Abstract
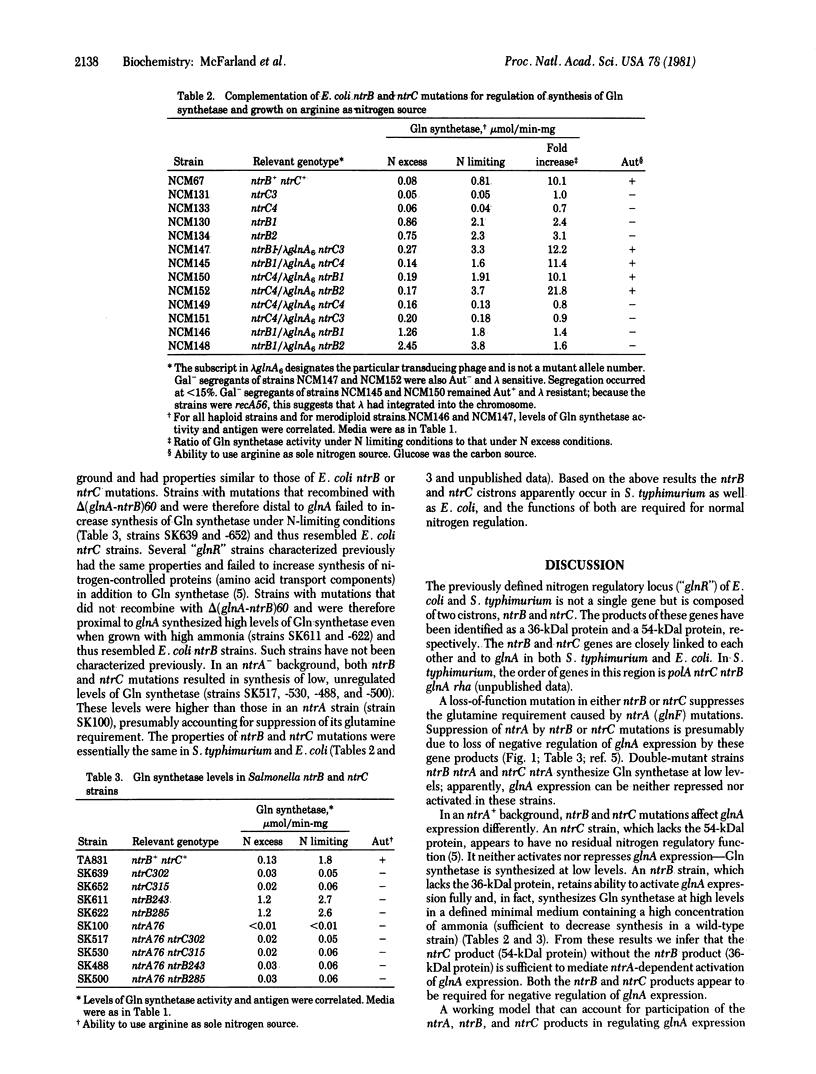
The nitrogen regulatory locus "glnR" of Escherichia coli and Salmonella typhimurium is composed of two cistrons, which we propose to call ntrB and ntrC (nitrogen regulation B and C). Frameshift mutations in ntrB and ntrC were isolated on a lambda phage that carries the E. coli ntrB and ntrC genes and the closely linked glnA gene, the structural gene encoding glutamine synthetase [L-glutamate:ammonia ligase (ADP-forming), EC 6.3.1.2]; mutations were selected as suppressors of glnF (which we propose to rename ntrA), a selection used previously to isolate glnR mutations. Phage DNA from one mutant (ntrB) failed to direct synthesis of a 36-kilodalton (kDal) protein whose synthesis was directed by DNA from the parent phage (ntrB+) in a coupled in vitro transcription/translation system. DNA from three other mutants (ntrC) failed to direct synthesis of a 54-kDal protein; DNA from two of these mutants instead directed synthesis of smaller proteins, 53 and 50 kDal, respectively. In all four cases, DNA from frameshift revertants directed synthesis of both the 36-kDal and 54-kDal proteins. These results suggested that ntrB and ntrC were separate genes which encoded 36-kDal and 54-kDal protein products, respectively. Frameshift mutations in ntrB and ntrC complemented each other with regard to regulation of glnA expression in vivo and growth on arginine as nitrogen source, another nitrogen-controlled phenotype; this confirmed that ntrB and ntrC are separate cistrons that encode diffusible products. The ntrB and ntrC genes were also defined in S. typhimurium. Studies of mutant strains provided information on the roles of the ntrB and ntrC products in activation and repression of glnA expression and raised the possibility that these products function as a protein complex in regulating expression of nitrogen-controlled genes.
Full text
PDF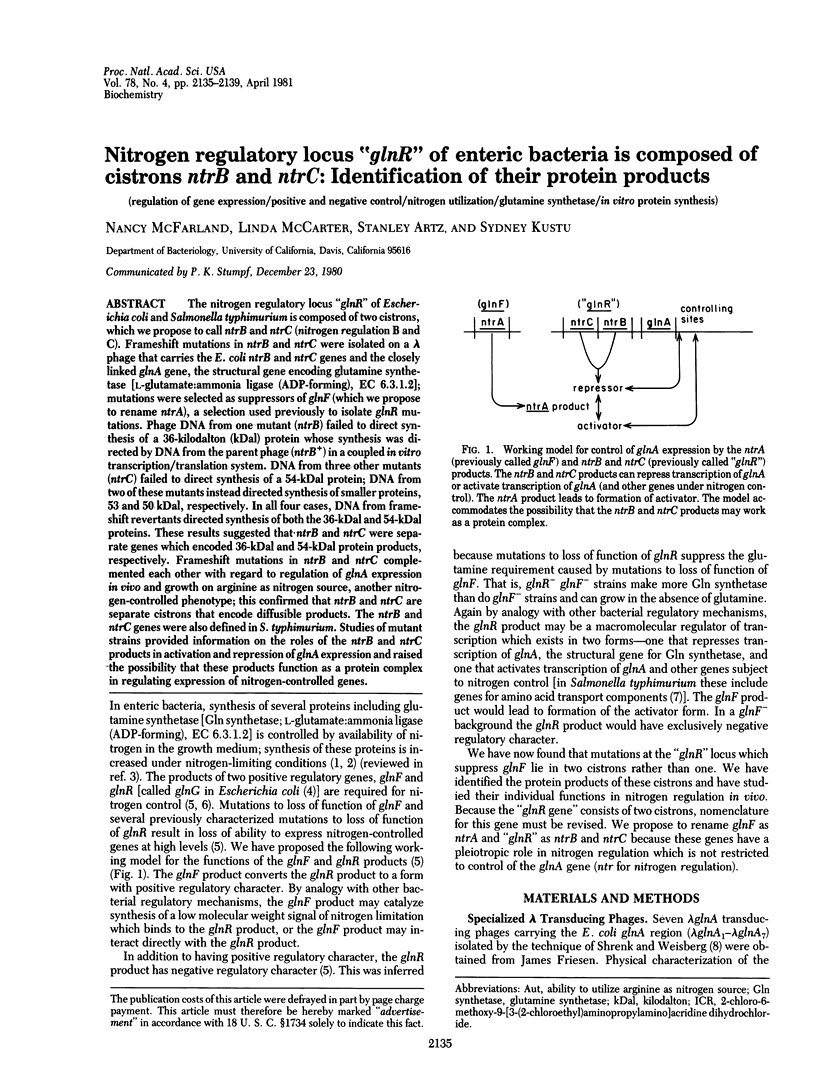



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ames G. F. Resolution of bacterial proteins by polyacrylamide gel electrophoresis on slabs. Membrane, soluble, and periplasmic fractions. J Biol Chem. 1974 Jan 25;249(2):634–644. [PubMed] [Google Scholar]
- Artz S. W., Broach J. R. Histidine regulation in Salmonella typhimurium: an activator attenuator model of gene regulation. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3453–3457. doi: 10.1073/pnas.72.9.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft S., Rhee S. G., Neumann C., Kustu S. Mutations that alter the covalent modification of glutamine synthetase in Salmonella typhimurium. J Bacteriol. 1978 Jun;134(3):1046–1055. doi: 10.1128/jb.134.3.1046-1055.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron L. S., Penido E., Ryman I. R., Falkow S. Behavior of coliphage lambda in hybrids between Escherichia coli and Salmonella. J Bacteriol. 1970 Apr;102(1):221–233. doi: 10.1128/jb.102.1.221-233.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J., Neumann C., Kustu S. Mutant strains (nit) of Salmonella typhimurium with a pleiotropic defect in nitrogen metabolism. J Bacteriol. 1976 Oct;128(1):86–98. doi: 10.1128/jb.128.1.86-98.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csonka L. N., Clark A. J. Construction of an Hfr strain useful for transferring recA mutations between Escherichia coli strains. J Bacteriol. 1980 Jul;143(1):529–530. doi: 10.1128/jb.143.1.529-530.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia E., Bancroft S., Rhee S. G., Kustu S. The product of a newly identified gene, gInF, is required for synthesis of glutamine synthetase in Salmonella. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1662–1666. doi: 10.1073/pnas.74.4.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustu S. G., McFarland N. C., Hui S. P., Esmon B., Ames G. F. Nitrogen control of Salmonella typhimurium: co-regulation of synthesis of glutamine synthetase and amino acid transport systems. J Bacteriol. 1979 Apr;138(1):218–234. doi: 10.1128/jb.138.1.218-234.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustu S. G., McKereghan K. Mutations affecting glutamine synthetase activity in Salmonella typhimurium. J Bacteriol. 1975 Jun;122(3):1006–1016. doi: 10.1128/jb.122.3.1006-1016.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustu S., Burton D., Garcia E., McCarter L., McFarland N. Nitrogen control in Salmonella: regulation by the glnR and glnF gene products. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4576–4580. doi: 10.1073/pnas.76.9.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magasanik B. Classical and postclassical modes of regulation of the synthesis of degradative bacterial enzymes. Prog Nucleic Acid Res Mol Biol. 1976;17:99–115. doi: 10.1016/s0079-6603(08)60067-7. [DOI] [PubMed] [Google Scholar]
- NEIDHARDT F. C., MAGASANIK B. Reversal of the glucose inhibition of histidase biosynthesis in Aerobacter aerogenes. J Bacteriol. 1957 Feb;73(2):253–259. doi: 10.1128/jb.73.2.253-259.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. Z., Gold L. M., Huang W. M. The identification of prereplicative bacteriophage T4 proteins. J Biol Chem. 1973 Aug 10;248(15):5499–5501. [PubMed] [Google Scholar]
- Ohtsubo E., Hsu M. T. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli: structure of F100, F152, and F8 and mapping of the Escherichia coli chromosomal region fep-supE-gal-attlambda-uvrB. J Bacteriol. 1978 Jun;134(3):778–794. doi: 10.1128/jb.134.3.778-794.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahel G., Tyler B. A new glnA-linked regulatory gene for glutamine synthetase in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4544–4548. doi: 10.1073/pnas.76.9.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrenk W. J., Weisberg R. A. A simple method for making new transducing lines of coliphage lambda. Mol Gen Genet. 1975;137(2):101–107. doi: 10.1007/BF00341676. [DOI] [PubMed] [Google Scholar]
- Stadtman E. R., Ginsburg A., Ciardi J. E., Yeh J., Hennig S. B., Shapiro B. M. Multiple molecular forms of glutamine synthetase produced by enzyme catalyzed adenylation and deadenylylation reactions. Adv Enzyme Regul. 1970;8:99–118. doi: 10.1016/0065-2571(70)90011-7. [DOI] [PubMed] [Google Scholar]
- Stadtman E. R., Smyrniotis P. Z., Davis J. N., Wittenberger M. E. Enzymic procedures for determining the average state of adenylylation of Escherichia coli glutamine synthetase. Anal Biochem. 1979 May;95(1):275–285. doi: 10.1016/0003-2697(79)90217-3. [DOI] [PubMed] [Google Scholar]
- Woolfolk C. A., Shapiro B., Stadtman E. R. Regulation of glutamine synthetase. I. Purification and properties of glutamine synthetase from Escherichia coli. Arch Biochem Biophys. 1966 Sep 26;116(1):177–192. doi: 10.1016/0003-9861(66)90026-9. [DOI] [PubMed] [Google Scholar]



