Abstract
Starch synthase (SS) I and IIIa are the first and second largest components of total soluble SS activity, respectively, in developing japonica rice (Oryza sativa L.) endosperm. To elucidate the distinct and overlapping functions of these enzymes, double mutants were created by crossing the ss1 null mutant with the ss3a null mutant. In the F2 generation, two opaque seed types were found to have either the ss1ss1/SS3ass3a or the SS1ss1/ss3ass3a genotype. Phenotypic analyses revealed lower SS activity in the endosperm of these lines than in those of the parent mutant lines since these seeds had different copies of SSI and SSIIIa genes in a heterozygous state. The endosperm of the two types of opaque seeds contained the unique starch with modified fine structure, round-shaped starch granules, high amylose content, and specific physicochemical properties. The seed weight was ∼90% of that of the wild type. The amount of granule-bound starch synthase I (GBSSI) and the activity of ADP-glucose pyrophosphorylase (AGPase) were higher than in the wild type and parent mutant lines. The double-recessive homozygous mutant prepared from both ss1 and ss3a null mutants was considered sterile, while the mutant produced by the leaky ss1 mutant×ss3a null mutant cross was fertile. This present study strongly suggests that at least SSI or SSIIIa is required for starch biosynthesis in rice endosperm.
Keywords: Amylopectin, amylose, endosperm, mutant, rice, starch synthase
Introduction
Starch, consisting of two homopolymers of the α-D-glucosyl unit, is one of the most important carbohydrates on earth. The branched amylopectin makes up 65–85% of starch weight, while basically linear amylose makes up the remainder. Four enzyme classes catalyse the reactions of starch biosynthesis: ADP-glucose pyrophosphorylase (AGPase), starch synthase (SS), starch branching enzyme (BE), and starch debranching enzyme (DBE) (Smith et al., 1997; Myers et al., 2000; Nakamura, 2002; Ball and Morell, 2003).
SS (EC 2.4.1.21) elongates α-glucans by adding glucose residues from ADP-glucose to the glucan non-reducing ends through α-1,4 glucosidic linkages. Of all the starch biosynthetic enzymes, SS has the largest number of isoforms. In rice, there are 10 SS isozymes divided into five types: SSI, SSII, SSIII, SSIV, and GBSS (granule-bound starch synthase I), with each enzyme having one, three, two, two, and two isozymes, respectively (Hirose and Terao, 2004). The SSI, SSIIa, SSIIIa, and GBSSI genes are highly expressed in developing endosperm (Hirose and Terao, 2004; Ohdan et al., 2005). SSI and SSIII(a) are the main isozymes associated with amylopectin biosynthesis in cereals, such as maize (Gao et al., 1998; Cao et al., 1999) and japonica rice (Fujita et al., 2006), and also in Arabidopsis leaves (Delvalle et al., 2005; Zhang et al., 2005). In contrast, SSII and SSIII are the primary isozymes present in potato tubers (Marshall et al., 1996; Edwards et al., 1999; Lloyd et al., 1999) and pea embryos (Craig et al., 1998). Clues concerning the specific function of each SS isozyme have been elucidated through biochemical and molecular studies together with mutant and transgenic plant analyses.
ss1 (Fujita et al., 2006) and ss3a (Fujita et al., 2007), mutants of the japonica rice cultivar Nipponbare, were isolated from a retrotransposon (Tos17) insertion line using a reverse genetics approach. These mutants have helped clarify the function of the SSI and SSIIIa isozymes. The rice ss3a mutant corresponds to the dull-1 mutant in maize (Mangelsdorf, 1947; Davis et al., 1955; Gao et al., 1998), with both mutants exhibiting similar endosperm starch phenotypes. With the exception of japonica rice (Fujita et al., 2006) and Arabidopsis (Delvalle et al., 2005), no SSI-defective mutants have been identified in other plant species to date.
The analysis of japonica rice defective in SSIIa (Nakamura et al., 2005) made it possible to identify and characterize the SSI isozyme. In japonica rice, the ss1 mutation had no effect on the size or shape of seeds and starch granules, or on the crystallinity of endosperm starch. However, the changes in amylopectin chain length distribution in four allelic mutants were positively correlated with the levels of SSI activity (0–25% of the wild type). These results show that SSI generates chains with a degree of polymerization (DP) 8–12 from short DP 6–7 chains emerging from the branch point in the A and B1 chains of amylopectin (Fujita et al., 2006). These findings were confirmed through in vitro experiments using recombinant rice SSI (Fujita et al., 2008).
In contrast, the rice ss3a mutant phenotypes differed significantly from those of wild-type plants (Fujita et al., 2007). First, the ss3a mutant seeds had a white core, and the starch granules were small, round, and relatively loosely packed into the endosperm cell. Secondly, the level of amylopectin B2–4 chains with DP ≥30 was reduced to ∼60% of that of the wild type. Thirdly, SSIIIa deficiency in the ss3a mutant resulted in an increase in SSI and GBSSI gene expression, resulting in a change of DP ≤30 in amylopectin chain length and a 1.5-fold increase in amylose content, respectively.
In most japonica cultivars, GBSSI activity is reduced by ∼10% due to GBSSI mutations (Sano, 1984; Isshiki et al., 1998). The japonica ss1 and ss3a mutants reported are triple mutants [ss1/ss2a/gbss1L (where the superscript L denotes leaky) and ss2a/ss3a/gbss1L, respectively] when compared with wild-type indica rice. However, in these mutants, the quantity of endosperm starch and seed weight are nearly the same as in the wild type. A specific SS isozyme deficiency results in an overlapping of chain elongation reactions with those of other SSs or other compensation mechanisms. Typically, the minor SS isozyme function is masked in wild-type plants due to the large number of SS isozymes present in higher plants. To study individual SS isozyme function, the overlapping activity of many other SS isozymes needs to eliminated.
The present study describes the generation of double-recessive deficient mutants of SSI and SSIIIa. The double-recessive mutants derived from a cross between null ss1 and null ss3a mutants were considered sterile. However, the heterozygous mutants produced fertile opaque seeds. In an attempt to obtain fertile double-recessive mutant lines, the null ss3a mutant and the leaky ss1 mutant were crossed. The pleiotropic effects on starch biosynthesis in the developing endosperm following loss or reduction of SSI and SSIIIa activity are also discussed.
Materials and methods
Plant materials
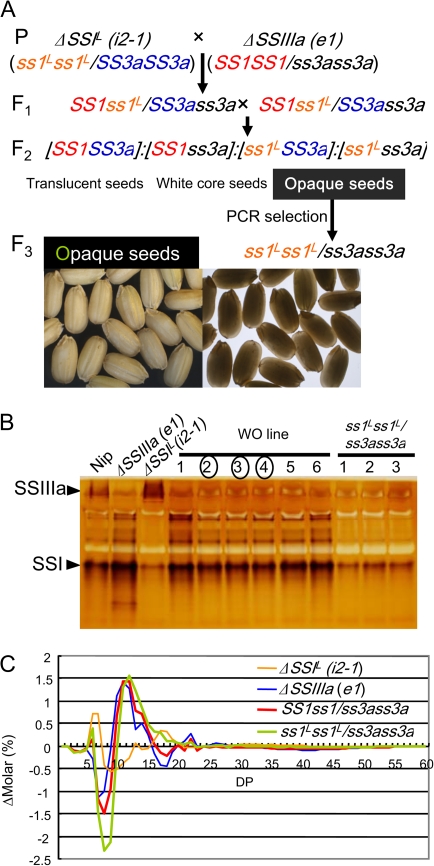
ss1 mutant lines ΔSSI containing null (e7) and leaky (i2-1) (Fujita et al., 2006), and the null ss3a mutant line ΔSSIIIa (e1, or ss3a-1 in Fujita et al., 2007) were used for generating ss1 and ss3a double mutant lines. The parental cultivar Nipponbare (Nip) was used as a control. ΔSSI (e7 and i2-1) was crossed with ΔSSIIIa (e1), and the resulting double heterozygotes (F1) were self-pollinated. Opaque seeds of the F2 generation were self-pollinated again. In the crosses between i2-1 and e1, double-recessive seeds of F2 opaque seeds were identified via PCR screening and self-pollinated (Fig. 8). PCR screening was conducted as described by Fujita et al. (2006, 2007). Rice plants were grown during the summer months in an experimental paddy field at Akita Prefectural University under natural environmental conditions.
Fig. 8.
Pedigree, seed morphology (A), native-PAGE/SS activity staining of developing endosperm (B), and differences in the amylopectin chain length distribution pattern (C) of ss1Lss1L/ss3ass3a after a cross between the leaky ss1 mutant (ΔSSI, i2-1) and the ss3a mutant (ΔSSIIIa, e1). The morphology of rice dehulled seeds was observed using a stereo-microscope with overhead light (left panel) and on a light box (right panels). Lane numbers with circles on the zymogram (B) show WO endosperm having the same level or a slightly lower SSI activity band than that of Nipponbare.
Native-PAGE/activity staining and enzyme assay
Native-PAGE/activity staining of DBE and BE was performed using the methods of Fujita et al. (1999) and Yamanouchi and Nakamura (1992), respectively. SS activity staining was performed on 7.5% (w/v) acrylamide slab gels containing 0.8% (w/v) oyster glycogen (G8751, Sigma) according to Nishi et al. (2001) with the addition of 0.5 M citrate in the reaction mixture. The assay for AGPase was performed as described by Nakamura et al. (1989).
Starch or amylopectin structure analysis
Extraction of starch from mature and developing rice endosperm for amylopectin chain length distribution assessment was performed according to Fujita et al. (2001). The chain length distributions of endosperm α-glucans were analysed using capillary electrophoresis as described by O'Shea and Morell (1996) and Fujita et al. (2001) in a P/ACE MDQ Carbohydrate System (Beckman Coulters, CA, USA).
Gel filtration chromatography of starches and amylopectin was performed according to Fujita et al. (2007, 2009) using a Toyopearl HW55S gel filtration column (300×20 mm) connected in series to three columns (300×20 mm) of Toyopearl HW50S.
Endosperm starch granule analysis
Rice seed α-glucan quantification, X-ray diffraction, rapid visco-analyser (RVA) analysis of the pasting properties of endosperm starch, differential scanning calorimetry (DSC) of the thermal properties of endosperm starch, and scanning electron microscopy (SEM) of starch granules (JEOL-5600) were performed as described previously (Fujita et al., 2003, 2006).
Protein extraction and quantification from mature endosperm
Preparation of soluble protein (SP), loosely bound protein (LBP), and tightly bound protein (TBP) from the mature endosperm was performed according to the methods of Fujita et al. (2006). The SSI and GBSSI protein amounts within these fractions were estimated by measuring the immunoblot band intensities with NIH image (ver. 5.5).
Results
Generation of opaque seeds from a cross between SSI- and SSIIIa-deficient mutant lines
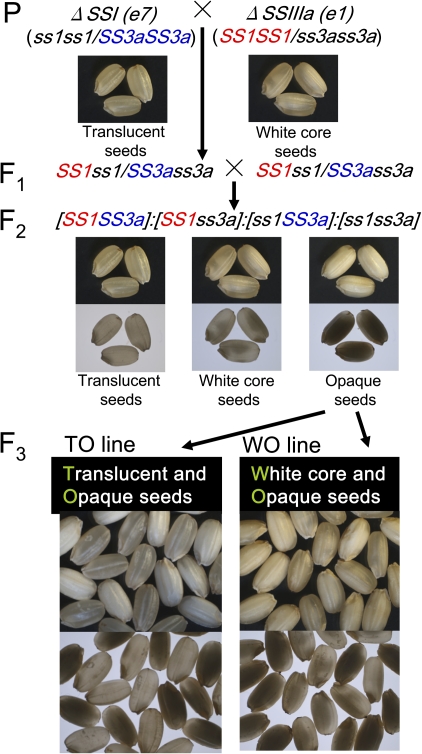
To understand the effects of the concurrent inactivation of the two major SS isozymes, the null ss1 mutant (e7) was crossed with the null ss3a mutant (e1). Morphological analysis showed that the ss1 seeds were translucent, whereas the ss3a seeds had a white core (Fig. 1; Fujita et al., 2006, 2007). Approximately 10% of the opaque seeds appeared in the F2 segregation from self-pollination of F1 seeds (Fig. 1; Supplementary Table S1 available at JXB online). F3 seeds resulting from the self-pollination of the F2 opaque seeds could be segregated into different seed morphologies, indicating that the opaque seeds were not double-recessive homozygous. Moreover, two types of F3 segregations appeared: one (TO line) was separated into translucent (T) and opaque (O) seeds, and the other (WO line) was separated into those with a white core (W) and those with opaque (O) seeds (Fig. 1). F3 to F5 seeds resulting from TO line opaque seed self-pollination were segregated into an approximate 1:1 ratio of translucent to opaque seeds. F3 to F4 seeds resulting from WO line opaque seed self-pollination also segregated into an approximate 1:1 ratio of white core to opaque seeds (Supplementary Table S1). Some empty seeds were found in F3 to F5 seeds and F3 to F4 seeds in both the TO and WO lines, respectively. However, the incidence rate of such phenomenon was dependent on the harvest year (Supplementary Table S1).
Fig. 1.
Pedigree of opaque seeds of the TO and WO lines and seed morphology. The morphology of rice dehulled seeds was observed using a stereo-microscope with overhead light (upper panels) and on a light box (lower panel). (This figure is available in colour at JXB online.)
Genotype and SS activity in the TO and WO lines
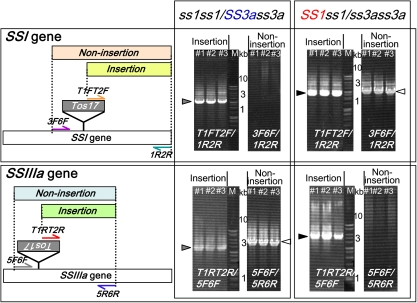
The genotypes of the ss1 and ss3a mutant lines can be confirmed by examining for the presence of the Tos17 insertion (Fujita et al., 2006, 2007). To determine the genotype of the TO and WO seeds with different morphologies, nested PCR was conducted with DNA from three independent seedlings from each line (Fig. 2). The DNA extracted from seedlings of TO opaque seeds was found to contain the homozygous recessive SSI gene and the heterozygous SSIIIa gene (ss1ss1/SS3ass3a). In contrast, the DNA extracted from WO opaque seeds was found to contain the heterozygous SSI gene and the homozygous recessive SSIIIa gene (SS1ss1/ss3ass3a). The genotypes of the TO translucent seeds and WO white core seeds were determined to be ss1ss1/SS3aSS3a and SS1SS1/ss3ass3, respectively, the same genotypes as those of the parent ss1 and ss3a mutants, respectively (data not shown). However, the genotype of the opaque seed endosperm (3n) was not identified in this study.
Fig. 2.
Determination of genotypes in opaque seeds of the TO and WO lines by nested PCR. The left figures show the site of Tos17 insertion and the position of the primer pairs. Horizontal half arrows show the binding sites for nested PCR primers used for genotype determination (T1FT2F, 1R2R, and 3F6F for the SSI gene, and T1RT2R, 5F6F, and 5R6R for the SSIIIa gene). Primer pairs are indicated below the photographs. ‘T1FT2F/1R2R’ means that the primer pair T1F/1R was used for the first PCR and T2F/2R for the second PCR. M, molecular markers. (This figure is available in colour at JXB online.)
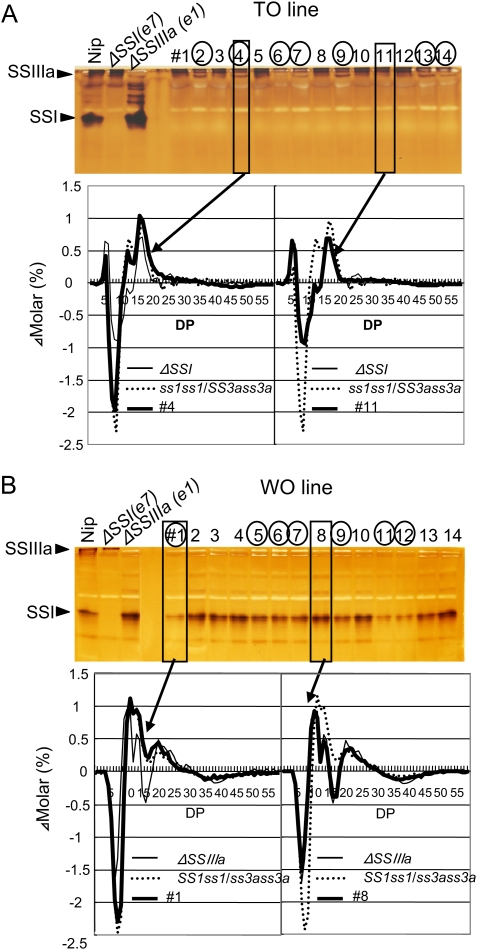
To detect SS activity in the endosperm during the development of self-pollinated ss1ss1/SS3ass3a seeds, 14 developing endosperm samples were randomly selected and the crude enzyme extract of each seed was subjected to native-PAGE/SS activity staining (Fig. 3A, upper panel).
Fig. 3.
Native-PAGE/SS activity staining of self-pollinated developing endosperm of ss1ss1/SS3ass3a in the TO line (A) and SS1ss1/ss3ass3a in the WO line (B) and their chain length distribution. Fourteen developing endosperm samples were randomly chosen, and the zymogram pattern of native-PAGE/SS activity staining was compared with those of Nipponbare, the ss1 mutant (ΔSSI, e7), and the ss3a mutant (ΔSSIIIa, e1). SSIIIa (A) or SSI (B) activity bands in 14 developing endosperm samples were divided into two groups, namely higher or lower activity bands, respectively, compared with Nipponbare. Lane numbers with circles indicate endosperm having the same or a slightly lower SSIIIa (A) or SSI (B) activity band, respectively, than that of Nipponbare. Chain length distributions of endosperm starch of a typical lower or higher activity band (boxed lanes) were analysed (bold lines) and compared with those of ΔSSI (thin line) and ss1ss1/SS3ass3a (dotted line) (A), or ΔSSIIIa (thin line) and SS1ss1/ss3ass3a (dotted line) (B). (This figure is available in colour at JXB online.)
Although the SSI activity band was absent in all the developing seeds, the SSIIIa activity bands were present. Notably, after evaluating >50 seeds, none of the developing ss1ss1/SS3ass3a endosperm samples had completely lost both SS activity bands (data not shown). As previously shown, the SSIIIa activity bands in the null ss1 mutants were slightly stronger than those of the wild type (Fujita et al., 2006). The SSIIIa activity bands of 14 developing ss1ss1/SS3ass3a endosperm samples were divided into two groups, namely those that exhibited higher activity (lanes #1, 3, 5, 8, 10, 11, and 12) and those that exhibited lower activity (lanes #2, 4, 6, 7, 9, 13, and 14) than that of Nipponbare (Fig. 3A, upper panel).
The same experiments were repeated in the developing seeds of SS1ss1/ss3ass3a. The SSIIIa activity bands were lost in all the developing seeds tested. However, the SSI activity bands remained. Notably, no self-pollinated SS1ss1/ss3ass3a seeds completely lost both SS activity bands among the >50 developing endosperm samples assessed (data not shown). As previously shown, the SSI activity bands in the null ss3a mutant are significantly stronger than those of the wild type (Fujita et al., 2007). The SSI activity bands in 14 developing endosperm samples of the self-pollinated SS1ss1/ss3ass3a line were divided into two groups, namely the higher group (#2, 3, 4, 8, 10, 13, and 14) and the same or slightly lower group (#1, 5, 6, 7, 9, 11, and 12) (Fig. 3B, upper panel).
These results suggest that either SSI or SSIIIa activity varies among the different lines having different copies of SSI and SSIIIa genes in a heterozygous state generated by the ss1×ss3a cross. A schematic representation of the levels of SSI and SSIIIa activities is shown in Table 1.
Table 1.
Summary of genotypes and characterization of starch-related phenotypes in various mutant lines and the wild type used in this study
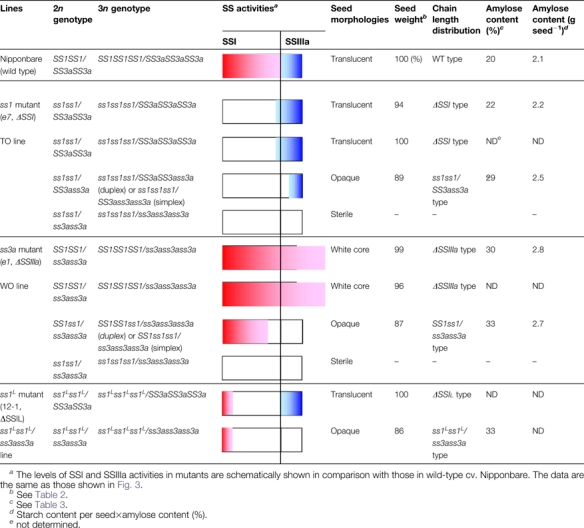 |
As no developing endosperm from the self-pollinated ss1ss1/SS3ass3a and SS1ss1/ss3ass3a lines was identified as having completely lost both SSI and SSIIIa activity, these results strongly suggest that double-recessive ss1ss1/ss3ass3a seeds are sterile and become empty in the hulls (Supplementary Table S1 at JXB online).
Characterization of TO and WO opaque seeds and endosperm starch
The ss1 and ss3a mutant seed weights were not significantly different from those of the wild type (Table 2; Fujita et al., 2006, 2007). The dehulled grain weights of ss1ss1SS3ass3a and SS1ss1ss3ass3a seeds were lower (89% and 87%, respectively) than those of the wild type and parent mutants (Table 2), although there was no significant difference between these lines and the wild type by t-test at P <0.05. The starch content of opaque seeds of both lines remained at >75% of that of the wild type (Table 2).
Table 2.
Dehulled grain weight and carbohydrate content
| Lines | Dehulled grain weight (mg) | Starch content | Soluble fraction (%) |
| Nipponbare | 20.6±0.3a (100)b | (100)b | 3.5±0.3c |
| ΔSSI | 19.3±0.2 (94) | (94.6) | 3.2±0.2 |
| ΔSSIIIa | 20.3±0.3 (99) | (85.8) | 4.4±1.4 |
| ss1ss1/SS3aSS3a | 20.5±0.3 (100) | – | – |
| ss1ss1/SS3ass3a | 18.2±0.3 (89) | (80.1) | 2.9±1.4 |
| SS1SS1/ss3ass3a | 19.8±0.2 (96) | – | – |
| SS1ss1/ss3ass3a | 18.0±0.2 (87) | (75.5) | 2.9±1.4 |
Mean ±SE of 20 seeds. No significant differences between Nipponbare and mutant lines by t-test at P <0.05.
Percentage of the wild type.
Mean ±SE of three seeds.
To characterize the fertile opaque seed phenotype, chain length distribution of the endosperm amylopectin was determined using capillary electrophoresis (Fig. 4). The chain length distribution patterns of ss1ss1/SS3ass3a and SS1ss1/ss3ass3a (Fig. 4A, B) were quite different from those of the parent mutant line and wild type (Fig. 4C), while those of ss1ss1/SS3aSS3a and SS1SS1/ss3ass3a showed the same pattern as seen in the ss1 and ss3a mutants, respectively (data not shown). At least 20 seeds of the ss1ss1/SS3ass3a line exhibited a nearly identical amylopectin chain length pattern, and the same result was also obtained with seeds of the SS1ss1/ss3ass3a line (data not shown), suggesting that there is no dosage effect between the simplex (ss1ss1ss1/SS3ass3ass3a or SS1ss1ss1/ss3ass3ass3a) and duplex (ss1ss1ss1/SS3aSS3ass3a or SS1SS1ss1/ss3ass3ass3a) in the 3n genotype in the endosperm.
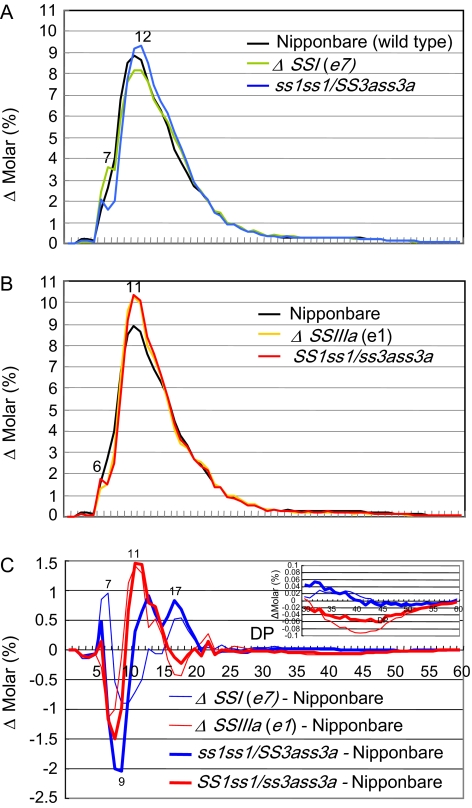
Fig. 4.
Amylopectin structure of ss1ss1/SS3ass3a and SS1ss1/ss3ass3a. (A) Chain length distribution patterns of endosperm amylopectin in the mature endosperm of ss1ss1/SS3ass3a, the ss1 mutant (e7, ΔSSI), and the wild-type parent cultivar Nipponbare. (B) Chain length distribution patterns of endosperm amylopectin in the mature endosperm of SS1ss1/ss3ass3a, the ss3a mutant (e1, ΔSSIIIa), and the wild-type parent cultivar Nipponbare. (C) Differences in the chain length distribution patterns of amylopectin in the mature endosperm of ss1ss1/SS3ass3a and SS1ss1/ss3ass3a and their parent mutant lines. The inset in C shows the magnification of the pattern in the range of chains with DP 30–60. The numbers on the plots are the DP values.
In the ss1ss1/SS3ass3a line, amylopectin chains with DP ≤10 significantly decreased, while those with DP 11–20 increased significantly when they were compared with the wild type (Fig. 4C). In the ss3a mutant, long amylopectin B2–4 chains of ∼DP 42 or 74 significantly decreased to ∼60% of wild-type levels (Fujita et al., 2007). The amylopectin of the SS1ss1/ss3ass3a line showed a decrease in long chains at ∼DP 42, similar to the ss3a mutant, although the extent of reduction was more pronounced in the ss3a mutant (Fig. 4C, inset). However, the decrease in short chains with DP ≤9 and the increase of those with DP 11–20 were more pronounced than in the ss3a mutant (Fig. 4C). The ss1ss1/SS3ass3a pattern was quite distinctive in comparison with the ss1 mutant, whereas the SS1ss1/ss3ass3a pattern was similar to that of the ss3a mutant (Fig. 4A, B). These phenotypes indicate that the additive reduction of SSIIIa activity affects the amylopectin fine structure more significantly than that of SSI. Furthermore, the SSIIIa heterozygous mutation is sufficient to alter amylopectin fine structure.
Following native-PAGE/SS activity staining, starch samples from the same developing endosperm samples were used for chain length distribution analysis. All endosperm samples displaying high SSIIIa activity had amylopectin chain length distributions very similar to that of the ss1 mutant, while endosperm samples with low SSIIIa activity showed patterns (Fig. 3A, lower panel, data not shown) similar to the endosperm of ss1ss1/SS3ass3a mature seeds (Fig. 4C). The amylopectin chain length distribution of all endosperm samples with high SSI activity bands had patterns similar to the ss3a mutant, while those from all endosperm samples with low SSI activity bands had similar patterns (Fig. 3B, lower panel, data not shown) to those of the endosperm of SS1ss1/ss3ass3a mature seeds (Fig. 4C). These results strongly suggest that a reduction of SS activity, as a result of heterozygous SSI and SSIIIa genes, leads to a change in amylopectin fine structure.
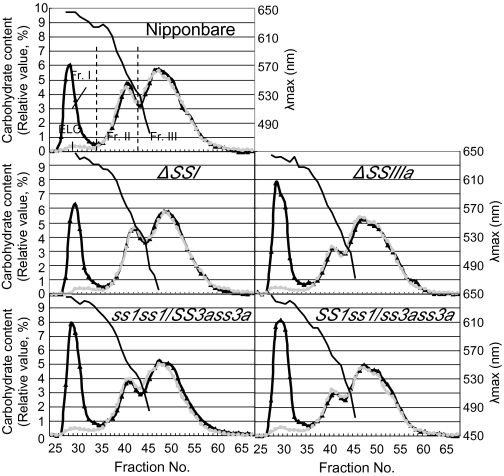
To analyse the starch structure and components in mature ss1ss1/SS3ass3a and SS1ss1/ss3ass3a seeds further, the isoamylolysates of endosperm starch and purified amylopectin were subjected to size exclusion chromatography using Toyopearl HW55S and HW50S gel filtration columns (Fig. 5). The λmax values of the α-glucan–iodine complex indicate that fraction I (Fr. 1) contained most, if not all, of the amylose. A small amount of fraction I was detected in purified amylopectin from Nipponbare endosperm starch (Fig. 5) and was designated an ‘extra-long chain’ (DP ≥500) amylopectin (Takeda et al., 1987; Horibata et al., 2004). Therefore, Fr. I from endosperm starch includes both true amylose and ‘extra-long chain’ content. The value obtained after subtraction of extra-long chain content from the apparent amylose content of starch is equivalent to the true amylose content of starch (Horibata et al., 2004). The proportion of each starch component was calculated based on the data shown in Fig. 5, and the results are shown in Table 3.
Fig. 5.
Size separation of debranched endosperm starch and purified amylopectin from ss1ss1/SS3ass3a and SS1ss1/ss3ass3a, ss1 mutant (ΔSSI, e7), ss3a mutant (ΔSSIIIa, e1), and wild-type Nipponbare by gel filtration chromatography through Toyopearl HW55S–HW50S columns. Elution profiles of isoamylase-debranched starch (black lines) and purified amylopectin (grey lines) are shown. Each fraction (Fr. I, II, and III) was divided into the valley of the carbohydrate content curve determined by the phenol sulphuric acid method.
Table 3.
The composition of carbohydrate content (weight %) in the fractions separated by gel filtration chromatography of endosperm starch and purified amylopectin
| Fr. Ia (%) | Fr. IIa (%) | Fr. IIIa (%) | III/II | TACb (%) | ||
| Nipponbare | Starchc | 19.9 | 23.3 | 56.8 | 2.4 | 17.8 |
| Amylopectin | 2.1d | 23.3 | 56.5 | 2.4 | ||
| ΔSSI | Starch | 22.3 | 21.2 | 56.5 | 2.7 | 19.7 |
| Amylopectin | 2.6 | 21.1 | 56.4 | 2.7 | ||
| ΔSSIIIa | Starch | 30.2 | 14.9 | 54.9 | 3.7 | 26.9 |
| Amylopectin | 3.3 | 15.3 | 56.4 | 3.7 | ||
| ss1ss1/ | Starch | 28.6 | 21.2 | 50.2 | 2.4 | 25.3 |
| SS3ass3a | Amylopectin | 3.3 | 22.9 | 45.8 | 2.0 | |
| SS1ss1/ | Starch | 32.5 | 15.8 | 51.7 | 3.3 | 29.4 |
| ss3ass3a | Amylopectin | 3.1 | 15.7 | 49.2 | 3.1 |
Each fraction (Fr. I, II, and III) was divided into the valley of the carbohydrate content curve determined by the phenol sulphuric acid method (Fig. 5).
True amylose content=apparent amylose content (Fr. I of starch) – extra-long chains (Fr. I of amylopectin).
Total carbohydrate content was 100%.
The areas for Fr. II and Fr. III of amylopectin were superimposed on those of the starch, and the amount of Fr. I of amylopectin (extra-long chain) was calculated.
The apparent amylose content of the ss1 mutant (22.3%) was slightly higher than that of Nipponbare (19.9%), while that of the ss3a mutant (30.2%) was significantly higher than that of the wild type (Fig. 5, Table 3; Fujita et al., 2007). On the other hand, the apparent amylose content of the ss1ss1/SS3ass3a (28.6%) and SS1ss1/ss3ass3a (32.5%) lines, showing lower SSIIIa and SSI activity than that of the ss1 and ss3a mutants, respectively, was higher than that of the parent mutants. The highest amylose content of rice cultivars, including indica and japonica rice lines, as measured by gel filtration with Toyopearl HW55S and HW50S columns, is ∼31–33% (Horibata et al., 2004; Inouchi et al., 2005), although the high-amylose maize BEIIb (amylose-extender) mutant has an apparent amylose content of 50–85% (Li et al., 2008). Therefore, the amylose content of SS1ss1/ss3ass3a in this study belongs to the highest group in rice. The extra-long chain in ss1ss1/SS3ass3a (3.3%) and SS1ss1/ss3ass3a (3.1%) lines and the ss3a mutant (3.3%) having no or low SSIIIa activity was higher than that of the ss1 mutant (2.6%) and the wild type (2.1%).
Fr. II included B2–4 long chains of amylopectin connecting tandem clusters of amylopectin, and Fr. III included short chains within one cluster of amylopectin. The ratio of Fr. III to Fr. II (III/II) amylopectin chains in ss3a mutant endosperm starch (3.7) was higher than that in Nipponbare endosperm starch (2.4), indicating that the ss3a mutant endosperm composition included fewer long amylopectin chains than Nipponbare (Table 3; Fujita et al., 2007). The III/II ratio of amylopectin chains in SS1ss1/ss3ass3a (3.3) was also higher than that of the wild type (2.4) but lower than that of the ss3a mutant. However, the ratio in ss1ss1/SS3ass3a (2.0) and in ss1 (2.7) was similar to that of the wild type. These results were consistent with the chain length distribution obtained by capillary electrophoresis (Fig. 4).
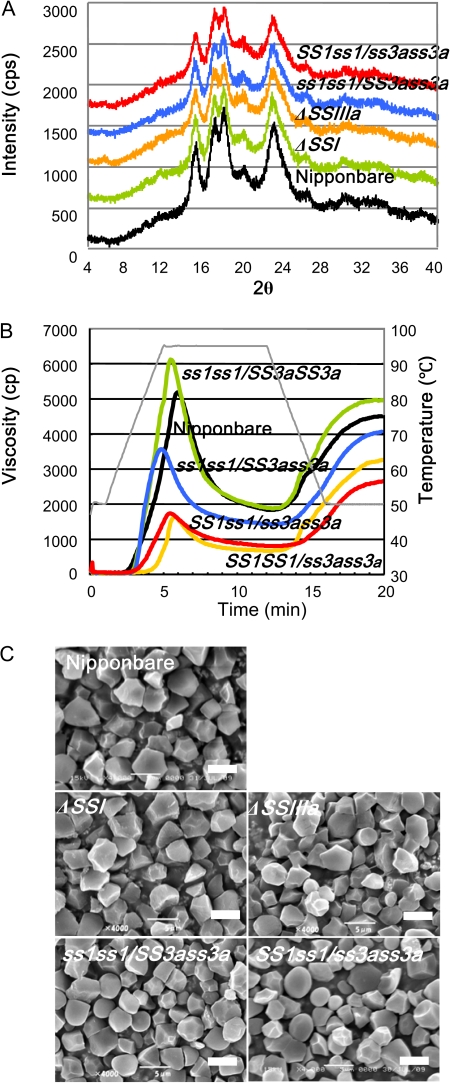
Cereal endosperm starch displays the A-type X-ray diffraction pattern, and potato starch displays the B-type diffraction pattern. The starch granules of ss1ss1/SS3ass3a, SS1ss1/ss3ass3a, and their mutant parents displayed the typical A-type X-ray diffraction pattern (Fig. 6A). When compared with wild-type starch, the height and sharpness of the major starch peaks were apparently lower in the mutant lines with particularly reduced SSIIIa activity. This result indicates that the degree of starch granule crystallinity in these mutant lines is reduced (Fig. 6A).
Fig. 6.
Characterization of the endosperm starch in the ss1ss1/SS3ass3a, SS1ss1/ss3ass3a, ss1 mutant (ΔSSI, e7), ss3a mutant (ΔSSIIIa, e1), and wild-type Nipponbare. (A) X-ray diffraction patterns of endosperm starch. The height and sharpness of major peaks show the degree of starch granule crystallinity. (B) Pasting properties of endosperm starch by rapid visco-analyser (RVA). The viscosity pattern shown is one of at least three replicates. The thin line indicates the change in temperature during measurement with an RVA. (C) Scanning electron micrographs of the endosperm starch granules. Bar=5 μm. (This figure is available in colour at JXB online.)
The pasting properties of the endosperm starch were analysed using an RVA (Fig. 6B). The paste viscosity of starch from SS1ss1/ss3ass3a and SS1SS1/ss3ass3a, which have the same genotype as the ss3a mutant, was dramatically lower than that of the starch from the wild type (Fig. 6B; Fujita et al., 2007). In contrast, the peak viscosity of endosperm starch from ss1ss1/SS3aSS3a, having the same genotype as the ss1 mutant, was higher than that of the wild type. The peak viscosity of endosperm starch from ss1ss1/SS3ass3a was lower than that of the wild type. These results reveal that endosperm viscosity is related to SSI and SSIIIa activity. A decrease in SSI and SSIIIa activity elevates and lowers the peak viscosity, respectively, of the endosperm starch.
To evaluate physicochemical properties of endosperm starch in ss1ss1/SS3ass3a and SS1ss1/ss3ass3a, the gelatinization temperature of the endosperm starch was analysed by DSC. The temperatures [for the onset (To), peak (Tp), and conclusion (Tc)] of endosperm starch gelatinization in the ss3a mutant or SS1SS1/ss3ass3a were slightly higher (0–3 °C) than those of the wild type, However, the temperatures of endosperm starch gelatinization in SS1ss1/ss3ass3a were 3–5 °C higher than in the wild type. In contrast, the temperatures in the ss1 mutant or ss1ss1/SS3aSS3a line were 5–9 °C higher than those in the wild type, while those in ss1ss1/SS3ass3a were significantly higher (8–10 °C) than those in the wild type (Table 4).
Table 4.
Thermal properties of endosperm starch as determined by DSC
| Lines | Toa (°C) | Tpb (°C) | Tcc (°C) | ΔHd (mJ mg−1) |
| Nipponbare | 47.0±0.8e | 55.3±0.1 | 62.4±0.4 | 12.1±3.2 |
| ΔSSI | 55.9±0.4 | 62.9±0.3 | 69.5±0.2 | 14.1±1.6 |
| ΔSSIIIa | 47.0±0.2 | 55.7±0.4 | 63.2±0.3 | 12.1±0.8 |
| ss1ss1/SS3aSS3a | 53.3±1.6 | 60.7±0.7 | 67.8±1.7 | 14.4±2.5 |
| ss1ss1/SS3ass3a | 57.4±0.6 | 64.1±0.1 | 70.6±0.2 | 18.1±1.7 |
| SS1SS1/ss3ass3a | 47.9±1.2 | 57.1±0.4 | 65.3±0.2 | 11.5±0.5 |
| SS1ss1/ss3ass3a | 50.5±0.7 | 60.2±0.1 | 66.7±0.2 | 12.5±1.4 |
Onset temperature.
Peak temperature.
Conclusion temperature.
Gelatinization enthalpy of starch.
Mean ±SE of three seeds.
To determine whether the reduction in SSI and SSIIIa activity affects starch granule morphology, SEM of purified starch granules was performed (Fig. 6C). Previous studies reported that the endosperm starch granules of the wild type and ss1 mutant formed similarly sized (∼5 μm) polygonal granules with sharp edges. In contrast, starch granules isolated from ss3a endosperm were slightly smaller and rounder than those of the wild type (Fig. 5B; Fujita et al., 2006, 2007). The starch from the ss1ss1/SS3ass3a and SS1ss1/ss3ass3a lines consisted of a substantially larger proportion of round granules compared with the ss3a mutant (Fig. 6C).
Pleiotropic effects of other enzymes related to starch biosynthesis in the TO and WO lines
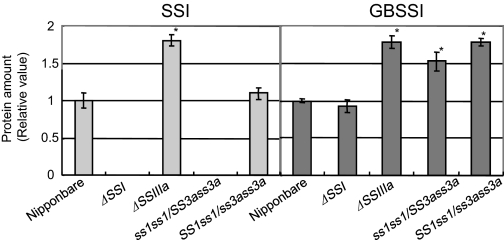
A reduction in SSIIIa activity leads to an increase in SSI and GBSSI gene expression. This results in structural and physicochemical alterations in the endosperm starch (Fujita et al., 2007). To assess the effect of SSI and SSIIIa deficiencies in the ss1ss1/SS3ass3a and SS1ss1/ss3ass3a lines quantitatively, SSI and GBSSI protein levels in mature seeds were estimated by immunoblot analysis (Fig. 7). SSI was found not only in the soluble fraction, but also in the starch granule-bound fraction of developing rice endosperm, whereas most GBSSI binds to starch granules (Fujita et al., 2006, 2007). The SSI and GBSSI proteins were separated into soluble protein+loosely bound protein (SP+LBP), and tightly bound protein (TBP) fractions, and the total amounts of SSI and GBSSI (SP+LBP+TBP) were estimated (Fig. 7). SSI protein was not detected in the ss1 mutant and ss1ss1/SS3ass3a. Although SS1ss1/ss3ass3a contains a heterozygous SSI gene, the SSI protein level in the seeds was comparable with that of the wild type (Fig. 7), although the SSI activity band detected by native-PAGE/SS activity staining was lower in intensity than the wild-type band (Fig. 3B). The GBSSI protein level was 1.8 times higher in the ss3a mutants than in the wild type. In contrast, GBSSI protein levels in the ss1 mutants were nearly the same as or slightly lower than in the wild type (Fig. 7). The GBSSI protein amount in the ss1ss1/SS3ass3a and SS1ss1/ss3ass3a lines was 1.5 and 1.8 times higher than in the wild type, respectively (Fig. 7).
Fig. 7.
Amount of SSI or GBSSI protein in the mature endosperm of ss1ss1/SS3ass3a, SS1ss1/ss3ass3a, ss1 mutant (ΔSSI, e7), ss3a mutant (ΔSSIIIa, e1), and wild-type Nipponbare. The total protein amount in the three fractions [SP (soluble protein)+LBP (loosely bound protein) and TBP (tightly bound protein) of SSI or GBSSI protein] was quantified by immunoblotting using antiserum raised against SSI or GBSSI (Fujita et al., 2006). The data are the mean ±SE of three seeds. Asterisks denote statistically significant differences between Nipponbare and mutant lines by t-test at P <0.05.
To evaluate pleiotropic effects caused by the deficiency and reduction in SSI and SSIIIa activity, the activities of other starch biosynthesis enzymes were measured in the developing seeds of self-pollinated ss1ss1/SS3ass3a and SS1ss1/ss3ass3a. Isoamylase, pullulanase, and phosphorylase (PHO), detected by native-PAGE/DBE activity staining, and BEI, BEIIa, and BEIIb, detected by native-PAGE/BE activity staining in developing endosperm revealed no obvious differences (Suppplementary Fig. S1A, B at JXB online). The AGPase activity of the rice ss1 mutant (Fujita et al., 2006) and the rice and maize ss3(a) mutant (Fujita et al., 2007; Singletary et al., 1997) was reported to be higher than that of the wild type. In this study, the AGPase activity of the ss1 and ss3a mutants and the developing endosperm in these mutants was found to be ∼1.5 times the wild-type level (Table 5). In comparison, the AGPase activity of the developing endosperm in ss1ss1/SS3ass3a and SS1ss1/ss3ass3a was ∼2.0 times the wild-type level (Table 5).
Table 5.
AGPase activities of developing endosperm
| Lines | AGPase activitya (μmol min−1 endosperm−1) |
| Nipponbare | 0.236±0.017 (100)b |
| ΔSSI | 0.325±0.034 (138) |
| ΔSSIIIa | 0.370±0.016* (157) |
| ss1ss1/SS3aSS3ac | 0.376±0.013* (159) |
| ss1ss1/SS3ass3ac | 0.479±0.034* (203) |
| SS1SS1/ss3ass3ac | 0.385±0.029* (163) |
| SS1ss1/ss3ass3ac | 0.462±0.009*(196) |
Mean ±SE of three seeds.
Percentage of the wild type.
Genotypes of developing endosperm of self-pollinated ss1ss1/SS3ass3a and SS1ss1/ss3ass3a were determined by the chain length distribution pattern of remaining starch after measurement of AGPase activities.
*Significant differences between Nipponbare and mutant lines by t-test at P <0.05.
Production of double homozygous recessive lines
The next generation of the self-pollinated ss1ss1/SS3ass3a line will segregate again into translucent and opaque seeds, while the SS1ss1/ss3ass3a line will segregate into white core and opaque seeds (Fig. 1). Fertilization of opaque seeds was possible due to remaining SSIIIa or SSI activity in the developing endosperm (Fig. 3A, B, upper panels). For these reasons, the leaky rice ss1 mutant line (i2-1), having one-sixth the activity of the wild type (Fujita et al., 2006), was crossed with the null ss3a mutant (e1) to obtain a double-recessive homozygous mutant. Opaque seeds of the F2 generation were selected by PCR using seedling DNA, and the seeds bearing double-recessive homozygous genotypes were self-pollinated (Fig. 8). Native-PAGE/SS activity staining of 14 randomly chosen developing endosperm samples of self-pollinated seeds did not detect any SSIIIa activity bands. However, a faint SSI band was detected at a level lower than that of SS1ss1/ss3ass3a and similar to that of i2-1 (Fig. 8B, Table 1; Suppplementary Fig. S1 at JXB online). Moreover, these self-pollinated seeds had an opaque morphology (Fig. 8A), indicating that the seeds are of the double-recessive homozygous mutant line (ss1Lss1L/ss3ass3a). The chain length distribution pattern of these opaque seeds exhibited a reduction in 7≤DP≤12 that was greater than that of the SS1ss1/ss3ass3a line (Fig. 8C). The ss1Lss1L/ss3ass3a seed weight was 86% that of the wild type, and the ss1Lss1L/ss3ass3a amylose content of endosperm starch was slightly higher (apparent amylose content: 33%) than that of SS1ss1/ss3ass3a (data not shown). These results are consistent with the hypothesis that double-recessive ss1ss1/ss3ass3a seeds, having no SSI and SSIIIa activity, are sterile. Moreover, the structure and physicochemical properties of the starch accumulated in the ss1Lss1L/ss3ass3a endosperm should be analysed in detail. It is possible that this starch may have potential uses in food and industrial applications. Therefore, the preparation of ss1 leaky mutants may prove to be useful tool to avoid the sterility of the seed producing starch with modified but invaluable functional properties.
Discussion
Significance of production of SS isozyme double mutant lines
Among starch biosynthesis enzymes, SS has the largest number of isozymes of all higher plants, indicating that the elongation of α-glucans by SS is essential for starch biosynthesis. Each isozyme plays a distinct role in starch biosynthesis, and the presence of multiple isozymes may allow for functional compensation when one isozyme is absent or defective. To elucidate the function of each SS isozyme, the elimination of other functionally overlapping SS isozymes is essential. A limited number of studies have been conducted with double or triple SS mutant lines in Arabidopsis (Szydlowski et al., 2009), antisense potato (ss2/ss3, ss3/gbss1; Edwards et al., 1999; Lloyd et al., 1999), and maize double mutants (sug2/dul1:ss2a/ss3 and wx/du1:gbss1/ss3; Wang et al., 1993a, b). Although leaf starch phenotypes in ss1/ss2/ss3 triple mutant lines of Arabidopsis were examined (Szydlowski et al., 2009), to our knowledge this is the first case of ss1-related multiple mutants. Here, the phenotypic characterization of storage starch in rice endosperm was characterized for the first time using various mutant lines defective in both SSI and SSIIIa genes.
In this study, three mutant lines of rice (ss1ss1/SS3ass3a, SS1ss1/ss3ass3a, and ss1Lss1L/ss3ass3a), exhibiting lower SS activities compared with ss1 or ss3a single mutants (Fujita et al., 2006 or 2007, respectively), were isolated. In total, seven lines displayed various levels of SSI and SSIIIa activities. These include the two heterozygous lines, the leaky double-recessive mutant line, the parent single mutant lines, and the wild-type parent lines (Table 1). These lines were of great use in quantitative analysis of the distinct and overlapping functions of SSI, SSIIIa, and other SS isozymes in rice endosperm. In most previous studies, the double mutant lines were prepared through crosses between the null mutant lines, making it impossible to evaluate the effects of SS activity reduction on starch phenotypes.
Effects of SSI and SSIIIa deficiency on starch accumulation and amylopectin fine structure in rice endosperm
The major SS isozymes responsible for amylopectin biosynthesis in rice endosperm are SSI, SSIIa, and SSIIIa. These SSs have distinct roles in α-1,4 glucan chain elongation in amylopectin (Fujita et al., 2006). The Arabidopsis ss4 mutant, the only mutant of the SSIV group (Roldán et al., 2007), had minor effects on amylopectin structure. Despite this, SSIV is considered important for the initiation of starch granules (Szydlowski et al., 2009). SSIIa plays an essential role in A and B1 chain synthesis by elongating short and intermediate chains of amylopectin clusters in rice amylopectin. However, the japonica type rice varieties such as cv. Nipponbare, used as the wild-type parent line in this study, are defective in the SSIIa gene (Nakamura et al., 2005).
The null ss1 mutant accumulated the same amount of starch as the wild type, despite significant changes in the chain length distribution of amylopectin. This suggests that SSI plays a specific role in the synthesis of very short chains by attacking the chains of DP 6 and 7 (Table 1; Fujita et al., 2006). These observations strongly suggest that deficiencies or serious reductions in the activity of the SSs (both SSI and SSIIa) responsible for A and B1 chain elongation in amylopectin clusters do not severely affect the starch accumulation in the endosperm. The most probable SS isozyme responsible for amylopectin synthesis in the ss1 mutant is SSIIIa (Table 1; Fujita et al., 2006, 2007). When SSI and SSIIa activities are deficient or significantly reduced, SSIIIa is predicted to elongate short chains in addition to long chains despite its preferential elongation of long B2–4 chains connecting multiple clusters of amylopectin.
The sterility of the double-recessive ss1 and ss3a mutant (Fig. 4, Table 1) strongly implicates that either SSI or SSIIIa is required for starch biosynthesis in rice endosperm. Other SS isozymes are unable to functionally complement these isozymes. In the present study, seven mutant and wild-type lines yielded fertile seeds with three different morphologies: transculent (normal), white core, and opaque seeds (Figs. 1, 9, Table 1). The ss1Lss1L/ss3ass3a and ss1ss1/SS3ass3a mutants produced fertile seeds regardless of an 85–90% reduction in SSI or SSIIIa activity (SSI+SSIIIa) compared with the wild type. These results show that SSI and SSIIIa can play the principal role in starch synthesis in rice endosperm, maintaining ∼60% of the normal starch production (Table 2, data not shown) and starch granular structure with semi-crystalline properties (Fig. 6A, C, Table 4) if either of them is present. Thus, SSI or SSIIIa is indispensable for starch biosynthesis in rice endosperm, and these isozymes can strongly compensate for each other when the counterpart is lacking. At the same time, observations of modified amylopectin structure in the ss1 and ss3a mutants (Fig. 4; Fujita et al., 2006, 2007) implies that their contribution to amylopectin fine structure cannot be completely complemented by each other.
When SS activity is assayed by measuring the incorporation of 14C from ADP-glucose into glucan, SSI activity is at least 50% of the total SS activity in developing rice endosperm (Fujita et al., 2007). The SSIIIa contribution to the total SS activity is not easy to assess due to the increase in SSI activity caused by the ss3a mutation. The native-PAGE activity staining method estimates the SSI and SSIIIa activities to be ∼60% and 30%, respectively, of the total SS activity. The remaining activity (∼10%) is accounted for by the other minor SS isozymes. These minor SSs corresponded to the unidentified SS activity bands observed by native-PAGE/SS activity staining of the fractions obtained by anion-exchange chromatography of extracts of the ss1 mutant (Fujita et al., 2007). All of the examined mutant endosperm contained minor SS activity bands of varying intensity on the native gels. However, no data on the substantial contribution of minor SS isozymes to starch synthesis are currently available.
Indirect effects of SSI and SSIIIa activity reduction or loss on amylose content
Amylose content is a major factor influencing the physicochemical properties of endosperm starch. Several SS mutant lines are known to accumulate high quantities of amylose in starch granules. These include maize ss2 (sug-2; Zhang et al., 2004) and ss3 (du1; Wang et al., 1993a, b), rice ss3a (Fujita et al., 2007), wheat ss2a (Yamamori et al., 2000), barley ss2 (Morell et al., 2003), and Arabidopsis ss2/ss3 (Zhang et al., 2008) mutants. Among the mutant lines used in this study, ss1, ss3a, ss1ss1/SS3ass3a, SS1ss1/ss3ass3a, and ss1Lss1L/ss3ass3a lines had higher amylose content (when expressed as a percentage in starch) than the wild type (Fig. 5, Table 3). On the other hand, it is possible that the increased amylose content of opaque seeds (ss1ss1/SS3ass3a, SS1ss1/ss3ass3a, and ss1Lss1L/ss3ass3a) was derived at least partly from a decrease in amylopectin biosynthesis caused by the extremely low SS activity levels in endosperm. To distinguish whether the increase in amylose was due to the increase in the synthesis of amylose or to the decrease in the synthesis of amylopectin, the amylose content was also expressed per seed (Table 1). The result shows that the former is the case for ss3a, ss1ss1/SS3ass3a, and SS1ss1/ss3ass3a mutant lines. The increase in amylose content could be explained by the increase in GBSSI. This is considered to be the case for the ss3a, ss1ss1/SS3ass3a, and SS1ss1/ss3ass3a mutant lines compared with the wild type (Fig. 7). Another possibility is that the increased supply of ADP-glucose by AGPase elevates GBSSI activity, which results in the enhanced amylose synthesis because GBSSI is known to have a higher Km for ADP-glucose than the other soluble SS isozymes (Clarke et al., 1999). The increase in AGPase activity was observed in the ss1, ss3a, ss1ss1/SS3ass3a, and SS1ss1/ss3ass3a mutant lines (Table 5), as reported in the maize ss3 mutant (Singletary et al., 1997). Higher AGPase levels (Table 5) and lower SS activities (Fig. 3) are also considered to increase plastidial ADP-glucose levels, resulting in increased amylose synthesis (Fig. 5, Table 1).
Genetic analysis and the reason for sterility
Endosperm has the 3n genotype. In principle, the genotype of SS1ss1/ss3ass3a in the self-pollinated line should have segregated into SS1SS1SS1/ss3ass3ass3a, SS1SS1ss1/ss3ass3ass3a, SS1ss1ss1/ss3ass3ass3a, and ss1ss1ss1/ss3ass3ass3a in WO endosperm at a ratio of 1:1:1:1 (Table 1). It is assumed that the SS1SS1SS1/ss3ass3ass3a and ss1ss1ss1/ss3ass3ass3a endosperm must have had the parent-type white core and sterile seed phenotypes, respectively. Since the empty (sterile) seeds accounted for much less than 50% of the total seeds of the self-pollinated products (Supplementary Table S1 at JXB online), it is unlikely that the SS1ss1ss1/ss3ass3ass3a (simplex) seed produces the empty seeds. Thus, it is assumed that the opaque seed genotype was SS1SS1ss1/ss3ass3ass3a (duplex) or SS1ss1ss1/ss3ass3ass3a (simplex). If this assumption is correct, the ratio of the white core, opaque seeds, and sterile seeds would have been 1:2:1, which differs from the observed ratio (Supplementary Table S1). The reason for this discrepancy remains unresolved, yet one likely possibility is that the simplex (SS1ss1ss1/ss3ass3ass3a) genotype could have become sterile under environmental stress conditions. The preliminary observation showed that the empty seed at the developmental stage contained a translucent liquid in the endosperm. However, the number of empty seeds varied depending on the harvest year (Supplementary Table S1). This indicates that both genetic and environmental factors affect seed sterility. Overall, starch biosynthesis in rice endosperm is closely related to fertilization of the seeds. More detailed studies such as with the reciprocal crossing between the F1 and the parent mutant are needed.
Conclusion
The present investigation utilized a variety of ss1 and ss3a rice mutant lines exhibiting different SS isozyme activities to demonstrate that SSI and SSIIIa play essential roles in starch production, ultimately enabling the fundamental fine structure of endosperm amylopectin. It is particularly noted that endosperm including either SSI or SSIIIa even at lower activity levels could accumulate up to ∼60% of the starch observed in the wild type. Since the japonica-type rice used in this study is defective in SSIIa, the contribution of SSIIa to the functional coordination between SSI and SSIIIa remains to be elucidated. The contribution of other starch synthetic enzymes, namely BE, DBE, or PHO, also cannot be completely excluded. However, in vitro assays revealed no significant changes in the activity of these enzymes (Supplementary Fig. S1 at JXB online). Recent studies revealed protein–protein interaction among SS and BE isozymes in endosperm from wheat (Tetlow et al., 2004, 2008) and maize (Hennen-Bierwagen et al., 2009). If SSI, SSIIa, and/or SSIIIa also associate with each other, and in addition with BE, DBE, and/or PHO, to form protein–protein complexes in rice endosperm, detailed analysis of ss1 and ss3a lines will provide new insights into the dynamic mechanism for starch biosynthesis regulation, as well as into the redundancy of SS isozymes in cereal endosperm.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Pleiotropic effect of the deficiency or reduction of SSI or SSIIIa activity on SS, DBE, BE, and PHO activity in the TO line, WO line, and ss1Lss1L/ss3ass3a, ss1 mutant (ΔSSI, e7, i2-1), ss3a mutant (ΔSSIIIa, e1), and wild type Nipponbare when measured by native-PAGE/activity staining of their developing endosperm.
Table S1. Segregation of F2 and F3 to F5 seeds of the TO and WO lines.
Acknowledgments
The authors are grateful to Dr Yasuhito Takeda and Dr Isao Hanashiro (Kagoshima University), Dr Kimiko Itoh (Niigata University), and Dr Sayuri Akuzawa (Tokyo University of Agriculture) for their participation in helpful discussions, and Dr Yoshiko Toyosawa and Yuko Nakaizumi (Akita Prefectural University) for technical support. This work was partially supported by the Program for the Promotion of Basic and Applied Research for Innovations in Bio-oriented Industry and a Grant-in-Aid for Scientific Research (B) (19380007).
Glossary
Abbreviations
- AGPase
ADP-glucose pyrophosphorylase
- GBSSI
granule-bound starch synthase I
- RT-PCR
reverse transcription PCR
References
- Ball SG, Morell MK. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annual Review of Plant Biology. 2003;54:207–233. doi: 10.1146/annurev.arplant.54.031902.134927. [DOI] [PubMed] [Google Scholar]
- Cao H, Imparl-Radosevich J, Guan H, Keeling PL, James MG, Myers AM. Identification of the soluble starch synthase activities of maize endosperm. Plant Physiology. 1999;120:205–215. doi: 10.1104/pp.120.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke BR, Denyer K, Jenner CF, Smith AM. The relationship between the rate of starch synthesis, the adenosine 5′-diphosphoglucose concentration and the amylose content of starch in developing pea embryos. Planta. 1999;209:324–329. doi: 10.1007/s004250050639. [DOI] [PubMed] [Google Scholar]
- Craig J, Lloyd JR, Tomlinson K, Barber L, Edwards A, Wang TL, Martin C, Hedley CL, Smith AM. Mutations in the gene encoding starch synthase II profoundly alter amylopectin structure in pea embryos. The Plant Cell. 1998;10:413–426. doi: 10.1105/tpc.10.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JH, Kramer HH, Whistler RL. Expression of the gene du in the endosperm of maize. Agronomy Journal. 1955;47:232–235. [Google Scholar]
- Delvallé D, Dumez S, Wattebled F, Roldán I, Planchot V, Berbezy P, Colonna P, Vyas D, Chatterjee M, Ball S, Mérida A, D'Hulst C. Soluble starch synthase I: a major determinant for the synthesis of amylopectin in Arabidopsis thaliana leaves. The Plant Journal. 2005;43:398–412. doi: 10.1111/j.1365-313X.2005.02462.x. [DOI] [PubMed] [Google Scholar]
- Edwards A, Fulton DC, Hylton CM, Jobling SA, Gidley M, Rossner U, Martin C, Smith AM. A combined reduction in activity of starch synthases II and III of potato has novel effects on the starch of tubers. The Plant Journal. 1999;17:251–261. [Google Scholar]
- Fujita N, Goto S, Yoshida M, Suzuki E, Nakamura Y. The function of rice starch synthase I expressed in E.coli. Journal of Applied Glycoscience. 2008;55:167–172. [Google Scholar]
- Fujita N, Hasegawa H, Taira T. The isolation and characterization of a waxy mutant of diploid wheat (Triticum monococcum L.) Plant Science. 2001;160:595–602. doi: 10.1016/s0168-9452(00)00408-8. [DOI] [PubMed] [Google Scholar]
- Fujita N, Kubo A, Francisco PB, Jr, Nakakita M, Harada K, Minaka N, Nakamura Y. Purification, characterization, and cDNA structure of isoamylase from developing endosperm of rice. Planta. 1999;208:283–293. doi: 10.1007/s004250050560. [DOI] [PubMed] [Google Scholar]
- Fujita N, Kubo A, Suh S-D, Wong K-S, Jane J-L, Ozawa K, Takaiwa F, Inaba Y, Nakamura Y. Antisense inhibition of isoamylase alters the structure of amylopectin and the physicochemical properties of starch in rice endosperm. Plant and Cell Physiology. 2003;44:607–618. doi: 10.1093/pcp/pcg079. [DOI] [PubMed] [Google Scholar]
- Fujita N, Toyosawa Y, Utsumi Y, Higuchi T, Hanashiro I, Ikegami A, Akuzawa S, Yoshida M, Mori A, Inomata K, Itoh R, Miyao A, Satoh H, Nakamura Y. Characterization of PUL-deficient mutants of rice (Oryza sativa L.) and the function of PUL on the starch biosynthesis in the rice endosperm. Journal of Experimental Botany. 2009;60:1009–1023. doi: 10.1093/jxb/ern349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, Nakamura Y. Function and characterization of starch synthase I using mutants in rice. Plant Physiology. 2006;140:1070–1084. doi: 10.1104/pp.105.071845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Kondo T, Saito K, Utsumi Y, Tokunaga T, Nishi A, Satoh H, Park JH, Jane JL, Miyao A, Hirochika H, Nakamura Y. Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiology. 2007;144:2009–2023. doi: 10.1104/pp.107.102533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Wanat J, Stinard PS, James MG, Myers AM. Characterization of dull1, a maize gene coding for a novel starch synthase. The Plant Cell. 1998;10:399–412. doi: 10.1105/tpc.10.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennen-Bierwagen TA, Lin Q, Grimaud F, Planchot V, Keeling PL, James MG, Myers AM. Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in high molecular weight complexes: a model for regulation of carbon allocation in maize amyloplasts. Plant Physiology. 2009;149:1541–1559. doi: 10.1104/pp.109.135293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Terao T. A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.) Planta. 2004;220:9–16. doi: 10.1007/s00425-004-1314-6. [DOI] [PubMed] [Google Scholar]
- Hizukuri S. Polymodal distribution of the chain lengths of amylopectins, and its significance. Carbohydrate Research. 1986;147:342–347. [Google Scholar]
- Horibata T, Nakamoto M, Fuwa H, Inouchi N. Structural and physicochemical characteristics of endosperm starches of rice cultivars recently bred in Japan. Journal of Applied Glycoscience. 2004;51:303–313. [Google Scholar]
- Inouchi N, Hibiu H, Li T, Horibata T, Fuwa H, Itani T. Structure and properties of endosperm starches from cultivated rice of Asia and other countries. Journal of Applied Glycoscience. 2005;52:239–246. [Google Scholar]
- Isshiki M, Morino K, Nakajima M, Okagaki RJ, Wessler SR, Izawa T, Shimamoto K. A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5' splice site of the first intron. The Plant Journal. 1998;15:133–138. doi: 10.1046/j.1365-313x.1998.00189.x. [DOI] [PubMed] [Google Scholar]
- Li L, Jiang H, Campbell M, Blanco M, Jane J-L. Characterization of maize amylose-extender (ae) mutant starches. Part I: relationship between resistant starch contents and molecular structures. Carbohydrate Polymer. 2008;74:396–404. [Google Scholar]
- Lloyd JR, Landschutze V, Kossmann J. Simultaneous antisense inhibition of two starch-synthase isoforms in potato tubers leads to accumulation of grossly modified amylopectin. Biochemial Journal. 1999;338:515–521. [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf PC. The inheritance of amylaceous sugary endosperm and its derivatives in maize. Genetics. 1947;32:448–458. doi: 10.1093/genetics/32.5.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J, Sidebottom C, Debet M, Martin C, Smith AM. Identification of the major starch synthase in the soluble fraction of potato tubers. The Plant Cell. 1996;8:1121–1135. doi: 10.1105/tpc.8.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell MK, Kosar-Hashemi B, Cmiel M, Samuel MS, Chandler P, Rahman S, Buleon A, Batey IL, Li Z. Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. The Plant Journal. 2003;34:173–185. doi: 10.1046/j.1365-313x.2003.01712.x. [DOI] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG. Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiology. 2000;122:989–997. doi: 10.1104/pp.122.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y. Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: rice endosperm as a model tissue. Plant and Cell Physiology. 2002;43:718–725. doi: 10.1093/pcp/pcf091. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Francisco BP, Jr, Hosaka Y, Satoh A, Sawada T, Kubo A, Fujita N. Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Molecular Biology. 2005;58:213–227. doi: 10.1007/s11103-005-6507-2. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Yuki K, Park SY, Ohya T. Carbohydrate metabolism in the developing endosperm of rice grains. Plant and Cell Physiology. 1989;30:833–839. [Google Scholar]
- Nishi A, Nakamura Y, Tanaka N, Satoh H. Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiology. 2001;127:459–472. [PMC free article] [PubMed] [Google Scholar]
- Ohdan T, Francisco PB, Jr, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. Journal of Experimental Botany. 2005;56:3229–3244. doi: 10.1093/jxb/eri292. [DOI] [PubMed] [Google Scholar]
- O'Shea MG, Morell MK. High resolution slab gel electrophoresis of 8-amino-1,3,6-pyrenetrisulfonic acid (APTS) tagged oligosaccharides using a DNA sequencer. Electrophoresis. 1996;17:681–686. doi: 10.1002/elps.1150170410. [DOI] [PubMed] [Google Scholar]
- Roldán I, Wattebled F, Mercedes Lucas M, Delvallé D, Planchot V, Jiménez S, Pérez R, Ball S, D'Hulst C, Mérida A. The phenotypeof soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control ofstarch granule formation. The Plant Journal. 2007;49:492–504. doi: 10.1111/j.1365-313X.2006.02968.x. [DOI] [PubMed] [Google Scholar]
- Sano Y. Differential regulation of waxy gene expression in rice endosperm. Theoretical and Applied Genetics. 1984;68:467–473. doi: 10.1007/BF00254822. [DOI] [PubMed] [Google Scholar]
- Singletary GW, Banisadr R, Keeling PL. Influence of gene dosage on carbohydrate synthesis and enzymatic activities in endosperm of starch-deficient mutants of maize. Plant Physiology. 1997;113:293–304. doi: 10.1104/pp.113.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Denyer K, Martin C. The synthesis of the starch granule. Annual Review of Plant Physiology and Molecular Biology. 1997;48:67–87. doi: 10.1146/annurev.arplant.48.1.67. [DOI] [PubMed] [Google Scholar]
- Szydlowski N, Ragel P, Raynaud S, Lucas MM, Roldán I, Montero M, Muńoz FJ, Ovecka M, Bahaji A, Planchot V, Pozueta-Romero J, D'Hulst C, Mérida A. Starch granule initiation in Arabidopsis requires the presence of either class IV or class II starch synthases. The Plant Cell. 2009;21:2443–2357. doi: 10.1105/tpc.109.066522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y, Hizukuri S, Juliano BO. Structures of rice amylopectins with low and high affinities for iodine. Carbohydrate Research. 1987;168:79–88. [Google Scholar]
- Tetlow IJ, Wait R, Lu Z, Akkasaeng R, Bowsher CG, Esposito S, Kosar-Hashemi B, Morell MK, Emes MJ. Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein–protein interactions. The Plant Cell. 2004;16:694–708. doi: 10.1105/tpc.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow IJ, Beisel KG, Cameron S, Makhmoudova A, Liu F, Bresolin NS, Wait R, Morell MK, Emes MJ. Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiology. 2008;146:1878–1891. doi: 10.1104/pp.108.116244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, White P, Pollak L, Jane J-L. Characterization of starch structures of 17 maize endosperm mutant genotypes with Oh43 inbred line background. Cereal Chemistry. 1993a;70:171–179. [Google Scholar]
- Wang YJ, White P, Pollak L, Jane J-L. Amylopectin and intermediate materials in starches from mutant genotypes of the Oh43 inbred line. Cereal Chemistry. 1993b;70:521–525. [Google Scholar]
- Yamamori M, Fujita S, Hayakawa K, Matsuki J, Yasui T. Genetic elimination of a starch granule protein, SGP-1, of wheat generates an altered starch with apparent high amylose. Theoretical and Applied Genetics. 2000;101:21–29. [Google Scholar]
- Yamanouchi H, Nakamura Y. Organ specificity of isoforms of starch branching enzyme (Q-enzyme) in rice. Plant and Cell Physiology. 1992;33:985–991. [Google Scholar]
- Zhang X, Colleoni C, Ratushna V, Sirghie-Colleoni M, James MG, Myers AM. Molecular characterization demonstrates that the Zea mays gene sugary2 codes for the starch synthase isoform SSIIa. Plant Molecular Biology. 2004;54:865–879. doi: 10.1007/s11103-004-0312-1. [DOI] [PubMed] [Google Scholar]
- Zhang X, Myers AM, James MG. Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis. Plant Physiology. 2005;138:663–674. doi: 10.1104/pp.105.060319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Szydlowski N, Delvallé D, D'Hulst C, James MG, Myers AM. Overlapping functions of the starch synthase SSII and SSIII in amylopectin biosynthesis in Arabidopsis. BMC Plant Biology. 2008;8:96–113. doi: 10.1186/1471-2229-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.