Abstract
Lipid domain formation induced by annexin was investigated in mixtures of phosphatidylcholine (PC), phosphatidylserine (PS), and cholesterol (Chol), which were selected to mimic the inner leaflet of a eukaryotic plasma membrane. Annexins are ubiquitous and abundant cytoplasmic, peripheral proteins, which bind to membranes containing PS in the presence of calcium ions (Ca2+), but whose function is unknown. Prompted by indications of interplay between the presence of cholesterol in PS/PC mixtures and the binding of annexins, we used Monte Carlo simulations to investigate protein and lipid domain formation in these mixtures. The set of interaction parameters between lipids and proteins was assigned by matching experimental observables to corresponding variables in the calculations. In the case of monounsaturated phospholipids, the PS-PC and PC-Chol interactions are weakly repulsive. The interaction between protein and PS was determined based on experiments of annexin binding to PC/PS mixtures in the presence of Ca2+. Based on the proposal that PS and cholesterol form a complex in model membranes, a favorable PS-Chol interaction was postulated. Finally, protein-protein favorable interactions were also included, which are consistent with observations of large, two-dimensional, regular arrays of annexins on membranes. Those net interactions between pairs of lipids, proteins and lipids, and between proteins are all small, of the order of the average kinetic energy. We found that annexin a5 can induce formation of large PS domains, coincident with protein domains, but only if cholesterol is present.
Introduction
Cholesterol (Chol) is one of the most abundant lipids in animal plasma membranes. Yet its role in membrane physiology remains to be understood. When cholesterol is mixed with saturated and unsaturated phospholipids, its preference for ordered, saturated acyl chains leads to the formation of liquid-ordered (Lo) and liquid-disordered (Ld) domains that coexist in the plane of the membrane (1–5). Mixtures of cholesterol with saturated phospholipids, such as phosphatidylcholine (PC) and especially sphingomyelin (SM), form particularly ordered bilayers (1,6,7), which are still liquid above the main phase transition of the phospholipid (8–10). If the PC is unsaturated, SM/PC/Chol mixtures may be good models for the outer leaflet of the cell membrane. Cholesterol has been suggested to organize the membrane through formation of liquid-ordered domains, which constitute the physical basis of lipid rafts (11–14).
The lipid composition of the inner leaflet of animal plasma membranes, however, is very different; it consists mainly of phosphatidylethanolamine (PE), phosphatidylserine (PS), and cholesterol (15–17). Mixtures of PE/PS/Chol have been much less studied than those of SM/PC/Chol. One important feature of PE/PS/Chol membranes, though, is that they do not form Lo phases or rafts (18). If the role of cholesterol in the outer leaflet is in membrane organization through compartmentalization into raft (Lo) and nonraft areas (11), what is its role in the inner leaflet?
We wanted to investigate the role of cholesterol in a system mimicking the inner leaflet. The model system chosen must be quantitatively well understood, in terms of lipid-lipid and lipid-protein interactions, so that meaningful predictions are possible. This knowledge is especially necessary because differences in interactions between lipids in membranes are generally small (4,19), and the effect of cholesterol may be subtle. Previously, we showed that addition of a peripheral membrane protein, the C2 domain of synaptotagmin I, induces lipid domain formation in the fluid state, in mixtures of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS). PS/protein domains result from coupling of a favorable protein-PS interaction with an unfavorable PC-PS interaction (20). We further conjectured that “one might also postulate that some proteins that have no apparent enzymatic or signaling activities, like the annexins, might function specifically to organize lipid domains upon which other proteins can bind” (20). Although they have no known enzymatic function, these membrane-binding proteins account for ∼2% of the cytoplasmic proteins.
The design of this investigation was prompted by a few observations: annexins show a preference for binding PS relative to PC in the presence of Ca2+ (21); cholesterol seems to influence the interaction of annexins with phospholipid bilayers (22,23); there is evidence for a preferential interaction of cholesterol with PS relative to PC, especially if the lipids have saturated or monounsaturated acyl chains (24–26) (if the lipid has a polyunsaturated acyl chain, the preference is reversed but very weak (27)); and annexins are prone to forming large, two-dimensional, regular arrays on membrane surfaces (28), suggesting that protein-protein interactions are also involved.
On this basis, we hypothesized that cholesterol may effectively enhance the PS-PC repulsion, leading to larger PS domains, even in the absence of protein. Then, in the presence of Ca2+, the annexins would induce formation of large domains in PC/PS/Chol mixtures. These mixtures resemble the lipid composition of the inner leaflet of eukaryotic plasma membranes, with the difference that the zwitterionic lipid PE is replaced by another zwitterionic lipid, PC. The replacement of PE by PC was motivated by practical considerations and is explained below. To model this system, we used Monte Carlo simulations of a simple lattice representing the lipid bilayer, to which proteins adsorb. The goal was to predict the conditions—composition, concentrations, and magnitudes of interactions between lipids and proteins—under which domains should be observed.
Methods
Monte Carlo simulations
Simulations were performed as previously described (5,20,29,30) using standard Monte Carlo methods (31–33). The lipid membrane was represented by a 100 × 100 triangular lattice with skew-periodic boundary conditions. Each site on the lattice is occupied by one phospholipid or cholesterol. To obtain equilibrium configurations, the lipids are exchanged by randomly selecting partners on the lattice, using a nonnearest-neighbor Kawasaki step (34). The proteins are initially placed in solution, in a virtual volume calculated to yield the correct lipid concentration, [L]. For a 100 × 100 lattice, which contains 104 lipids, this volume is 107[L]/NA (Avogadro's number), thus yielding [L] = 1 mM. The total number of proteins in the system was 103, thus yielding a concentration of 100 μM. The proteins are allowed to bind to the lattice and, when bound, occupy a 19-site hexagon, superimposed on the lipids. On the surface, a protein is allowed to move its center to another lattice site (randomly chosen), or to desorb back into solution. The choice between these two types of moves is aleatory. Steric overlap of proteins is not allowed. In addition, acceptance or rejection of all attempted moves, for both lipids and proteins, is based on the Metropolis criterion (35) with a move probability that depends exponentially on the interaction free energy change, using a random number (36) for the decision. The simulations included a preequilibration period of 2 × 104 Monte Carlo cycles followed by a period of 1–2 × 106 acquisition cycles, which were found to be more than sufficient to reach equilibrium, as judged by the evolution of domain sizes and protein binding. Simulations in larger lattices, of 200 × 200 and 300 × 300 sites, which correspond to an increase of up to an order of magnitude in system size, yielded equivalent results (Fig. S1 and Fig. S2 in the Supporting Material). Six interaction parameters are used, three involving the annexin and three involving only the lipids. Both a5-PC and a5-Chol interactions are represented by a membrane binding free energy, , the a5-PS preferential interaction is represented by εP, and the protein-protein interaction, by εA. The three lipid-lipid, unlike nearest-neighbor AB interaction parameters are of the form ωAB = εAB – 1/2(εAA + εBB), where the εij represent the contact (nearest-neighbor) interaction between the lipids i and j, and A, B = PC, PS, or Chol (4).
Results and Discussion
The experimental model system simulated
The experimental model system consisted of bilayers of PC/PS/Chol mixtures and the peripheral, membrane-binding protein annexin a5. We believe PC/PS/Chol mixtures should capture the essence of the lipid component of the inner leaflet of a eukaryotic plasma membrane, with respect to the properties investigated here, although this is a working hypothesis at this point. PC was used instead of PE because the phase transition temperatures of the PE (37,38) are not suitable for the experiments designed to obtain the protein-lipid interactions. At first sight, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) appears like the most obvious choice to mimic the PE component of the biological membrane. However, the gel-to-fluid transition of POPE occurs at 25°C and the POPS transition, at 12°C. Therefore, a PE/PS 60:40 mixture is very near phase separation, at room temperature, a drastic and unphysiological condition that must be avoided for subtler effects to be observed. Dioleoyl-sn-glycero-3-phosphoethanolamine has a more convenient gel-to-fluid transition temperature, –16°C, but its fluid-to-hexagonal II phase transition occurs at 10°C, rendering it useless at room temperature for these experiments. (POPE would not suffer from this complication, with a fluid-to-hexagonal II transition at 72°C.) In using POPC instead of POPE, we could also take advantage of our previous work on PC/PS mixtures (20,39) and of the knowledge of most interaction parameters with high confidence.
Previously, we examined binding of annexin a5 to large unilamellar vesicles composed of PC/PS in the presence of varying concentrations of the lanthanide ion Tb3+, which was a mimic for Ca2+ (21). More recently, annexin a5 binding to PC/PS membranes was reexamined, but in the presence of its physiological ligand, Ca2+. (J. W. Gauer, K. J. Knutson, J. R. Murphy, S. R. Jaworski, and A. Hinderliter, unpublished data). The equilibrium binding constants for annexin a5 obtained are now summarized. In aqueous solution, binding of Ca2+ to annexin a5 was described by a model with five identical and independent sites, with a binding constant K0 = 3.0 × 103 M–1. In the presence of PC/PS 60:40 membranes (large unilamellar vesicles), the binding isotherm included two types of sites. The first two sites were of high affinity, with binding constant K1a = 1 × 105 M–1. The second three sites were of low affinity, similar to those in solution, with K1b = 5.6 × 103 M–1. Measuring binding of annexin a5 to PC/PS 60:40 in the presence of 1 mM Ca2+ yielded an apparent binding constant = 5.7 × 104 M–1, which is given by
| (1) |
With the simplifying approximation of K0 ≈ K1b, a self-consistent thermodynamic cycle was constructed, from which the binding constant of annexin a5 to PC/PS 60:40, in the absence of Ca2+, was calculated to be KL = 50 M–1.
Interaction parameters for the Monte Carlo simulations
Simulations of PC/PS membranes, with and without Chol, were performed in the absence and in the presence of the peripheral protein annexin a5, using standard Monte Carlo methods (5,20,29,33). The lipid membrane was simulated as a 100 × 100 triangular lattice, each site representing a phospholipid or a cholesterol molecule. The annexins were added from a virtual volume representing the aqueous solution above the membrane. When bound, each protein covers the area of a 19-site hexagon on the lattice (20). The proteins associate with the lipid lattice with binding constants derived from experiment, which depends on Ca2+ concentration and membrane composition. In all simulations, the number of proteins in the system was 103, which corresponds to a solution concentration of 100 μM, but the number bound was ≈50–100 proteins. The number of lipids was 104, which corresponds to a concentration of 1 mM.
Binding of a5 to POPC was too weak to be reliably measured. In the Monte Carlo simulations, we assumed a sufficiently small free energy of interaction, , which corresponds to a dissociation constant KD = 1 M. The same interaction was assumed between annexin a5 and Chol (no preference over PC). Then, based on the experimental binding constants to PC/PS 60:40, we determined εP which represents the free energy by which annexin a5 binds better to a PS lipid, located underneath the protein, than to a PC lipid. This parameter was determined by varying its value in Monte Carlo simulations of annexin a5 binding to a PC/PS 60:40 lattice, in the limit of very few proteins bound (to minimize excluded volume interactions), until the fraction of bound a5 matched the value calculated from the experimentally derived binding constants, in the presence of various Ca2+ concentrations, at room temperature (295 K). We found that εP = – 375, – 445, and – 470 cal/mol-lipid in the presence of 20, 100, and 200 μM Ca2+, respectively. All simulations reported here were performed with εP = – 445 cal/mol, corresponding to 100 μM Ca2+.
In addition to these parameters, three lipid-lipid, unlike nearest-neighbor interaction parameters were necessary. They are defined by ωAB = εAB – 1/2(εAA + εBB) and represent the excess free energy of contacts between the three possible unlike A-B lipid pairs (PS-PC, PS-Chol, and PC-Chol) over the average of the interactions between each set of like pairs (4). The PS-PC interaction (ωPS–PC) was set to +240 cal/mol, the value we previously found to provide the best match between experimental and simulated pyrene excimer/monomer emission ratios (20). Measurements of Ca2+ activity in combination with simulations have shown that PS and PC do not mix randomly, but form separate clusters, despite the electrostatic repulsion between PS headgroups (41). Consistent with those observations, molecular dynamics simulations indicated a net favorable PS-PS interaction in mixtures with PC (42), in the presence of Ca2+, corresponding to ωPS–PC ≈ + 200 to +350 cal/mol at room temperature (4). This is probably because the high hydrogen bonding capacity of the PS headgroups overrides the electrostatic repulsion, at least in the presence of Ca2+. Note that the interaction parameters used in our simulations correspond to experimental conditions that include Ca2+.
The PC-Chol interaction (ωPC–Chol) was set to +200 cal/mol, a value previously found to yield very good agreement between Monte Carlo simulations and fluorescence (Förster) energy transfer data in SM/POPC/Chol mixtures (29). This value was also obtained from measurements of lipid partitioning determined by isothermal titration calorimetry (4,43). However, the PC-Chol interaction depends on the type of PC and on the state of the bilayer. Cholesterol and dipalmitoylphosphatidylcholine (DPPC) interact favorably in the Lo phase, thus ωDPPC–Chol < 0 (4,5,44). The same applies to interactions with dimyristoylphosphatidylcholine (DMPC) and distearoylphosphatidylcholine (DSPC) in the Lo phase (4). In the Ld phase, however, the interaction between saturated PC and Chol is essentially zero (4,44). PC with polyunsaturated acyl chains, on the other hand, interact very unfavorably with Chol (45–47), because the large conformational space of the unsaturated acyl chains (48,49) is restricted next to cholesterol molecules, resulting in an entropic penalty (4). The interaction of monounsaturated PC (POPC) with Chol must lie somewhere in between those of polyunsaturated and saturated PC in the Ld phase (50). Thus ωPOPC–Chol ≈ + 100 to +200 cal/mol (29) appears correct, at least at low cholesterol concentration (Ld), which is the regime used here (10 mol% Chol). Now probably, the POPC-Chol interaction becomes more favorable at high cholesterol concentration, so that ωPOPC–Chol ≈ 0 in POPC/Chol 70:30 (4).
Recent Monte Carlo simulations of the phase diagram of DSPC/dioleoylphosphatidylcholine (DOPC)/Chol (51) employed (at room temperature and in our notation) ωDSPC–Chol = –760, ωDOPC–Chol = –590, and ωDSPC–DOPC = +320 cal/mol. The phospholipids were simulated as linked dimers (51), so the values are not directly comparable, but they would actually be larger in absolute value if translated into the simpler model we use. The DSPC/DOPC/Chol system, however, is quite different from POPS/POPC/Chol and even from SM/POPC/Chol, because its behavior is dominated by the very unfavorable DSPC-DOPC interaction (4,51,52). A favorable DSPC-Chol interaction is in qualitative agreement with experiment, which, however, yielded much smaller absolute values, ωDSPC–Chol ≈ –250 cal/mol close to room temperature (4). The negative value of the DOPC-Chol interaction (51), on the other hand, appears to fall outside the pattern, in the Ld phase, but may be correct at high cholesterol concentration. We suspect that fits of similar quality to the ternary DSPC/DOPC/Chol phase diagram are possible using a smaller absolute value for ωDSPC–Chol and ωDOPC–Chol > 0 but small, in the low-cholesterol regions. Those simulations are probably not very sensitive to the exact values of these two interactions, as long as their difference remains constant, because of the dominant effect of ωDSPC–DOPC.
The PS-PC and PC-Chol interactions used in our simulations are weakly repulsive. In contrast, the PS-Chol interaction was set to ωPS–Chol = –350 cal/mol, which is favorable and identical to that previously used for the SM-Chol interaction (29). This choice was based on observations that suggested the formation of a PS-Chol complex in lipid monolayers (24), as proposed for SM-Chol (53). That SM-Chol and PS-Chol interactions may be of similar magnitude is further suggested by the observation that PS can compete with SM for Chol (54). Formation of a complex or a preferential association requires a favorable interaction, hence ωPS–Chol < 0. Smaller absolute values for both SM-Chol and PS-Chol interactions, but still negative (favorable) compared to POPC-Chol, are calculated from partitioning experiments that use cyclodextrin (26). The value of –350 cal/mol was used as a starting point, but the effect of ωPS–Chol on domain size is examined below in detail.
Finally, we allowed for the possibility that annexins interact favorably when adjacent to each other on the membrane surface. Evidence for a favorable interaction between the proteins comes from the observation of ordered two-dimensional arrays of annexins on membranes with clear hexagonal packing (28). The corresponding Gibbs free energy is represented by the interaction parameter εA = –0.68 kcal/mol. This value was chosen because it is the minimum necessary to produce small protein clusters in PC/PS 60:40 (Fig. 1 D and Fig. 2, light gray bars), which were observed experimentally by fluorescence microscopy (55). Fig. 2 shows the effect of varying εA from –0.65 kcal/mol (black bars) to –0.68 kcal/mol (light gray) to –0.70 kcal/mol (dark gray). This variation of εA results in going from very small clusters (dominated by single proteins) to very large ones (essentially one large domain) over a range of only 50 cal/mol. If εA ≤ –0.8 kcal/mol (more negative), complete agglutination of the protein on the membrane results, which was not observed experimentally (55).
Figure 1.
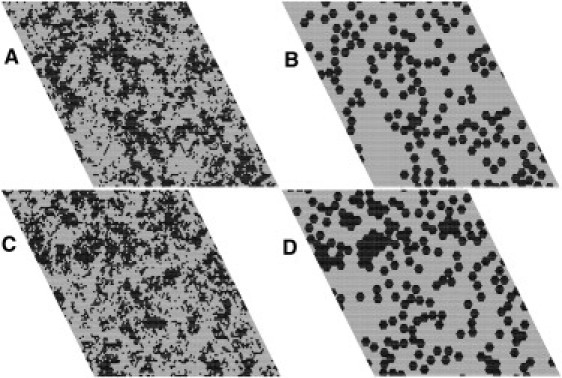
Snapshot of a simulation of PC/PS 60:40 in the presence of annexin. (A) Lipid and (B) protein without protein-protein interactions (εA = 0); (C) lipid and (D) protein with a protein-protein interaction of εA = –0.68 kcal/mol. Lipid (A and C): PS = black, PC = gray. Protein (B and D): black hexagons on lipid matrix (gray).
Figure 2.
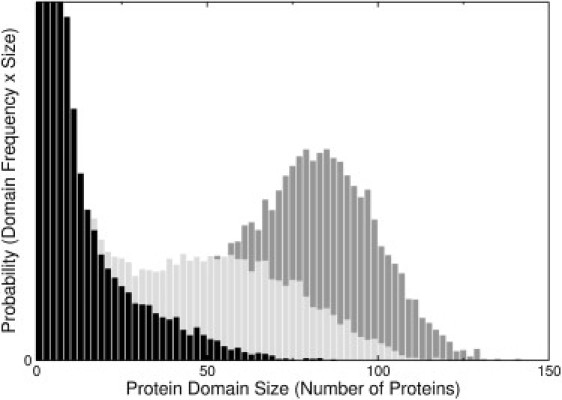
Protein domain size distributions in PC/PS 60:40 (number of proteins in domains of a given size) as a function of the protein-protein interaction parameter, εA: black, –0.65 kcal/mol; light gray, –0.68 kcal/mol; dark gray, – 0.70 kcal/mol. The ordinate scale is truncated for clarity.
Lipid and protein domain formation
We will now describe the most significant results of these Monte Carlo simulations. First, we simulated the effect of replacing 10% of the PC with Chol in a PC/PS 80:20 membrane. As found previously (20), in the absence of Chol, the distribution of PS in the PC matrix is close to random (Fig. 3 A). In PC/PS/Chol 70:20:10, however, small PS/Chol domains form (Fig. 3 B). When annexin a5 is added to PC/PS 80:20, no significant change in PS domain distribution occurs, whether or not the proteins interact favorably with each other (Fig. 3 C). Even with favorable protein-protein interactions, the protein distribution on PC/PS 80:20 is also essentially random (Fig. 3 D). Similarly, in PC/PS 60:40 no phase separation occurs, and even small protein clusters are only observed in the presence of attractive protein-protein interactions (Fig. 1).
Figure 3.
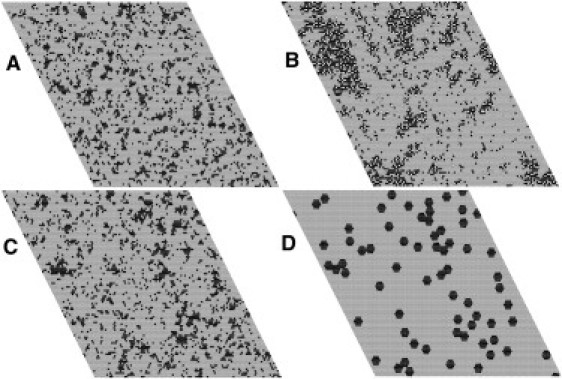
Snapshots of Monte Carlo simulations of PC/PS 80:20 (A) and PC/PS/Chol 70:20:10 (B) in the absence of annexin; PC/PS 80:20 lipid (C) and protein (D) in the presence of annexin with a protein-protein interaction parameter εA = –0.68 kcal/mol. The PS-Chol interaction is ωPS–Chol = –350 cal/mol. Lipid: PS, black; PC, gray; Chol, white. Protein: black hexagons on lipid matrix (gray).
When annexin a5 is added to PC/PS/Chol 70:20:10, however, larger PS/Chol domains form, even without protein-protein interactions (Fig. 4 A). If the proteins interact favorably with each other the PS/Chol domains increase dramatically in size (εA = –0.68 kcal/mol, Fig. 4 C), concomitant with formation of large protein domains on the membrane surface (Fig. 4 D). Such large protein domains do not form if the proteins interact only by hard core repulsions (εA = 0, Fig. 4 B).
Figure 4.
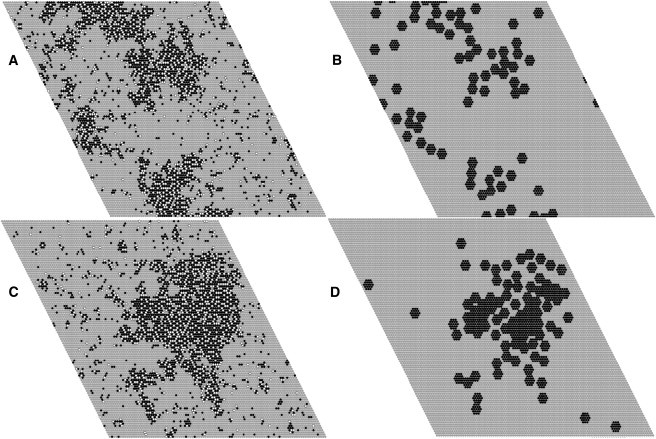
Snapshots of simulations of PC/PS/Chol 70:20:10 in the presence of annexin. (A) Lipid and (B) protein, without protein-protein interactions (εA = 0); (C) lipid and (D) protein, with a protein-protein interaction εA = –0.68 kcal/mol. Lipid: PS, black; PC, gray; Chol, white. Protein: black hexagons on lipid matrix (gray).
A more quantitative representation of these observations is provided by the distribution functions of the PS domains, shown in Fig. 5. In the absence of Chol, even with proteins bound on the membrane, the PS domain distribution is dominated by very small clusters (Fig. 5, black). Similarly, in the absence of protein but in the presence of Chol, the PS domain size distribution shows essentially an exponential decay in size, albeit less pronounced (Fig. 5, light gray). Well-defined, large PS domains are only observed if Chol and annexin are both present, and especially so if the proteins interact favorably with each other (Fig. 5, dark gray).
Figure 5.
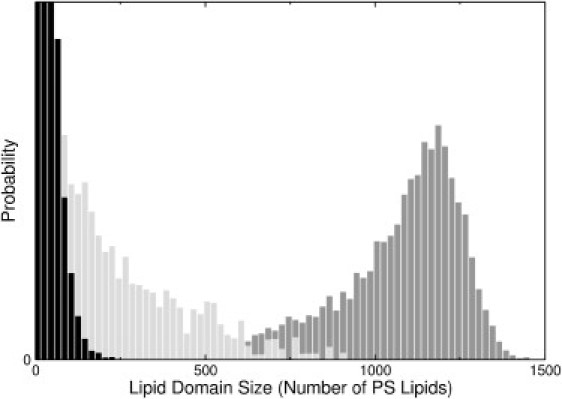
PS domain size distributions (number of PS lipids in domains of a given size). Black, PC/PS 80:20 in the presence of a5 with a protein-protein interaction εA = –0.68 kcal/mol; this corresponds to Fig. 3C. Light gray, PC/PS/Chol 70:20:10 without protein and ωPS–Chol = –350 cal/mol; this corresponds to Fig. 3B. Dark gray, PC/PS/Chol 70:20:10 in the presence of a5, with εA = –0.68 kcal/mol and ωPS–Chol = –350 cal/mol; this corresponds to Fig. 4C. The ordinate axis gives the product of domain size by the number of domains of that size, thus representing how much PS is found in a domain of a given size; the ordinate scale is arbitrary and is truncated for clarity.
It is known from experiment that the primary effect of cholesterol is not on annexin binding, which is similar in PC/PS and PC/PS/Chol at fixed PS content in the presence of Ca2+ (22,23) (also, S. Jaworski and A. Hinderliter, unpublished data). Rather, cholesterol appears to affect lipid-lipid interactions in the membrane. Therefore, it is important to determine the effect of ωPS–Chol on domain formation, which is shown in Fig. 6 (A, ωPS–Chol = –250; B, –300; C, –350; D, –400 cal/mol). The corresponding protein domains coincide with the PS domains (Fig. S3). These results show that if ωPS–Chol ≥ –300 cal/mol (less negative) the large domains dissipate (although this can be compensated by slightly more favorable protein-protein interactions). The effect of the PS-Chol interaction is cast in quantitative terms in Fig. 7, which shows the PS domain size distributions for the same four values of ωPS–Chol (increasingly negative from left to right).
Figure 6.
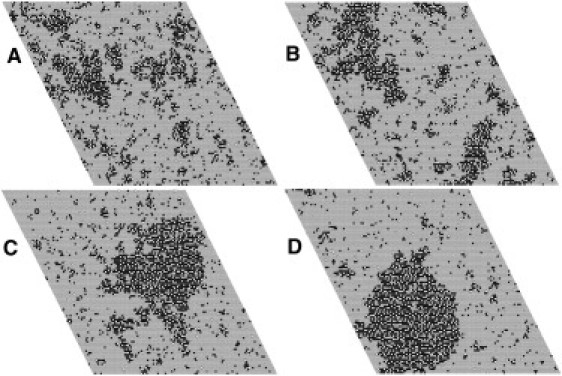
Effect of varying the PS-Chol interaction (ωPS–Chol) on PS domain sizes in PC/PS/Chol 70:20:10, in the presence of annexin a5. (A) ωPS–Chol = –250 cal/mol, (B) –300 cal/mol, (C) –350 cal/mol, and (D) –400 cal/mol. The other parameters are the same as in Fig. 4. PS, black; PC, gray; Chol, white.
Figure 7.
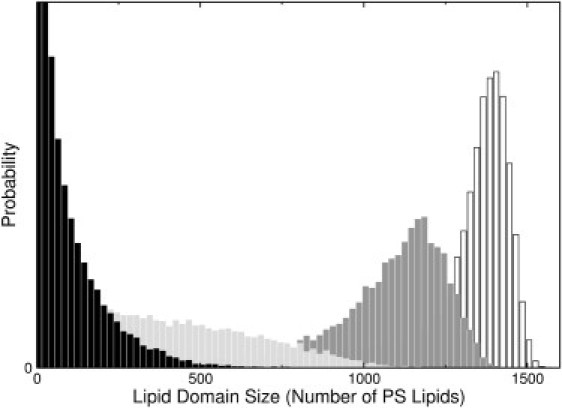
Effect of varying the PS-Chol interaction on the PS domain size distributions in PC/PS/Chol 70:20:10 (number of PS lipids in domains of a given size). The four distributions represent the same conditions as panels A–D in Fig. 6. Black (A) ωPS–Chol = –250 cal/mol; Light Gray (B) –300 cal/mol; Dark Gray (C) –350 cal/mol; White (D) –400 cal/mol. The ordinate axis gives the product of domain size by the number of domains of that size, thus representing how much PS is found in domains of different sizes; the ordinate scale is arbitrary and is truncated for clarity.
These results do not contradict the observation that cholesterol does not induce formation of Lo domains in bilayers modeling the plasma membrane inner leaflet (18), because our simulations indicate that large domains do not form in POPC/POPS/Chol mixtures in the absence of annexin (Fig. 3 B). It would be interesting, however, to determine if probes designed to detect domains based on partitioning between ordered and disordered phases (18) could detect domains in POPE/POPS/Chol in the presence of annexin.
Magnitudes of interactions and physical interpretation
It is well established that annexins bind to PS-containing membranes in the presence of Ca2+. We used experimentally measured binding affinities of annexin a5 for Ca2+ and for PC/PS membranes to set the free energies of interaction of annexin a5 with PC and PS in the Monte Carlo simulations. The preference of a5 for PS over PC means that the preferential interaction of the protein with PS contributes an additional free energy of εP = –445 cal/mol of lipid to its binding to the membrane surface, in the presence of 100 μM Ca2+. Because this is not a very large energy, addition of annexin does not lead to appreciable PS clustering in PC/PS lattices (Fig. 3, C and D).
However, adding annexin to PC/PS/Chol mixtures resulted in formation of PS domains that were especially large if a favorable protein-protein interaction was also included in the Monte Carlo simulations (Fig. 4 C; Fig. 5, dark gray bars). The largest domains that can form in a 100 × 100 lattice are of course limited by the size of the lattice. In the presence of favorable protein-protein interactions, there is essentially one annexin/PS/Chol domain that scales with the lattice size (Fig. S1). In very large systems, we expect this domain to grow to sizes observable experimentally by fluorescence microscopy.
The free energies of interaction used in the Monte Carlo simulations are small. They are approximately of the magnitude of the thermal energy (RT) for protein-protein interactions, about RT/2 for lipid-lipid interactions, and just slightly above RT/2 for protein-lipid preferential interactions. Yet these small interactions are sufficient to induce formation of large lipid and protein domains in a PC/PS/Chol lattice (Fig. 4, C and D). Those large domains, however, are predicted to occur in a membrane only in the presence of cholesterol and if ωPS–Chol is ≤–350 cal/mol (more negative). Otherwise, even with a favorable protein-protein interaction and a preference of annexin a5 for PS over PC, no large lipid domains form in PC/PS/Chol membranes.
The physical reason why cholesterol is essential for the large domains to form was previously discussed for the SM/POPC/Chol system (4). The negative ωPS–Chol increases the likelihood of observing PS-Chol complexes. Because the interaction of POPC with PS and Chol is unfavorable, a POPC molecule adjacent to the complex is repelled by both, therefore even more strongly. The value of –350 cal/mol was previously used for the SM-Chol interaction, which resulted in very large SM/Chol domains in mixtures with POPC (4,29). The difference between these two ternary lipid mixtures (without protein) arises because the SM-PC interaction (+300 cal/mal) in SM/POPC/Chol (29) is significantly more unfavorable than the PS-PC interaction (+240 cal/mol) in POPC/POPS/Chol, so POPC is repelled more strongly by the “complex” in the former case.
It appears that large domains form in these mixtures if the combination of the three interaction parameters is such that the sum of the two repulsive minus the attractive one is at least ≈ +850 cal/mol, at room temperature (4,5). In PC/PS/Chol this corresponds to ωPS–PC + ωPC–Chol – ωPS–Chol = 790 cal/mol, which is close but not enough (Fig. 3 B). The protein-PS interaction coupled with the protein-protein interaction provides the small additional driving force for large PS-rich domains to appear. This role of cholesterol—not so much in forming rigid lipid rafts, but rather in providing an additional, if subtle, contribution to clustering of PS domains in the inner leaflet of the plasma membrane—is clearly suggested by the present results.
Acknowledgments
This work was supported by Research Corporation Cottrel College Science Award 7622 (P.F.A.) and National Institutes of Health grant GM064443 (A.H.).
Supporting Material
References
- 1.Ipsen J.H., Karlström G., Zuckermann M.J. Phase equilibria in the phosphatidylcholine-cholesterol system. Biochim. Biophys. Acta. 1987;905:162–172. doi: 10.1016/0005-2736(87)90020-4. [DOI] [PubMed] [Google Scholar]
- 2.Feigenson G.W., Buboltz J.T. Ternary phase diagram of dipalmitoyl-PC/dilauroyl-PC/cholesterol: nanoscopic domain formation driven by cholesterol. Biophys. J. 2001;80:2775–2788. doi: 10.1016/S0006-3495(01)76245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veatch S.L., Keller S.L. Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys. J. 2003;85:3074–3083. doi: 10.1016/S0006-3495(03)74726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almeida P.F.F. Thermodynamics of lipid interactions in complex bilayers. Biochim. Biophys. Acta. 2009;1788:72–85. doi: 10.1016/j.bbamem.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Almeida P.F. A simple thermodynamic model of the liquid-ordered state and the interactions between phospholipids and cholesterol. Biophys. J. 2011;100:420–429. doi: 10.1016/j.bpj.2010.12.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vist M.R., Davis J.H. Phase equilibria of cholesterol/dipalmitoylphosphatidylcholine mixtures: 2H nuclear magnetic resonance and differential scanning calorimetry. Biochemistry. 1990;29:451–464. doi: 10.1021/bi00454a021. [DOI] [PubMed] [Google Scholar]
- 7.Quinn P.J., Wolf C. The liquid-ordered phase in membranes. Biochim. Biophys. Acta. 2009;1788:33–46. doi: 10.1016/j.bbamem.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Rubenstein J.L., Smith B.A., McConnell H.M. Lateral diffusion in binary mixtures of cholesterol and phosphatidylcholines. Proc. Natl. Acad. Sci. USA. 1979;76:15–18. doi: 10.1073/pnas.76.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almeida P.F.F., Vaz W.L.C., Thompson T.E. Lateral diffusion in the liquid phases of dimyristoylphosphatidylcholine/cholesterol lipid bilayers: a free volume analysis. Biochemistry. 1992;31:6739–6747. doi: 10.1021/bi00144a013. [DOI] [PubMed] [Google Scholar]
- 10.Kahya N., Scherfeld D., Schwille P. Probing lipid mobility of raft-exhibiting model membranes by fluorescence correlation spectroscopy. J. Biol. Chem. 2003;278:28109–28115. doi: 10.1074/jbc.M302969200. [DOI] [PubMed] [Google Scholar]
- 11.Almeida P.F.F., Pokorny A., Hinderliter A. Thermodynamics of membrane domains. Biochim. Biophys. Acta. 2005;1720:1–13. doi: 10.1016/j.bbamem.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 13.Jacobson K., Mouritsen O.G., Anderson R.G. Lipid rafts: at a crossroad between cell biology and physics. Nat. Cell Biol. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- 14.Pike L.J. The challenge of lipid rafts. J. Lipid Res. 2009;50(Suppl):S323–S328. doi: 10.1194/jlr.R800040-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Op den Kamp J.A.F. Lipid asymmetry in membranes. Annu. Rev. Biochem. 1979;48:47–71. doi: 10.1146/annurev.bi.48.070179.000403. [DOI] [PubMed] [Google Scholar]
- 16.Cullis P.R., Hope M.J. Physical properties and functional roles of lipids in membranes. In: Vance D.E., Vance J.E., editors. Biochemistry of Lipids and Membranes. Benjamin-Cummings; Menlo Park, CA: 1985. pp. 28–33. [Google Scholar]
- 17.Zachowski A. Phospholipids in animal eukaryotic membranes: transverse asymmetry and movement. Biochem. J. 1993;294:1–14. doi: 10.1042/bj2940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang T.Y., Silvius J.R. Cholesterol does not induce segregation of liquid-ordered domains in bilayers modeling the inner leaflet of the plasma membrane. Biophys. J. 2001;81:2762–2773. doi: 10.1016/S0006-3495(01)75919-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loew S., Hinderliter A., May S. Stability of protein-decorated mixed lipid membranes: the interplay of lipid-lipid, lipid-protein, and protein-protein interactions. J. Chem. Phys. 2009;130:045102. doi: 10.1063/1.3063117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinderliter A., Almeida P.F.F., Biltonen R.L. Domain formation in a fluid mixed lipid bilayer modulated through binding of the C2 protein motif. Biochemistry. 2001;40:4181–4191. doi: 10.1021/bi0024299. [DOI] [PubMed] [Google Scholar]
- 21.Almeida P.F.F., Sohma H., Hinderliter A. Allosterism in membrane binding: a common motif of the annexins? Biochemistry. 2005;44:10905–10913. doi: 10.1021/bi050474g. [DOI] [PubMed] [Google Scholar]
- 22.Ayala-Sanmartin J., Henry J.P., Pradel L.A. Cholesterol regulates membrane binding and aggregation by annexin 2 at submicromolar Ca(2+) concentration. Biochim. Biophys. Acta. 2001;1510:18–28. doi: 10.1016/s0005-2736(00)00262-5. [DOI] [PubMed] [Google Scholar]
- 23.Ayala-Sanmartin J. Cholesterol enhances phospholipid binding and aggregation of annexins by their core domain. Biochem. Biophys. Res. Commun. 2001;283:72–79. doi: 10.1006/bbrc.2001.4748. [DOI] [PubMed] [Google Scholar]
- 24.Radhakrishnan A., Anderson T.G., McConnell H.M. Condensed complexes, rafts, and the chemical activity of cholesterol in membranes. Proc. Natl. Acad. Sci. USA. 2000;97:12422–12427. doi: 10.1073/pnas.220418097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leventis R., Silvius J.R. Use of cyclodextrins to monitor transbilayer movement and differential lipid affinities of cholesterol. Biophys. J. 2001;81:2257–2267. doi: 10.1016/S0006-3495(01)75873-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niu S.-L., Litman B.J. Determination of membrane cholesterol partition coefficient using a lipid vesicle-cyclodextrin binary system: effect of phospholipid acyl chain unsaturation and headgroup composition. Biophys. J. 2002;83:3408–3415. doi: 10.1016/S0006-3495(02)75340-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huster D., Arnold K., Gawrisch K. Influence of docosahexaenoic acid and cholesterol on lateral lipid organization in phospholipid mixtures. Biochemistry. 1998;37:17299–17308. doi: 10.1021/bi980078g. [DOI] [PubMed] [Google Scholar]
- 28.Richter R.P., Him J.L.K., Brisson A.R. On the kinetics of adsorption and two-dimensional self-assembly of annexin A5 on supported lipid bilayers. Biophys. J. 2005;89:3372–3385. doi: 10.1529/biophysj.105.064337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frazier M.L., Wright J.R., Almeida P.F. Investigation of domain formation in sphingomyelin/cholesterol/POPC mixtures by fluorescence resonance energy transfer and Monte Carlo simulations. Biophys. J. 2007;92:2422–2433. doi: 10.1529/biophysj.106.100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jerala R., Almeida P.F.F., Biltonen R.L. Simulation of the gel-fluid transition in a membrane composed of lipids with two connected acyl chains: application of a dimer-move step. Biophys. J. 1996;71:609–615. doi: 10.1016/S0006-3495(96)79261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heimburg T. Wiley-VCH; Weinheim, Germany: 2007. Thermal Biophysics of Membranes. [Google Scholar]
- 32.Sugár I.P., Biltonen R.L., Mitchard N. Monte Carlo simulations of membranes: phase transition of small unilamellar dipalmitoylphosphatidylcholine vesicles. Methods Enzymol. 1994;240:569–593. doi: 10.1016/s0076-6879(94)40064-4. [DOI] [PubMed] [Google Scholar]
- 33.Binder K., Heermann D.W. 3rd ed. Springer; New York: 1997. Monte Carlo Simulation in Statistical Physics. [Google Scholar]
- 34.Kawasaki K. Kinetics of Ising models. In: Domb C., Green M.S., editors. Vol. 2. Academic Press; New York: 1972. pp. 443–501. (Phase Transitions and Critical Phenomena). [Google Scholar]
- 35.Metropolis N., Rosenbluth A.W., Teller E. Equation of state calculations by fast computing machines. J. Chem. Phys. 1953;21:1087–1092. [Google Scholar]
- 36.Press W.H., Teukolsky S.A., Flannery B.P. 2nd ed. Cambridge University Press; Cambridge: 1994. Numerical Recipes in FORTRAN: The Art of Scientific Computing. [Google Scholar]
- 37.Silvius J.R. Thermotropic phase transitions of pure lipids in model membranes and their modification by membrane proteins. In: Jost P.C., Griffith O.H., editors. Vol. 2. Wiley-Interscience; New York: 1982. pp. 239–281. (Lipid-Protein Interactions). [Google Scholar]
- 38.Cevc G., editor. Phospholipids Handbook. Marcel Dekker; New York: 1993. [Google Scholar]
- 39.Hinderliter A., Biltonen R.L., Almeida P.F.F. Lipid modulation of protein-induced membrane domains as a mechanism for controlling signal transduction. Biochemistry. 2004;43:7102–7110. doi: 10.1021/bi036334t. [DOI] [PubMed] [Google Scholar]
- 40.Reference deleted in proof.
- 41.Huang J., Swanson J.E., Feigenson G.W. Nonideal mixing of phosphatidylserine and phosphatidylcholine in the fluid lamellar phase. Biophys. J. 1993;64:413–425. doi: 10.1016/S0006-3495(93)81382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodríguez Y., Mezei M., Osman R. Association free energy of dipalmitoylphosphatidylserines in a mixed dipalmitoylphosphatidylcholine membrane. Biophys. J. 2007;92:3071–3080. doi: 10.1529/biophysj.106.089078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsamaloukas A., Szadkowska H., Heerklotz H. Nonideal mixing in multicomponent lipid/detergent systems. J. Phys. Condens. Matter. 2006;18:S1125–S1138. doi: 10.1088/0953-8984/18/28/S02. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J., Cao H., Regen S.L. Cholesterol-phospholipid association in fluid bilayers: a thermodynamic analysis from nearest-neighbor recognition measurements. Biophys. J. 2006;91:1402–1406. doi: 10.1529/biophysj.106.084152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harroun T.A., Katsaras J., Wassall S.R. Cholesterol hydroxyl group is found to reside in the center of a polyunsaturated lipid membrane. Biochemistry. 2006;45:1227–1233. doi: 10.1021/bi0520840. [DOI] [PubMed] [Google Scholar]
- 46.Brzustowicz M.R., Cherezov V., Wassall S.R. Molecular organization of cholesterol in polyunsaturated membranes: microdomain formation. Biophys. J. 2002;82:285–298. doi: 10.1016/S0006-3495(02)75394-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bakht O., Pathak P., London E. Effect of the structure of lipids favoring disordered domain formation on the stability of cholesterol-containing ordered domains (lipid rafts): identification of multiple raft-stabilization mechanisms. Biophys. J. 2007;93:4307–4318. doi: 10.1529/biophysj.107.114967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feller S.E., Gawrisch K., MacKerell A.D., Jr. Polyunsaturated fatty acids in lipid bilayers: intrinsic and environmental contributions to their unique physical properties. J. Am. Chem. Soc. 2002;124:318–326. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 49.Feller S.E., Gawrisch K. Properties of docosahexaenoic-acid-containing lipids and their influence on the function of rhodopsin. Curr. Opin. Struct. Biol. 2005;15:416–422. doi: 10.1016/j.sbi.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 50.Silvius J.R. Role of cholesterol in lipid raft formation: lessons from lipid model systems. Biochim. Biophys. Acta. 2003;1610:174–183. doi: 10.1016/s0005-2736(03)00016-6. [DOI] [PubMed] [Google Scholar]
- 51.Dai J., Alwarawrah M., Huang J. Simulation of the lo-ld phase boundary in DSPC/DOPC/cholesterol ternary mixtures using pairwise interactions. J. Phys. Chem. B. 2011;115:1662–1671. doi: 10.1021/jp110243v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsamaloukas A., Szadkowska H., Heerklotz H. Thermodynamic comparison of the interactions of cholesterol with unsaturated phospholipid and sphingomyelins. Biophys. J. 2006;90:4479–4487. doi: 10.1529/biophysj.105.080127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radhakrishnan A., McConnell H.M. Condensed complexes in vesicles containing cholesterol and phospholipids. Proc. Natl. Acad. Sci. USA. 2005;102:12662–12666. doi: 10.1073/pnas.0506043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garg S., Tang J.X., Naumann C.A. Actin-induced perturbation of PS lipid-cholesterol interaction: a possible mechanism of cytoskeleton-based regulation of membrane organization. J. Struct. Biol. 2009;168:11–20. doi: 10.1016/j.jsb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 55.Vats K., Knutson K., Sheets E.D. Peripheral protein organization and its influence on lipid diffusion in biomimetic membranes. ACS Chem. Biol. 2010;5:393–403. doi: 10.1021/cb900303s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


