Abstract
The HAP complex occurs in many eukaryotic organisms and is involved in multiple physiological processes. Here it was found that in Picea wilsonii, HAP5 (PwHAP5), a putative CCAAT-binding transcription factor gene, is involved in pollen tube development and control of tube orientation. Quantitative real-time reverse transcription-PCR showed that PwHAP5 transcripts were expressed strongly in germinating pollen and could be induced by Ca2+. Overexpression of PwHAP5 in pollen altered pollen tube orientation, whereas the tube with PwHAP5RNAi showed normal growth without diminishing pollen tube growth. Furthermore, PwFKBP12, which encodes an FK506-binding protein (FKBP) was screened and a bimolecular fluorescence complementation assay performed to confirm the interaction of PwHAP5 and PwFKBP12 in vivo. Transient expression of PwFKBP12 in pollen showed normal pollen tube growth, whereas the tube with PwFKBP12RNAi bent. The phenotype of overexpression of HAP5 on pollen tube was restored by FKBP12. Altogether, our study supported the role of HAP5 in pollen tube development and orientation regulation and identified FKBP12 as a novel partner to interact with HAP5 involved in the process.
Keywords: Growth orientation, Picea wilsonii, pollen tube, PwFKBP12, PwHAP5
Introduction
The CCAAT box, one of the most ubiquitous promoter elements in eukaryote genomes (Bucher, 1990; Maity and de Crombrugghe, 1998; Mantovani, 1999), occurs in 30% of all eukaryotic genes (Bucher, 1990; Wuarin et al., 1990). It can be recognized by a highly specific heteromeric factor called the HAP (for histone- or haem-associated protein) complex, also known as CBF (for CCAAT-binding factor) or NF-Y (for nuclear factor Y) (Mantovani, 1999), which is an evolutionarily conserved transcription factor that occurs in a wide range of eukaryotes from yeast to humans. The HAP complex in mammals and plants (known as NF-Y in mammals) includes three subunits: NF-YA (CBF-B or HAP2), NF-YB (CBF-A or HAP3), and NF-YC (CBF-C or HAP5), which are required for DNA binding of the complex and are sufficient for transcriptional activity (Maity and de Crombrugghe, 1998; Mantovani, 1999). In yeast, the HAP complex is composed of four subunits: HAP2, HAP3, HAP4, and HAP5 (McNabb and Pinto, 2005). In contrast to yeast and mammals, in which a single gene generally encodes each subunit, plants have greatly expanded subunit classes. For example, in Arabidopsis, 10, 11, and 13 genes encode the HAP2, HAP3, and HAP5 subunits, respectively (Riechmann et al., 2000), and rice has 10 HAP2 genes, 11 HAP3 genes, and 7 HAP5 genes (Thirumurugan et al., 2008). Thus, the large number of HAP2/HAP3/HAP5 heterotrimer combinations in plants provides the potential for the HAP complex to be recruited into a wide range of processes and play diverse roles in gene transcription in higher plants (Edwards et al., 1998). However, relatively little is known about the biological function of the HAP complex in plants compared with its role in yeast and mammals, which has been extensively analysed (Pinkham and Guarente, 1985; Dang et al., 1996; Mantovani, 1999).
A growing body of evidence indicates that individual plant HAP subunits function in numerous physiological processes, including embryogenesis and seed maturation (Lotan et al., 1998; Kwong et al., 2003; Lee et al., 2003; Yazawa and Kamada, 2007; Yamamoto et al., 2009), chloroplast biogenesis (Miyoshi et al., 2003), meristem growth (Combier et al., 2006), and stress responses (Nelson et al., 2007; Liu and Howell, 2010). The first identified plant HAP gene, LEAFY COTYLEDON1 (LEC1), in Arabidopsis and its most closely related subunit, LEC1-LIKE (L1L), which is similar to AtHAP3 subunits, controls embryogenesis and seed maturation via interaction with ABA-response element (ABRE)-binding factor, bZIP67 (Lotan et al., 1998; Kwong et al., 2003; Lee et al., 2003; Yamamoto et al., 2009). Similarly, C-LEC1 in carrot was shown to be a functional orthologue of LEC1 that regulates gene expression during carrot embryo development (Yazawa and Kamada, 2007). In particular, HAP subunits are involved in flowering regulation, and changes in member activities can influence flowering time (Ben-Naim et al., 2006; Wenkel et al., 2006; Cai et al., 2007; Chen et al., 2007; Kumimoto et al., 2008). Wenkel et al. (2006) revealed that the CCT-domain protein CO forms a heterotrimeric CO–AtHAP3–AtHAP5 complex and functions in the photoperiodic flowering pathway. AtHAP3b promotes flowering by enhancing the expression of key flowering time genes in the photoperiod-regulated flowering pathway, but is not involved in flowering promoted by gibberellin or vernalization (Cai et al., 2007; Chen et al., 2007; Kumimoto et al., 2008). Subsequently, Wei et al. (2010) identified DTH8 in rice, which encodes a putative HAP3 subunit and appears to play an important role as a novel suppressor in the signal network of photoperiodic flowering, as well as regulation of plant height and yield potential (Wei et al., 2010). Most recently, HAP5 subunits have been shown to be required for CONSTANS-mediated, photoperiod-dependent flowering and involved in the regulation of photosynthesis-related genes in plants (Kumimoto et al., 2010). Despite the many reports concerning the roles of HAP subunits in flowering time regulation, whether they function in other plant sexual reproduction processes, such as pollen tube development, is not known.
During sexual reproduction in flowering plants, pollen grains land on the stigma of the pistil and germinate into a cylindrical tube, which penetrates the stigma, grows through the transmitting tract, and orients towards the ovule to deliver the sperm cell for fertilization. Thus, pollen germination and tube growth are key steps in the success of fertilization. Tube growth and sperm delivery in conifers are fundamentally different from those in angiosperms (Singh, 1978). Pollen tubes of coniferous species, which represent a major evolutionary divergence in the development of the male gametophytes (Lazzaro et al., 2005), with little information regarding this development available, grow more slowly than angiosperm pollen tubes; they also tend to ramify, and lack a tip-to-base zonation of organelles (Knuiman et al., 1996), implying that the development and fertilization mechanisms of the tubes of the two types of plant differ.
In this study, a gene encoding the HAP5 transcription factor in Picea wilsonii Mast. was isolated and characterized. The expression pattern of PwHAP5 was examined and it was demonstrated that PwHAP5 was induced by Ca2+, and that overexpression of PwHAP5 in pollen altered the orientation of pollen tube growth. Moreover, a novel HAP5-interacting protein, an FK506-binding protein (FKBP12), was identified, which interacts with the C-terminal, N-terminal, and full-length sequence of PwHAP5 in vivo to regulate pollen tube development. These results provide a novel function for this protein in the control of pollen tube orientation by interaction with PwHAP5.
Materials and methods
Plant material
Cones with mature pollen were collected in mid-April 2008 from mature P. wilsonii Mast. trees in the Beijing Botanical Garden, Institute of Botany, Chinese Academy of Sciences, and were dried overnight at room temperature. The dry pollen was stored at –80 °C until use.
In vitro P. wilsonii pollen germination
P. wilsonii pollen grains stored at –80 °C were resuscitated by transfer to 4 °C for 12 h, and then to room temperature for another 2 h. The resuscitated pollen was cultured in standard liquid pollen germination and tube growth medium [12% sucrose, 0.03% Ca(NO3)2, 0.01% H3BO3, and 5 mM citrate-phosphate buffer, pH 5.8]. Pollen grains were incubated in small dishes at 25±1 °C in a saturated atmosphere (100% relative humidity) and experimentally sampled at 6, 12, 18, 24, 30, and 36 h after germination.
RNA extraction
For RNA isolation, the plant tissues were harvested separately, frozen in liquid nitrogen, and stored at –80 °C until use. Total RNA from germinating pollen was isolated using Trizol reagent (Gibco-BRL, Grand Island, NY, USA) according to the manufacturer's instructions. Total RNA from other tissues was extracted using standard cetyltrimethylammonium bromide (CTAB) extraction and lithium chloride precipitation as described previously (Chang et al., 1993). The quality and quantity of RNA was assessed by agarose gel electrophoresis and UV spectroscopy, respectively.
cDNA library construction and screening
Poly(A) RNA (0.5 μg) isolated from P. wilsonii pollen incubated in Ca2+-stressed medium (0.1% Ca2+ concentration) for 12 h was used to synthesize first-strand cDNA, which was then amplified via long-distance PCR according to the manufacturer's protocol (SMART™ cDNA Library Construction Kit; Clontech, Palo Alto, CA, USA). The original cDNA library, estimated to contain 5×105 independent recombinants, was amplified with Escherichia coli XL-Blue before screening. The cDNA library was screened by differential hybridization (one hybridization with a Ca2+-untreated pollen cDNA probe and another with a Ca2+-treated pollen cDNA probe) as described previously (Wu et al., 2004). One cDNA clone, PwHAP5, is described in this study.
RT-PCR and quantitative real-time RT-PCR
Total RNA extracted from each tissue was treated with DNase I to remove genomic DNA, after which the cDNA was synthesized. Reverse transcription (RT) reactions were performed using M-MLV reverse transcriptase (Promega, Madison, WI, USA) with oligo(dT) primers. Each sample was assayed in triplicate. The PwHAP5 gene was amplified using the gene-specific primers 5′-ATCAGCAGCAGCCCACAA-3′ and 5′-ATCCTCGTCTGCCTTCAT-3′. An elongation factor α gene (EF1-α) was amplified as an internal control in the RT-PCRs, and the primer sequence used was as previously described (Yu et al., 2009).
For quantitative RT-PCR analysis, SYBR Green Real-Time PCR Master Mix (Toyobo, Osaka, Japan) and Chromo 4 Real-Time PCR Detector (Bio-Rad, Hercules, CA, USA) were used. Quantitative real-time RT-PCR analysis of PwHAP5 was performed using the specific primers of the PwHAP5 gene, 5′-GGTAACCAGATGAGGGAG-3′ and 5′-TCTTGTTCTCCTCGGTGT-3′. EF1-α mRNA expression was amplified as an internal control for real-time analysis, using the specific primers 5′-AACTGGAGAAGGAACCCAAG-3′ and 5′-AACGACCCAATGGAGGATAC-3′. At least two different biological replicates were used for all RT-PCR determinations, which were performed with 30 cycles of denaturation, annealing, and extension steps for each sample. Quantitative PCR experiments were repeated independently three times.
Determination of RNAi specificity
In the experiments reported here, the specificity of RNAi was assured using a modification of the method of Miki and Shimamoto (2004). To test transient suppression of gene expression caused by direct introduction of the RNAi vector into P. wilsonii pollen, the pFGCLat52-GFP vector, which carries a 300-bp fragment of the green fluorescent protein (GFP) gene, was generated. The GFP–RNAi construct, Lat52–GFP, as a target gene and Lat52–CHERRY as a control for bombardment efficiency were co-bombarded. The results shown in Supplementary Fig. S1 (available at JXB online) indicate that the bombarded Lat52–GFP gave sufficient expression of GFP protein, which was evidenced by the green fluorescence in the pollen. When the GFP–RNAi construct was co-bombarded with Lat52–GFP, strong silencing of the green fluorescence was observed, while no difference in the expression of the control CHERRY gene was observed (Supplementary Fig. S1 at JXB online). These results suggested that the RNAi vector based on pFGCLat52 was useful for transient suppression of gene function in P. wilsonii pollen. Meanwhile, a 386-bp fragment for PwHAP5RNAi and a 294-bp fragment for PwFKBP12RNAi, which were conserved at a very low level among members of the PwHAP5 family and PwFKBP family, respectively, were used in the experiment in order that the sequences used as RNAi trigger had enough specificity.
RNAi vector constructs
To achieve the objective of pollen-specific expression, the 35S promoter in pFGC5941 was replaced by the Lat52 promoter to form the pFGCLat52 vector. To construct the PwHAP5RNAi and PwFKBP12RNAi vector, two fragments (post-blast) were obtained. The first was a 386-bp fragment, conservative to PwHAP5 and amplified from cDNA of P. wilsonii pollen using the primer pair: 5′-TCTAGAGGCGCGCCTGATCTCTGCAGAGGCACC-3′ (bold italics represent the XbaI–AscI site) and 5′-GGATCCATTTAAATTACGTCCTCAAAGAACCC-3′ (bold italics represent the BamHI–SwaI site). The second was a 294-bp fragment conservative to PwFKBP12 amplified from cDNA of P. wilsonii pollen using primer pair 5′-TCTAGAGGCGCGCCTGAGGATCTGTCTCAGA-3′ (bold italics represent the XbaI–AscI site) and 5′-GGATCCATTTAAATGTTTGCTGGCAATATC-3′ (bold italics represent the BamHI–SwaI site). The fragments were digested with SwaI and AscI and inserted on the left side (5′ end) of the chalcone synthase intron from Petunia hybrida in pFGCLat52, and further digested with BamHI and XbaI and inserted on the right side (3′ end) of the chalcone synthase intron, thus completing the two RNAi vectors.
Particle bombardment-mediated transient expression in pollen
Mature P. wilsonii pollen grains were used in a transient expression experiment, using the particle bombardment procedure described in ‘In vitro P. wilsonii pollen germination’, above. Microprojectile bombardment was performed using the helium-driven PDS-1000–He biolistic system (Bio-Rad). Tungsten particles (1.1 μm) were coated with plasmid DNA according to the manufacturer's recommendation (Bio-Rad; Sanford et al., 1993). The pollen grains were incubated for 12 h or 24 h before observation under a confocal microscope (LSM 510 META; Zeiss), with 100 tubes of each sample counted and measured. Pollen grains were considered to be germinated when the pollen tube length was greater than the diameter of the pollen grain (Rodriguez-Riano and Dafni, 2000). Fluorescence and transmitted light images were recorded using the fluorescence microscope.
Y2H assays
The full-length coding region (F), plus a fragment carrying residues 1–77 (N77) and another carrying residues 92–221 (C130) of PwHAP5, were fused to the plasmid pGBKT7 and transformed into Saccharomyces cerevisiae strain AH109 with the empty pGADT7-Rec vector. The transformants of all three fragments plus the empty pGADT7-Rec vector can grow on SD/–Trp–Leu–His–Ade medium. F, N77, and C130 were used as bait to screen a P. wilsonii pollen cDNA library constructed in the pGADT7-Rec vector. The cDNA library was transformed into yeast strain AH109 with F, N77, and C130. Y2H assays were performed according to standard methods (Chien et al., 1991). Positive clones were selected on SD/–Trp–Leu–His–Ade medium. After confirmation using the 5-bromo-4-chloro-3-indoyl-α-D-galactoside (X-α-Gal) test and retransformation, the inserts were sequenced. In addition, pGADT7-Rec and pGADT7-PwFKBP12 were transformed into yeast strain AH109, with the empty pGBDT7 vector as control. The expression of the third reporter gene lacZ was followed by measuring at OD420 the accumulation of the product metabolized by β-galactosidase, with o-nitrophenyl β-D-galactopyranoside (o-NPG; Sigma, St Louis, MO, USA) as substrate.
Bimolecular fluorescence complementation (BiFC) assays
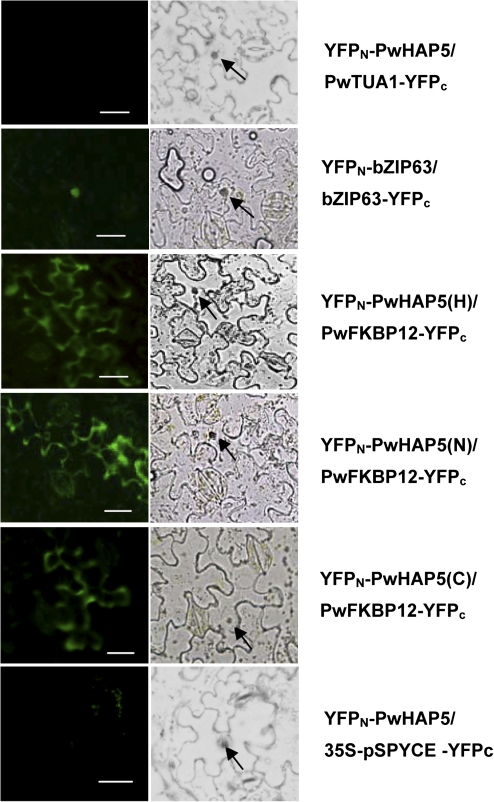
BiFC assays were performed as described by Guo et al. (2009). cDNA without a termination codon encoding PwHAP5 was cloned into pSPYNE-35S, and the cDNAs encoding PwFKBP12 were cloned into pSPYCE-35S. Both the cDNAs encoding PwTUA1 (P. wilsonii α-tubulin protein) in pSPYCE-35S and the empty vector 35S-pSPYCE were used as negative controls, and the bZIP63-pSPYNE-35S and bZIP63-pSPYCE-35S vectors were used as positive controls (Walter et al., 2004). Thus, the plasmids pUC-SPYNE-PwHAP5 and pUC-SPYCE-PwFKBP12 were expressed as PwHAP5–yellow fluorescent protein (YFP)N and PwFKBP12–YFPC fusion proteins. These vectors were introduced into Agrobacterium tumefaciens strain GV3101. For infiltration of Nicotiana benthamiana, the P19 protein of tomato bushy stunt virus was used to suppress gene silencing. The A. tumefaciens strains were grown overnight in YEB media containing appropriate antibiotic selections. Cells were pelleted at 4000 g, resuspended in infiltration medium (10 mM MgCl2, 10 mM MES, 150 mM acetosyringone), and incubated for at least 2 h at room temperature. Co-infiltration of A. tumefaciens strains containing the BiFC constructs and the P19 silencing plasmid was carried out at an OD600 of 0.7:0.7:1.0. Resuspended cells were infiltrated into leaves of 4-week-old N. benthamiana plants as described previously (Voinnet et al., 2003; Walter et al., 2004). After 2 d, epidermal cell layers of tobacco leaves were assayed for fluorescence under a fluorescence microscope (BX51 model 7.3; Olympus). These data clearly indicated both that PwFKBP12 is an interaction partner of PwHAP5 in vivo and that the bimolecular interaction takes place in the cytoplasm.
Results
Isolation and characterization of the cDNA clone encoding HAP5 from P. wilsonii
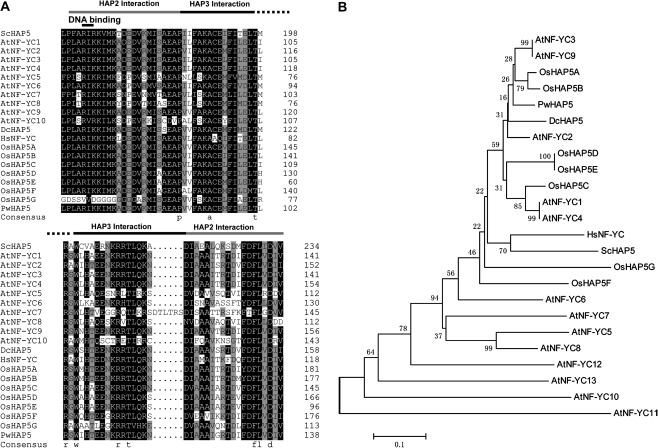
The putative PwHAP5 cDNA clone was isolated from a P. wilsonii subtractive cDNA library of pollen after a 12-h incubation in Ca2+-stressed medium (0.1% Ca2+ concentration) via differential hybridization screening to identify genes involved in pollen development. The resulting 988-nucleotide PwHAP5 cDNA, with an ATG initiation codon at position 64 and a termination codon at position 667 (TGA), contained an open reading frame (ORF) encoding a 202-amino-acid polypeptide with a calculated molecular weight of 22.44 kDa and a predicted pI of 6.20. The full cDNA sequence was submitted to GenBank as accession number EU579453.1. Like the majority of HAP5 (NF-YC) proteins, the PwHAP5 protein contained the required conservative amino acids. The predicted PwHAP5 amino acid sequence contained a fragment (∼53 residues) with strong similarity to the conserved domain of HAP5 subunits in other organisms (Fig. 1A). Of these 53 residues, 10 were absolutely conserved among all HAP5 subunits. The protein also contained NF-YA/NF-YB (HAP2/HAP3) interaction domains, as well as a DNA-binding domain similar to previously characterized NF-YCs (Yazawa and Kamada, 2007).
Fig. 1.
HAP5 gene of P. wilsonii. (A) Alignment of the HAP5 proteins, sequences correspond to the conserved regions in HAP5 proteins across various lineages. Dc, Daucus carota; Hs, Homo sapiens; Os, Oryza sativa; Sc, Saccharomyces cerevisiae. Note that the HAP2 interaction domain extends across two separate regions. The DNA-binding domain in HAP5 consists of the two amino acids AR (found in most HAP5 homologues). (B) Phylogenetic tree of P. wilsonii HAP5 (PwHAP5) and other HAP5 proteins previously characterized. A neighbor-joining tree based on the deduced amino acid sequences of the conserved domains in HAP5s. This bootstrap consensus tree was based on 1000 replicates. Numbers on nodes are bootstrap values. The accession numbers in GenBank and sources of the protein are as follows: AtNF-YC1(At3g48590), AtNF-YC2(At1g56170), AtNF-YC3(At1g54830), AtNF-YC4(At5g63470), AtNF-YC5 (At5g50490), AtNF-YC6(At5g50480), AtNF-YC7(At5g50470), AtNF-YC8(At5g27910), AtNF-YC9(At1g08970), AtNF-YC10(At1g07980), AtNF-YC11(At3g12480), AtNF-YC12(At5g38140), AtNF-YC13(At5g43250) from Arabidopsis thaliana; DcHAP5(AB104612) from D. carota; HsNF-YC(U78774) from H. sapiens; OsHAP5A(AB288041), OsHAP5B(AB288042), OsHAP5C(AB288043), OsHAP5D(AB288044), OsHAP5E(AB288045), OsHAP5F(AB288046), OsHAP5G(AB288047) from O. sativa; ScHAP5(U19932) from S. cerevisiae.
To examine evolutionary relationships among HAP5 proteins, a phylogenetic analysis based on the amino acid sequences of the conserved domains was conducted. This showed that PwHAP5 and AtNF-YC3/9, which are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis (Kumimoto et al., 2010), and OsHAP5A/B were in the same clade and were highly conserved (Fig. 1B).
Expression pattern of PwHAP5 in different tissues and germinating pollen at different developmental stages
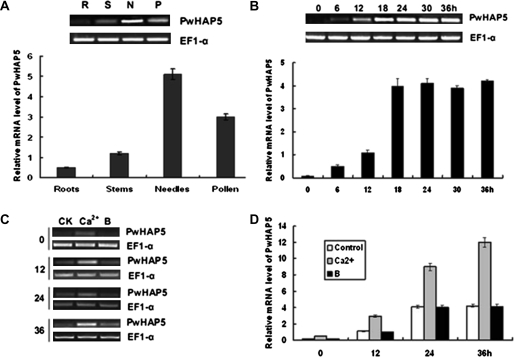
To obtain the profile of PwHAP5 expression patterns, total RNA was isolated from needles, stems, roots, and from incubated germinating P. wilsonii pollen mixtures at 6-h intervals and transcript accumulation examined using RT-PCR and quantitative real-time RT-PCR analyses. PwHAP5 transcripts were expressed strongly in needles, germinating pollen, and stems, but less in roots (Fig. 2A). Among the various tissues, needles had the highest PwHAP5 transcription level. PwHAP5 expression was further examined during pollen development. As shown in Fig. 2B, PwHAP5 expression was first detected in pollen 6 h post-incubation (germination only). It increased gradually, reaching a maximum 18 h post-incubation. Transcription levels remained at this same high level during the late stages (24, 30, and 36 h post-incubation). The PwHAP5 expression level in boron-stressed (0.1% H3BO3 concentration) and Ca2+-stressed medium (0.1% Ca2+ concentration) was also analysed during various pollen tube developmental stages. PwHAP5 was induced by Ca2+, but not by boron, during all of the tested stages (Fig. 2C).
Fig. 2.
Expression of PwHAP5 in different tissues and in developing pollen tubes of P. wilsonii. (A) Tissue-specific expression of PwHAP5 in P. wilsonii. Total RNA was isolated from needles, stems, roots, and pollen (incubated after 0, 6, 12, 18, and 24 h). Above the graph is shown semi-quantitative RT-PCR analysis of expression of PwHAP5. The EF1-α gene was amplified as an internal control. The graph shows quantification of PwHAP5 expression. Quantitative real-time RT-PCR was performed using PwHAP5-specific primers. Column heights represent expression levels relative to the EF1-α gene. The values are means ±SD (n = 3) from three independent experiments. (B) Expression of PwHAP5 in pollen at different development stages (incubated after 0, 6, 12, 18, 24, 30, and 36 h). Above the graph is shown semi-quantitative RT-PCR analysis of expression of PwHAP5; the graph represents the quantification of PwHAP5 expression. The control and values are as in (A). (C) Semi-quantitative RT-PCR analysis of expression of PwHAP5 in pollen 0, 12, 24, and 36 h after incubation, induced by 0.1% (w/v) Ca2+ or 0.1% (w/v) H3BO3. (D) Quantification of PwHAP5 expression in P. wilsonii pollen at the same intervals after incubation as in (C). The control and values are as in (A).
The effects of PwHAP5 on pollen tube growth
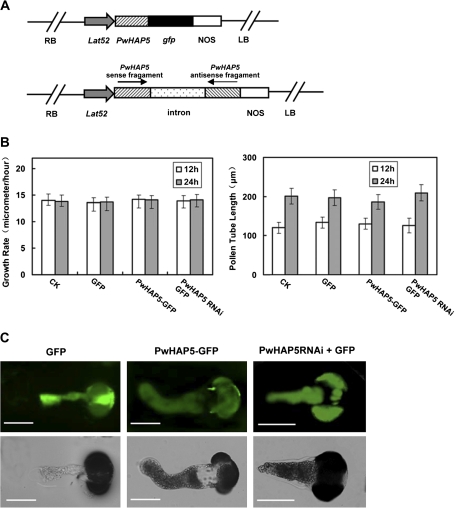
To further examine the function of the PwHAP5 gene in pollen tube development, an overexpression vector and an RNAi expression vector of PwHAP5 were constructed (Fig. 3A). The vectors were used to transiently transform P. wilsonii pollen by microprojectile bombardment. A reporter gene encoding GFP was fused to PwHAP5 to visualize transiently transformed pollen. The Lat52 promoter was used to drive the pollen-specific expression of transgenes in these studies (Twell et al., 1990). Considering that GFP fused to HAP5RNAi vector directly could affect gene silencing, PwHAP5RNAi was transiently co-expressed under the pollen-specific promoter Lat52 (Twell et al., 1990) with Lat52::GFP as an indicator. No significant difference was observed in pollen germination percentage and tube length between the samples transformed with fusion proteins and those left untreated (Fig. 3B) suggesting that neither the microprojectile bombardment itself nor the fusion proteins had any significant effect on pollen germination and pollen tube length. The orientation of P. wilsonii pollen tube growth was altered when PwHAP5 was overexpressed in the pollen tube. However, the pollen tube with PwHAP5RNAi showed normal growth (Fig. 3C). This phenotype was observed in most of the transformed pollen tubes although some tip bending was occasionally observed in untreated pollen tubes after prolonged culture (Table 1).
Fig. 3.
Transient expression of PwHAP5 alters the orientation of P. wilsonii pollen tube growth. (A) Diagram of plant expression vectors. The pollen-specific expression promoter Lat52 was used to drive the expression of PwHAP5 in pollen tube. (B) The effect of PwHAP5 on pollen germination and tube length using microprojectile bombardment. After 12 h and 24 h of incubation, the percentage germination and tube length of pollen bombarded with the GFP-only, PwHAP5 overexpression, or RNAi construct showed no significant difference compared with the untreated control pollen (P<0.05). Error bars indicate averages from the number of pollen tubes measured (n≥50). (C) Transient expression of PwHAP5 in P. wilsonii pollen tube growth. PwHAP5–GFP represents overexpression of PwHAP5 in P. wilsonii pollen tube. PwHAP5RNAi+GFP represents interference of PwHAP5. GFP was used as control. The data were obtained from three independent experiments, and every condition was tested three times. Bars 20 μm.
Table 1.
Phenotype analysis of transformed pollen tubes
| Transformed vectors | Bent | Unbent | Bent/unbent ratio |
| CK | 1 | 49 | 1 : 49 |
| GFP | 1 | 48 | 1 : 48 |
| PwHAP5–GFP | 46 | 4 | 11.5 : 1 |
| PwHAP5 (RNAi)+GFP | 2 | 48 | 1 : 24 |
| PwHAP5–CHERRY | 45 | 4 | 11.25 : 1 |
| PwFKBP12(RNAi)+GFP | |||
| PwFKBP12–GFP | 2 | 51 | 1 : 25.5 |
| PwHAP5–CHERRY | 2 | 47 | 1 : 23.5 |
| PwFKBP12–GFP | |||
| PwHAP5(RNAi)+CHERRY | 1 | 48 | 1 : 48 |
| PwFKBP12–GFP | |||
| PwFKBP12(RNAi)+GFP | 48 | 2 | 24 : 1 |
| PwHAP5(RNAi)+CHERRY | 0 | 50 | |
| PwFKBP12(RNAi)+GFP |
PwHAP5 protein interacts with PwFKBP12
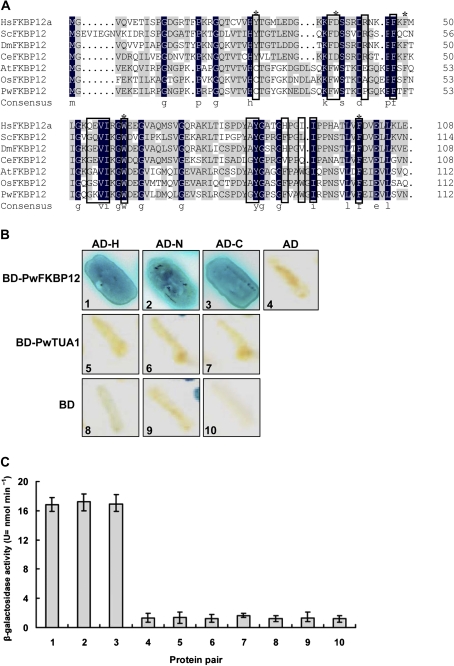
Y2H screening was performed to identify proteins that interact with PwHAP5. The PwHAP5 protein contained HAP2/HAP3 interaction domains and a DNA-binding domain. To examine which domain is crucial to the function of PwHAP5 and its interaction with other proteins, PwHAP5 was split into two fragments. The bait was constructed by fusing 77 N-terminal amino acids (N), 130 C-terminal amino acids (C), and full-length PwHAP5 (H) to the pGBDKT7 vector to screen the cDNA library from P. wilsonii pollen. A total of 6×106 colonies were screened and 35 positive clones corresponding to eight cDNAs were identified (data not shown). Among the eight clones, the 5-15/3-11 clone was highly homologous to AtFKBP12 (FK506-binding protein) in Arabidopsis, and it was named PwFKBP12. The full cDNA sequence of PwFKBP12 was submitted to GenBank under accession number GQ5140630. As shown in Fig. 4A, PwFKBP12 conserves three of the five residues with strongest influence over catalytic activity in mammalian FKBP12 (DeCenzo et al., 1996; Tradler et al., 1997), as well as a cysteine pair (Cys26 and Cys80) that is unique to the plant FKBP12 isoforms and was vital for interaction with calcineurin in vitro (Xu et al., 1998).
Fig. 4.
Interaction of PwHAP5 with FKBP12. (A) Amino acid alignment of FKBP12 isoforms from different species. H. sapiens (Hs; GenBank protein accession number AAI19733), S. cerevisiae (Sc; AAA03564), Drosophila melanogaster (Dm; AAF57582), Caenorhabditis elegans (Ce; CAA22330), A. thaliana (At; AAB57847), and O. sativa (Os; NP_001048188.1) aligned with P. wilsonii (Pw). Asterisks above residues denote residues that are important for peptidyl prolyl cis/trans isomerase activity in mammalian FKBP12 (DeCenzo et al., 1996; Tradler et al., 1997). Residues in boxes are involved in FK506 and/or rapamycin binding in mammalian FKBP12 (Van Duyne et al., 1993). (B) The interactions were assayed in the GAL4 (a regulator of galactose-induced genes) Y2H system to re-examine the interactions of full-length PwHAP5 (H), N77 (N), and C130 (C) proteins with the pGBDKT7 vector alone (BD) and with PwFKBP12. As a control, combinations of the pGADT7-Rec vector (AD) with the pGBDKT7 vector (BD) and PwTUA1 are also shown. Yeast cells expressing AD–H, AD–N, AD–C, or AD alone and each of BD fusion proteins or BD alone were grown on medium-selective plates (SD/–Leu–Trp–His–Ade). The blue represents growth and interaction between PwHAP5 and PwFKBP. (C) Liquid β-galactosidase assay using o-NPG as a substrate. β-galactosidase activity is expressed in U (=nmol min−1). The values displayed are the average β-galactosidase activities for three individual double transformants, with standard deviations indicated by error bars.
Protein interactions between N/C/H and PwFKBP12 were further confirmed by analysing growth on selective medium, followed by measuring true β-galactosidase activity. Growth of the N–PwFKBP12, C–PwFKBP12, and H–PwFKBP12 combinations, but no growth of the control combinations was observed (Fig. 4B). β-Galactosidase activities of the N/C/H fusion proteins were nearly 20 times higher than those of the controls (Fig. 4C), indicating specific interaction between PwHAP5 and PwFKBP12.
In vivo detection of the interaction between PwHAP5 and PwFKBP12
Next a BiFC assay was performed (Walter et al., 2004) in a tobacco transient expression system (Voinnet et al., 2003) to confirm the interaction of PwHAP5 and PwFKBP12 in vivo. PwFKBP12 was fused with YFPC (SPYCE), and the full length (H) of the PwHAP5 protein was fused with YFPN (SPYNE). Fluorescence from YFP in transgenic tobacco epidermis transformed with PwHAP5(H)–YFPN and PwFKBP12–YFPC was observed throughout the cytoplasm, but not in the nucleus (Fig. 5). The same results were obtained in transgenic tobacco epidermis transformed with PwHAP5 N77 or C130 and PwFKBP12–YFPc (Fig. 5). The negative controls (empty vectors or PwTUA1–YFPC) produced no or only background fluorescence and the positive control (YFPN-bZIP63/bZIP63–YFPC) produced fluorescence only in the nucleus, showing the specificity of the BiFC assays.
Fig. 5.
In vivo interaction of PwHAP5 with PwFKBP in the BiFC system. BiFC in A. tumefaciens-infiltrated tobacco (N. benthamiana) leaves was visualized by laser confocal microscopy. The laser-scanning confocal microscopy images show fluorescence (indicated by YFP) and merged images of double transformed tobacco leaves with the YFPN–PwHAP5 (full-length PwHAP5; H), N77 (N), and C130 (C), respectively) and PwFKBP12–YFPC fusions (YFPN–PwHAP5/ PwFKBP12–YFPC). PwHAP5 interacts with PwFKBP12 in the cytoplasm, and does not interact with the negative control PwTUA1 or empty vector. The YFP fluorescence of the positive control vectors, bZIP63-pSPYNE-35S and bZIP63-pSPYCE-35S, was detected only in the nucleus. Arrows indicate the nucleus. Bar 25 μm.
The effect of PwHAP5 and PwFKBP12 on pollen tube growth
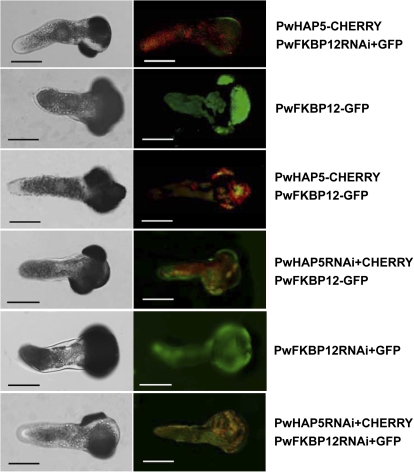
To further clarify the function of the association of PwHAP5 and PwFKBP12 in pollen tube growth, RNAi and overexpression vectors of PwHAP5 and PwFKBP12 fused to CHERRY and GFP, respectively, were constructed. The overexpression and RNAi vectors were used to transform P. wilsonii pollen transiently alone or in combination via microprojectile bombardment. Considering that GFP/CHERRY fused to PwHAP5RNAi/PwFKBP12RNAi vector directly could affect gene silencing, PwHAP5RNAi/PwFKBP12RNAi was transiently co-expressed under the pollen-specific promoter Lat52 (Twell et al., 1990) with Lat52::GFP/CHERRY as an indicator. As expected, the pollen tube overexpressing PwHAP5 or with PwFKBP12RNAi showed bent growth (Fig. 6, Table 1), which was consistent with that observed in Fig. 3. However, neither the transient expression of PwFKBP12 alone nor the co-expression of PwHAP5 and PwFKBP12 in pollen altered pollen tube growth orientation and the pollen tubes showed normal growth (Figs 3, 6). Furthermore, the pollen tube with PwHAP5RNAi (whether PwFKBP12 overexpression or RNAi) also showed normal growth (Figs 3, 6). No significant difference was observed in the pollen germination percentage or tube growth rate between samples transformed with GFP or CHERRY and those left untreated, suggesting that neither microprojectile bombardment itself nor GFP or CHERRY had a significant effect on pollen germination and pollen tube growth in P. wilsonii (Fig. 3; Yu et al., 2009).
Fig. 6.
Transient expression of PwHAP5 or PwFKBP12 in P. wilsonii pollen tube. The overexpression vectors PwHAP5 or PwFKBP12 fused to CHERRY or GFP, respectively, were constructed and expressed transiently alone or used to co-transform P. wilsonii pollen by microprojectile bombardment. The pollen-specific expression promoter Lat52 was used to drive the expression of PwHAP5 or PwFKBP12 in pollen tube. Microscopic analysis of pollen tube expression was performed 24 h after gene transfer of PwFKBP12 and PwHAP5 in pollen. Transient expression of PwHAP5 alone affects P. wilsonii pollen tube growth orientation, whereas both transient expression of PwFKBP12 and co-expression of PwHAP5 and PwFKBP12 do not. Left panel: transmitted light images. Right panel: fluorescence images. The yellow fluorescence is the overlay of GFP and CHERRY proteins. The data were obtained from three independent experiments, and each condition was tested three times. Bars, 20 μm.
Discussion
Plant sexual reproduction is a key determinant of crop yield, and although the HAP gene family and its orthologues have been shown to be implicated in flowering time regulation, knowledge of the function of plant HAP genes in pollen tube growth is limited. In this report, strong evidence is provided that HAP5, which encodes a CCAAT-binding transcription factor and interacts with FKBP12 in cytoplasm, is involved in pollen tube growth.
PwHAP5 is highly expressed in germinating pollen and involved in pollen tube growth
This study confirms that PwHAP5, an orthologous P. wilsonii transcription factor, has an important role in pollen tube development and orientation regulation. This represents a novel function of HAP5 in the HAP plant gene family. The protein identified contained NF-YA/NF-YC (HAP2/HAP5) interaction domains, as well as a DNA-binding domain similar to previously characterized NF-YCs (Yazawa and Kamada, 2007). PwHAP5 was highly conserved with AtNF-YC3/9 and OsHAP5A/B and in the same clade (Fig. 1B). RT-PCR and quantitative real-time RT-PCR analyses showed that PwHAP5 is highly expressed in needle and germinating pollen and maintains a high level of expression throughout pollen tube development (Fig. 2). Furthermore, it was shown that the direction of tube growth is altered by particle bombardment-mediated transient overexpression of PwHAP5. However, the pollen tube with PwHAP5RNAi showed normal growth, without diminishing pollen tube growth (Fig. 3). It is noteworthy that neither overexpression nor RNAi of PwHAP5 influences pollen germination percentage and tube growth rate (Fig. 3). This is in contrast to PwTUA1, a pollen-specific tubulin gene cloned from P. wilsonii pollen, overexpression of which greatly increases pollen germination percentage and pollen tube elongation (Yu et al., 2009). These results extend the functions of HAP5, which is implicated not only in the control of flowering time (Kumimoto et al., 2010), but also pollen tube growth orientation regulation.
Interestingly, distinct from the phenotype of overexpressed ROP GTPases, which act as key molecular switches controlling tip growth in Arabidopsis pollen tubes previously reported (Li et al., 1999), in which pollen tubes ballooned or were furcated by the ectopic accumulation of ROP proteins, overexpression of PwHAP5 altered only the direction of pollen tube growth (Fig. 3), suggesting a more pivotal role of HAP5 in pollen tube growth orientation regulation than in tube polarity maintenance compared with ROP GTPases.
PwFKBP12 interacts with PwHAP5 to regulate pollen tube growth orientation
FKBPs, which are receptors of the immunosuppressive drug FK506, are referred to as immunophilins and are found in all living organisms (Geisler and Bailly, 2007). Although historically linked to immunosuppression and proline bond rotation, the physiological importance of FKBPs far surpasses FK506 binding and general protein folding, extending to cellular processes, including signal transduction, chloroplast function, DNA transcription, protein trafficking, apoptosis, and fertility (Heitman et al., 1992; Luan et al., 1994; Gopalan et al., 2004; Kang et al., 2008; Meiri et al., 2010).
In this study, PwFKBP12 was screened via a Y2H system using PwHAP5 as bait (Fig. 4). Yeast cell growth and β-galactosidase activity were not observed in the control combinations of the pGADT7-Rec vector (AD) with the pGBDKT7 vector (BD) and PwTUA1, suggesting specific interaction between PwHAP5 and PwFKBP12. Unfortunately, HAP2 or HAP3 subunit cannot be screened via a Y2H system using the full length, N-terminus, or C-terminus of PwHAP5 as bait. The possibility cannot be excluded that the accumulation of PwHAP2 or PwHAP3 was less compared with that of PwFKBP12 during the stage of P. wilsonii pollen adopted in this study. The function of PwFKBP12 interaction with PwHAP5 in pollen tube development was investigated further. Transient expression of PwFKBP12 in pollen showed normal pollen tube growth whether PwHAP5 overexpression or RNAi or not, in contrast to the alteration of pollen tube growth direction by PwHAP5 overexpression (Figs 3, 6). The pollen tube with PwFKBP12RNAi bent, whereas PwHAP5RNAi alone has normal growth. Importantly, the overexpression phenotype of HAP5 in pollen tubes is restored by FKBP12 overexpression, as shown by the normal growth observed when PwHAP5 and PwFKBP12 were co-expressed in pollen (Fig. 6). These data provide clear evidence that PwFKBP12 interacts with PwHAP5 to regulate pollen tube growth orientation. Notably, neither PwFKBP12 or PwHAP5 overexpression or RNAi nor the co-expression or the RNAi of the two genes affected the pollen germination percentage or pollen tube growth rate (data not shown).
The BiFC assay in a tobacco transient expression system demonstrated that the interaction of PwHAP5 and PwFKBP12 in vivo occurred mainly in the cytoplasm (Fig. 5). Arabidpsis FKBP12 was reported to be localized in the cytoplasm (Geisler and Bailly, 2007). This study also supports the localization of PwFKBP12 in the cytoplasm (Fig. 6). However, the localization of different HAP subunits may differ in different cells or physiological processes. Frontini et al. (2004) reported that NF-YA (HAP2) and NF-YB (HAP3) are nuclear proteins, whereas NF-YC (HAP5) localizes to both cytoplasmic and nuclear compartments and its nuclear localization is determined by the interaction with its heterodimerization partner NF-YB. Interestingly, the compartmentalization of NF-YC transcription factors is a dynamic process and regulated under different physiological processes (Frontini et al., 2004; Liu and Howell, 2010). Kahle et al. (2005) found that only the NF-YB/NF-YC (HAP3/HAP5) dimer, but not the monomeric components, are recognized by importin 13 and are imported into the nucleus. In the present study, the HAP3 homologue could not be obtained by Y2H screening using PwHAP5 (H/N/C) as bait, which possibly explains in part why fluorescence representing PwHAP5 was not observed in the nucleus. However, localization of HAP5 in the nucleus is not excluded.
Potential mechanisms of FKBP12 interaction with HAP5 in pollen tube growth
Some FKBPs are reported to be related to Ca2+ (Timerman et al., 1993, 1995) and involved in sexual reproduction in plants (Kurek et al., 2002). Kurek et al. (2002) found that wheat FKBP73 could bind calmodulin via the CaM-binding domain and that the deletion of the CaM-binding and TPR domains resulted in male-sterile plants, suggesting a role for FKBP73 in the plant sexual reproduction process. This study uncovered a novel function of FKBP12 in pollen tube development interaction with HAP5 in the control of pollen tube growth direction. In mammals, FKBP12 is tightly bound to the calcium release channel/ryanodine receptor of skeletal muscle terminal cisternae of the sarcoplasmic reticulum (Timerman et al., 1993, 1995). Kurek et al. (2002) showed that FKBP12–FK56 complexes can bind to and inhibit the activity of calcineurin, a Ca2+ or calmodulin-dependent protein phosphatase. PwHAP5 was induced by Ca2+, but not by boron, during pollen tube development (Fig. 2C), suggesting that PwHAP5 is a Ca2+-responsive gene. Since Ca2+ is a key factor for pollen tube growth and maintenance of polar growth (Li et al., 1999), it is likely that PwFKBP12 is involved in the plant sexual reproduction process via interaction with the Ca2+-responsive gene HAP5 or probably modulating Ca2+ or Ca2+-related proteins or enzymes involved in pollen tube development and guidance in plants. Meanwhile, considering that FKBPs may function as protein-folding chaperones, the possibility cannot be excluded that PwFKBP12 binds to other proteins to function in the process. Nevertheless, the hypothesis and the accurate signalling pathway related to HAP5 and FKBP12 involved in pollen tube development and orientation need detailed investigation in the future. However, the identification of PwHAP5 and PwFKBP12 in P. wilsonii presents an opportunity to identify downstream genes regulated by HAP5 mediating pollen tube guidance and fertilization, since pollen tube growth direction in the ovary determines ovule targeting and sperm delivery to the female gametophyte.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Transient suppression of GFP gene expression in P. wilsonii pollen by the pFGCLat52 -based vector.
Acknowledgments
The authors thank Professor Jörg Kudla (University of Münster, Germany) for the BiFC vector, Professor David Baulcombe (Sainsbury Laboratory, UK) for p19 protein, and Dr J. Haseloff (MRC Laboratory of Molecular Biology, Cambridge, UK) for the GFP construct, pBINmGFP5-ER. They also thank Professor Dapeng Zhang (Tsinghua University) for the kind suggestions and other anonymous scientists for the critical revision in language of the manuscript. This work was supported by a grant from Agricultural Ministry of China (Grant no. 2008ZX08009-003 and 2009ZX08009-062B to LYZ) and the Foundation for National Natural Science (Grant No. 30700646 to LYZ).
Glossary
Abbreviations
- ABRE
abscisic acid response element
- BiFC
bimolecular fluorescence complementation
- CHSA
chalcone synthase
- CTAB
cetyltrimethylammonium bromide
- EF1-α
elongation factor α
- GFP
green fluorescent protein
- HAP
histone (or haem)-associated protein
- o-NPG
o-nitrophenyl β-D-galactopyranoside
- ORF
open reading frame
- RT
reverse transcription
- X-α-Gal
5-bromo-4-chloro-3-indoyl-α-D-galactoside
- YFP
yellow fluorescent protin
References
- Ben-Naim O, Eshed R, Parnis A, Teper-Bamnolker P, Shalit A, Coupland G, Samach A, Lifschitz E. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. The Plant Journal. 2006;46:462–476. doi: 10.1111/j.1365-313X.2006.02706.x. [DOI] [PubMed] [Google Scholar]
- Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. Journal of Molecular Biology. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- Cai X, Ballif J, Endo S, Davis E, Liang M, Chen D, DeWald D, Kreps J, Zhu T, Wu Y. A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiology. 2007;145:98–105. doi: 10.1104/pp.107.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Purgear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter. 1993;11:117–121. [Google Scholar]
- Chen NZ, Zhang XQ, Wei PC, Chen QJ, Ren F, Chen J, Wang XC. AtHAP3b plays a crucial role in the regulation of flowering time in Arabidopsis during osmotic stress. Journal of Biochemistry and Molecular Biology. 2007;40:1083–1089. doi: 10.5483/bmbrep.2007.40.6.1083. [DOI] [PubMed] [Google Scholar]
- Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proceedings of the National Academy of Sciences, USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier JP, Frugier F, de Billy F, et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes and Development. 2006;20:3084–3088. doi: 10.1101/gad.402806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang VD, Bohn C, Bolotin-Fukuhara M, Daignan-Fornier B. The CCAAT box-binding factor stimulates ammonium assimilation in Saccharomyces cerevisiae, defining a new cross-pathway regulation between nitrogen and carbon metabolisms. Journal of Bacteriology. 1996;178:1842–1849. doi: 10.1128/jb.178.7.1842-1849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCenzo MT, Park ST, Jarrett BP, Aldape RA, Futer O, Murcko MA, Livingston DJ. FK506-binding protein mutational analysis: defining the active-site residue contributions to catalysis and the stability of ligand complexes. Protein Engineering. 1996;9:173–180. doi: 10.1093/protein/9.2.173. [DOI] [PubMed] [Google Scholar]
- Edwards D, Murray JA, Smith AG. Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiology. 1998;117:1015–1022. doi: 10.1104/pp.117.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frontini M, Imbriano C, Manni I, Mantovani R. Cell cycle regulation of NF-YC nuclear localization. Cell Cycle. 2004;3:217–222. [PubMed] [Google Scholar]
- Geisler M, Bailly A. Tete-a-tete: the function of FKBPs in plant development. Trends in Plant Science. 2007;12:465–473. doi: 10.1016/j.tplants.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Gopalan G, He Z, Balmer Y, Romano P, Gupta R, Heroux A, Buchanan BB, Swaminathan K, Luan S. Structural analysis uncovers a role for redox in regulating FKBP13, an immunophilin of the chloroplast thylakoid lumen. Proceedings of the National Academy of Sciences, USA. 2004;101:13945–13950. doi: 10.1073/pnas.0405240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YH, Yu YP, Wang D, Wu CA, Yang GD, Huang JG, Zheng CC. GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytologist. 2009;183:62–75. doi: 10.1111/j.1469-8137.2009.02838.x. [DOI] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. Proline isomerases at the crossroads of protein folding, signal transduction, and immunosuppression. New Biologist. 1992;4:448–460. [PubMed] [Google Scholar]
- Kahle J, Baake M, Doenecke D, Albig W. Subunits of the heterotrimeric transcription factor NF-Y are imported into the nucleus by distinct pathways involving importin beta and importin 13. Molecular and Cellular Biology. 2005;25:5339–5354. doi: 10.1128/MCB.25.13.5339-5354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang CB, Hong Y, Dhe-Paganon S, Yoon HS. FKBP family proteins: immunophilins with versatile biological functions. Neurosignals. 2008;16:318–325. doi: 10.1159/000123041. [DOI] [PubMed] [Google Scholar]
- Knuiman B, Win de AHN, Pierson ES, Geurts H, Kengen HMP, Derksen J. Development and cellular organization of Pinus sylvestris pollen tubes. Sexual Plant Reproduction. 1996;9:93–101. [Google Scholar]
- Kumimoto RW, Adam L, Hymus GJ, Repetti PP, Reuber TL, Marion CM, Hempel FD, Ratcliffe OJ. The nuclear factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta. 2008;228:709–723. doi: 10.1007/s00425-008-0773-6. [DOI] [PubMed] [Google Scholar]
- Kumimoto RW, Zhang Y, Siefers N, Holt 3rd. NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. The Plant Journal. 2010;63:379–391. doi: 10.1111/j.1365-313X.2010.04247.x. [DOI] [PubMed] [Google Scholar]
- Kurek I, Dulberger R, Azem A, Tzvi BB, Sudhakar D, Christou P, Breiman A. Deletion of the C-terminal 138 amino acids of the wheat FKBP73 abrogates calmodulin binding, dimerization and male fertility in transgenic rice. Plant Molecular Biology. 2002;48:369–381. doi: 10.1023/a:1014023329807. [DOI] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. The Plant Cell. 2003;15:5–18. doi: 10.1105/tpc.006973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro MD, Cardenas L, Bhatt AP, Justus CD, Phillips MS, Holdaway-Clarke TL, Hepler PK. Calcium gradients in conifer pollen tubes; dynamic properties differ from those seen in angiosperms. Journal of Experimental Botany. 2005;56:2619–2628. doi: 10.1093/jxb/eri256. [DOI] [PubMed] [Google Scholar]
- Lee H, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proceedings of the National Academy of Sciences, USA. 2003;100:2152–2156. doi: 10.1073/pnas.0437909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lin Y, Heath RM, Zhu MX, Yang Z. Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. The Plant Cell. 1999;11:1731–1742. doi: 10.1105/tpc.11.9.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. The Plant Cell. 2008;20:2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. The Plant Cell. 2010;22:782–796. doi: 10.1105/tpc.109.072173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- Luan S, Albers MW, Schreiber SL. Light-regulated, tissue-specific immunophilins in a higher plant. Proceedings of the National Academy of Sciences, USA. 1994;91:984–988. doi: 10.1073/pnas.91.3.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity SN, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends in Biochemical Sciences. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- McNabb DS, Pinto I. Assembly of the Hap2p/Hap3p/Hap4p/Hap5p-DNA complex in Saccharomyces cerevisiae. Eukaryotic Cell. 2005;4:1829–1839. doi: 10.1128/EC.4.11.1829-1839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiri D, Tazat K, Cohen-Peer R, Farchi-Pisanty O, Aviezer-Hagai K, Avni A, Breiman A. Involvement of Arabidopsis ROF2 (FKBP65) in thermotolerance. Plant Molecular Biology. 2010;72:191–203. doi: 10.1007/s11103-009-9561-3. [DOI] [PubMed] [Google Scholar]
- Miki D, Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant and Cell Physiology. 2004;45:490–495. doi: 10.1093/pcp/pch048. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Ito Y, Serizawa A, Kurata N. OsHAP3 genes regulate chloroplast biogenesis in rice. The Plant Journal. 2003;36:532–540. doi: 10.1046/j.1365-313x.2003.01897.x. [DOI] [PubMed] [Google Scholar]
- Nelson DE, Repetti PP, Adams TR, et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proceedings of the National Academy of Sciences, USA. 2007;104:16450–16455. doi: 10.1073/pnas.0707193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkham JL, Guarente L. Cloning and molecular analysis of the HAP2 locus: a global regulator of respiratory genes in Saccharomyces cerevisiae. Molecular and Cellular Biology. 1985;5:3410–3416. doi: 10.1128/mcb.5.12.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Riano T, Dafni A. A new procedure to asses pollen viability. Sexual Plant Reproduction. 2000;12:241–244. [Google Scholar]
- Sanford JC, Smith FD, Russell JA. Optimizing the biolistic process for different biological applications. Methods in Enzymology. 1993;217:483–509. doi: 10.1016/0076-6879(93)17086-k. [DOI] [PubMed] [Google Scholar]
- Singh H. Handbuch der Pflanzenanatomie. Vol. 10. Berlin: Gebrutber Borntraeger; 1978. Embryology of Gymnosperms. part 2. [Google Scholar]
- Thirumurugan T, Ito Y, Kubo T, Serizawa A, Kurata N. Identification, characterization and interaction of HAP family genes in rice. Molecular Genetics and Genomics. 2008;279:279–289. doi: 10.1007/s00438-007-0312-3. [DOI] [PubMed] [Google Scholar]
- Timerman AP, Ogunbumni E, Freund E, Wiederrecht G, Marks AR, Fleischer S. The calcium release channel of sarcoplasmic reticulum is modulated by FK-506-binding protein. Dissociation and reconstitution of FKBP-12 to the calcium release channel of skeletal muscle sarcoplasmic reticulum. Journal of Biological Chemistry. 1993;268:22992–22999. [PubMed] [Google Scholar]
- Timerman AP, Wiederrecht G, Marcy A, Fleischer S. Characterization of an exchange reaction between soluble FKBP-12 and the FKBP.ryanodine receptor complex. Modulation by FKBP mutants deficient in peptidyl-prolyl isomerase activity. Journal of Biological Chemistry. 1995;270:2451–2459. doi: 10.1074/jbc.270.6.2451. [DOI] [PubMed] [Google Scholar]
- Tradler T, Stoller G, Rucknagel KP, Schierhorn A, Rahfeld JU, Fischer G. Comparative mutational analysis of peptidyl prolyl cis/trans isomerases: active sites of Escherichia coli trigger factor and human FKBP12. FEBS Letters. 1997;407:184–190. doi: 10.1016/s0014-5793(97)00345-1. [DOI] [PubMed] [Google Scholar]
- Twell D, Yamaguchi J, McCormick S. Pollen-specific gene expression in transgenic plants: coordinate regulation of two different tomato gene promoters during microsporogenesis. Development. 1990;109:705–713. doi: 10.1242/dev.109.3.705. [DOI] [PubMed] [Google Scholar]
- Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. Journal of Molecular Biology. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant Journal. 2003;33:949–956. doi: 10.1046/j.1365-313x.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. The Plant Journal. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- Wei X, Xu J, Guo H, Jiang L, Chen S, Yu C, Zhou Z, Hu P, Zhai H, Wan J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiology. 2010;153:1747–1758. doi: 10.1104/pp.110.156943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. The Plant Cell. 2006;18:2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CA, Yang GD, Meng QW, Zheng CC. The cotton GhNHX1 gene encoding a novel putative tonoplast Na(+)/H(+) antiporter plays an important role in salt stress. Plant and Cell Physiology. 2004;45:600–607. doi: 10.1093/pcp/pch071. [DOI] [PubMed] [Google Scholar]
- Wuarin J, Mueller C, Schibler U. A ubiquitous CCAAT factor is required for efficient in vitro transcription from the mouse albumin promoter. Journal of Molecular Biology. 1990;214:865–874. doi: 10.1016/0022-2836(90)90341-I. [DOI] [PubMed] [Google Scholar]
- Xu Q, Liang S, Kudla J, Luan S. Molecular characterization of a plant FKBP12 that does not mediate action of FK506 and rapamycin. The Plant Journal. 1998;15:511–519. doi: 10.1046/j.1365-313x.1998.00232.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Kagaya Y, Toyoshima R, Kagaya M, Takeda S, Hattori T. Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. The Plant Journal. 2009;58:843–856. doi: 10.1111/j.1365-313X.2009.03817.x. [DOI] [PubMed] [Google Scholar]
- Yazawa K, Kamada H. Identification and characterization of carrot HAP factors that form a complex with the embryo-specific transcription factor C-LEC1. Journal of Experimental Botany. 2007;58:3819–3828. doi: 10.1093/jxb/erm238. [DOI] [PubMed] [Google Scholar]
- Yu Y, Li Y, Li L, Lin J, Zheng C, Zhang L. Overexpression of PwTUA1, a pollen-specific tubulin gene, increases pollen tube elongation by altering the distribution of α-tubulin and promoting vesicle transport. Journal of Experimental Botany. 2009;60:2737–2749. doi: 10.1093/jxb/erp143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.