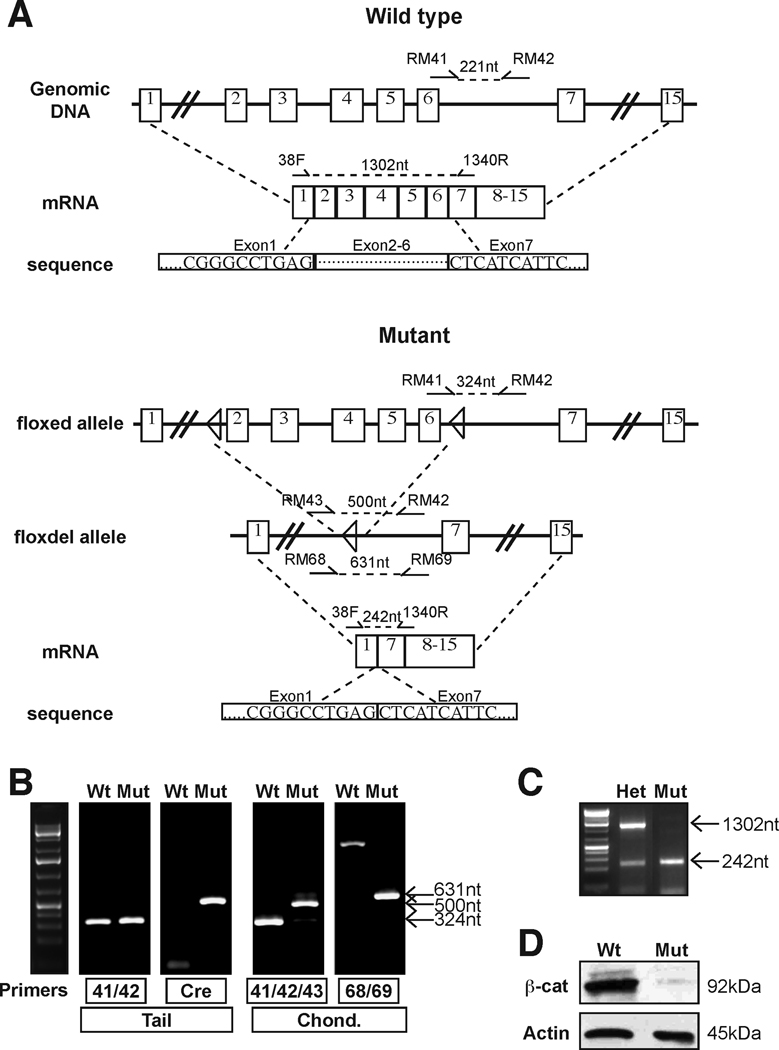
Figure 2. Mutant chondrocytes lack full-length β-catenin.
(A) Schematic detailing the genomic DNA and resulting mRNA for wild type and Ctnnb1 mutant loci (based on (Brault et al., 2001)). Numbered boxes represent exons and triangles show the loxP recombination sites. Primers for genotyping (RM41, RM42, RM43, RM68, RM69) and for RT-PCR (38F and 1340R) are noted along with the lengths of the predicted amplicons. Partial DNA sequences of predicted transcripts are shown. (B) Genotyping of tail DNA identified Ctnnb1flox/flox (wild type, Wt; no Cre) and Ctnnb1flox/flox; cre (mutant, Mut) specimen. Analysis of DNA from isolated growth plate chondrocytes of these specimens revealed only amplicons from the recombined locus (500 and 631nt bands) in Mut but not in Wt. In the Wt mice RM 43/42 and RM68/69 are sufficiently far apart to prevent amplification. (C) RT-PCR analysis of chondrocyte cDNA from a heterozygote (Ctnnb1flox/+; cre) reveals both the full-length β-catenin (1302nt) and a truncated (242nt) transcript, whereas mutant (Ctnnb1flox/flox; cre) chondrocytes express only the truncated transcript. (D) By Western blot analysis, wild type chondrocytes (Ctnnb1flox/+ or Ctnnb1+/+; cre+/−) express high levels of full-length β-catenin (92kDa), which is lacking in mutant chondrocytes.
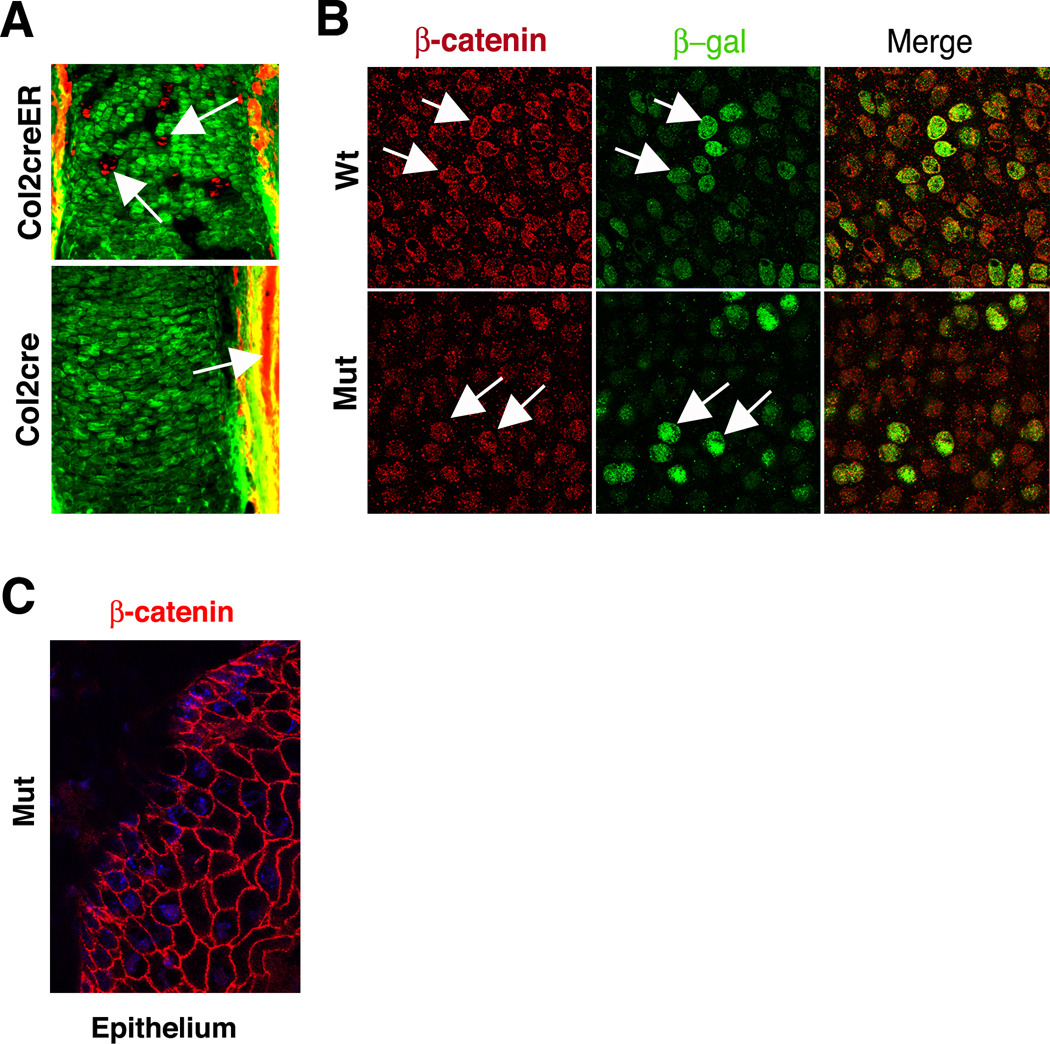
Figure 3. Efficient recombination results in the absence of β-catenin in cartilage.
(A) Cre-mediated recombination is efficient in cartilage. (Top) Epifluorescence illumination of tissue sections from embryos exposed to tamoxifen shows that the reporter locus can readily distinguish chondrocytes in which recombination occurred (green) from those in which recombination is absent (red, arrows). (Bottom) The Col2a1cre line used to generate Ctnnb1 mutants is highly efficient at promoting recombination of the reporter locus, as evidenced by the predominance of green fluorescence and the lack of red fluorescence. As expected, cells in the surrounding soft tissue lack recombination (red, arrows). (B) Immunofluorescence detection in sections of wild type cartilage demonstrates strong plasma membrane localization of β-catenin (arrows). Only some cells that express β-catenin show increased TOPGAL activity (green), as determined using an anti-β-galactosidase antibody. By contrast, Ctnnb1 mutant cartilage shows only background levels of cytoplasmic anti-β-catenin signal and not cortical localization, but retains strong TOPGAL transcription (arrows). (C) Robust expression of β-catenin (red) at the plasma membrane is found in the epithelium surrounding Ctnnb1 mutant cartilage. The nucleus is stained with DAPI (blue).
Cartilage lacking β-catenin has grossly normal architecture and maturation profiles
To test for potential effects of genetic background, we next asked if the phenotype of this Ctnnb1 mutant was consistent with previous publications (Akiyama et al., 2004; Hill et al., 2005). Histological staining of the cellular architecture revealed that the tissue is grossly normal (Fig 4A) and in situ hybridization demonstrated that mutant chondrocytes undergo normal phases of maturation (Fig 4B). However, the cell density was reduced in mutant cartilage, consistent with known roles for β-catenin in promoting cell proliferation (Fig 4A) (Dickinson et al., 1994; Olson and Papkoff, 1994). In addition, the least mature chondrocytes comprised a greater proportion of the growth plate in mutant than in wild type (Fig 4C), suggesting a delay in the initiation of chondrocyte maturation. Moreover, hypertrophic chondrocytes and other undefined cell types persist in the presumptive primary ossification center, consistent with a delay in the final steps of chondrocyte maturation. These hypertrophic chondrocytes might explain in part the increased number of apoptotic cells in mutant cartilage as shown by immunofluorescence for cleaved caspase-3 (Fig 4B). Collectively, these data demonstrate that β-catenin activity has primary roles in regulating cell proliferation and the terminal maturation of chondrocytes as previously described.
Figure 4. β-catenin mutant growth plates have normal tissue architecture and delayed maturation.
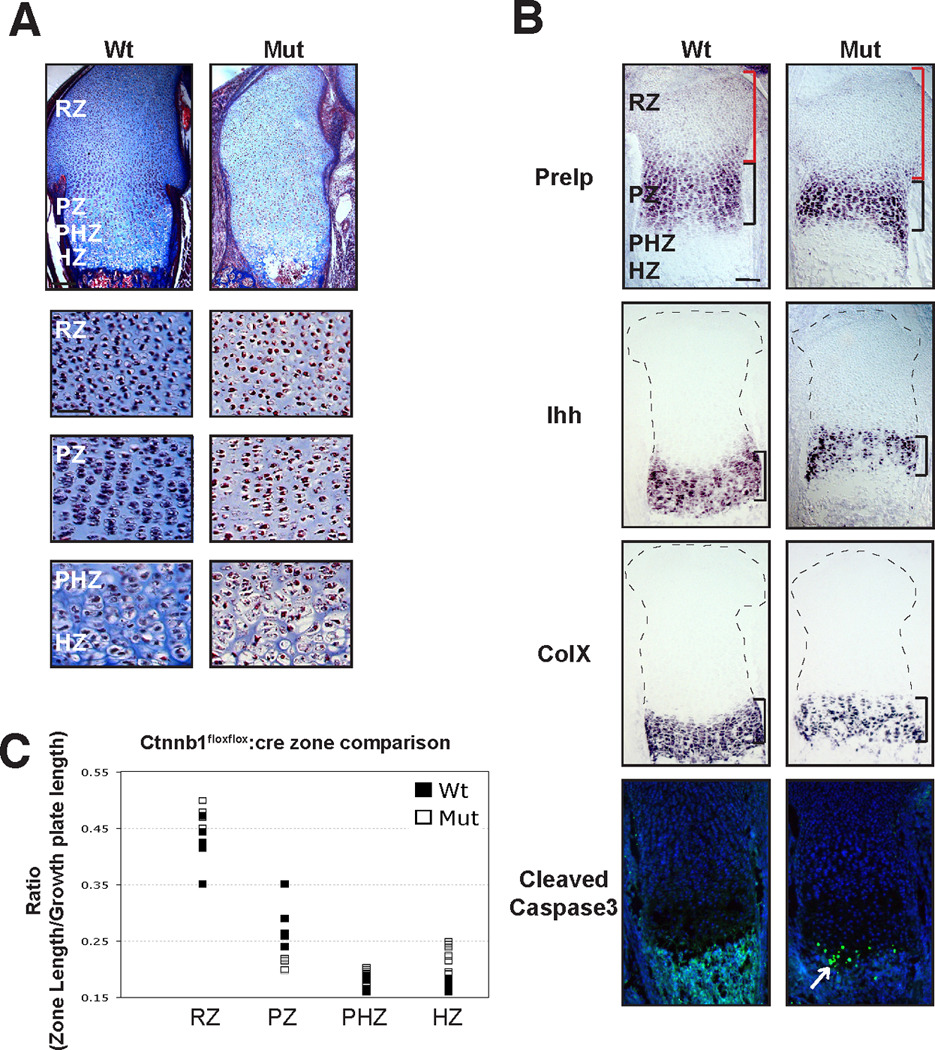
Sections of 18.5 dpc distal femur were stained with Masson’s Trichrome (A) or processed for in situ hybridization (B; Prelp, Ihh, ColX) or for immunofluorescence (B; cleaved caspase 3). (A). β-catenin mutant cartilage shows similar cellular architecture to wild type in all maturation zones. Differences in chondrocyte density between wild type and mutant samples were noted. (B) Mutant chondrocytes transit through normal phases of maturation indicated by expression of Prelp (proliferative zone), Ihh (prehypertrophic zone), and ColX (hypertrophic zone). Cleaved caspase 3 antibody staining shows an increase in apoptotic cells in hypertrophic chondrocytes of β-catenin mutant growth plates. (C) In β-catenin mutant growth plates, the resting zone is significantly longer (measurement denoted with red bracket) and the prehypertrophic and hypertrophic zones are slightly longer than in wild type embryos. In contrast, the proliferative zone is shorter in mutants than in wild type embryos (n=5). Data are reported as a ratio of the zone length to the total growth plate length. Scale bar equals 200 µm in low magnification panels in (A) and all panels in (B). Scale bar equals 100 µm in high magnification panels in (A).
Truncated β-catenin can enhance reporter activity
Even though full-length β-catenin protein was not detected by Western blot, the presence of a truncated transcript in mutant cartilage (Fig 2C) suggested the possibility of a gain of Ctnnb1 function. We first asked if the mutant transcript could generate a stable protein product. Western blot analysis of lysates from 293T cells expressing the flox delete transcript with or without a heterologous Kozak sequence revealed the presence of a truncated protein, suggesting that translation can be initiated at an internal ATG codon (Fig 5A). Next, we used a transient reporter assay to determine if expression of the truncated transcript could promote transcriptional activation of the β-catenin dependent firefly luciferase reporter Super 8xTOPFLASH (TOPFLASH) or the negative control reporter Super 8xFOPFLASH (FOPFLASH) that contains mutated TCF/LEF binding sites (Fig 5B). In all conditions, signals generated from FOPFLASH were never greater than those produced from TOPFLASH stimulated by GFP, demonstrating the dependence of the response on TCF/LEF binding to the reporter cassette (data not shown). Expression of full-length β-catenin or Tcf-4 promoted activation of TOPFLASH. In contrast, TOPFLASH was not activated by the truncated cDNA, Lef1, or dominant negative Tcf-4 alone. However, the truncated molecule (with or without the Kozak sequence) significantly increased TOPFLASH activity when co-transfected with full-length β-catenin or Tcf-4 (Fig 5B and data not shown). TOPFLASH activity required Tcf function in all conditions. Thus, the mutant allele potentially encodes a molecule that functions in the β-catenin/TCF pathway.
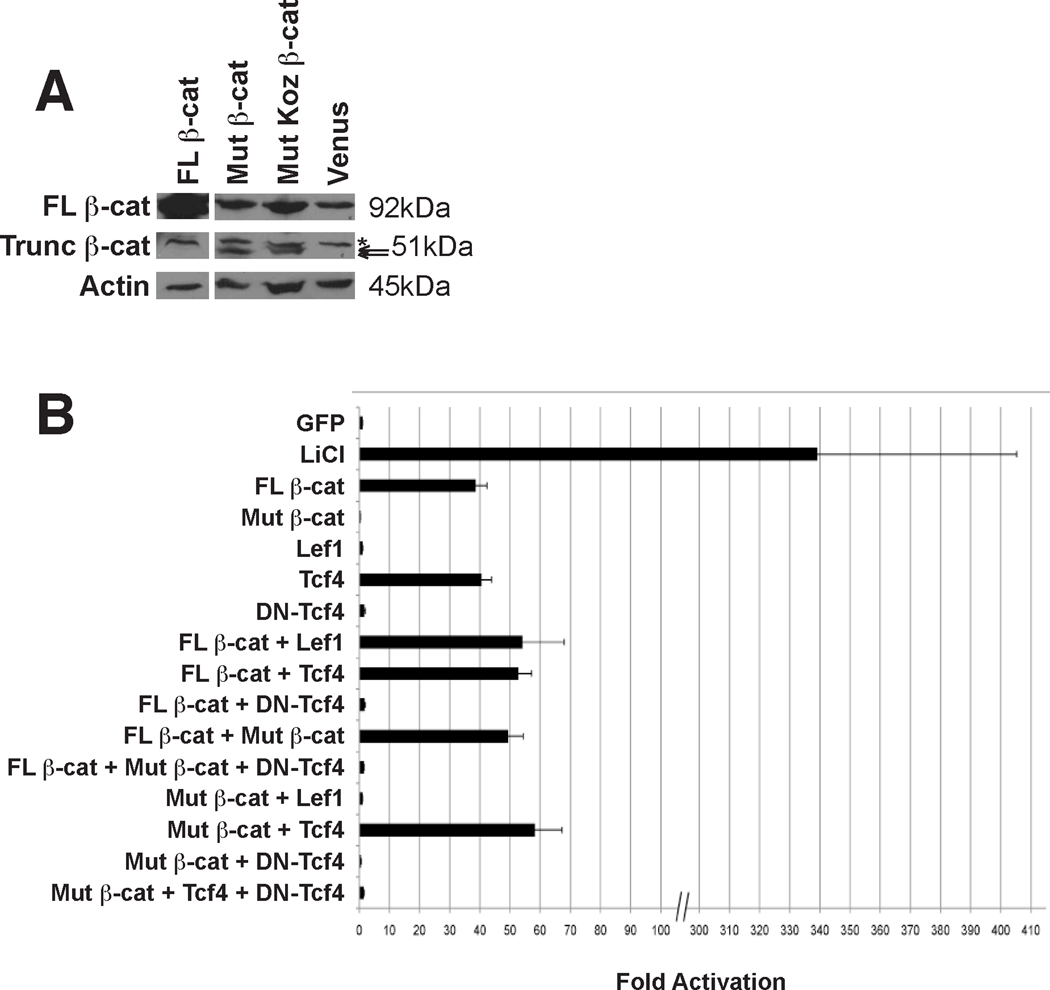
Figure 5. Truncated β-catenin can enhance reporter activity.
(A) Western blot analysis of 293T cell lysates using an anti-β-catenin antibody detects full-length β-catenin (FL β-cat, 92kDa) as well as a smaller protein in cells transfected with mutant β-catenin (Trunc β-cat) that matches the predicted size for a message with (51.6 kDa) or without (49.7 kDa, predicted size based on initiation using the first methionine, aa328, in exon7) a heterologous Kozak sequence. A weak background band at >51kDa is present in all lysates (asterisk). (B) Cultures of 293T cells were transfected with TOPFLASH, CMV-Renilla, and expression vectors for the indicated proteins. Fold activation was calculated as a ratio of signals [firefly luciferase/Renilla luciferase] for the experimental samples normalized by the ratio obtained for the GFP control. Error bars report the confidence intervals for each experimental condition. Mut β-catenin (exons 7 – 15) does not have activity alone, but confers greater activity when combined with FL β-catenin. Co-transfection of FL β-catenin with either Lef1 or Tcf4 increases activity, and the combination of Mut β-catenin with Tcf4, but not Lef1, confers greater activity. Dominant negative (DN)-Tcf4 suppresses activity in all combinations. Treatment with the GSK3β inhibitor LiCl shows maximal stimulation of the reporter in 293T cells.
Discussion
Ctnnb1flox/flox produces a null mutation when recombined in cartilage
Cre-mediated recombination of the conditional Ctnnb1flox/flox allele deletes exons 2–6 and generates a presumed null mutation by virtue of removing the initiator methionine encoded by exon 2, although this assumption was never explicitly tested (Brault et al., 2001). Here we show that cartilage-specific recombination of the Ctnnb1flox/flox allele results in the efficient deletion of exons 2–6 and the loss of full-length β-catenin protein.
The presence of a stable transcript from the recombined locus posed a concern due to the potential to generate a protein product via translation initiation from a methionine codon located downstream of exon 6 or in an alternative reading frame. Expression of this mutant transcript with or without an in-frame Kozak sequence to initiate translation generated stable protein as determined by Western blot analysis. However, expression of these truncated transcripts did not activate the TOPFLASH reporter, demonstrating that the protein products alone are not activators of β-catenin dependent transcription.
By contrast, co-transfection of the mutant construct with full-length β-catenin or Tcf-4 potentiated reporter activation. The fact that the endogenous truncated transcript encodes a protein that has a lower apparent molecular mass than the truncated transcript containing a 5’ heterologous Kozak sequence strongly suggests that translation initiates from an in-frame methionine (e.g. M328 in exon 7) downstream of exon 6 and thus the relevant functional domain is in the C-terminus of β-catenin. Initiation at M328 would result in a molecule that lacks the N-terminal phosphorylation sites that target β-catenin for degradation, but retains three armadillo (ARM) protein-protein interaction motifs and the C-terminal transactivation domain. As such, the mutant molecule might function as a dominant activator of Wnt signaling target genes. For example, truncated β-catenin could interact with Tcf-4 and potentiate its function on the promoter. However, the absence of intrinsic transcriptional activation properties in luciferase reporter assays is consistent with a mutant protein that interferes with the function of cellular antagonists of β-catenin activity. One potential target is the APC/GSK3β/Axin complex. Our studies using LiCl provide strong evidence that the level of endogenous GSK3β activity in 293T cells is sufficient to repress transcriptional activation even in the presence of overexpressed Ctnnb1. Thus, competitive inhibition of the interaction of full-length β-catenin with the APC/GSK3β/Axin complex could provide one mechanism for the potentiation of β-catenin mediated transcription by the truncated protein. Alternatively, truncated β-catenin could interact with Chibby or ICAT, two proteins that bind the C-terminus of β-catenin and inhibit the formation of β-catenin-Tcf transcriptional complexes (Daniels and Weiss, 2002; Takemaru et al., 2003; Gottardi and Gumbiner, 2004; Xing et al., 2008). In this model, the mutant β-catenin molecule would act as a competitive inhibitor of these antagonists. Regardless, the mutant transcript results in a gain of function only in the presence of an activated pathway, which is absent in mutant cartilage, and therefore the recombined locus produces a null allele in this context.
TOPGAL activity can be independent of canonical Wnt signaling
TOPFLASH is a widely regarded as faithful reporter of β-catenin/Tcf activity in vitro. TOPGAL, the in vivo equivalent of TOPFLASH, is strongly expressed in regions of the early embryo and the skin known to be sites of β-catenin activity. However, during cartilage development, robust expression of β-galactosidase from the TOPGAL transgene in Ctnnb1 mutant cartilage demonstrates that the reporter locus is activated independent of β-catenin in vivo. Similar observations were made for the retina where TOPGAL expression was observed in cells presumed to be mutant for β-catenin (Fuhrmann et al., 2009). What is not certain from our data is whether the observed TOPGAL activity is constitutive or is regulated by a β-catenin independent signaling event. Moreover, due to limited recombination at the earliest developmental stages, we do not know if the staining patterns in newly formed cartilage are a composite of β-catenin dependent and independent regulation, or if a switch between dependent and independent mechanisms occurs during development.
Why is the tight regulation of the parent reporter construct not maintained in vivo? Fuhrmann et al suggests that the reason lies in β-catenin-independent regulators of TCF/LEF, which have recently been described (Reviewed in (Carlsson et al., 1993; Hsu et al., 1998; Labbe et al., 2000; Arce et al., 2006)). Although consistent with our luciferase reporter studies, the presence of these regulators does not explain the patchy expression of TOPGAL in cartilage and in other tissues. This activity pattern could result from position effects due to endogenous enhancer or promoter activity at the integration site. However, in cartilage, TOPGAL activity is often observed in columns of proliferative chondrocytes that are generated by clonal expansion (Ahrens et al., 2009). These results suggest that the salt and pepper activity pattern is not stochastic in nature and instead derives from an epigenetic event that occurs in progenitors of the resting zone and regulates transgene activation throughout chondrocyte maturation. Alternatively, these findings might imply regulation of transgene activation by the cell cycle since columnar chondrocytes show partly coordinated cell cycle activity. Similar explanations could account for the observation of small groups of β-galactosidase positive cells in Ctnnb1 mosaic retinas (Fuhrmann et al., 2009).
Novel genetic tools require stringent validation processes
Transcriptional reporters of cell signaling activity and conditional mouse mutants are rapidly becoming important tools for mechanistic studies of development and disease. For example, two reporter lines for β-catenin activity, TOPGAL (DasGupta and Fuchs, 1999) and BATGAL (Maretto et al., 2003), have been used in more than 100 published studies. As with all reagents, the utility of these tools depends on the level of confidence that a particular result has a discreet and predictable cause. Although TOPGAL transgene activity is often interpreted as evidence for canonical Wnt signaling, our studies and the work of others unambiguously demonstrate that activity is observed in the absence of β-catenin, the key mediator of canonical Wnt signaling. Thus, without tissue-specific loss of function controls, TOPGAL activity cannot be used as an indicator of endogenous canonical Wnt signaling. Moreover, the high basal (uninduced) activity in some tissues greatly reduces the dynamic range and therefore limits the utility of the reporter for semi-quantitative studies of Wnt signaling. The results of our work highlight the need for stringent validation of genetic resources to ensure the proper interpretation of experimental results. First, while in vitro analysis is useful for the development of reporter cassettes, in vivo analyses are required to validate transgenic reporters. Second, all in vivo reporter mouse lines should be analyzed using both gain and loss of function tests. Third, due to concerns about epigenetic and position effects, in vivo reporter cassettes integrated at distinct sites should be used to test for variability in response. Moreover, our work demonstrates that recombination of conditional loci may produce truncated transcripts that have the potential to generate translation products that have off-target effects. Thus, it is not sufficient to demonstrate the absence of wild type protein when a stable mutant transcript is present. In such cases, thorough analysis of potential translation products and associated functions should be performed.
Experimental Procedures
Unless stated otherwise, all standard chemical reagents were obtained from Sigma-Aldrich (St. Louis, MO).
Mouse strains & animal care
The mouse strains used are B6.129-Ctnnb1tm2Kem/KnwJ (Brault et al., 2001), Col2a1::cre (Ovchinnikov et al., 2000), Col2a1::creER (Nakamura et al., 2006), (Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (tdTomato; Jackson Laboratory) and Tg(Fos-lacZ)34Efu/J (TOPGAL; Jackson Laboratory) (DasGupta and Fuchs, 1999). Where indicated, tamoxifen injections were performed as previously described (Nakamura et al., 2006). Animal care and use was in accordance with NIH guidelines and was approved by the Animal Care and Use Committee of Northwestern University. The morning after mating is designated as 0.5 days post coitum (dpc).
DNA Isolation and Genotyping
Genomic DNA was collected from tail snips or from cartilage by cell lysis in buffer containing proteinase K (Roche Diagnostics, Indianapolis, IN), isopropanol precipitation, and reconstitution in Tris-EDTA buffer. β-catenin alleles were analyzed as described (Brault et al., 2001). Briefly primers RM41, RM42, and RM43 were used to detect wild type (221nt), floxed (324nt), and floxdel (500nt) β-catenin alleles, and recombination was confirmed using primers RM68 and RM69 (631nt). Cre recombinase was detected using CreF 5’ - TAA AAT GTC CAA TTT ACT GAC CGT AC – 3’ and CreR 5’ - CTG GCA ATT TCG GCT ATA CGT AAC AC – 3’. TOPGAL mice were identified by genotyping with LacZ primers as described (Jackson Laboratories, Bar Harbor, ME).
Chondrocyte Isolation
Growth plate cartilage from the distal femur and the proximal tibia were dissected free from articular cartilage and the perichondrium/periosteum, and then homogenized in DNA lysis buffer (100mM Tris-Cl, pH 7.5, 100mM NaCl, 50mM EDTA, 0.5% SDS containing 250 µg/ml proteinase K), Trizol (Invitrogen, Carlsbad, CA), or standard RIPA buffer.
RNA Isolation, RT-PCR, and Cloning
RNA was isolated from chondrocytes using Trizol (Invitrogen, Carlsbad, CA). First strand cDNA synthesis was produced using the SMARTer cDNA Synthesis kit (Clontech, Mountain View, CA). Specific targets were detected by PCR amplification using Platinum Taq polymerase and Platinum Pfx polymerase (Invitrogen, Carlsbad, CA) in conjunction with the following primers: β-cat 38F 5’ – GTC AGC TCG TGT CCT GTG AA – 3’ and β-cat 1340R 5’ – GAG CAG ACA GAC AGC ACC TT – 3’ amplified a 1302nt and 350nt region. The truncated mutant band was cut from the gel and column purified (Qiagen, Valencia, CA) and cloned into pGEM (Promega, Madison, WI). For luciferase assays, a truncated form of β-catenin (exons 7–15) with or without a Kozak sequence at the 5’ end was cloned from full-length β-catenin into pCDNA3.1 using the following primers: NheI β-catenin Exon1/7 5’ GCT AGC CGG GCC TGA GCT CAT CAT TCT GGC CAG TGG or NheI Kozak β-catenin Exon1/7 5’ – GCT AGC GCC ACC ATG GGC GGG CCT GAG CTC ATC ATT CTG GCC AGT GG – 3’ and β-catenin 2673R 5’ – GGA GCG GCC GCC CAC ACT GGC TTT TAT AAG CTT TC – 3’. All subcloned DNA fragments were verified by DNA sequencing.
Western Analysis
Isolated chondrocytes or 293T cell lysates were homogenized in RIPA buffer containing Compleat® protease inhibitors (Roche Diagnostics, Indianapolis, IN) then sonicated. Total protein was recovered by chloroform methanol precipitation (Wessel and Flugge, 1984) and reconstituted in 1X NuPAGE® loading buffer (Invitrogen, Carlsbad, CA). Proteins were separated on NuPAGE 4–15% gels (Invitrogen, Carlsbad, CA) and electro-blotted to PVDF membranes (Millipore, Inc., Billerica, MA). Membranes were blocked in 5% nonfat dried milk, incubated sequentially in primary then secondary antibodies, and developed using the PicoWest or FemtoWest chemiluminescence kits (Thermo Fischer Scientific Inc., Rockford, IL). The following antibodies were used for Western analysis: anti-β-catenin (1:500, R&D Systems, Minneapolis, MN or Santa Cruz Biotechnology, Inc, Santa Cruz, CA), anti-β-actin (1:500, Cell Signaling, Danvers, MA), and HRP-conjugated secondary antibodies (Cell signaling, Danvers, MA).
Histology
To analyze cell morphology, 18.5 dpc limbs were dissected, fixed in Bouin’s Fixative (Ricca Chemical Company, Texas) at 4°C overnight, dehydrated through an ethanol series followed by xylenes, embedded in paraffin (Paraplast X-tra, Tyco-Healthcare). Sections were stained with Masson’s Trichrome using a standard protocol.
Tissue preparation, immunofluorescence, and in situ hybridization
For immunofluorescence (IF) and in situ hybridization (ISH) analysis, tissue was fixed in 4% paraformaldehyde (PFA), equilibrated in 30% sucrose/PBS, frozen in O.C.T. (Tissue-Tek, Torrance, CA), sectioned (12 µm), and mounted on Superfrost Plus glass slides (VWR International, Inc.). For IF, sections were permeabilized in 1% Triton X-100, blocked in 10% FBS before incubating sequentially with the following primary and secondary antibodies: anti-cleaved caspase-3 (1:500, Cell Signaling), anti-β-galactosidase (1:2,000, a gift from R. Holmgren) (Sisson et al., 2006) (Zhang et al., 1994), anti-β-catenin (1:100; BD Biosciences, San Jose, CA), and anti-rabbit Alexa 488 (1:500, Invitrogen, Carlsbad, CA). Where indicated, tissue was counter-stained with 4’,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich).
In situ hybridization analysis was performed as previously described (Ahrens et al., 2009). For quantitative analysis, the length of the expression domain was measured along the longitudinal axis of the cartilage and calculated as a ratio over the total length of the growth plate (articular surface to the mineralized region).
Luciferase Assay
293T cells were transfected with 100ng 8XSuperTOPFLASH or 8X SuperFOPFLASH (gifts from R. Moon, University of Washington), 3 ng CMV-Renilla, 300, 450, or 900 ng pcDNA3-human-beta-catenin (provided by E. Fearon, through Addgene, Inc., plasmid 16828), pcDNA-hTcf4 (provided by C. Gottardi, Northwestern University), pCS2+Lef1 (provided by R. Moon through Addgene, Inc., plasmid 16709), pcDNA/Myc DeltaN TCF4 (provided by B. Vogelstein through Addgene, Inc., plasmid 16513), and 300, 450, or 900 ng pCAG-GFP (provided by C. Cepko through Addgene, Inc., plasmid 11150), pcDNA3.1-mutant β-catenin or mutant β-catenin + Kozak in 24-well plates using Fugene HD transfection reagent (Roche Diagnostics). After 24 hours, cell lysates were assayed with the Dual Luciferase kit (Promega, Madison, WI).
Acknowledgements
We thank A.K. Hadjantonakis, Robert Holmgren, Randall Moon, and Susan Mackem for providing reagents. This work was supported by the Cellular Molecular Basis of Disease Training Grant (MJA), the National Institutes of Health/NIAMS (AR054857), and by the National Center for Research Resources (NCRR).
Footnotes
The authors do not have conflicts of interest associated with this work.
References
- Ahrens MJ, Li Y, Jiang H, Dudley AT. Convergent extension movements in growth plate chondrocytes require gpi-anchored cell surface proteins. Development. 2009;136:3463–3474. doi: 10.1242/dev.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR, McCrea PD, de Crombrugghe B. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 2004;18:1072–1087. doi: 10.1101/gad.1171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- Carlsson P, Waterman ML, Jones KA. The hLEF/TCF-1 alpha HMG protein contains a context-dependent transcriptional activation domain that induces the TCR alpha enhancer in T cells. Genes Dev. 1993;7:2418–2430. doi: 10.1101/gad.7.12a.2418. [DOI] [PubMed] [Google Scholar]
- DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Dickinson ME, Krumlauf R, McMahon AP. Evidence for a mitogenic effect of Wnt-1 in the developing mammalian central nervous system. Development. 1994;120:1453–1471. doi: 10.1242/dev.120.6.1453. [DOI] [PubMed] [Google Scholar]
- Fuhrmann S, Riesenberg AN, Mathiesen AM, Brown EC, Vetter ML, Brown NL. Characterization of a transient TCF/LEF-responsive progenitor population in the embryonic mouse retina. Invest Ophthalmol Vis Sci. 2009;50:432–440. doi: 10.1167/iovs.08-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R. Lack of beta-catenin affects mouse development at gastrulation. Development. 1995;121:3529–3537. doi: 10.1242/dev.121.11.3529. [DOI] [PubMed] [Google Scholar]
- Han C, Yan D, Belenkaya TY, Lin X. Drosophila glypicans Dally and Dally-like shape the extracellular Wingless morphogen gradient in the wing disc. Development. 2005;132:667–679. doi: 10.1242/dev.01636. [DOI] [PubMed] [Google Scholar]
- Hartmann C, Tabin CJ. Wnt-14 plays a pivotal role in inducing synovial joint formation in the developing appendicular skeleton. Cell. 2001;104:341–351. doi: 10.1016/s0092-8674(01)00222-7. [DOI] [PubMed] [Google Scholar]
- Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8:727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Hsu SC, Galceran J, Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Labbe E, Letamendia A, Attisano L. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways. Proc Natl Acad Sci U S A. 2000;97:8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dudley AT. Noncanonical frizzled signaling regulates cell polarity of growth plate chondrocytes. Development. 2009;136:1083–1092. doi: 10.1242/dev.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci U S A. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon RT, Bowerman B, Boutros M, Perrimon N. The promise and perils of Wnt signaling through beta-catenin. Science. 2002;296:1644–1646. doi: 10.1126/science.1071549. [DOI] [PubMed] [Google Scholar]
- Nakamura E, Nguyen MT, Mackem S. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev Dyn. 2006;235:2603–2612. doi: 10.1002/dvdy.20892. [DOI] [PubMed] [Google Scholar]
- Olson DJ, Papkoff J. Regulated expression of Wnt family members during proliferation of C57mg mammary cells. Cell Growth Differ. 1994;5:197–206. [PubMed] [Google Scholar]
- Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26:145–146. [PubMed] [Google Scholar]
- Sisson BE, Ziegenhorn SL, Holmgren RA. Regulation of Ci and Su(fu) nuclear import in Drosophila. Dev Biol. 2006;294:258–270. doi: 10.1016/j.ydbio.2006.02.050. [DOI] [PubMed] [Google Scholar]
- Spater D, Hill TP, O'Sullivan RJ, Gruber M, Conner DA, Hartmann C. Wnt9a signaling is required for joint integrity and regulation of Ihh during chondrogenesis. Development. 2006;133:3039–3049. doi: 10.1242/dev.02471. [DOI] [PubMed] [Google Scholar]
- Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Yang Y, Topol L, Lee H, Wu J. Wnt5a and Wnt5b exhibit distinct activities in coordinating chondrocyte proliferation and differentiation. Development. 2003;130:1003–1015. doi: 10.1242/dev.00324. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ungar A, Fresquez C, Holmgren R. Ectopic expression of either the Drosophila gooseberry-distal or proximal gene causes alterations of cell fate in the epidermis and central nervous system. Development. 1994;120:1151–1161. doi: 10.1242/dev.120.5.1151. [DOI] [PubMed] [Google Scholar]