Abstract
The voltage-dependent anion channel (VDAC), a major outer mitochondrial membrane protein, is thought to play an important role in energy production and apoptotic cell death in mammalian systems. However, the function of VDACs in plants is largely unknown. In order to determine the individual function of plant VDACs, molecular and genetic analysis was performed on four VDAC genes, VDAC1–VDAC4, found in Arabidopsis thaliana. VDAC1 and VDAC3 possess the eukaryotic mitochondrial porin signature (MPS) in their C-termini, while VDAC2 and VDAC4 do not. Localization analysis of VDAC–green fluorescent protein (GFP) fusions and their chimeric or mutated derivatives revealed that the MPS sequence is important for mitochondrial localization. Through the functional analysis of vdac knockout mutants due to T-DNA insertion, VDAC2 and VDAC4 which are expressed in the whole plant body are important for various physiological functions such as leaf development, the steady state of the mitochondrial membrane potential, and pollen development. Moreover, it was demonstrated that VDAC1 is not only necessary for normal growth but also important for disease resistance through regulation of hydrogen peroxide generation.
Keywords: Arabidopsis thaliana, defence response, mitochondrial porin signature, mitochondrial membrane potential, pollen germination, voltage-dependent anion channel
Introduction
The voltage-dependent anion channel (VDAC), a highly conserved major mitochondrial outer membrane protein, is present in all eukaryotic species examined (Godbole et al., 2003). It is thought to regulate metabolite transport between mitochondria and the cytoplasm in both physiological and pathological conditions (Kroemer et al., 2007). VDAC is encoded by a small gene family; Saccharomyces cerevisiae has two VDAC isoforms, and mammals, including mice and human, have three isoforms (for a review, see Kusano et al., 2009). In plants, at least three isoforms of VDAC have been reported in Oryza sativa (rice) and Nicotiana tabacum (tobacco) (Roosens et al., 2000; Al Bitar et al., 2002, 2003; Tateda et al., 2009), and five different VDACs are present in Arabidopsis thaliana, Lotus japonicus, and Medicago truncatula (Clausen et al., 2004; Wandrey et al., 2004; Lee et al., 2009). Plants thus appear to contain more VDAC isoforms than yeast and mammals. Nevertheless, information regarding plant VDACs is fragmented, and individual functions and the physiological significance of all VDAC isoforms in one plant species have not been described yet (Kusano et al., 2009). This prompted the investigation of all of the VDAC members from one plant species. The VDAC family in A. thaliana was selected for examination because Arabidopsis is a model plant and its entire genomic sequence has been published (Arabidopsis Genome Initiative, 2000).
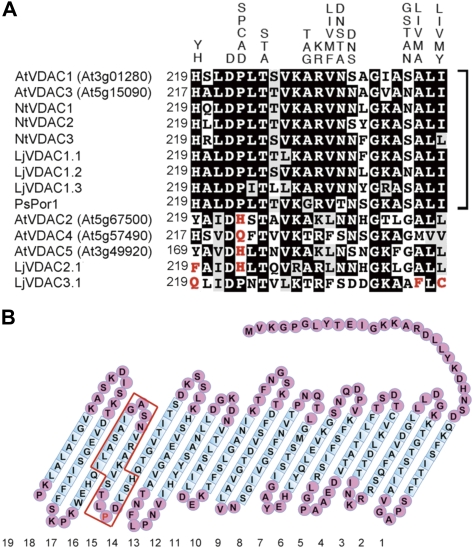
Some plant VDAC members, which are most distantly related to yeast and animal VDACs, also possess a 23 amino acid long eukaryotic mitochondrial porin signature (MPS) (PROSITE Pattern-ID:PS00558) in their C-terminal regions: [YH]-X(2)-D-[SPCAD]-X-[STA]-X(3)-[TAG]-[KR]-[LIVMF]-[DNSTA]-[DNS]-X(4)-[GSTAN]-[LIVMA]-X-[LIVMY], where X represents any amino acid, whereas the other plant members have sequences which do not match this motif (Fig. 1A). Interestingly, phylogenetic analysis of VDACs isolated from several dicotyledonous plant species are clearly classified into two groups; one carrying the conserved MPS motif and the other carrying a divergent MPS motif (Tateda et al., 2009). This suggests that the VDAC members that have canonical MPS motifs have been conserved during evolution of the plant kingdom, and that the members with diverse MPS sequences emerged by mutation. However, the functional significance of the conserved MPS and its mutated forms remains to be determined (Wandrey et al., 2004; Young et al., 2007).
Fig. 1.
Amino acid sequence alignment of MPS motif regions of plant VDACs and the predicted secondary structure of Arabidopsis VDAC1. (A) Comparison of MPS regions of VDACs from dicot plants. The alternative residues of the MPS motif are displayed above the aligned amino acid sequences by one-letter code. Identical and similar residues are highlighted with black and grey backgrounds, respectively. VDAC members which contain the conserved MPS motifs are indicated by square brackets. The residues which diverged from the conserved MPS motif are marked in red. The AGI codes or accession numbers used were AtVDAC1 (At3g01280), AtVDAC2 (At5g67500), AtVDAC3 (At5g15090), AtVDAC4 (At5g57490), AtVDAC5 (At3g49920), NtVDAC1 (AB286176), NtVDAC2 (AB286177), NtVDAC3 (AB286178), LjVDAC1.1 (AY316737), LjVDAC1.2 (AY316738), LjVDAC1.3 (AY316739), LjVDAC2.1 (AY316740), LjVDAC3.1 (AY316741), and PsPor1 (Z25540). At, Arabidopsis thaliana; Nt, Nicotiana tabacum; Lj, Lotus japonicus; Ps, Pisum sativum. (B) The secondary structure of VDAC1, as predicted by the program FORTE (http://www.cbrc.jp/forte/). Nineteen transmembrane (TM) domains were predicted for Arabidopsis VDAC1. The sequence enclosed in red corresponds to the MPS. The proline residue at amino acid 223 described in the text is marked in red.
Early studies on VDACs were mainly focused on its isoforms in animals. Functional analysis of animal VDAC members revealed that they play an important role in apoptosis by participating in the release of cytochrome c (Tsujimoto and Shimizu, 2002). Moreover, overexpression of rice VDAC in the Jurkat T-cell line induced apoptosis, which can be blocked by Bcl-2 (Godbole et al., 2003). In plants, VDAC was identified as a one of the hypersensitive response (HR) marker genes (Lacomme and Roby, 1999; Swidzinski et al., 2004). Since then, however, almost all reports described the isolation and the expressional characterization of the VDAC isoforms (for a review, see Kusano et al., 2009). Recently, it was demonstrated that Nicotiana benthamiana VDACs are involved in non-host resistance against Pseudomonas cichorii (Tateda et al., 2009). Moreover, based on the observation that hydrogen peroxide (H2O2) production in VDAC-silenced plants infected with P. cichorii was significantly reduced compared with that in control plants and that H2O2 production increased if VDAC expression was enhanced in a dexamethazone-inducible manner, it was proposed that H2O2 is involved in the VDAC-mediated defence pathway (Tateda et al., 2009).
In this study, the molecular and genetic characterization of all VDAC genes present in A. thaliana was performed through subcellular localization, histochemical monitoring of promoter–β-glucosidase (GUS) activity, and functional analysis of T-DNA knockout mutants. Moreover, the defence potential of vdac1 and vdac3 knockout plants against bacterial pathogens was examined. Based on these results, the possible functions of individual Arabidopsis VDAC members in vegetative and reproductive growth, and in pathogen defence are discussed.
Materials and methods
Plant materials and growth conditions
Two wild-type (WT) A. thaliana plants (accessions Columbia and Wassilewskija) and their T-DNA insertion lines were used in this work. The seeds were surface sterilized in a solution of 1% sodium hypochlorite and 0.1% Tween-20 for 15 min, and then washed extensively with sterilized water. The seeds were incubated in a growth chamber at 22 °C with a 16 h light/8 h dark photoperiod on 1/2 MS (Murashige and Skoog, 1962) agar medium containing 1% sucrose. Plants were also grown on soil in a growth room under the same light regime at 22 °C.
Transgenic plants carrying VDAC promoter-–GUS constructs
Herein, At3g01280, At5g67500, At5g15090, At5g57490, and At3g49920 are described as VDAC1, VDAC2, VDAC3, VDAC4, and VDAC5, respectively. The regions encompassing the VDAC1–VDAC4 promoters were amplified by PCR using the corresponding primer pairs (see Supplementary Table S1A available at JXB online) and A. thaliana genomic DNA as a template. The promoter lengths used for promoter–GUS constructs are also shown in Supplementary Table S1A. The amplified and sequence-verified fragments, termed VDAC1-pro, VDAC2-pro, VDAC3-pro, and VDAC4-pro, respectively, were digested with XbaI or SalI and BamHI restriction enzymes. The resulting fragments were subcloned into the corresponding sites of the pBI101 vector, yielding VDAC1pro-GUS, VDAC2pro-GUS, VDAC3pro-GUS, and VDAC4pro-GUS, respectively. These recombinant vectors were introduced into Agrobacterium tumefaciens strain GV3101, which was then used to produce transgenic A. thaliana seeds by means of the floral dip method (Zhang et al., 2006).
Histochemical GUS staining assays
Plant samples were treated with cold 90% acetone for 15 min on ice, and washed twice with 100 mM phosphate buffer (pH 7.0). The samples were soaked in GUS staining solution [0.5 mg ml−1 X-gluc, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, 0.1% Triton X-100, 100 mM phosphate buffer (pH 7.0), 10 mM EDTA] and subjected to weak negative pressure using a vacuum pump. The samples were further incubated in the same solution for 16 h, fixed in 70% ethanol, and then observed by light microscopy.
Construction of green fluorescent protein (GFP) fusion plasmids and microscopic observation in onion cells
The coding regions of VDAC1–VDAC4 were amplified by PCR. The VDAC1-m2 and VDAC2-M1 fragments were amplified by three-step PCR, and those of VDAC1-P223H and VDAC2-H223P by two-step PCR with the corresponding primer pairs (Supplementary Table S1B). The fragments encoding VDAC1, VDAC3, VDAC4, VDAC1-m2, and VDAC1P-223H were digested with XhoI and KpnI, while those encoding VDAC2, VDAC2-M1, and VDAC2-H223P were digested with XbaI and KpnI. The resulting fragments were subcloned into the corresponding sites of the pGFP-2 vector, yielding pAtVDAC1-GFP, pAtVDAC2-GFP, pAtVDAC3-GFP, pAtVDAC4-GFP, pAtVDAC1-m2-GFP, pAtVDAC2-M1-GFP, pAtVDAC1-P223H-GFP, and pAtVDAC2-H223P-GFP, respectively.
Constructs containing two mitochondrial markers, pmtGFP and pmtmCherry, were generated. The fragments encoding intact mCherry and truncated F0F1ATPase [nucleotide positions 1 (the first nucleotide of the start codon) to 228, abbreviated in this study as ‘mt’] were amplified by PCR with the appropriate primer pairs. The fragment encoding F0F1ATPase (1–228) was digested with XhoI and KpnI, and cloned into the corresponding sites of the pGFP-2 vector, resulting in pmtGFP. The mCherry-encoded fragment was verified by DNA sequencing. The GFP portion of pmtGFP was replaced with the KpnI and SacI double-digested mCherry-encoded fragment, yielding pmtmCherry. The resulting GFP and mCherry constructs were delivered into onion bulbs by particle bombardment. After incubating the bulbs at 22 °C for 16 h in darkness, the epidermal layers were peeled off and observed with a fluorescence microscope (BX61, Olympus).
Introduction of GFP fusion constructs into A. thaliana protoplasts and microscopic observation
Arabidopsis protoplasts were prepared from 4-week-old plant leaves and transformed with the GFP fusion constructs using 40% polyethylene glycol (PEG) solution as described previously (Yoo et al., 2007). Briefly, the protoplasts were incubated with the fusion constructs in 40% PEG solution at 23 °C for 30 min, washed with W5 solution, and further incubated overnight in darkness at 23 °C. The protoplasts were observed with a fluorescence microscope, and the z-stack images were deconvolved using Lumina Vision OL software (Mitani Corp. Ltd, Tokyo, Japan).
Evaluation of mitochondrial membrane potential (MMP)
Protoplasts, prepared as described above, were treated with Rhodamine 123 (1 μg ml−1 dissolved in W5 solution) for 5 min at room temperature, and the protoplasts were gently washed with W5 solution three times (Zhang and Xing, 2008). The protoplasts were observed with a fluorescence microscope as described in the previous section, and the fluorescence intensity of the images was detected using Lumina Vision OL.
Identification of Arabidopsis lines bearing T-DNA insertions in VDAC genes
The vdac1-3 (SALK_039833), vdac1-5 (SALK_058473C), vdac1-6 (SALK_011520C), vdac2-2 (SAIL_726_H02), vdac3-3 (SAIL_238_A01), and vdac4-2 (SALK_023908) T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center (Ohio State University, USA) (Alonso et al., 2003). The vdac3-2 line (FLAG_368B09, accession Wassilewskija) was obtained from the Institute Jean-Pierre Bourgin (Institut National de la Recherche Agronomique, France) (Samson et al., 2002). The homozygous lines were identified by PCR using the respective gene-specific primer and the left border primer of the T-DNA. The primers listed in Supplementary Table S2 at JXB online were used to confirm VDAC expression and the T-DNA insertion in the vdac mutants.
Trypan blue staining
Trypan blue staining was performed as described (Bowling et al., 1997; Mackey et al., 2002). Briefly, leaves were soaked in the dye solution consisting of a 2:1 ratio of 100% ethanol and lactic acid–phenol–trypan blue solution (25% phenol, 25% lactic acid, 25% glycerin, and 0.5 mg ml−1 trypan blue), and heated at 95 °C for 2 min. After staining, leaves were washed several times with 2.5 g ml−1 chloral hydrate solution.
Reverse transcription-PCR (RT-PCR) and real-time RT-PCR analyses
Total RNA fractions extracted using a Sepasol(R)-RNA I Super-Kit (Nacalai Tesque, Kyoto, Japan) were treated with recombinant DNase I (RNase-free) (Takara, Shiga, Japan). The RNA fraction was heat-denatured at 95 °C for 5 min, and then subjected to reverse transcription at 42 °C for 60 min using ReverTra Ace (TOYOBO, Otsu, Japan) and oligo(dT) primers. RT-PCR was performed as described (Takahashi et al., 2004). Constitutively expressed A. thaliana Act2, which served as an internal control for the assays, and PR1, PR2, and PR5 were amplified by PCR using the primers listed in Supplementary Table S3 at JXB online.
Real-time RT-PCR was performed using the StepOne™ Real time PCR system (Applied Biosystems, Foster City, CA, USA) with the FastStart Universal SYBR Green Master (ROX) (Roche, Switzerland). The primers used were designed by Primer Express Software Version 3.0 (Applied Biosystems). The primers listed in Supplementary Table S2B at JXB online were used for the two-step real-time RT-PCR that was performed with the following program: one cycle of 94 °C for 10 min, followed by 40 cycles of 94 °C for 15 s and 60 °C for 60 s. The Arabidopsis gene encoding cap-binding protein 20 (CBP20) was used as an internal control for the assays. The amount of cDNA was calculated relative to the signals of a standard dilution of the respective PCR products using StepOne™ v2.1 (Applied Biosystems).
In vitro pollen germination assay
The in vitro pollen germination assay was performed as described (Fan et al., 2001). Briefly, pollen was placed on an agar plate [1% (w/v) agar] containing germination buffer [50 mM MES (pH 5.8), 1 mM KCl, 10 mM CaCl2, 0.8 mM MgSO4, 0.01% (w/v) H3BO3, 18% (w/v) sucrose], incubated at 22 °C at 100% humidity for 12 h, and observed by light microscopy.
Infiltration of Pseudomonas syringae pv. tomato DC3000 and its derivative strains into Arabidopsis leaves and bacterial growth assays
Pseudomonas syringae pv. tomato (Pst) DC3000 and Pst DC3000 (avrRpt2) were provided by Dr J. Dangl (North Carolina University). Bacteria were grown overnight at 28 °C in LB medium containing 50 μg ml−1 rifampicin (for Pst DC3000), or rifampicin and kanamycin (50 μg ml−1 each) [for Pst DC3000 (avrRpt2)]. After centrifugation, bacterial cells were resuspended in 10 mM MgCl2 solution to 5×106 colony-forming units (cfu) ml−1 for subsequent real-time RT-PCR and cell death assay analyses, or to 1×105 cfu ml−1 for subsequent bacterial growth analysis. The resuspended bacterial culture solutions were infiltrated into Arabidopsis leaves using a needleless syringe and further incubated for a certain period of time. The number of viable bacterial cells in leaf discs was assayed.
Measurement of H2O2 content in Arabidopsis leaves
H2O2 content was measured using 2′,7′-dichlorofluorescin diacetate (DCFH-DA), as described (Sanchez et al., 1990; Tateda et al., 2009). Briefly, 20 μM DCFH-DA dissolved in distilled water was infiltrated into the plant leaf samples, which were then incubated in darkness at room temperature for 5 min. Then, leaf discs were punched out using a cork borer (inner diameter, 7 mm). The fluorescence of the samples was measured with a LAS-1000 plus Luminescent Image Analyzer (Fujifilm, Tokyo), and quantified using Multi Gauge Ver. 3.0 software (Fujifilm, Tokyo).
Statistical analysis
The data analysis was performed using the statistical tools (Student's t-test) of Microsoft Excel software.
Results
The five VDAC isoforms present in A. thaliana
Five Arabidopsis VDAC isoforms, VDAC1 (At3g01280), VDAC2 (At5g67500), VDAC3 (At5g15090), VDAC4 (At5g57490), and VDAC5 (At3g49920), which exhibit high identity (ranging from 68% to 50%), can be extracted from the GenBank database. VDACs are known as β-strand-rich proteins (Abrecht et al., 2000; Colombini, 2009). Using the FORTE program (Tomii and Akiyama, 2004; http://www.cbrc.jp/forte), the VDAC1–VDAC4 proteins were, like human VDACs, predicted to contain 19 transmembrane (TM) β-strand domains (Fig. 1B and data not shown). VDAC5 is assumed to encode the truncated protein that lacks the sixth to ninth TM domains (data not shown). The eukaryotic MPS found in VDACs (Young et al., 2007) is positioned in the 15–17th TM domains (Fig. 1B). Of the five Arabidopsis VDACs, VDAC1 and VDAC3 contain the canonical MPS motif in their C-terminal region, whereas VDAC2, VDAC4, and VDAC5 contain divergent MPS sequences (Fig. 1A).
The next step was to try detect the VDAC transcripts from A. thaliana accession Col-0. Of the five VDAC genes, it was not possible to isolate a sequence identical to the VDAC5 sequence published in the public database. The fragments amplified from reverse-transcribed cDNAs were three kinds of splicing variants of VDAC5 and their deduced amino acid sequences were truncated (data not shown). It was thus assumed that VDAC5 is a pseudo-gene. Henceforth, all of the VDAC genes present in Arabidopsis were characterized, excluding VDAC5.
Subcellular localization of VDACs
The subcellular localization of Arabidopsis VDACs in plant cells was examined using constructs consisting of GFP fused to the C-terminal regions of VDAC1–VDAC4. These VDAC–GFP fusion constructs were delivered into onion epidermal cells by particle bombardment or into protoplasts prepared from Arabidopsis leaves by PEG treatment (Fig. 2). VDAC1–GFP and VDAC3–GFP were exclusively detected in mitochondria (Fig. 2A). In contrast, the green fluorescence signals of VDAC2–GFP and VDCA4–GFP were detected not only in mitochondria, but also in another subcellular area (Fig. 2A). The mitochondrial targeting of the VDAC–GFP constructs was confirmed by a mitochondria-localized fluorescence fusion protein consisting of the truncated F0F1ATPase γ-subunit and intact fluorescent mCherry protein delivered into onion epidermal cells (Fig. 2B). As previously mentioned, VDAC1 and VDAC3 contain the conserved MPS sequence in their C-termini, whereas VDAC2 and VDAC4 do not (Fig. 1A). It was assumed that the canonical MPS sequence is important for strict targeting to mitochondria. To test this possibility, the following constructs were generated: the MPS regions (amino acid positions 219–241) of VDAC1 and VDAC2 were named M1 and m2, respectively, and the M1 of VDAC1 was replaced with m2, resulting in VDAC1-m2–GFP (Fig. 2C). Conversely, the m2 of VDAC2 was replaced with M1, resulting in VDAC2-M1–GFP (Fig. 2C). The chimeric constructs were similarly introduced into Arabidopsis protoplasts. GFP fluorescence of VDAC1-m2–GFP was observed in a pattern similar to VDAC2–GFP or VDAC4–GFP, whereas VDAC2-M1–GFP localized primarily to mitochondria, as did VDAC1–GFP or VDAC3–GFP (Fig. 2D). Next, experiments were carried out to try to establish whether a specific amino acid residue of M1 determines the strict mitochondrial localization of VDAC1. Based on a comparison of the conserved MPS sequences with those of M1 and m2, it was hypothesized that the proline at amino acid position 223 of M1 is a possible candidate residue for determining mitochondrial localization (Fig. 1). The corresponding residue of m2 is histidine. Thus, a construct that encoded VDAC1 with the m2-specific residue at position 223 (i.e. VDAC1P223H–GFP) and one that encoded VDAC2 with the M1-specific residue at position 223 (i.e. VDAC2H223P–GFP) were generated by means of site-directed mutagenesis. As expected, VDAC1P223H had the properties of VDAC2 in terms of cellular localization, whereas VDAC2H223P localized primarily to mitochondria, as did VDAC1 (Fig. 2D).
Fig. 2.
Subcellular localization of VDAC–GFP fusion proteins and their derivative proteins in Arabidopsis mesophyll protoplasts and onion epidermal cells. (A) Subcellular localization of VDAC–GFP fusion proteins in Arabidopsis protoplasts. Left, GFP fluorescence; middle, autofluorescence emitted by chlorophyll; right, merged GFP and autofluorescence images. mtGFP is a mitochondrial marker. Bars=10 μm. (B) Subcellular localization of VDAC–GFP fusion proteins in onion epidermal cells. As a reference, mitochondria-targeted F0F1ATPase-mCherry (mtmCherry) was co-bombarded into onion cells. Left, GFP fluorescence; middle, mtmCherry fluorescence; right, merged GFP and mtmCherry images. mtGFP is a mitochondrial marker. Bars=50 μm. (C) Schematic diagram of intact VDAC1 and 2, and of the chimeric proteins produced. In VDAC1, the conserved MPS motif (referred to as M1 in this study) spans from amino acid residue 219 to 241. VDAC2 contains the divergent sequence (referred to as m2 in this study). Swapped chimeras of the MPS regions of VDAC1 and VDAC2 were constructed. In VDAC1-m2, the M1 region of VDAC1 was replaced with the m2 region of VDAC2. In VDAC2-M1, the m2 region of VDAC2 was replaced with M1. (D) Subcellular localization of VDAC1 and VDAC2 derivatives in Arabidopsis protoplasts. In addition to VDAC1-m2 and VDAC2-M1 fusion constructs, mutagenized VDAC1P223H and VDAC2H223P fusion constructs were expressed in Arabidopsis protoplasts. Left, GFP fluorescence; middle, autofluorescence emitted by chlorophyll; right, merged GFP and autofluorescence images. Bars=10 μm.
Spatiotemporal analysis of the expression of four VDAC genes
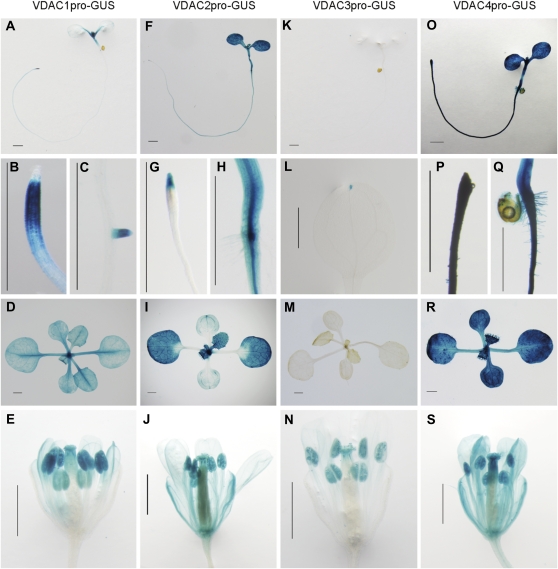
As a step toward determining the functional sites of the four VDAC genes in planta, transgenic Arabidopsis plants carrying the respective VDAC promoter–GUS constructs were generated, and the spatiotemporal expression pattern of the reporter gene was evaluated. The VDAC1 promoter was active in shoot meristems and the surrounding regions (Fig. 3A), corresponding to the root meristematic zone, but not in root caps (Fig. 3B), lateral roots (Fig. 3C), the entire leaf area (Fig. 3D), and the stigma and anthers (Fig. 3E). The activity of the VDAC2 promoter was constitutively elevated in the above-ground parts of the plant (Fig. 3F) and in the root tips (Fig. 3G) and steles (Fig. 3H). The activity was detected in whole leaves of the plants which had developed further (Fig. 3I) and in whole flower organs, including sepals (Fig. 3J). VDAC3 promoter activity was scarcely detected in leaves, except for the leaf tips (Fig. 3K–M), and rather strong activity was detected in the anthers and stigma (Fig. 3N). The VDAC4 promoter was constitutively active in whole seedlings (Fig. 3O) and in almost all organs (Fig. 3P–S).
Fig. 3.
Histochemical localization of GUS activity in transgenic Arabidopsis plants containing VDAC1–VDAC4 promoter–GUS constructs. (A–E) VDAC1pro-GUS, (F–J) VDAC2pro-GUS, (K–N) VDAC3pro-GUS, (O–S) VDAC4pro-GUS. GUS activity was assayed in: (A), (F), (K), and (O) 7-day-old seedlings; (B), (C), (G), (H), (L), (P), and (Q) root and leaf portions (at high magnification) of (A), (F), (K) and (O). (D), (I), (M), and (R) show developing leaves of 14-day-old plantlets; (E), (J), (N) and (S) show flower organs of 42-day-old plants. Bars=1 mm.
Characterization of VDAC T-DNA insertion mutant lines
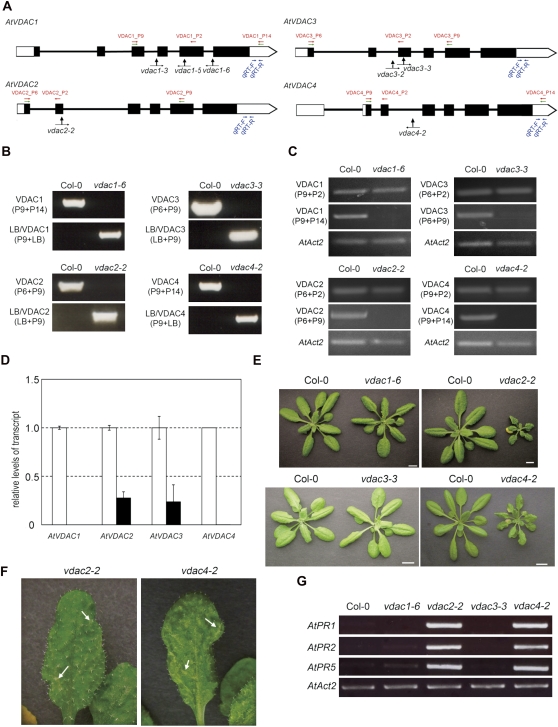
To dissect the function of VDAC genes genetically, four T-DNA insertion alleles of VDAC1, one of VDAC2, two of VDAC3, and one of VDAC4 were investigated (Fig. 4A). Of these, five lines (i.e. vdac1-6, vdac2-2, vdac3-2, vdac3-3, and vdac4-2) were obtained as homozygotes (Fig. 4B–E), whereas vdac1-3 and vdac1-5 were only obtained as heterozygotes. The level of the respective VDAC transcripts in the homozygous T-DNA insertion mutant plant was measured by RT-PCR using primer pairs which correspond to the 5′ region of the T-DNA insertion sites and which span the T-DNA insertion sites, respectively. VDAC transcripts were detected when the 5′-side primer pairs were used, but null or reduced levels of the transcripts were detected when the primers spanning the T-DNA insertion sites were used (Fig. 4C). Moreover, the RNA expression levels were quantified using the 3′-side primers of T-DNA insertions by quantitative real-time PCR. VDAC1 or VDAC4 expression was totally suppressed in vdac1-6 or vdac4-2 plants, and VDAC2 or VDAC3 expression decreased to 27% or 23% in vdac2-2 or vdac3-3, respectively, compared with that of the WT (Fig. 4D).
Fig. 4.
Characterization of A. thaliana VDAC T-DNA insertion mutants. (A) Schematic structures of VDAC1, VDAC2, VDAC3, and VDAC4, the positions and orientations of the primers used in PCR, and the positions of the T-DNA insertions. Filled boxes, exons; open regions, untranslated regions; black lines, introns. Green arrows, positions of primers used in B; red arrows, positions of primers used in C; blue arrows, positions of primers used in D. Black arrows indicate the positions of T-DNA insertions in vdac1-3, vdac1-5, vdac1-6, vdac2-2, vdac3-2, vdac3-3, and vdac4-2. (B) Genomic PCR analysis of vdac T-DNA insertion mutants. The primers used are described in parentheses. (C) RT-PCR of VDAC transcripts in vdac mutant leaves. The primers used are described in parentheses; lower, AtAct2 primer pairs. (D) Quantification of VDAC transcripts in vdac mutant leaves. White column, Col-0 plants; black column, T-DNA insertion vdac mutants. This experiment was replicated three times. (E) Phenotypes of 4-week-old vdac mutant plants grown at 22 °C. Bars=1 cm. (F) Phenotypes of rosette leaves of the mutants. Arrows indicate the dotted pale green area. (G) RT-PCR analysis of three PR genes. AtAct2 is used as a loading control.
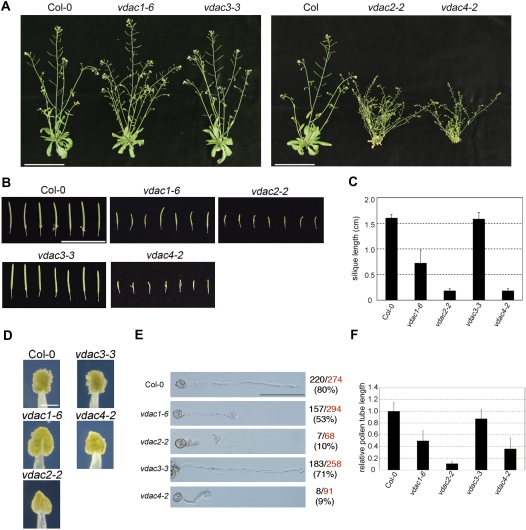
The growth rates of vdac1-6 and vdac3-3 are comparable with those of the WT (Col-0) but, interestingly, two of the mutants (vdac2-2 and vdac4-2) exhibited retarded growth rates and had a miniature phenotype compared with the WT throughout development (Fig. 4E). In addition, both mutants (vadc2-2, and vdac4-2) showed a lesion mimic phenotype (Fig. 4F). Furthermore, up-regulation of the pathogenesis-related genes PR1, PR2, and PR5 was observed in vdac2-2 and vdac4-2, but not in vdac1-6 and vdac3-3, under normal conditions (Fig. 4G).
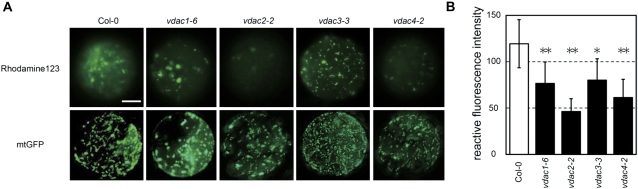
VDAC is known as an outer mitochondrial membrane component of the permeability transition (PT) pore, and the PT pore participates in the regulation of the mitochondrial membrane potential (MMP) (Kusano et al., 2009). To establish why these mutant plants exhibited diverse phenotypes, the MMP of the protoplasts prepared from the mutant plants was examined using Rhodamine123. In all of the vdac mutants tested, the MMP values were lower (mean value of vdac1-6, 64%; vdac2-2, 39%; vdac3-3, 67%; vdac4-2, 51%) than those of the WT (Fig. 5A, B). Loss of function of VDAC2 or VDAC4 resulted in a greater loss of MMP values than did loss of function of VDAC1 or VDAC3 (Fig. 5A, B).
Fig. 5.
Detection of mitochondrial membrane potential (MMP) in the protoplasts prepared from vdac mutant plants. (A) MMP in the protoplasts prepared from vdac mutant plants. Upper row, protoplasts isolated from the rosette leaves of 24-day-old plants, treated with Rhodamine123 for 5 min, and observed by fluorescence microscopy; lower row, mitochondria were visualized by green fluorescence derived from mtGFP. Bar=10 μm. (B) Quantification of MMP based on the fluorescence intensity of Rhodamine123. Significant differences between the WT and mutant samples are indicated by * (P <0.05) and ** (P <0.01). These experiments were replicated three times.
Possible function of VDAC genes during the reproductive stages
Self-crossing of the heterozygotes of vdac1-3 and vdac1-5 did not result in a vdac1/vdac1 genotype in the progeny (Table 1). In vdac1-6, the T-DNA was inserted in the most 3′-distal region of VDAC1 of all the insertion mutants tested, and the homozygous vdac1-6 line could set seeds in siliques that were slender compared with those of the WT (Col-0) (Fig. 6A–C). Moreover, the pollen germination rate of vdac1-6 was 27% lower compared with the WT, and germinated pollen tube length was also affected in vdac1-6 (Fig. 6E, F). Apparently, vdac1-6 and vdac3-3 did not show any growth retardation compared with the WT, whereas vdac2-2 and vdac4-2 grew at half the rate, taking at least a few weeks more to reach the flowering stage (Fig. 6A). The siliques of the vdac2-2 and vdac4-2 homozygous lines were short compared with those of the WT (Fig. 6B, C), but they did not set fertile seeds. Anther maturation in the vdac2-2 and vdac4-2 homozygous lines appeared to be disturbed, and their anthers contained fewer pollen grains than those of the WT (Fig. 6D). Furthermore, the germination rate of pollen obtained from vdac2-2 and vdac4-2 was reduced about one-eighth and germinated pollen tubes were also drastically shorter than those of the WT (Fig. 6E, F).
Table 1.
Segregation analysis of vdac1 mutations by PCR-based genotyping of the progeny derived from self-fertilization
| Genotype | Line name | Progeny genotype |
% WT | ||
| VDAC1/VDAC1 | VDAC1/vdac1 | vdac1/vdac1 | |||
| VDAC1/VDAC1 | Col-0 | 58 | 0 | 0 | 100.00 |
| VDAC1/vdac1 | vdac1-3 | 105 | 45 | 0 | 66.67 |
| VDAC1/vdac1 | vdac1-5 | 67 | 33 | 0 | 67.00 |
Fig. 6.
Characterization of A. thaliana vdac mutants during the flowering stage. (A) Phenotypes of flowering plants of WT, vdac1-6, vdac2-2, vdac3-3, and vdac4-2 grown at 22 °C. Left, 6-week-old WT, vdac1-6, and vdac3-3 plants; right, 6-week-old plant of the WT and 10-week-old plant of vdac2-2 and vdac4-2. Bar=10 cm. (B) Morphology of siliques produced by WT and vdac mutant plants. Bar=2 cm. (C) Silique length of WT and vdac mutant plants. (D) Anthers at the maturation stage. Bar=0.2 mm. (E) In vitro pollen germination assay. Images show typical germinated pollen. Bar=100 μm. The observed total numbers and the germination rate of pollen are shown on the right side of the panels. Red, observed total number; black, germinated number; parentheses, germination rate. (F) Relative pollen tube growth of germinated pollen in vdac1-6, vdac2-2, vdac3-3, and vdac4-2 compared with that of Col-0. The ratio of pollen tube length of vdac mutant seeds relative to that of Col-0 was displayed.
Defence response in vdac1-6 and vdac3-3 plants against bacterial pathogens
To examine the relationship between VDAC function and defence activity against the bacterial pathogen Pst, the expression profiles of VDAC genes during pathogen infection were analysed using quantitative RT-PCR. All four VDAC members were up-regulated upon infection with the bacterial pathogen. The relative transcript levels of VDAC1 and VDAC3 were enhanced to a greater degree when plants were infected with the avirulence pathogen, Pst DC3000 (avrRpt2), than with the virulence pathogen, Pst DC3000 (Fig. 7A). Next, the growth of Pst DC3000 and its derivative, Pst DC3000 (avrRpt2), was tested in the VDAC loss-of-function mutant plants. Here, two mutant plants were used, vdac1-6 and vdac3-3, since the other two mutants, vdac2-2 and vdac4-2, were too small to infiltrate with the bacterial solution and constitutively expressed PR genes (Fig. 4E, G). The host defence activity against Pst DC3000 (avrRpt2), but not against Pst DC3000, was compromised in vdac1-6 plants compared with control plants at 48 h post-inoculation (Fig. 7B, C). This phenomenon was not observed in vdac3-3 plants (Fig. 7B, C). This result indicates that VDAC1 is involved in plant defence against the avirulence pathogen Pst DC3000 (avrRpt2). Next, the production of H2O2 in vdac1-6 plants upon Pst DC3000 (avrRpt2) infection was compared with that in the pathogen-infected WT. The amount of H2O2 in vdac1-6 plants was less than half of that produced by the WT at 9 h post-inoculation (Fig. 7D). When Arabidopsis WT plants were challenged with Pst DC3000 (avrRpt2), the HR was induced and cell death was observed around the inoculation sites. Therefore, experiments were carried out to examine whether similar cell death occurred when vdac1-6 and vdac3-3 leaves were infiltrated with Pst DC3000 (avrRpt2) (inoculation size, 5×106 cfu ml−1). While cell death was observed in the majority of WT leaves by 20–24 h post-bacterial inoculation, cell death was seldom observed in vdac1-6 plants at the same time points (Fig. 7E). In the WT, 58 leaves exhibited clear cell death (C-CD; Fig. 7E, left), 21 leaves exhibited weak cell death (W-CD), and two leaves showed no cell death (N-CD) symptoms. The corresponding ratio in vdac1-6 was six (C-CD), to 60 (W-CD; Fig. 7E, right), to 16 (N-CD). In contrast, this phenomenon was not observed in vdac3-3 plants (Fig. 7F). These findings suggest that the loss of function of VDAC1 suppresses the activation of cell death.
Fig. 7.
Involvement of VDAC1 in the defence response against bacterial pathogens. (A) Quantitative expression analysis of VDAC1–VDAC4 after inoculation with 5×106 cfu ml−1 Pst DC3000 or Pst DC3000 (avrRpt2). White, light grey, and dark grey columns, control plants treated with MgCl2 solution (for 3, 6, and 9 h, respectively); light blue, turquoise, and royal blue columns, plants inoculated with Pst DC3000 (3, 6, and 9 h post-inoculation); pale pink, pink, and red columns, plants inoculated with Pst DC3000 (avrRpt2) (3, 6, and 9 h post-inoculation). Bacterial growth of Pst DC3000 (avrRpt2) (B) and Pst DC3000 (C) in Col-0 (white column), vdac1-6 (black column), and vdac3-3 (grey column) plants. Leaves were inoculated with 1×105 cfu ml−1 bacteria. Asterisks (**P <0.01) indicate a significant difference in bacterial growth between the WT and vdac1-6 hosts. (D) H2O2 production in vdac1-6 leaves upon Pst DC3000 (avrRpt2) infection. White column, Col-0 leaves; black column, vdac1-6 leaves. The leaves were infiltrated with 5×106 cfu ml−1 Pst DC3000 (avrRpt2). The average fluorescence intensity emitted by the respective control plants 3 h after bacterial infiltration was set at 1. (E) HR-like phenotype (upper panel) and trypan blue staining (lower panel) of rosette leaves at 20 h after inoculation with Pst DC3000 (avrRpt2) in the WT and vdac1-6. The leaves were infiltrated with 5×106 cfu ml−1 Pst DC3000 (avrRpt2). The ratio of phenotypes [clear cell death (C):weak cell death (W):no cell death (N)] is shown below the photos and the representative phenotypes are denoted in red. These experiments were replicated more than three times. (F) Trypan blue staining of rosette leaves at 20 h after inoculation with Pst DC3000 (avrRpt2) in the WT and vdac3-3. The same conditions were used as in E. These experiments were replicated three times.
Discussion
Subcellular localization of VDACs and the role of MPS
Whereas VDAC1 and VDAC3 contain a canonical MPS motif and are exclusively localized to mitochondria (Fig. 2A, B), VDAC2 and VDAC4 have divergent sequences in the corresponding region and are localized not only in mitochondria but also in other unidentified cellular regions (Fig. 2A, B). The Pisum sativum (pea) VDAC isoform, which bears a conserved MPS motif (Fig. 1A), was detected primarily in mitochondria (Clausen et al., 2004). Similar to the situation in Arabidopsis, three of the five VDACs present in L. japonicus contain well-conserved MPS motifs (Wandrey et al., 2004), whereas the remaining two contain divergent sequences in the corresponding region (Fig. 1A). Wandrey et al. (2004) further analysed the cellular localization of VDACs in L. japonicus cells using anti-VDAC antibody and demonstrated that they are present not only in mitochondria, but also in numerous small vesicles at the cell periphery. To our knowledge, correlations between the cellular localization of VDAC proteins and the presence or absence of an MPS motif have not been examined carefully. To address this issue, a swapping experiment between the MPS regions and the residual regions of VDAC1 (bearing the conserved MPS) and VDAC2 (containing a divergent MPS) was performed. The resulting VDAC1-m2 exhibited the same subcellular localization as that of VDAC2 and VDAC4, whereas the localization of VDAC2-M1 was the same as that of VDAC1 and VDAC3 (Fig. 2A, D), suggesting that the conserved MPS motif restricts VDAC localization to mitochondria.
Function of four VDACs, VDAC1–VDAC4, in plant growth
It was not possible to generate homozygous vdac1/vdac1 plants containing the vdac1-3 or vdac1-5 mutant allele (Table 1). Homozygous mutant seeds were obtained for vdac1-6, in which the T-DNA was inserted into the most 3' region of VDAC1. These results suggest that VDAC1 is more important than the other VDAC members for mitochondrial porin formation. The vdac1-5 and vdac1-6 alleles may encode truncated proteins that include regions 1–121 and 1–175, respectively, of the intact VDAC protein (275 amino acids long). Using the program FORTE, 19 TM domains were predicted for VDAC1. The VDAC1-6 truncated protein, which is predicted to include only the first to the 12th TM regions, retains partial activity, as the MMP, pollen germination, and pollen tube growth were affected in vdac1-6 (Figs. 5B, 6E, F). VDAC1/vdac1 (vdac1-3 or vdac1-5) was self-crossed, and the genotypes of the progeny were analysed. As seen in Table 1, the ratio of VDAC1/VDAC1 to VDAC1/vdac1 exceeds 2:1. These results suggest that the gametes bearing the vdac1-3 or vdac1-5 allele are unstable. Thus, VDAC1 is essential for growth.
VDAC3 promoter activity was rather weak and was detected only in the leaf tips of seedlings, shoot meristems, and anthers (Fig. 3). In the vdac3-3 knockout mutant, the growth phenotype does not differ from the WT and, consistent with this observation, the MMP is only 30% less than that in the WT. Furthermore, no changes in the transcript levels of the other VDAC members were observed following VDAC3 disruption (data not shown). Therefore, it is concluded that VDAC3 is not essential for plant growth.
The T-DNA insertion mutants of VDAC2 and VDAC4 exhibited severe growth retardation, which may be explained by their low MMPs. The vdac2-2 and vdac4-2 single knockout mutants exhibited clear phenotypes, even though the expression of VDAC4 in vdac2-2 and that of VDAC2 in vdac4-2 increased 2-fold (data not shown). This result suggests that VDAC2 cannot complement a defect in VDAC4, and vice versa. VDAC2 and VDAC4 have distinct functions, and both proteins may play important roles in energy production and plant growth.
All of the VDAC promoters were active in floral organs, particularly in the anthers and stigma (Fig. 3). Interestingly, the pollen produced by vdac2-2 and vdac4-2 homozygous lines failed to germinate (Fig. 6E, F), and these plants could not set fertile seeds. Heterozygous vdac1-3 and vdac1-5 could not form homozygous mutant seeds (Table 1). Moreover, the vdac1-6 homozygote could set seeds, but with very low efficiency (data not shown). Therefore, in addition to their roles in normal plant growth, VDAC1, VDAC2, and VDAC4 play important roles in the reproductive stage. For these reasons, so far it has not been possible to obtain the complementation lines such as vdac1-6/VDAC1, vdac2-2/VDAC2, and vdac4-2/VDAC4. In keeping with this, it is worth noting that knockout of VDAC3 in male mice results in defects in sperm motility (Craigen and Graham, 2008).
Defensive role of VDAC1 against avirulent bacterial pathogen
Lee et al. (2009) reported that four isoforms of Arabidopsis VDAC genes, corresponding to VDAC1–VDAC4 in the present study, were up-regulated upon inoculation with the virulent pathogen, Pst DC3000. Here a similar result was obtained (Fig. 7A). In addition, it was observed that VDAC1–VDAC4 were further up-regulated in Arabidopsis plants in response to inoculation with Pst DC3000 carrying the avirulence factor gene avrRpt2 (Fig. 7A). When vdac1-6 and vdac3-3 plants were inoculated with Pst DC3000 (avrRpt2), the host defence of vdac1-6, but not that of vdac3-3, was compromised relative to that of WT plants (Fig. 7B). Consistent with this result, H2O2 production was reduced and HR cell death was suppressed in vdac1-6 plants upon inoculation with Pst DC3000 (avrRpt2) (Fig. 7D, E). At present, it is not known how VDAC(s) regulate H2O2 production. One possibility is that the source of H2O2 production is the mitochondrion itself. The newly synthesized VDAC protein(s) during avirulence pathogen infection alter the composition of the mitochondrial outer membrane, which may trigger production of reactive oxygen species, including H2O2. Based on the result obtained, the following model is proposed: Arabidopsis plants carrying the R gene, RPS2, sense the effector protein, avrRpt2, and activate the defence signalling pathway (Jones and Dangl, 2006) in which VDAC1 is involved. The downstream component of the VDAC1-mediated defence pathway is H2O2. It should be emphasized that HR cell death was abrogated in vdac1-6 plants, suggesting that VDAC1 is involved in avirulent pathogen-triggered HR cell death. Nicotiana benthamiana VDAC genes are involved in plant defence against non-host bacterial pathogens, possibly through H2O2 production soon after sensing the pathogen (Tateda et al., 2009). To date, the data indicate that some VDAC member(s) play a key role in triggering HR cell death and/or in mounting a defence reaction against non-host and avirulent bacterial pathogens. Therefore, VDAC2 and VDAC4 might contribute to disease resistance through the regulation of H2O2 generation like VDAC1. In order to determine the function of VDAC2 and VDAC4 in defence response, an attempt was made to analyse the bacterial resistance activity in vdac2-2 and vdac4-2 plants. However, it was not possible to test the contribution of these plants in disease resistance because the mature rosette leaves of these two mutants were too small to be infiltrated with the bacterial solution. It is worth rememebering that several PR genes were constitutively expressed even under the non-stress conditions, suggestive of enhanced innate immunity. Plant VDAC expression is modulated not only by biotic stresses, but also by various abiotic stresses (Kusano et al., 2009). Recently, Yan et al. (2009) described that the transgenic Arabidopsis plants suppressing AtVDAC2 expression showed an abscisic acid (ABA)-insensitive phenotype, suggesting the presence of cross-talk between VDAC and ABA signalling.
In summary, here various pieces of novel information about Arabidopsis VDACs are presented. VDAC1 is associated with mitochondria and its disruption leads to lethality. This VDAC also functions in the reproductive stage and is involved in defence reactions against avirulent pathogens. VDAC3 expression is rather low compared with that of the other VDAC members and its knockout mutant does not show any clear phenotype. Therefore, it was not possible to investigate its function further in this study. Disruption of VDAC2 and VDAC4 functions reduces the MMP of the host cells and severely retards host growth. Interestingly, the spatiotemporal expression and cellular localization of VDAC2 and VDAC4 are quite similar, but they clearly have distinct functions. VDAC2 and VDAC4 also have important roles in the reproductive stage of development.
Supplementary data
Supplementary data are available at JXB online.
Table S1. (A) The primers and promoter length used for VDAC promoter–GUS constructs. (B) The primers used for VDAC–GFP fusion constructs, the pmtmCherry construct, domain-swapped constructs between VDAC1 and VDAC4, and the site-directed mutation constructs.
Table S2. (A) Primers used for RT-PCR to analyse the expression of VDAC genes. (B) Primers used for real-time PCR to analyse the expression of VDAC genes and AtCBP20. (C) Primers used to confirm T-DNA insertion in the vdac mutants. (D) Combination of the primers used in PCR for analysing T-DNA insertion.
Table S3. Primers used for RT-PCR to detect Arabidopsis PR1, PR2, PR5, and Act2.
Acknowledgments
We thank N.-H. Chua and J.L. Dangl for providing vectors and bacterial strains. We also thank the Arabidopsis Biological Resource Center, Nottingham Arabidopsis Stock Centre, and Institute Jean-Pierre Bourgin for providing the Arabidopsis mutant seeds. Thomas Berberich is acknowledged for critically reading the manuscript. This work was supported in part by Grant-in-Aids from the Japan Society for the Promotion of Science to CT (21-6801), TK (21380063), and YT (21780087).
Glossary
Abbreviations
- DCFH-DA
2',7'-dichlorofluorescin diacetate
- GUS
β-glucuronidase
- HR
hypersensitive response
- MMP
mitochondrial membrane potential
- MPS
mitochondrial porin signature
- PTP
permeability transition pore
- TM
transmembrane
- VDAC
voltage-dependent anion channel
References
- Abrecht H, Goormaghtigh E, Ruysschaert J-M, Homblé F. Structure and orientation of two voltage-dependent anion-selective channel isoforms. Journal of Biological Chemistry. 2000;275:40992–40999. doi: 10.1074/jbc.M006437200. [DOI] [PubMed] [Google Scholar]
- Al Bitar F, Roosens N, Boxtel JV, Dewaele E, Jacobs M, Homblé F. Expression of the rice vdac isoform2: histochemical localization and expression level. Biochimica et Biophysica Acta. 2002;1579:133–141. doi: 10.1016/s0167-4781(02)00532-8. [DOI] [PubMed] [Google Scholar]
- Al Bitar F, Roosens N, Smeyers M, Vauterin M, Boxtel JV, Jacobs M, Homblé F. Sequence analysis, transcriptional and posttranscriptional regulation of the rice vdac family. Biochimica et Biophysica Acta. 2003;1625:43–51. doi: 10.1016/s0167-4781(02)00590-0. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dongag X. The cpr5 mutant of Arabidopsis expresses both NPRl-dependent and NPRl-independent resistance. The Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen C, Ilkavets I, Thompson R, Philippar K, Vojta A, Möhlmann T, Neuhaus E, Fulgosi H, Soll J. Intracellular localization of VDAC proteins in plants. Planta. 2004;220:30–37. doi: 10.1007/s00425-004-1325-3. [DOI] [PubMed] [Google Scholar]
- Colombini M. The published 3D structure of the VDAC channel: native or not? Trends in Biochemical Sciences. 2009;34:382–389. doi: 10.1016/j.tibs.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Craigen WJ, Graham BH. Genetic strategies for dissecting mammalian and Drosophila voltage-dependent anion channel functions. Journal of Bioenergetics and Biomembranes. 2008;40:207–212. doi: 10.1007/s10863-008-9146-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L-M, Wang Y-F, Wang H, Wu W- H. In vitro Arabidopsis pollen germination and characterization of the inward potassium currents in Arabidopsis pollen grain protoplasts. Journal of Experimental Botany. 2001;52:1603–1614. [PubMed] [Google Scholar]
- Godbole A, Varghese J, Sarin A, Mathew MK. VDAC is a conserved element of death pathways in plant and animal systems. Biochimica et Biophysica Acta. 2003;1642:87–96. doi: 10.1016/s0167-4889(03)00102-2. [DOI] [PubMed] [Google Scholar]
- Jones DG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiological Reviews. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- Kusano T, Tateda C, Berberich T, Takahashi Y. Voltage-dependent anion channels: their roles in plant defense and cell death. Plant Cell Reports. 2009;28:1301–1308. doi: 10.1007/s00299-009-0741-z. [DOI] [PubMed] [Google Scholar]
- Lacomme C, Roby D. Identification of new early markers of the hypersensitive response in Arabidopsis thaliana. FEBS Letters. 1999;459:149–153. doi: 10.1016/s0014-5793(99)01233-8. [DOI] [PubMed] [Google Scholar]
- Lee SM, Hoang MHT, Han HJ, Kim HS, Lee K, Kim KE, Kim DH, Lee SY, Chung WS. Pathogen inducible voltage-dependent anion channel (AtVDAC) isoforms are localized to mitochondria membrane in Arabidopsis. Molecules and Cells. 2009;27:321–327. doi: 10.1007/s10059-009-0041-z. [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF, Wii A, Dangl JF. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Roosens N, Al Bitar F, Jacobs M, Homble F. Characterization of a cDNA encoding a rice mitochondrial voltage-dependent anion channel and its gene expression studied upon plant development and osmotic stress. Biochimica et Biophysica Acta. 2000;1463:470–476. doi: 10.1016/s0005-2736(99)00246-1. [DOI] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, Caboche M, Lecharny A. FLAGdb/FST: a database for mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Research. 2002;30:94–97. doi: 10.1093/nar/30.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez FA, Santema JS, Hilhorst R, Visser AJ. Fluorescence detection of enzymatically formed hydrogen peroxide in aqueous solution and in reversed micelles. Analytical Biochemistry. 1990;187:129–132. doi: 10.1016/0003-2697(90)90429-d. [DOI] [PubMed] [Google Scholar]
- Swidzinski JA, Leaver CJ, Sweetlove LJ. A proteomic analysis of plant programmed cell death. Phytochemistry. 2004;65:1829–1838. doi: 10.1016/j.phytochem.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Berberich T, Yamashita K, Uehara Y, Miyazaki A, Kusano T. Identification of tobacco HIN1 and two closely related genes as spermine-responsive genes and their differential expression during the Tobacco mosaic virus-induced hypersensitive response and during leaf- and flower-senescence. Plant Molecular Biology. 2004;54:613–622. doi: 10.1023/B:PLAN.0000038276.95539.39. [DOI] [PubMed] [Google Scholar]
- Tateda C, Yamashita K, Takahashi F, Kusano T, Takahashi Y. Plant voltage-dependent anion channels are involved in host defense against Pseudomonas cichorii and in Bax-induced cell death. Plant Cell Reports. 2009;28:41–51. doi: 10.1007/s00299-008-0630-x. [DOI] [PubMed] [Google Scholar]
- Tomii K, Akiyama Y. FORTE: a profile–profile comparison tool for protein fold recognition. Bioinformatics. 2004;20:594–595. doi: 10.1093/bioinformatics/btg474. [DOI] [PubMed] [Google Scholar]
- Tsujimoto S, Shimizu S. The voltage-dependent anion channel: an essential player in apoptosis. Biochimie. 2002;84:187–193. doi: 10.1016/s0300-9084(02)01370-6. [DOI] [PubMed] [Google Scholar]
- Wandrey M, Trevaskis B, Brewin N, Udvardi MK. Molecular and cell biology of a family of voltage-dependent anion channel porins in Lotus japonicus. Plant Physiology. 2004;134:182–193. doi: 10.1104/pp.103.031484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, He H, Tong S, Zhang W, Wang J, Li X, Yang Y. Voltage-dependent anion channel 2 of Arabidopsis thaliana (AtVDAC2) is involved in ABA-mediated early seedling development. International Journal of Molecular Sciences. 2009;10:2476–2486. doi: 10.3390/ijms10062476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S-D, Cho Y-H, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- Young MJ, Bay D, Hauser G, Court DA. The evolutionary history of mitochondrial porins. BMC Evolutionary Biology. 2007;7:31–52. doi: 10.1186/1471-2148-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xing D. Methyl jasmonate induces production of reactive oxygen species and alterations in mitochondrial dynamics that precede photosynthetic dysfunction and subsequent cell death. Plant and Cell Physiology. 2008;49:1092–1111. doi: 10.1093/pcp/pcn086. [DOI] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nature Protocols. 2006;1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.