Abstract
Studies of rice protein expression have increased considerably with the development of rice functional genomics. In order to obtain reliable expression results in western blotting, information on appropriate reference proteins is necessary for data normalization. To date, no published study has identified and systematically validated reference proteins suitable for the investigation of rice protein expression. In this study, nine candidate proteins were selected and their specific antibodies were obtained through immunization of rabbits with either recombinant proteins expressed in Escherichia coli or synthesized peptides. Western blotting was carried out to detect the expression of target proteins in a set of 10 rice samples representing different rice tissues/organs at different developmental stages. The expression stability of the proteins was analysed using geNorm and Microcal Origin 6.0 software. The results indicated that heat shock protein (HSP) and elongation factor 1-α (eEF-1α) were the most constantly expressed among all rice proteins tested throughout all developmental stages, while the proteins encoded by conventional internal reference genes fluctuated in amount. Comparison among the profiling of translation and transcription [expressed sequence tags (EST) and massively parallel signature sequencing (MPSS)] revealed that a correlation existed. Based on the standard curves derived from the antigen–antibody reaction, the concentrations of HSP and eEF-1α proteins in rice leaves were ∼0.12%. Under the present experimental conditions, the lower limits of detection for HSP and eEF-1α proteins in rice were 0.24 ng and 0.06 ng, respectively. In conclusion, the reference proteins selected in this study, and the corresponding antibodies, can be used in qualitative and quantitative analysis of rice proteins.
Keywords: Antibody-based proteomics, rice (Oryza sativa L.), reference gene, reference protein, western blotting
Introduction
Housekeeping genes refer to the essential genes widely expressed in vivo. These genes are responsible for most basic metabolic processes and they tend to be constitutively expressed. Therefore, housekeeping genes are usually used as reference genes when comparing the relative expression levels of different samples. In biological experiments, the selection and use of appropriate reference genes to normalize experimental results is necessary to ensure accuracy and reliability. The use of inappropriate reference genes, on the other hand, may lead to relatively large errors in a significant proportion of samples (Vandesompele et al., 2002; Huggett et al., 2005).
Ideally, reference genes should have a constant level of expression across all tissue samples under various experimental conditions (Suzuki et al., 2000). However, most reference genes display constant expression levels only under certain circumstances. For example, the commonly used reference genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH), actin, and tubulin, are not suitable as reference genes under several conditions (Thellin et al., 1999; Selvey et al., 2001; Lee et al., 2002; Gutierrez et al., 2008). Thus, selecting a suitable reference gene and using it under the appropriate conditions is critical to experimental design.
Rice (Oryza sativa L.) is one of the most important cereal crops, serving as a staple food for almost half of the world's population. It is also an important model monocot plant used for genetic and molecular studies. Based on the success of rice genome sequencing programmes (Goff et al., 2002; Yu et al., 2002), a number of whole-genome microarray systems have been established, accelerating the investigation of the rice transcriptome (Wei et al., 2009a; Wang et al., 2010). Recent studies of transcription in important biological processes, including heterosis (Wei et al., 2009a), growth and development (Jain, 2009), and stress response (Liu et al., 2008; Fujino and Matsuda, 2010), have identified several constitutive genes, providing information essential to the selection of reference genes.
Ubiquitin, elongation factor-1α (eEF-1α), and 18S and 25S rRNA are presently the most commonly used rice reference genes (Kim et al., 2003; Jain et al., 2006). The phosphatase 2A coatomer subunits and the ubiquitin-conjugating enzyme have been used for Arabidopsis thaliana (Czechowski et al., 2005). The cell division control gene, ADP-ribosylation factor, and an RNase L inhibitor-like gene have been used for wheat (Paolacci and Tanzarella, 2009), SKIP16, UKN1, and UKN2 for soybean (Jian et al., 2008), CAC, TIP41, and SAND for tomato (Exposito-Rodriguez et al., 2008), and eEF-1α for potato (Nicot et al., 2005).
The success of rice genome projects has encouraged further research on rice proteomics. A number of differentially expressed proteins in biological processes, such as rice growth (Zhao et al., 2005; Yang et al., 2007; Kim et al., 2009), stress response (Imin et al., 2004; Ali and Komatsu, 2006; Hashimoto and Komatsu, 2007; Wei et al., 2009b; Chi et al., 2010; Pandey et al., 2010), and callus differentiation (Yin et al., 2007), were identified by using the combination of two-dimensional gel electrophoresis and mass spectrometry (2DGE-MS), making rice the ‘cornerstone’ of cereal crops for proteomics research (Agrawal et al., 2009). On the basis of proteomics, in-depth data mining will reveal post-translational modifications, and the integration of proteomics and other ‘-omics’ may help to elucidate the function of target proteins.
The next step is to define the function of the proteins identified in the Rice Genome Project (Wang et al., 2006; Xiang et al., 2006). Western blotting, the method most commonly used for investigation of protein expression, is sensitive, specific, and convenient. Obviously, reference proteins are essential for normalizing western blotting data, as they are used as internal controls when adjusting the various factors involved in a given experiment. The use of Ponceau S staining (Romero-Calvo et al., 2010) and proteins encoded by commonly used reference genes, such as actin, tubulin, and GAPDH has been widely reported in the literature (Kim et al., 2003; Jain et al., 2006). However, no evidence has been presented to confirm constant protein expression encoded by these reference genes. This, in turn, may affect the accuracy of studies on protein expression (Ferguson et al., 2005). To date, no systematic identification and validation of rice reference proteins has been reported.
In this study, nine antibodies against candidate reference proteins were generated in order to detect the expression of target proteins in a set of 10 samples representing different rice tissues/organs at different developmental stages. The results indicate that heat shock protein (HSP) and eEF-1α have more stable protein expression than actin, tubulin, and GAPDH. Thus, HSP and eEF-1α are more suitable for use as reference proteins in rice protein research. The concentration of HSP and eEF-1α proteins in rice and their lower limits of detection were also examined. Taken together, the results provided supporting data for the application of antibodies in both qualitative and quantitative analyses of rice proteins.
Materials and methods
Biological materials
In this study, cDNA libraries, expression vectors pET-30a (Novagen) and pET30a-GST, a glutathione S-transferase (GST) tag-containing version of pET30a, created in our laboratory (Cao et al., 2010), and bacterial strains (DH5α, BL21, and ER2566) were used. Restriction enzymes BamHI, XhoI, EcoRI, HindIII, T4 DNA ligase, and Ex Taq DNA polymerase were purchased from TAKARA.
Rice samples
Four kinds of rice samples were collected and used for western blotting analysis in this study: (i) 10 samples from the seedling (shoot and root), tillering (leaf and stem), booting (flag leaf and young panicle), flowering (flag leaf and panicle), and filling stages (flag leaf and seed) of rice variety 93-11 (Oryza sativa L.); (ii) seven leaf samples collected at 4 h intervals starting at 12 pm within a single day; (iii) eight samples from leaves of the 4021-3, homozygous transgenic rice line with the bacterial blight resistance gene Xa21 (Xiang et al., 2006), inoculated with the incompatible Philippine race 6 of Xanthomonas oryzae pv. oryzae (Xoo) at 0, 1, 2, 4, and 8 h, and 1, 3, and 5 d; and (iv) four samples from wheat, maize, cotton, and A. thaliana leaves during the growing period. All materials were frozen using liquid nitrogen and stored at –70 °C until use.
Antigenic peptide prediction and primer design
BEPITOPE software (Odorico and Pellequer, 2003) was used to predict antigenic fragments from which those which were unique in the rice genome, once verified by BLASTP, were chosen as the antigen to generate specific antibodies against target proteins. PrimerCE software (Cao et al., 2010) was used to design the primers based on the coding sequence (CDS) downloaded from the TIGR database (http://rice.plantbiology.msu.edu/) as shown below. The underlined letters are restriction enzyme recognition sites. The detailed positions in full-length cDNA and the peptide sequences are listed in Table 1. Os09g30418.1F, 5′-CGGGATCCTTCGCCTTCCAGGCCGAGAT-3′; Os09g30418.1R, 5′-CCGCTCGAGCTCCTCAAGGTATTCCAGCTGA-3′; Os05g04510.1F, 5′-GGAATTCCTTGGCGCTCGTCTTACGGAGG-3′; Os05g04510.1R, 5′-CCCAAGCTTGCCACTAGCAACAATGCTCTTGG-3′; Os06g46770.2F, 5′-GGAATTCATGCAGATCTTTGTGAAGACCC-3′; Os06g46770.2R, 5′-CCCAAGCTTCCTGAGCCTGAGCACAAGGTG-3′; Os03g08020.1F, 5′-GGAATTCAAGAACGTTGCGGTGAAGG-3′; Os03g08020.1R, 5′-CCCAAGCTTTCATTTCTTCTTGGCGGCAG-3′; Os03g50890.1F, 5′-GGAATTCACCATTGGTGCTGAGCGTTTC-3′; Os03g50890.1R, 5′-CCCAAGCTTTTAGAAGCATTTCCTGTGCACAAT-3′.
Table 1.
Rice reference protein candidates
| Gene name | Locus number | Annotation | Mol. wt (kDa) | Antigen | Fragment length (position)/peptide sequence |
| HSP | Os09g30418.1 | Heat shock protein | 94 | Expressed protein | 182 amino acids (8–189) |
| UBQ | Os06g46770.2 | Polyubiquitin containing 7 ubiquitin monomers | 60 | Expressed protein | 150 amino acids (1–150) |
| TUB | Os01g59150.1 | Tubulin beta-4 chain | 50 | Synthesized peptide | QYQDATADEEGEYEDEEQQ |
| eEF-1α | Os03g08020.1 | Elongation factor 1-α | 49 | Expressed protein | 148AA(301–448) |
| eIF-4α | Os02g05330.1 | Eukaryotic initiation factor 4-α | 47 | Synthesized peptide | DAKHYDSKMQELLHQGDNEE |
| SAMS | Os05g04510.1 | S-Adenosylmethionine synthetase 1 | 43 | Expressed protein | 150 amino acids (151–300) |
| ACT | Os03g50890.1 | Actin-1 | 42 | Expressed protein | 128 amino acids(251–378) |
| GAPDH | Os04g40950.1 | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic | 37 | Synthesized peptide | DLVSTDFQGDNRSSIFDAKAGI |
| UBC | Os02g42314.2 | Ubiquitin-conjugating enzyme E2 | 18 | Synthesized peptide | PDSPLNCDSGNLLRSGDIRGY |
Gene cloning, protein expression, and protein purification
PCR was carried out using plasmids from the rice cDNA libraries as templates. The indicated restriction enzymes were used to digest the amplicons and vectors, which were then gel purified. Next, the ligated products were transformed into E. coli DH5α, and the recombinants were verified using sequence analysis (Beijing Genomics Institute, Beijing, China). The recombinants were transformed into the E. coli expression strain ER2566 or BL21, and cultured overnight in LB medium supplemented with kanamycin (50 μg ml−1) at 37 °C. Cultures were diluted 1:100 with fresh Luria–Bertani medium (LB medium) supplemented with kanamycin (50 μg ml−1) and 1% glucose, and cultured at 37 °C to OD600 0.6–0.8. Next, isopropyl-β-d-thiogalactopyranoside (IPTG; 0.4 μM) was added for 3 h to induce the expression of fusion proteins. The bacterial cells were harvested, ruptured by using sonication, and purified by nickel column chromatography. The target proteins were then separated by using SDS–PAGE and stained with Coomassie blue.
Antibody generation
The polyclonal antibodies were generated by immunizing healthy rabbits using the purified fusion proteins or the synthesized peptides as antigens. The protein conjugations, immunizations, and antiserum purifications were carried out by BPI (Beijing Protein Innovation Co., Ltd, Beijing, China).
Extraction of rice proteins and determination of their concentration
Rice tissue was ground into a fine powder in liquid nitrogen. An 800 μl aliquot of extraction buffer [62.5 mM TRIS-HCl (pH 7.4), 10% glycerol, 0.1% SDS, 2 mM EDTA, 1 mM phenylmethylsulphonyl fluoride (PMSF), 5% (v/v) β-mercaptoethanol] was added to each 300mg powder sample. The mixture was vortexed and then chilled on ice for 10 min. Samples were centrifuged at 12 000 rpm for 10min at 4 °C, and the supernatant was collected and stored at –70 °C. The protein concentrations of the rice samples were determined using the Bradford method (Bradford, 1976). An equal amount of rice protein was loaded and separated by SDS–PAGE and then stained by Coomassie blue.
Western blotting and signal quantification analysis
Equal amounts of rice protein from different tissues/organs were separated using SDS–PAGE and electrotransferred to a PVDF membrane (Millipore Corporation, Bedford, MA, USA) at 100 V for 60 min. The membrane was immersed in 5% non-fat milk in a TTBS solution [0.2 M TRIS-HCl (pH 7.6), 1.37 M NaCl, 0.1% Tween-20] for 1h at room temperature. The proteins were incubated with the polyclonal antibodies in 5% non-fat milk in a TTBS solution for 3 h at room temperature and subjected to three 5 min rinses in a TTBS solution. The membrane was then incubated with a horseradish peroxidase-conjugated goat anti-rabbit antibody (Zhongshan Goldenbridge Biotechnology Co., Ltd, Beijing, China) for 1 h at room temperature, and subjected to three 5 min rinses in a TTBS solution. The blot was developed with a SuperECL Plus kit (Applygen, Beijing, China), and the signal was exposed with X-ray film.
The images were scanned and the intensity of each band was captured using an ImageMaster 2D Platinum version 5.0 (GE Healthcare Amersham Bioscience). The intensity of each band was standardized as a percentage of the total intensity and the results were referred to as a relative volume that represents the relative expression abundance of the gene in the samples tested. The relative expression abundance was used to evaluate protein expression stability. Western blotting and quantification analysis were performed in at least three biological replications.
Analysis of protein expression stability
The protein expression stability was evaluated by geNorm v.3.5 (http://medgen.ugent.be/∼jvdesomp/genorm/) (Vandesompele et al., 2002) and Microcal Origin 6.0 software (Microcal Software, Northampton, MA, USA). The relative protein expression values were imported into geNorm, which calculates the gene expression stability [M value or the mean of the standard variation of a given candidate gene relative to all other genes in the given set of samples (Murthi et al., 2008; Silveira et al., 2009)] to rank proteins in various tissue samples. The lower the M value, the more stable the protein expression. In addition, the relative values of protein expression were imported into Microcal Origin 6.0, which generates a box plot to estimate the expression stability of potential reference proteins.
Generation of western blotting standard curves and determination of the concentration of reference proteins in rice
The concentration of recombinant proteins was plotted against the western blotting signal generated by the corresponding antibodies in order to estimate the linear range of detection and the lower limits of detection of the recombinant proteins. Then, a series of diluted recombinant proteins and total rice proteins were assayed in the same western blotting membrane to generate the standard curve. The standard curve was used to calculate the protein concentration and percentage of reference proteins in rice. A series of dilutions of total rice proteins were also analysed using western blotting in order to determine the lower limits of detection for the rice reference proteins.
Transcriptional analysis of rice reference genes
Transcriptional data on rice leaf, root, stem, panicle, and seed were downloaded from the MPSS (http://mpss.udel.edu/rice/) (Nakano et al., 2006) and EST databases (http://www.ncbi.nlm.nih.gov/projects/dbEST/) (Boguski et al., 1993). The transcriptional level was divided into four grades based on the intensity of the expression signal in order to compare it with the western blotting results.
Results
Selection of candidate reference proteins
The following nine candidate reference proteins were selected from among the reference genes commonly used in RT-PCR and the constitutive genes recently identified in microarray analyses: HSP, eEF-1α, ACT (actin-1), TUB (tubulin β-4 chain) and GAPDH (Jain, 2009), UBQ (polyubiquitin containing seven ubiquitin monomers) (Wang et al., 2010), SAMS (S-adenosylmethionine synthetase 1) (Peleman et al., 1989; Rosic et al., 2010), eIF-4α (eukaryotic initiation factor 4α), and UBC (ubiquitin-conjugating enzyme E2) (Jain et al., 2006). Table 1 summarizes the names, accession numbers, annotations, molecular weights, immunogen types, amplification fragments, and peptide sequences. Annotations for the genes used in this study were downloaded from TIGR Rice Genome Pseudomolecules Release 5 (Ouyang et al., 2007).
Gene cloning, protein expression, and antibody generation
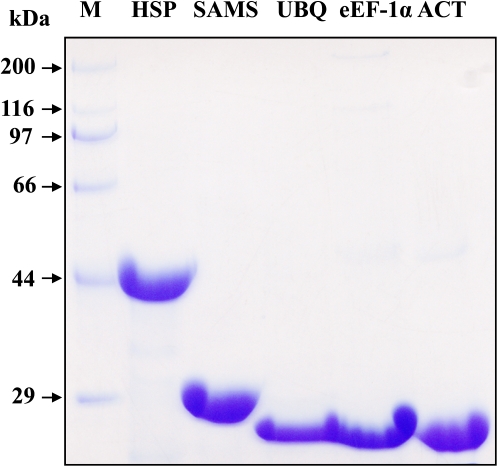
Using plasmid DNA isolated from the rice cDNA libraries as templates, the genes were amplified by PCR with gene-specific primers. Based on the results (data not shown), the sizes of the PCR products are consistent with their predicted length. The amplicons were introduced into the expression vector and the fusion proteins were induced by adding IPTG. The fusion proteins were expressed in the soluble fraction. The purity of the five expressed proteins separated by SDS–PAGE was >90% with concentrations of 1–1.5 mg ml−1 (Fig. 1), fulfilling the requirement for generating antibodies. Therefore, the purified proteins, along with the synthesized peptides, were used to immunize healthy rabbits to produce the antibodies. The specificity of the antibodies was validated using an enzyme-linked immunosorbent assay (ELISA) and western blotting (data not shown).
Fig. 1.
Escherichia coli-expressed reference protein fragments. Target genes were amplified by PCR and cloned into the expression vector, and the recombinant constructs were transformed into an E. coli strain for protein expression. The fusion proteins were purified and then separated by SDS–PAGE. The gel was stained by Commassie blue. (This figure is available in colour at JXB online.)
Expression profiling for the reference proteins
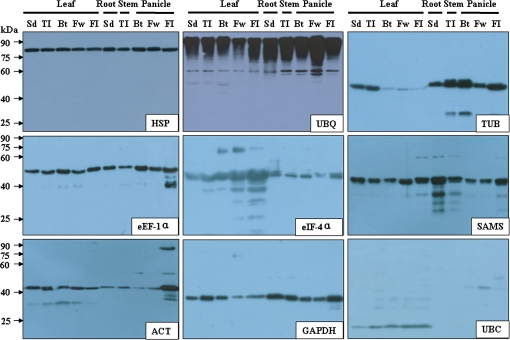
A total of 10 samples at five developmental stages were collected from rice plants growing in a field. The total proteins were extracted and the concentrations were determined. Equal amounts of protein (5 μg) were separated using SDS–PAGE. The proteins were transferred to a PVDF membrane and immunological detection was performed using the generated polyclonal antibodies.
All antibodies detected a major band at a position close to the predicted molecular weights (Fig. 2). According to western blotting analysis, both HSP and eEF-1α proteins were uniformly expressed in all samples and thus were constitutively expressed proteins. UBQ protein was also constitutively expressed. It should be noted that a batch of signal bands was detected at a higher molecular weight (>90kDa) position, implicating that a protein ubiquitination phenomena might have been detected. The TUB protein was expressed at a lower level in the leaf at the booting, flowering, and filling stages, but at a higher level in other tissues. The eIF-4α protein was expressed at a higher level in leaves. The SAMS protein was constitutively expressed in all samples. The ACT protein was expressed in all tissue samples, but had a higher abundance in the panicle at the filling stage. The GAPDH protein was constitutively expressed, while the UBC protein was only expressed in leaves. In summary, the levels of the tested proteins remained constant during at least part of the rice growing process; the behaviour pattern of each, however, varied.
Fig. 2.
Western blotting detection of rice reference proteins in tissue lysates. Sd, seedling stage; TI, tillering stage; Bt, booting stage; Fw, flowering stage; FI, filling stage. (This figure is available in colour at JXB online.)
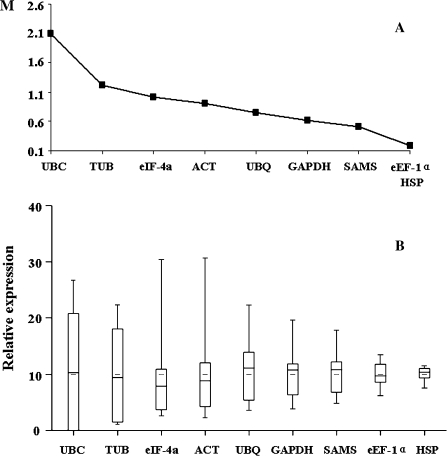
Expression stability of the reference proteins
In order to identify the proteins which were constantly expressed during rice development, stability was analysed using geNorm and Microcal Origin 6.0, and the proteins were ranked based on the results. The ranking of the reference proteins, from the least to the most stable, is as follows: UBC, TUB, eIF-4α, ACT, UBQ, GAPDH, SAMS, HSP, and eEF-1α (Fig. 3A). Microcal Origin 6.0 was used to represent the expression stability of the reference proteins visually. The box plot shows the minimum, maximum, and median expression levels of the reference proteins (Fig. 3B).The most stable proteins in all samples were HSP and eEF-1α, with coefficients of variation of 13% and 21%, respectively. In conclusion, HSP and eEF-1α were identified as the most stable reference proteins in all rice developmental stages. In contrast, the abundance of the proteins encoded by commonly used reference genes were considerably more variable, limiting their use as internal controls.
Fig. 3.
Expression stability of reference proteins in rice tissue lysates. (A) Expression stability analysed by geNorm. (B) Expression stability analysed by Microcal Origin 6.0.
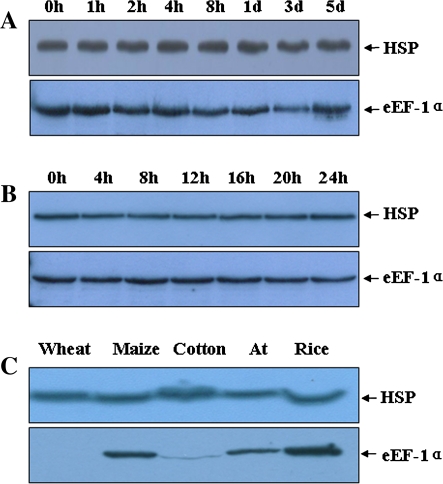
Application range of the reference proteins
In order to determine the application range of the reference proteins under different conditions, western blotting was carried out for rice leaves inoculated with an incompatible bacterial blight strain (Fig. 4A) as well as for those that were harvested at different times during a single day (Fig. 4B); the results indicated that the HSP and eEF-1α proteins were expressed constantly. In addition, leaves of wheat, maize, cotton, and A. thaliana were analysed, showing that the expression level of the anti-HSP antibody-detected signal remained constant, while that of the eEF-1α antibody-detected signal varied dramatically (Fig. 4C). Furthermore, the expression of HSP at different developmental stages for five elite rice varieties (PA64s, Te-qing, Zhenshan 97B, Guang-Lu- Ai 4, and Ai-jiao-nan-te) was also investigated and protein expression was found to be stable (XW and GL, unpublished data). These results demonstrate that the identified reference proteins can be applied under a wide range of conditions.
Fig. 4.
The application of HSP and eEF-1α as reference proteins. (A) Western blotting analysis using protein samples isolated from rice leaves inoculated with incompatible Xoo strains at the time points indicated. (B) Western blotting analysis using protein samples isolated from rice leaves at the intervals indicated, starting at 12 pm on a single day. (C) Western blotting analysis using protein samples isolated from wheat, maize, cotton, Arabidopsis thaliana (At), and rice leaves during the growing period. (This figure is available in colour at JXB online.)
Concentration of HSP and eEF-1α in rice
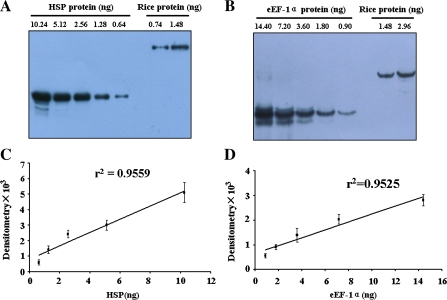
Western blotting was used to probe a series of fold dilutions of recombinant protein with the corresponding antibodies. The results indicated that the linear ranges of detection for recombinant HSP and eEF-1α were 0.41–8.2 ng and 1.5–24 ng, respectively. The lower limits of detection for the recombinant HSP and eEF-1α proteins were 0.41 ng and 0.15 ng, respectively (data not shown). A series of diluted recombinant proteins and total rice proteins were blotted in the linear ranges (Fig. 5A, B), and the standard curves were plotted (Fig. 5C, D). Based on the standard curves, the concentrations of HSP and eEF-1α reference proteins in rice were calculated, showing that 6 ng of HSP (0.12%) and 6 ng of eEF-1α (0.12%) proteins were in the total 5 μg of rice proteins. A series of fold dilutions of rice total protein were also analysed by western blotting and the lower limits of detection for the HSP and eEF-1α reference proteins in rice were 0.24 ng and 0.06 ng, respectively.
Fig. 5.
Determination of the concentration of HSP and eEF-1α proteins in rice. (A) Western blotting detection of diluted recombinant HSP and total rice proteins. (B) Western blotting detection of diluted recombinant eEF-1α and total rice proteins. (C) Standard curve of recombinant HSP protein versus the western blotting signal. (D) Standard curve of recombinant eEF-1α protein versus the western blotting signal. (This figure is available in colour at JXB online.)
Transcriptional analysis of the rice reference gene
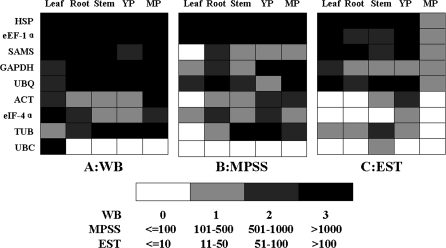
Both HSP and eEF-1α were identified as being constitutively expressed at the transcriptional level in a systematic analysis of the whole-genome microarray of rice (Jain, 2009). In order to understand further the transcription of the reference proteins, 97 978 37 signature sequences of candidate genes in rice leaf, root, stem, young panicle, and mature panicle were downloaded from the MPSS database (Supplementary Table S1 available at JXB online). A total of 571 800 ESTs of rice leaf, root, stem, young panicle, and mature panicle were downloaded from the TIGR database, and the ESTs that correspond to target genes were chosen by BLAST. The results are listed in Supplementary Table S2. The qualitative comparison among the protein expression, MPSS, and EST data suggested a correlation among the three. The MPSS and EST data also indicate that the transcriptions of HSP and eEF-1α genes were constant to a certain extent; however, those of ACT and TUB were more variable (Fig. 6).
Fig. 6.
Qualitative comparison of reference gene transcription and translation. YP, young panicle; MP, mature panicle. (A) Relative signal intensity detected by western blotting. (B) Relative signature sequence numbers detected by MPSS. (C) Relative EST number.
Discussion
In analysing protein expression, the use of the correct reference proteins has a direct impact on the reliability of the results. Thus, the identification of reference proteins and the validation of their application range are quite important. In this study, a set of nine candidate proteins were selected and western blotting analysis was carried out in order to detect their expression in a set of 10 samples representing different rice tissues/organs at different developmental stages. GeNorm and Microcal Origin 6.0 software were used to analyse the expression stability of the tested proteins. The results indicate that HSP and eEF-1α are the most stable rice reference proteins.
GeNorm operates on the principle that the expression ratio of two ideal reference genes is identical in all samples. In other words, changes in the expression ratio of two candidate reference genes indicate a decline in the stability of one or both of the genes. The gene expression stability was ranked by M value, and the two genes with the lowest M value, and therefore the most stable expression, were selected for normalization. This algorithm has been widely used for the identification of reference genes. Microcal Origin 6.0 calculates the minimum, maximum, and median expression level of reference proteins and enables researchers to compare the expression stability visually by examining the extent of variation evident in the box plot. Although the ranks generated by the two approaches differ in this study, both HSP and eEF-1α were shown to be the most stable reference proteins. In addition, both HSP and eEF-1α provided a wide range of applications as they were expressed constantly in rice materials resistant to bacterial blight, taken at different times during a single day, taken from different plants, and taken from different rice varieties.
HSPs are highly conserved among different organisms, and play important roles in protein folding, assembly, transportation, and degradation. Based on their molecular weight, HSPs can be grouped into five families: HSP100, HSP90, HSP70, HSP60, and small HSP (Schmitt et al., 2007). Because of their importance in eukaryotes, they are usually constitutively expressed and thus are used as internal controls. For example, a member of the HSP60 family has been used as a reference protein for liver tissues and hepatocellular carcinoma (Sun et al., 2009). The HSP identified as a reference protein in this study has a predicted mol. wt of 94kDa and thus belongs to the HSP90 family. Members of the HSP90 family were reported to be involved in resistance gene-mediated innate immune responses in plants (Hubert et al., 2003; Takahashi et al., 2003; Liu et al., 2004). Most of the HSPs are highly expressed. HSP90α and HSP90β, two major members of this family, account for 1–2% of all cytosolic proteins (Sreedhar et al., 2004).
eEF-1α is a protein factor that plays an important role in activating the elongation of amino acid chains in ribosomes and in regulating protein synthesis. eEF-1α is highly expressed and widely present in eukaryotic cells; for example, it accounts for 5% of the total protein in wheat embryos (Browning et al., 1990). eEF-1α is usually encoded by multiple genes to ensure its sufficient and reliable translation under stress conditions. For example, two copies of eEF-1α are in charge of translation in soybeans (Aguilar et al., 1991), and four copies are found in both the A. thaliana (Axelos et al., 1989) and rice (Kidou and Ejiri, 1998) genomes. In this study, HSP and eEF-1α proteins were found to be highly expressed in rice, each accounting for 0.12% of total rice proteins.
The western blotting results showed that most of the commonly used housekeeping genes, such as ACT and TUB, displayed considerable expression variability at the translation level. In fact, ACT and TUB have also displayed unstable expression at the transcription level. For instance, a preliminary study showed that the expression of ACT and GAPDH varied up to 2-fold among different cultivars of rice (Jain et al., 2006). Another study showed that UBQ5 and eEF-1α were the most reliable reference genes for normalization of real-time PCR data in rice, while ACT2 and β-TUB varied considerably in different rice tissues/organs (Zhang et al., 2008). Recently, 25 novel reference genes have been identified in a systematic analysis of the whole-genome microarray data for the various stages of vegetative and reproductive development in rice (Jain, 2009). Among the identified novel reference genes, HSP, the best internal control gene in the present study, was ranked as the first candidate. The eEF-1α gene, ranked second (Os03g08010) and fifth (Os03g08050) in the list, have the same amino acid sequence as the gene used in the present study. In the same study, the authors also analysed the relative expression levels of some conventional housekeeping genes, such as ACT and TUB, that have the same locus number as the genes in the present study. All these genes exhibited highly variable expression patterns in various developmental stages, a result consistent with the findings of the present study. Another systematic analysis of the genome-wide dynamic transcriptome throughout the life cycle of rice plants also identified 19 novel internal control genes (Wang et al., 2010). Among them, the UBQ (Os06g46770) ranked ninth has the same locus number as the gene used in the present study, and also had stable expression at the translation level. The qualitative comparison among protein expression, MPSS, and EST data also showed that HSP and eEF-1α were uniformly expressed during both translation and transcription, while ACT and TUB varied at these levels. These results provided further evidence for the existence of a correlation between transcription and translation.
Most of the antibodies generated in this study showed acceptable specificity and sensitivity. The lower limits of detection for HSP and eEF-1α in rice are ∼0.24ng and 0.06ng, respectively. In order to obtain more uniform antibodies with stable performance, the recombinant HSP protein was used to generate a monoclonal antibody. The results of the western blotting analysis demonstrated the consistency of monoclonal and polyclonal antibodies. The monoclonal HSP antibody routinely used in our laboratory showed stable performance (data not shown). In addition, the UBQ antibody can detect a ladder at higher molecular weight positions, and thus may be used to detect protein ubiquitination if the specificity can be validated.
Western blotting is widely performed to reveal the protein expression profile and the function of target proteins, a method that has been considered as an antibody-based proteomics (AbP) strategy (Uhlen and Ponten, 2005; Uemura et al., 2009). Compared with the conventional 2DE-MS-based strategy, AbP is easy to perform, more sensitive, and provides intuitive and quantitative results. By using this strategy, researchers can systematically investigate the target protein or protein families involved in a specific biological process. With the accumulation of rice protein-specific antibodies, the AbP strategy will be applied in more laboratories, contributing to a larger scope to the interpretation of the function of proteins and the mechanisms of biological processes.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Number of candidate gene signature sequences downloaded from the MPSS database.
Table S2. Number of candidate gene ESTs downloaded from the EST database.
Acknowledgments
The authors are grateful to Mr Yang Liu for critical reading of the manuscript. This work was supported by grants from The National Natural Sciences Foundation of China (30670175, 30730007) and 973 projects (2007CB109201, 2006CB910105).
Glossary
Abbreviations
- ACT
actin-1eEF-1αelongation factor 1-αeIF-4αeukaryotic initiation factor 4α
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase, cytosolicLB mediumLuria–Bertani medium
- MPSS
massively parallel signature sequencing
- SAMS
S-adenosylmethionine synthetase 1
- TUB
tubulin β-4 chain
- UBC
ubiquitin-conjugating enzyme E2
- UBQ
polyubiquitin containing seven ubiquitin monomers
- Xoo
Xanthomonas oryzae pv. oryzae
References
- Agrawal GK, Jwa NS, Rakwal R. Rice proteomics: ending phase I and the beginning of phase II. Proteomics. 2009;9:935–963. doi: 10.1002/pmic.200800594. [DOI] [PubMed] [Google Scholar]
- Aguilar F, Montandon PE, Stutz E. Two genes encoding the soybean translation elongation factor eEF-1 alpha are transcribed in seedling leaves. Plant Molecular Biology. 1991;17:351–360. doi: 10.1007/BF00040630. [DOI] [PubMed] [Google Scholar]
- Ali GM, Komatsu S. Proteomic analysis of rice leaf sheath during drought stress. Journal of Proteome Research. 2006;5:396–403. doi: 10.1021/pr050291g. [DOI] [PubMed] [Google Scholar]
- Axelos M, Liboz T, Bardet C, Le Van Thai A, Curie C, Lescure B. The gene family encoding the Arabidopsis thaliana translation elongation factor-1A: molecular cloning, characterization and expression. Molecular and General Genetics. 1989;219:106–112. doi: 10.1007/BF00261164. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Lowe TM, Tolstoshev CM. dbEST—database for ‘expressed sequence tags’. Nature Genetics. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Browning KS, Humphreys J, Hobbs W, Smith GB, Ravel JM. Determination of the amounts of the protein synthesis initiation and elongation factors in wheat germ. Journal of Biological Chemistry. 1990;265:17967–17973. [PubMed] [Google Scholar]
- Cao Y, Sun J, Zhu J, Li L, Liu G. PrimerCE: designing primers for cloning and gene expression. Molecular Biotechnology. 2010;46:113–117. doi: 10.1007/s12033-010-9276-3. [DOI] [PubMed] [Google Scholar]
- Chi F, Yang P, Han F, Jing Y, Shen S. Proteomic analysis of rice seedlings infected by Sinorhizobium meliloti 1021. Proteomics. 2010;10:1861–1874. doi: 10.1002/pmic.200900694. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito-Rodriguez M, Borges AA, Borges-Perez A, Perez JA. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biology. 2008;8:131. doi: 10.1186/1471-2229-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson RE, Carroll HP, Harris A, Maher ER, Selby PJ, Banks RE. Housekeeping proteins: a preliminary study illustrating some limitations as useful references in protein expression studies. Proteomics. 2005;5:566–571. doi: 10.1002/pmic.200400941. [DOI] [PubMed] [Google Scholar]
- Fujino K, Matsuda Y. Genome-wide analysis of genes targeted by qLTG3-1 controlling low-temperature germinability in rice. Plant Molecular Biology. 2010;72:137–152. doi: 10.1007/s11103-009-9559-x. [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Mauriat M, Guenin S, et al. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnology Journal. 2008;6:609–618. doi: 10.1111/j.1467-7652.2008.00346.x. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Komatsu S. Proteomic analysis of rice seedlings during cold stress. Proteomics. 2007;7:1293–1302. doi: 10.1002/pmic.200600921. [DOI] [PubMed] [Google Scholar]
- Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, Shirasu K, Dangl JL. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO Journal. 2003;22:5679–5689. doi: 10.1093/emboj/cdg547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes and Immunity. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- Imin N, Kerim T, Rolfe BG, Weinman JJ. Effect of early cold stress on the maturation of rice anthers. Proteomics. 2004;4:1873–1882. doi: 10.1002/pmic.200300738. [DOI] [PubMed] [Google Scholar]
- Jain M. Genome-wide identification of novel internal control genes for normalization of gene expression during various stages of development in rice. Plant Science. 2009;176:702–706. [Google Scholar]
- Jain M, Nijhawan A, Tyagi AK, Khurana JP. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochemical and Biophysical Research Communications. 2006;345:646–651. doi: 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- Jian B, Liu B, Bi Y, Hou W, Wu C, Han T. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Molecular Biology. 2008;9:59. doi: 10.1186/1471-2199-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidou S, Ejiri S. Isolation, characterization and mRNA expression of four cDNAs encoding translation elongation factor 1A from rice (Oryza sativa L.) Plant Molecular Biology. 1998;36:137–148. doi: 10.1023/a:1005960721762. [DOI] [PubMed] [Google Scholar]
- Kim BR, Nam HY, Kim SU, Kim SI, Chang YJ. Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnology Letters. 2003;25:1869–1872. doi: 10.1023/a:1026298032009. [DOI] [PubMed] [Google Scholar]
- Kim ST, Wang Y, Kang SY, Kim SG, Rakwal R, Kim YC, Kang KY. Developing rice embryo proteomics reveals essential role for embryonic proteins in regulation of seed germination. Journal of Proteome Research. 2009;8:3598–3605. doi: 10.1021/pr900358s. [DOI] [PubMed] [Google Scholar]
- Lee PD, Sladek R, Greenwood CM, Hudson TJ. Control genes and variability: absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Research. 2002;12:292–297. doi: 10.1101/gr.217802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang M, Duan J, Wu K. Gene expression analysis of germinating rice seeds responding to high hydrostatic pressure. Journal of Plant Physiology. 2008;165:1855–1864. doi: 10.1016/j.jplph.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Liu Y, Burch-Smith T, Schiff M, Feng S, Dinesh-Kumar SP. Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. Journal of Biological Chemistry. 2004;279:2101–2108. doi: 10.1074/jbc.M310029200. [DOI] [PubMed] [Google Scholar]
- Murthi P, Fitzpatrick E, Borg AJ, Donath S, Brennecke SP, Kalionis B. GAPDH, 18S rRNA and YWHAZ are suitable endogenous reference genes for relative gene expression studies in placental tissues from human idiopathic fetal growth restriction. Placenta. 2008;29:798–801. doi: 10.1016/j.placenta.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Nakano M, Nobuta K, Vemaraju K, Tej SS, Skogen JW, Meyers BC. Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Research. 2006;34:D731–D735. doi: 10.1093/nar/gkj077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, Evers D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. Journal of Experimental Botany. 2005;56:2907–2914. doi: 10.1093/jxb/eri285. [DOI] [PubMed] [Google Scholar]
- Odorico M, Pellequer JL. BEPITOPE: predicting the location of continuous epitopes and patterns in proteins. Journal of Molecular Recognition. 2003;16:20–22. doi: 10.1002/jmr.602. [DOI] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Research. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A, Rajamani U, Verma J, Subba P, Chakraborty N, Datta A, Chakraborty S, Chakraborty N. Identification of extracellular matrix proteins of rice (Oryza sativa L.) involved in dehydration-responsive network: a proteomic approach. Journal of Proteome Research. 2010;6:3443–3464. doi: 10.1021/pr901098p. [DOI] [PubMed] [Google Scholar]
- Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M. Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Molecular Biology. 2009;10:11. doi: 10.1186/1471-2199-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleman J, Boerjan W, Engler G, Seurinck J, Botterman J, Alliotte T, Van Montagu M, Inze D. Strong cellular preference in the expression of a housekeeping gene of Arabidopsis thaliana encoding S-adenosylmethionine synthetase. The Plant Cell. 1989;1:81–93. doi: 10.1105/tpc.1.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Calvo I, Ocon B, Martinez-Moya P, Suarez MD, Zarzuelo A, Martinez-Augustin O, de Medina FS. Reversible Ponceau staining as a loading control alternative to actin in Western blots. Analytical Biochemistry. 2010;401:318–320. doi: 10.1016/j.ab.2010.02.036. [DOI] [PubMed] [Google Scholar]
- Rosic NN, Pernice M, Rodriguez-Lanetty M, Hoegh-Guldberg O. 2010. Validation of housekeeping genes for gene expression studies in Symbiodinium exposed to thermal and light stress. Marine Biotechnology (NY) (in press) [DOI] [PubMed]
- Schmitt E, Gehrmann M, Brunet M, Multhoff G, Garrido C. Intracellular and extracellular functions of heat shock proteins: repercussions in cancer therapy. Journal of Leukocyte Biology. 2007;81:15–27. doi: 10.1189/jlb.0306167. [DOI] [PubMed] [Google Scholar]
- Selvey S, Thompson EW, Matthaei K, Lea RA, Irving MG, Griffiths LR. Beta-actin—an unsuitable internal control for RT-PCR. Molecular and Cellular Probes. 2001;15:307–311. doi: 10.1006/mcpr.2001.0376. [DOI] [PubMed] [Google Scholar]
- Silveira ED, Alves-Ferreira M, Guimaraes LA, da Silva FR, Carneiro VT. Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biology. 2009;9:84. doi: 10.1186/1471-2229-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedhar AS, Kalmar E, Csermely P, Shen YF. Hsp90 isoforms: functions, expression and clinical importance. FEBS Letters. 2004;562:11–15. doi: 10.1016/s0014-5793(04)00229-7. [DOI] [PubMed] [Google Scholar]
- Sun S, Yi X, Poon RT, Yeung C, Day PJ, Luk JM. A protein-based set of reference markers for liver tissues and hepatocellular carcinoma. BMC Cancer. 2009;9:309. doi: 10.1186/1471-2407-9-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29:332–337. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Casais C, Ichimura K, Shirasu K. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2003;100:11777–11782. doi: 10.1073/pnas.2033934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E. Housekeeping genes as internal standards: use and limits. Journal of Biotechnology. 1999;75:291–295. doi: 10.1016/s0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- Uemura N, Nakanishi Y, Kato H, Nagino M, Hirohashi S, Kondo T. Antibody-based proteomics for esophageal cancer: identification of proteins in the nuclear factor-kappaB pathway and mitotic checkpoint. Cancer Science. 2009;100:1612–1622. doi: 10.1111/j.1349-7006.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Ponten F. Antibody-based proteomics for human tissue profiling. Molecular and Cellular Proteomics. 2005;4:384–393. doi: 10.1074/mcp.R500009-MCP200. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002 doi: 10.1186/gb-2002-3-7-research0034. 3, RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xie W, Chen Y, Tang W, Yang J, Ye R, Liu L, Lin Y, Xu C, Xiao J, Zhang Q. A dynamic gene expression atlas covering the entire life cycle of rice. The Plant Journal. 2010;61:752–766. doi: 10.1111/j.1365-313X.2009.04100.x. [DOI] [PubMed] [Google Scholar]
- Wang YS, Pi LY, Chen X, et al. Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. The Plant Cell. 2006;18:3635–3646. doi: 10.1105/tpc.106.046730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Hu W, Lin Q, Cheng X, Tong M, Zhu L, Chen R, He G. Understanding rice plant resistance to the brown planthopper (Nilaparvata lugens): a proteomic approach. Proteomics. 2009b;9:2798–2808.. doi: 10.1002/pmic.200800840. [DOI] [PubMed] [Google Scholar]
- Wei G, Tao Y, Liu G, et al. A transcriptomic analysis of superhybrid rice LYP9 and its parents. Proceedings of the National Academy of Sciences, USA. 2009a;106:7695–7701. doi: 10.1073/pnas.0902340106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Cao Y, Xu C, Li X, Wang S. Xa3, conferring resistance for rice bacterial blight and encoding a receptor kinase-like protein, is the same as Xa26. Theoretical and Applied Genetics. 2006;113:1347–1355. doi: 10.1007/s00122-006-0388-x. [DOI] [PubMed] [Google Scholar]
- Yang P, Li X, Wang X, Chen H, Chen F, Shen S. Proteomic analysis of rice (Oryza sativa) seeds during germination. Proteomics. 2007;7:3358–3368. doi: 10.1002/pmic.200700207. [DOI] [PubMed] [Google Scholar]
- Yin L, Tao Y, Zhao K, Shao J, Li X, Liu G, Liu S, Zhu L. Proteomic and transcriptomic analysis of rice mature seed-derived callus differentiation. Proteomics. 2007;7:755–768. doi: 10.1002/pmic.200600611. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang X, Zhou G. A one-step real time RT-PCR assay for quantifying rice stripe virus in rice and in the small brown planthopper (Laodelphax striatellus Fallen) Journal of Virological Methods. 2008;151:181–187. doi: 10.1016/j.jviromet.2008.05.024. [DOI] [PubMed] [Google Scholar]
- Zhao C, Wang J, Cao M, Zhao K, Shao J, Lei T, Yin J, Hill GG, Xu N, Liu S. Proteomic changes in rice leaves during development of field-grown rice plants. Proteomics. 2005;5:961–972. doi: 10.1002/pmic.200401131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.