Abstract
Background
We tested the hypothesis that genetic variation in thrombotic and inflammatory pathways is independently associated with long-term mortality following coronary artery bypass grafting (CABG).
Methods and Results
Two separate cohorts of patients undergoing CABG at a single institution were examined, and all-cause mortality between 30 days and 5 years after the index CABG was ascertained from the National Death Index. In a discovery cohort of 1018 patients, a panel of 90 single nucleotide polymorphisms (SNPs) in 49 candidate genes was tested in Cox proportional hazard models to identify clinical and genomic multivariate predictors of incident death. After adjustment for multiple comparisons and clinical predictors of mortality, the homozygote minor allele of a common variant in the thrombomodulin (THBD) gene (rs1042579) was independently associated with significantly increased risk of all-cause mortality (HR 2.26; 95%CI, 1.31–3.92; p=0.003). Six tag SNPs in the THBD gene, one of which (rs3176123) in complete linkage disequilibrium with rs1042579, were then assessed in an independent validation cohort of 930 patients. Following multivariate adjustment for the clinical predictors identified in the discovery cohort and multiple testing, the homozygote minor allele of rs3176123 independently predicted all-cause mortality (HR 3.6; 95%CI, 1.67–7.78; p=0.001).
Conclusion
In two independent cardiac surgery cohorts, linked common allelic variants in the THBD gene are independently associated with increased long-term mortality risk following CABG, and significantly improve the classification ability of traditional postoperative mortality prediction models.
Keywords: CABG, cardiac surgery, genetic association, longitudinal cohort study
Several risk stratification tools have been developed to estimate the perioperative mortality risk for patients undergoing coronary artery bypass graft (CABG) surgery. The most widely used of these instruments, the European System for Cardiac Operative Risk Evaluation (EuroSCORE), has been validated for both short- and long-term morbidity and mortality.1–3 Population-based instruments remain limited, however, in their ability to predict death for specific individuals. Although coronary artery disease, myocardial infarction and cardiovascular mortality have all been shown to be heritable conditions in ambulatory population-based cohorts,4 evidence for a genetic basis of increased mortality following surgical myocardial revascularization remains limited.5–7 We therefore used two prospectively enrolled cohorts of patients undergoing CABG with cardiopulmonary bypass (CPB) to test the coupled hypotheses that specific DNA sequence variants in candidate genes involved in inflammation, thrombosis and endothelial dysfunction are independently associated with postoperative long-term mortality, and improve the predictive ability of the logistic EuroSCORE.
Methods
The study design and reporting of the results follows the recently published “Strengthening the Reporting of Genetic Association Studies” (STREGA) recommendations.8
Description of Study Cohorts
Patients enrolled in this study were participants in the PEGASUS (Perioperative Genetics and Safety Outcomes Study), an ongoing Institutional Review Board-approved prospective longitudinal study at Duke University Medical Center (DUMC), and provided informed consent. The present substudy targeted two separate cohorts of patients undergoing CABG surgery using CPB after 1989, in whom longitudinal follow-up was conducted through the Duke Clinical Research Institute (DCRI). Patients were excluded from enrollment in PEGASUS if they had a history of renal failure, active liver disease, bleeding disorders, autoimmune diseases, or immunosuppressive therapy. A standardized fentanyl/isoflurane anesthetic was administered to all patients. Perfusion support consisted of nonpulsatile CPB (30–32°C), crystalloid prime, pump flow rates > 2.4 L/min per m2, cold blood cardioplegia, α-stat blood gas management, serial hematocrits ≥ 0.18 while on CPB, and activated clotting times > 450 seconds. All patients received antifibrinolytic infusions with either aprotinin or aminocaproic acid.
Data Collection and End-point Definition
Data regarding patient demographics, preoperative comorbidities, medications, intraoperative variables and postoperative outcomes was collected and curated during the index hospitalization through the Duke Information System for Cardiovascular Care, an integral part of the Duke Databank for Cardiovascular Diseases. Longitudinal follow-up for patients from both the discovery and validation cohorts was conducted by the DCRI Follow-up Services Group, which is responsible for collecting annual follow-up data on death and nonfatal endpoints for the Duke Databank for Cardiovascular Diseases. The annual surveys collect data on general health, hospitalizations, myocardial infarction, stroke, cardiac problems, and medication use. Patients are surveyed 6 months after the index procedure and yearly thereafter by means of a mailed, self-administered questionnaire; non-responders are surveyed by telephone. The rate of response is 95% for mortality data, and there is an annual search for of the National Death Index for patients who are lost to follow-up (2%) or who have asked to be withdrawn (3%). Information on death is collected through next-of-kin interviews, reviews of hospital discharge summaries, death certificates and the National Death Index. For the purposes of this analysis, long-term mortality is defined as all-cause death occurring more than 30 days and up to 5 years following the index CABG surgery. Exclusion of early (30-day) mortality from analyses is justified by the known differences between early and late death hazard functions following CABG.
The discovery cohort was comprised of 1822 ethnically diverse subjects who underwent CABG with CPB at DUMC between 1989 and 2002. For patients who underwent more than one surgery within that period, only data from the first surgery were included. Subjects were excluded if genotypes or phenotypic data was missing (N=250), or censored if they died within the first 30 postoperative days (N=52) or later than 5-years postoperatively (N=502), yielding 1018 patients who met eligibility criteria and were included in the analysis. The validation cohort included 951 subjects of self-reported European ancestry, who underwent CABG with CPB at DUMC between 1985 and 2007. Of those, 21 lacked genotype or phenotypic data, yielding 930 analyzed patients.
Candidate Genes and Polymorphism Selection
Forty-nine candidate genes involved in inflammation, thrombosis and endothelial dysfunction were selected a priori based on systematic reviews of the literature, previous gene expression studies,9, 10 and expert opinion. Single nucleotide polymorhism (SNP) selection was based on the SeattleSNPs Variation Discovery Resource (http://pga.gs.washinton.edu/) using data for subjects of European descent. Ninety-six single nucleotide polymorphisms (SNPs) were selected in these candidate genes with an emphasis on common (minor allele frequency >5%) and functionally important variants, and genotyped in the discovery cohort. A list of the candidate genes and polymorphism studied is provided in Supplemental Table 1.
In the validation cohort we took advantage of all tag SNPs represented on the Illumina Human610-Quad BeadChip located in candidate genes demonstrating significance in the discovery cohort. These were tested in the replication cohort in order to more completely capture common allelic variation in the respective genes. A list of the SNPs examined in the validation cohort is provided in Supplemental Table 2.
Genotype Analysis
Genomic DNA was isolated from whole blood using standard procedures. In the discovery cohort genotyping was performed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry on a Sequenom system (Sequenom, San Diego, CA) at a core facility (Agencourt Bioscience Corporation, Beverly MA). Primers used and polymorphism details can be found in Supplemental Table 3. Sequenom raw data was analyzed with the SpectroTyper 3.1 software (Sequenom, San Diego, CA), with spectra and cluster plots checked by visual inspection of intensity plots and manual curation of genotype calls. Genotyping accuracy was validated at > 99% by scoring a panel of 6 SNPs in 100 randomly selected patients using an ABI 3700 capillary sequencer (Applied Biosystems, Foster City, CA).
Patients in the validation cohort were genotyped using the Human610-Quad BeadChip (Illumina, Inc., San Diego, CA) at the Duke Genomic Analysis Facility. Analysis of the Illumina raw data was done with the Illumina BeadStudio GenCall 6.3.0 using a low GenCall score cutoff of 0.15, followed by individual examination of all intensity plots with manual curation of genotype calls.
Statistical Analysis
Categorical and continuous demographic, clinical and procedural characteristics were compared between groups using Pearson chi-square and Wilcoxon rank sum tests, respectively. The discovery and validation cohorts were first analyzed separately and then combined. Genetic association testing was performed using PLINK (v1.07)11. Hardy-Weinberg equilibrium was evaluated using an exact test in survivors; 6 SNPs departed from HWE and were excluded. After application of genotype quality control criteria, a two-stage analysis strategy was used for polymorphism selection in the discovery cohort.12 Allelic associations with long-term mortality were first assessed univariately using χ2 tests for each of the remaining 90 SNPs. The association tests were performed using additive (homozygote major allele versus heterozygote versus homozygote minor allele), dominant (homozygote major allele versus heterozygote plus homozygote minor allele), and recessive (homozygote minor allele versus heterozygote plus homozygote major allele) models of inheritance for each polymorphism to avoid assumptions regarding inheritance modes. Because of the number of comparisons performed, family-wise error rates were estimated for 90 SNP comparisons within each model by permutation.13
Polymorphisms demonstrating significant univariate association with mortality in the discovery cohort (permutation-adjusted p-values <0.05) were further analyzed in the validation cohort. Because not every SNP tested in the discovery cohort was present on the Human610-Quad BeadChip, representative ‘proxy’ SNPs in complete linkage disequilibrium (r2=1) with discovery SNPs in the genes of interest were tested for univariate association with long-term mortality in the validation cohort using χ2 tests. Bonferroni-adjusted p-values < 0.05 were considered significant.
Within the discovery cohort, covariates with a 2-tailed nominal p<0.05 in univariate analyses were used to fit a multivariable logistic regression model of all-cause mortality. The logistic EuroSCORE1 was calculated for each patient to summarize preoperative and procedural factors that influence perioperative mortality. Definitions of the 17 variables comprising the EuroSCORE risk model are presented in Supplemental Table 4. Additional demographic, intraoperative and medication use variables were added to the logistic regression equation using forward stepwise variable selection guided by Akaike’s Information Criterion (AIC). Self-reported ethnicity and year of surgery were also tested as covariates in the model. To estimate the independent prognostic utility of genetic information on 5-year all-cause mortality, SNPs univariately significant in replicated analyses were used to construct a clinico-genomic model in the discovery cohort. The predictive value of adding the SNP to the clinical model was assessed by comparing the area under the receiver operator characteristic (ROC) curves (C-statistic), as well as 3 global measures of model fit: the likelihood ratio test, AIC and Bayesian information criterion (BIC). Furthermore, in order to take advantage of time-to-event information within the dataset, a Cox proportional hazard model was constructed using the variables from the clinico-genomic model to estimate covariate-adjusted hazard ratios (HRs) and 95% confidence intervals (CIs) associated with individual SNPs in the discovery cohort.
The final clinico-genomic multivariable logistic regression and Cox proportional hazard models constructed in the discovery cohort were tested in the independent validation cohort using the same SNPs if available or ‘proxy’ SNPs in complete linkage disequilibrium with the SNP of interest, as described above.
Finally, the discovery and validation cohort were combined to increase statistical power. The net reclassification improvement (NRI) and integrated discrimination improvement (IDI) were calculated to assess the models’ respective abilities to classify survivors from non-survivors.14, 15 Multivariate logistic regression, survival analysis, ROC curve generation, and reclassification analysis were conducted using R (v2.12.2, http://www.r-project.org).
Results
Characteristics of the discovery and validation cohorts, stratified by postoperative mortality, are shown in Table 1. In the discovery cohort (N=1018), mortality was observed in 183 patients (17.9%). Of the 90 candidate SNPs examined, one demonstrated significant association with long-term mortality, namely rs1042579 within THBD, with family-wise empirical (FWER) p-values corrected for multiple comparisons by Bonferroni testing of 0.017. In producing the clinical covariate model, multivariate analysis identified the logistic EuroSCORE as strongly associated with long-term mortality (Table 2). Of the traditional clinical and procedural variables not included in the EuroSCORE, forward variable selection resulted in three additional statistically significant independent predictors of long-term mortality, namely diabetes, beta-blocker use, and statin use at hospital discharge. Specifically, intraoperative aprotinin use was not significant in multivariable analyses. Self-reported ethnicity was not significant in either univariate or multivariate analyses (p=0.74). After adjusting for demographic, clinical and procedural predictors of mortality, the THBD rs1042579 (in recessive inheritance model) remained significant with a hazard ratio (95% CI) of 2.26 (1.31–3.92). Addition of rs1042579 genotype status to the clinical logistic regression model improved the area under the ROC curve from 0.747 to 0.754, and improved the predictive value as assessed with 3 global measures of model fit (Table 2). Of note, none of the variables tested was significantly different by THBD genotype. Specifically, the mean (SD) of the number of grafts was 2.9 (0.9) in homozygote TT patients versus 3.1 (0.9) in the other patients (p=0.16), suggesting that the observed association with mortality is independent of the extent of surgical revascularization.
Table 1.
Demographic, Clinical and Procedural Characteristics of the Discovery and Validation Cohorts
| Discovery Cohort | Validation Cohort | |||||
|---|---|---|---|---|---|---|
| Characteristics | Survived (n=835) |
Died (n=183) |
P* | Survived (n=831) |
Died (n=99) |
P* |
| Demographics | ||||||
| Age (years) | 61 (11) | 67 (10) | < 0.001 | 64 (10) | 67 (10) | < 0.001 |
| Gender (% female) | 29 | 49 | 0.004 | 23 | 34 | 0.01 |
| Medical history | ||||||
| Diabetes (%) | 32 | 56 | <0.001 | 30 | 33 | 0.43 |
| Peripheral vascular disease (%) | 14 | 25 | 0.008 | 11 | 23 | < 0.001 |
| Preoperative LVEF (%) | 52 (13) | 47 (15) | < 0.001 | 56 (14) | 50 (17) | < 0.001 |
| Procedural factors | ||||||
| CBP duration (min) | 111 (55) | 125 (50) | 0.019 | 113 (37) | 141 (70) | < 0.001 |
| Intraoperative aprotinin (%) | 8 | 16 | <0.001 | 15 | 18 | 0.14 |
| Number of grafts | 3.16(0.9) | 3.04(0.8) | 0.025 | 2.25(1.2) | 2.5(1.4) | 0.38 |
| IMA use (%) | 93 | 92 | 0.57 | 97 | 93 | 0.06 |
| Logistic EuroSCORE | 3.3 [1.8–6.4] | 6.3 [2.9–14.7] | < 0.001 | 3.2 [1.9–5.7] | 6.1 [2.9–13.8] | < 0.001 |
| Medications at hospital discharge | ||||||
| Aspirin (%) | 93 | 91 | 0.2 | 91 | 94 | 0.38 |
| Statin (%) | 53 | 42 | 0.009 | 44 | 40 | 0.53 |
| Beta-blocker (%) | 87 | 74 | <0.001 | 88 | 77 | 0.03 |
| Year of surgery | 0.84 | 0.7 | ||||
| Genotype | ||||||
| THBD rs1042579 (discovery cohort only)a (%) | <0.001# | |||||
| CC | 69 | 66 | ||||
| CT | 28 | 25 | ||||
| TT | 3 | 9 | ||||
| THBD rs3176123 (validation cohort only)b (%) | 0.019# | |||||
| GG | 65 | 60 | ||||
| GT | 31 | 29 | ||||
| TT | 4 | 11 | ||||
Values expressed as mean (SD), median [interquartile range], or as %.
Wilcoxon rank-sum (continuous variables); Pearson χ2 (categorical variables).
Probability values refer to a univariate 2-degrees-of-freedom genotypic model. LVEF, left ventricular ejection fraction; CPB, cardiopulmonary bypass; THBD, thrombomodulin.
the minor allele frequency of rs1042579 in the discovery cohort was 0.17;
the minor allele frequency of rs3176123 in the validation cohort was 0.2.
Table 2.
Multivariable Models of 5-year All-Cause Mortality Using Clinical, Procedural and Genotypic Information in the Discovery and Validation Cohorts
| Model variables | HR (95% CI) | Predictor P |
Model P |
Model AIC |
Model BIC |
Model -2LLR |
C-statistic |
|---|---|---|---|---|---|---|---|
| I. Discovery cohort (N=1018) | |||||||
| Logistic EuroSCORE | 2.12 (1.71–2.63) | <0.001 | |||||
| Diabetes | 2.26 (1.67–3.05) | <0.001 | |||||
| CPB duration (min) | 1.1 (0.97–1.25) | 0.13 | |||||
| Number of grafts | 0.85 (0.72–1.01) | 0.06 | |||||
| Intraoperative aprotinin | N/A | 0.51 | |||||
| Discharge beta-blocker | 0.58 (0.41–0.82) | 0.002 | |||||
| Discharge statin | 0.67 (0.5–0.92) | 0.01 | <0.0001 | 822a | 856 a | 808 a | 0.747 a |
| rs1042579 recessive (two copies of minor allele) | 2.26 (1.31–3.92) | 0.003 | <0.0001 | 813b | 852 b | 797 b | 0.754 b |
| II. Validation cohort (N=930) | |||||||
| Logistic EuroSCORE | 1.92 (1.3–2.85) | <0.001 | |||||
| Diabetes | 0.96 (0.51–1.79) | 0.9 | |||||
| CPB duration (min) | 1.52 (1.18–1.96) | 0.004 | |||||
| Number of grafts | 1.1 (0.7–1.7) | 0.42 | |||||
| Discharge beta-blocker | 0.78 (0.39–1.56) | 0.25 | |||||
| Discharge statin | 0.59 (0.31–1.14) | 0.21 | <0.0001 | 298 a | 326 a | 284 a | 0.703 a |
| rs3176123 recessive (two copies of minor allele) | 3.6 (1.67–7.78) | 0.001 | <0.0001 | 291 b | 32 b | 275 b | 0.732 b |
The predictive values of adding the THBD SNP rs1045279 or rs3176123 recessive genotypes (2 copies of the minor allele) to the baseline clinical models of all-cause mortality are shown in the discovery and validation cohorts, respectively. P values expressed using Wald test; HR, hazard ratio; CI, confidence interval; CPB, cardiopulmonary bypass; AIC, Akaike information criterion; BIC, Bayes information criterion - AIC and BIC reward the model for covariates that contribute significantly while penalizing for each additional variable (lower numbers signify better model fit);-2LLR, negative two log likelihood ratio;
values based on clinical multivariable model only;
values based on combined clinico-genetic model.
In the validation cohort (N=930), mortality was observed in 99 patients (10.6%). A total of 6 tagging SNPs were genotyped in the THBD gene (Supplemental Table 2). Univariate association testing in the validation cohort revealed the THBD rs3176123 SNP in a recessive inheritance model to be significantly associated with long-term mortality (Bonferroni-adjusted p-value < 0.01). When included in the Cox proportional hazard clinical model developed in the discovery cohort, THBD rs3176123 remained significant, with a hazard ratio (95%CI) of 3.6 (1.67–7.78). Addition of rs3176123 to the clinical logistic regression model improved the area under the ROC curve from 0.703 to 0.732, as well as the 3 global measures of model fit (Table 2). Importantly, the two THBD variants rs1042579 (in the discovery cohort) and rs3176123 (in the validation cohort) are known to be in complete linkage disequilibrium (Supplemental Figures 1 and 2), and therefore genotype information at these loci may be used interchangeably. Survival curves for both the discovery and validation cohort analyses are presented in Figure 1.
Figure 1.
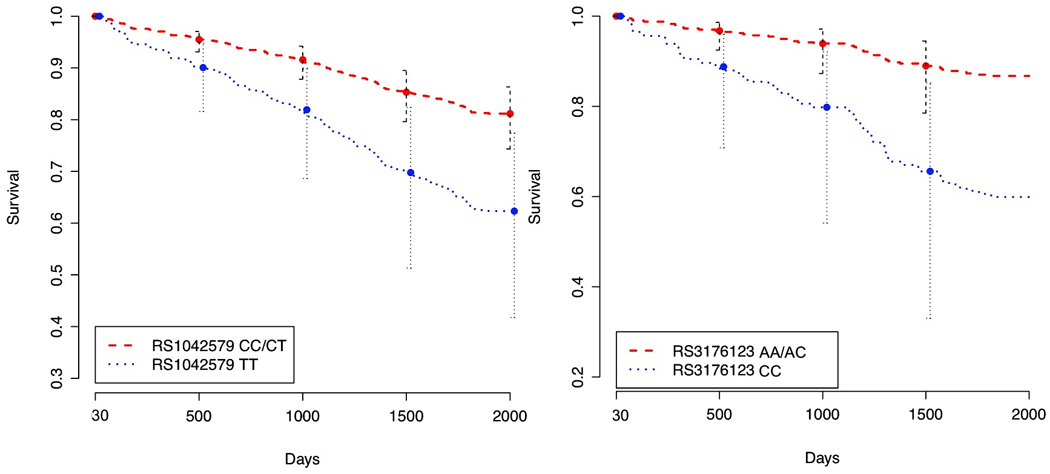
Long-term survival by THBD genotype in discovery and validation cohorts, after adjustment for clinical covariates in Cox proportional hazards models (baseline EuroSCORE, CPB duration, diabetes, number of grafts, and beta-blocker and statin use at hospital discharge). Shown are survival curves depending on the number of copies of risk alleles in rs1042579 or rs3176123 (two copies versus one or no copies) in the two cohorts, respectively.
The discovery and validation cohorts were combined for the multivariate analyses, and baseline demographics of the combined cohort can be seen in Supplemental Table 5 along with univariate associations with long-term mortality. There were a total of 282 deaths in the combined cohort during the follow-up period (14.4%). Global measures of model fit and discrimination for the combined cohort are presented in Supplemental Table 6. The resulting covariate-adjusted HR (95% CI) for patients homozygous for the recessive THBD genotype was 2.43 (1.57, 3.74). The C-statistic for the final clinico-genomic model was 0.74, compared with 0.725 for the clinical covariate model, suggesting a slight improvement in discriminatory accuracy (Supplemental Table 6, Supplemental Figure 3). Furthermore, the full genomic model’s ability to classify patients at risk improved significantly with a Net Reclassification Improvement of 0.194 (p=0.005) and an Integrated Discrimination Improvement of 0.022 (p = 0.01) when compared to the clinical covariate model. We then examined the reclassification of individual patients between risk categories by addition of THBD genotype. After defining three risk categories with cohort-specific clinical model mortality risk between 0–5%, 6–13% and >13%, we observed 96 patients correctly reclassified from the incorrect risk group to the correct risk group, and 73 patients who were incorrectly reclassified from the correct risk group to the incorrect risk group, by the addition of THBD genotype to the model, for a net gain of 23 correct reclassifications.
Discussion
In a prospective longitudinal follow-up study we found that common genetic variants in thrombomodulin are associated with long-term mortality after surgical revascularization, independent of traditional risk factors. Thrombomodulin (THBD) is an important endothelial anticoagulant protein that activates protein C, resulting in inactivation of factor Va and factor VIII, and reduction of thrombin formation. The protein, encoded by an intronless gene, is a ubiquitous endothelial-specific type I transmembrane receptor that binds thrombin and accelerates thrombin-mediated activation of protein C.16 In addition to its anticoagulant properties, THBD is also independently involved in anti-inflammatory responses, including innate immunity and complement regulation,17 mediated in part by activation of protein C as well as accelerating inactivation of complement-derived anaphylatoxins.18 The expression of THBD gene reflects endothelial activation and damage.19 Low concentrations of soluble THBD, especially when present along with elevated soluble intercellular adhesion molecule 1 (ICAM1), predispose to adverse cardiovascular events in the ambulatory population,20, 21 which formed the basis for its inclusion among the candidate genes to be studied in the context of adverse postoperative cardiovascular outcomes.
Whether THBD gene variants act as independent risk factors for adverse cardiovascular outcomes remains unclear. Several genetic studies have implicated polymorphisms within THBD to be associated with coronary artery disease,22 myocardial infarction,23 as well as composite incident cardiovascular events in non-surgical populations,24 and our findings suggest the participation of thrombomodulin in altered long-term survival of cardiac surgical patients as well. The two SNPs reported in our discovery and validation cohort analyses (rs1042579 and rs3176123, respectively)have been previously reported in high linkage disequilibrium in subjects of European(r2=1.0 Supplemental Figure 1 and 25) and Asian descent (r2=0.93),26 and for technical reasons the latter was genotyped in the validation cohort because the information they provide is similar. However, we further estimated pairwise linkage disequilibrium between the two variants in this study using a subcohort of N=421 patients genotyped at both loci; the calculated D’=0.98, r2=0.97 (Supplemental Figure 2). THBD rs1042579 is encoding for a non-synonymous aminoacid substitution (Ala473Val) in the sixth EGF-like domain of the gene, a region responsible for thrombin binding and activation of protein C. THBD rs3176123 is located in the 3’-untranslated region of the gene, and may affect mRNA stability. Both have been previously associated with plasma soluble thrombomodulin levels,25 suggesting they are functionally important. Recent reports suggest that expression of thrombomodulin on monocytes, in addition to plasma soluble levels, is associated with outcomes following CABG.26 Thus, future clinical utility of these findings could stem from identification of novel therapeutic targets for postoperative thromboprophylaxis based on THBD genetic variants.
While several models have been developed to estimate mortality risk following cardiac surgery, they are limited in their ability to predict death for specific individuals. We found that the addition of genetic information resulted in significant improvements in the ROC, the NRI, and the IDI when compared to the clinical model. All three measures suggest improved discriminatory ability of the model, thereby conferring more precise information for patients and providers evaluating the risks and benefits of CABG surgery.
The major limitation of genetic association studies is the potential for false-positive findings. We have attempted to minimize this by analyzing a relatively large population of patients in a prospective cohort design, thereby reducing the selection bias encountered in case-control genetic association studies. By selecting a limited subset of biologically relevant candidate genes, we were able to reduce the number of comparisons made during the first stage of analysis. We then repeated the analyses in an independent validation cohort to further minimize the likelihood of false-positive findings. Only SNPs from genes that were significantly associated with mortality in both the discovery and validation cohorts were incorporated into the final combined analysis. Taking advantage of the known local linkage disequilibrium structure in the THBD gene, we genotyped rs3176123 as a proxy for rs1042579, and used it in validating the risk prediction model for all-cause mortality. However, although the two variants are tightly linked, their correlation is not perfect (10% of the homozygous rs1042579 TT genotype may be the rs3176123 AC genotype, Supplemental Figure 2). Although this suggests that using the two variants interchangeably in the recessive model is not error free, it is unlikely that it would significantly confound the test statistics reported here. Previous studies suggest that the observed SNPs are putatively functional based on location within the gene and altered plasma thrombomodulin levels; we did not however conduct functional studies to further elucidate the potential biological effects of the SNPs analyzed. Specifically, we did not characterize the association between THBD variants and incidence of vein graft failure in this cohort. Finally, we found no race effect during multivariable regression modeling, and genomic control analysis found no evidence of population stratification within this data set.
In conclusion, genetic variants in thrombomodulin are independently associated with an increased risk of long-term all-cause mortality following surgical coronary revascularization, improving the predictive ability of the EuroSCORE. Although these findings lack immediate clinical utility, they delineate a potentially important genetic role in the etiology of adverse postoperative outcomes. Further research into understanding how genetic variation affects postoperative outcomes may inform antithrombotic therapies, postoperative surveillance, or lead to other novel approaches to improving care in cardiac surgical patients.
Supplementary Material
Acknowledgments
Funding Sources
These studies were supported in part by grants from the National Institutes of Health R01-HL075273 and R01-HL092071 (MVP), R01-AG09663 and HL54316 (MFN), and American Heart Association 0256342U and 9951185U (JPM), 9970128N (MFN) and 0120492U (MVP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Presented at the American Heart Association Scientific Sessions, Chicago, IL, November 14–17 2010
Journal Subject Codes: [109] Clinical genetics, [146] Genomics, [36] CV surgery: coronary artery disease
Disclosures
None.
References
- 1.Nashef SA, Roques F, Hammill BG, Peterson ED, Michel P, Grover FL, Wyse RK, Ferguson TB. Validation of European System for Cardiac Operative Risk Evaluation (EuroSCORE) in North American cardiac surgery. Eur J Cardiothorac Surg. 2002;22:101–105. doi: 10.1016/s1010-7940(02)00208-7. [DOI] [PubMed] [Google Scholar]
- 2.Nilsson J, Algotsson L, Hoglund P, Luhrs C, Brandt J. Comparison of 19 pre-operative risk stratification models in open-heart surgery. Eur Heart J. 2006;27:867–874. doi: 10.1093/eurheartj/ehi720. [DOI] [PubMed] [Google Scholar]
- 3.De Maria R, Mazzoni M, Parolini M, Gregori D, Bortone F, Arena V, Parodi O. Predictive value of EuroSCORE on long term outcome in cardiac surgery patients: a single institution study. Heart. 2005;91:779–784. doi: 10.1136/hrt.2004.037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnett DK, Baird AE, Barkley RA, Basson CT, Boerwinkle E, Ganesh SK, Herrington DM, Hong Y, Jaquish C, McDermott DA, O'Donnell CJ. Relevance of genetics and genomics for prevention and treatment of cardiovascular disease: a scientific statement from the American Heart Association Council on Epidemiology and Prevention, the Stroke Council, and the Functional Genomics and Translational Biology Interdisciplinary Working Group. Circulation. 2007;115:2878–2901. doi: 10.1161/CIRCULATIONAHA.107.183679. [DOI] [PubMed] [Google Scholar]
- 5.Muehlschlegel JD, Liu KY, Perry TE, Fox AA, Collard CD, Shernan SK, Body SC. Chromosome 9p21 variant predicts mortality after coronary artery bypass graft surgery. Circulation. 2010;122:S60–S65. doi: 10.1161/CIRCULATIONAHA.109.924233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Volzke H, Engel J, Kleine V, Schwahn C, Dahm JB, Eckel L, Rettig R. Angiotensin I-converting enzyme insertion/deletion polymorphism and cardiac mortality and morbidity after coronary artery bypass graft surgery. Chest. 2002;122:31–36. doi: 10.1378/chest.122.1.31. [DOI] [PubMed] [Google Scholar]
- 7.Zotz RB, Klein M, Dauben HP, Moser C, Gams E, Scharf RE. Prospective analysis after coronary-artery bypass grafting: Platelet GP IIIa polymorphism (HPA-1b/PlA2) is a risk factor for bypass occlusion, myocardial infarction, and death. Thromb Haemost. 2000;83:404–407. [PubMed] [Google Scholar]
- 8.Kubisz P, Stasko J, Holly P, Ivankova J, Pullmann R. More on: thrombomodulin gene polymorphisms or haplotypes as potential risk factors for venous thromboembolism: a population-based case-control study. J Thromb Haemost. 2005;3:2825–2827. doi: 10.1111/j.1538-7836.2005.01655.x. [DOI] [PubMed] [Google Scholar]
- 9.Ruel M, Bianchi C, Khan TA, Xu S, Liddicoat JR, Voisine P, Araujo E, Lyon H, Kohane IS, Libermann TA, Sellke FW. Gene expression profile after cardiopulmonary bypass and cardioplegic arrest. J Thorac Cardiovasc Surg. 2003;126:1521–1530. doi: 10.1016/s0022-5223(03)00969-3. [DOI] [PubMed] [Google Scholar]
- 10.Tomic V, Russwurm S, Moller E, Claus RA, Blaess M, Brunkhorst F, Bruegel M, Bode K, Bloos F, Wippermann J, Wahlers T, Deigner HP, Thiery J, Reinhart K, Bauer M. Transcriptomic and proteomic patterns of systemic inflammation in on-pump and off-pump coronary artery bypass grafting. Circulation. 2005;112:2912–2920. doi: 10.1161/CIRCULATIONAHA.104.531152. [DOI] [PubMed] [Google Scholar]
- 11.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. Plink: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoh J, Wille A, Zee R, Cheng S, Reynolds R, Lindpaintner K, Ott J. Selecting SNPs in two-stage analysis of disease association data: a model-free approach. Ann Hum Genet. 2000;64:413–417. doi: 10.1046/j.1469-1809.2000.6450413.x. [DOI] [PubMed] [Google Scholar]
- 13.Good PI. Permutation tests : a practical guide to resampling methods for testing hypotheses. New York: Springer; 2000. [Google Scholar]
- 14.Pencina MJ, D'Agostino RB, Sr, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30:11–21. doi: 10.1002/sim.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pencina MJ, D'Agostino RB, Sr, D'Agostino RB, Jr, Vasan RS. Evaluating the added predictive ability of a new marker: From area under the roc curve to reclassification and beyond. Stat Med. 2008;27:157–172. doi: 10.1002/sim.2929. discussion 207-112. [DOI] [PubMed] [Google Scholar]
- 16.Esmon C. Do-all receptor takes on coagulation, inflammation. Nat Med. 2005;11:475–477. doi: 10.1038/nm0505-475. [DOI] [PubMed] [Google Scholar]
- 17.Weiler H. Mouse models of thrombosis: thrombomodulin. Thromb Haemost. 2004;92:467–477. doi: 10.1160/TH04-05-0307. [DOI] [PubMed] [Google Scholar]
- 18.Delvaeye M, Noris M, De Vriese A, Esmon CT, Esmon NL, Ferrell G, Del-Favero J, Plaisance S, Claes B, Lambrechts D, Zoja C, Remuzzi G, Conway EM. Thrombomodulin mutations in atypical hemolytic-uremic syndrome. N Engl J Med. 2009;361:345–357. doi: 10.1056/NEJMoa0810739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morange PE, Simon C, Alessi MC, Luc G, Arveiler D, Ferrieres J, Amouyel P, Evans A, Ducimetiere P, Juhan-Vague I. Endothelial cell markers and the risk of coronary heart disease: the Prospective Epidemiological Study of Myocardial Infarction (PRIME) study. Circulation. 2004;109:1343–1348. doi: 10.1161/01.CIR.0000120705.55512.EC. [DOI] [PubMed] [Google Scholar]
- 20.Salomaa V, Matei C, Aleksic N, Sansores-Garcia L, Folsom AR, Juneja H, Chambless LE, Wu KK. Soluble thrombomodulin as a predictor of incident coronary heart disease and symptomless carotid artery atherosclerosis in the Atherosclerosis Risk in Communities (ARIC) study: a case-cohort study. Lancet. 1999;353:1729–1734. doi: 10.1016/s0140-6736(98)09057-6. [DOI] [PubMed] [Google Scholar]
- 21.Wu KK, Aleksic N, Ballantyne CM, Ahn C, Juneja H, Boerwinkle E. Interaction between soluble thrombomodulin and intercellular adhesion molecule-1 in predicting risk of coronary heart disease. Circulation. 2003;107:1729–1732. doi: 10.1161/01.CIR.0000064894.97094.4F. [DOI] [PubMed] [Google Scholar]
- 22.Wu KK, Aleksic N, Ahn C, Boerwinkle E, Folsom AR, Juneja H. Thrombomodulin Ala455Val polymorphism and risk of coronary heart disease. Circulation. 2001;103:1386–1389. doi: 10.1161/01.cir.103.10.1386. [DOI] [PubMed] [Google Scholar]
- 23.Ireland H, Kunz G, Kyriakoulis K, Stubbs PJ, Lane DA. Thrombomodulin gene mutations associated with myocardial infarction. Circulation. 1997;96:15–18. doi: 10.1161/01.cir.96.1.15. [DOI] [PubMed] [Google Scholar]
- 24.Auro K, Alanne M, Kristiansson K, Silander K, Kuulasmaa K, Salomaa V, Peltonen L, Perola M. Combined effects of thrombosis pathway gene variants predict cardiovascular events. PLoS Genet. 2007;3:e120. doi: 10.1371/journal.pgen.0030120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugiyama S, Hirota H, Kimura R, Kokubo Y, Kawasaki T, Suehisa E, Okayama A, Tomoike H, Hayashi T, Nishigami K, Kawase I, Miyata T. Haplotype of thrombomodulin gene associated with plasma thrombomodulin level and deep vein thrombosis in the Japanese population. Thromb Res. 2007;119:35–43. doi: 10.1016/j.thromres.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 26.Tsai CS, Tsai YT, Lin CY, Lin TC, Huang GS, Hong GJ, Lin FY. Expression of thrombomodulin on monocytes is associated with early outcomes in patients with coronary artery bypass graft surgery. Shock. 2010;34:31–39. doi: 10.1097/SHK.0b013e3181d494c4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


