Abstract
Mannitol is a putative osmoprotectant contributing to salt tolerance in several species. Arabidopsis plants transformed with the mannose-6-phosphate reductase (M6PR) gene from celery were dramatically more salt tolerant (at 100 mM NaCl) as exhibited by reduced salt injury, less inhibition of vegetative growth, and increased seed production relative to the wild type (WT). When treated with 200 mM NaCl, transformants produced no seeds, but did bolt, and exhibited less chlorosis/necrosis and greater survival and dry weights than the WT. Without salt there were no M6PR effects on growth or phenotype, but expression levels of 2272 genes were altered. Many fewer differences (1039) were observed between M6PR and WT plants in the presence of salt, suggesting that M6PR pre-conditioned the plants to stress. Previous work suggested that mannitol is an osmoprotectant, but mannitol levels are invariably quite low, perhaps inadequate for osmoprotectant effects. In this study, transcriptome analysis reveals that the M6PR transgene activated the downstream abscisic acid (ABA) pathway by up-regulation of ABA receptor genes (PYL4, PYL5, and PYL6) and down-regulation of protein phosphatase 2C genes (ABI1 and ABI2). In the M6PR transgenic lines there were also increases in transcripts related to redox and cell wall-strengthening pathways. These data indicate that mannitol-enhanced stress tolerance is due at least in part to increased expression of a variety of stress-inducible genes.
Keywords: Abscisic acid, cell wall, mannitol, mannose-6-phosphate reductase, osmoprotectant, redox, salt tolerance
Introduction
Mannitol is a primary photosynthetic product found in 70 higher plant families and many marine algae (Bieleski, 1982). In addition to its role as a primary photosynthetic product and translocated carbohydrate, in many species mannitol also appears to function in osmotic stress tolerance by serving as a compatible solute or osmoprotectant (Bohnert and Jensen, 1996). Several lines of evidence support these latter roles. Celery (Apium graveolens L.), which is a major mannitol producer, is quite salt tolerant, and mannitol biosynthesis in celery is increased by salt stress (Everard et al., 1994; Pharr et al., 1995). Plants transgenic for mannitol-related genes have shown increases in stress tolerance, particularly salt tolerance (Tarczynski et al., 1992; Thomas et al., 1995; Chaturvedi et al., 1997; Karakas et al., 1997; Sheveleva et al., 1998; Hu et al., 2005). These plants were all transformed with a bacterial catabolic NAD-dependent mannitol-1P dehydrogenase which ordinarily converts mannitol-1P to fructose-6P. In an alternative approach (utilized here), plants were transformed with the celery gene for mannose-6P reductase (M6PR) that usually converts mannose-6P to mannitol-1P as part of the path to mannitol biosynthesis (Zhifang and Loescher, 2003).
When Arabidopsis plants, which ordinarily do not contain mannitol, were transformed with the celery M6PR gene they accumulated substantial amounts of mannitol and were also quite salt tolerant, as assessed by analyses of fresh and dry weight (Zhifang and Loescher, 2003). Further study showed that the presence of the M6PR transgene maintained photosystem II and carboxylation efficiencies and thus protected photosynthesis against salt-related damage to the chloroplasts (Sickler et al., 2007). None of these studies, regardless of the transgene used, investigated transcriptomic changes, and the exact mechanisms by which mannitol exerts its protective effects in transgenic plants are still not clear. At the levels found in most transgenic plants, a role as a compatible solute would require primary accumulation in the cytosol; similarly, a role as an osmoprotectant would require accumulation in the cytosol, chloroplasts, and nucleus (Shen et al., 1997a, b). It seems unlikely that there are specific mannitol transporters as would be needed to maintain high cytosolic concentrations in transgenic plants that do not normally produce mannitol, and non-specific transporters would probably be compromised by competition from other substrates.
There are other results that suggest possible additional mechanisms responsible for the protective effects. Many species that ordinarily lack mannitol are still equipped to deal with it metabolically. Sequence analysis of the cDNA of the celery mannitol dehydrogenase (MTD) gene encoding the catabolic enzyme that oxidizes mannitol to mannose, revealed that this gene is the same as the ELI3 (Elicitor-Activated Gene 3) pathogenesis-related (PR) gene that has been described in numerous plants (Williamson et al., 1995). ELI3 is up-regulated in response to pathogen attack (Kiedrowski et al., 1992) and to treatment with the pathogen response signal molecule, salicylic acid (Chen et al., 1993; Yalpani et al., 1993).
Despite the unknowns, because only one or two genes are required, creating transgenic plants for mannitol biosynthesis is an attractive approach to stress tolerance, and particularly salt tolerance, which is a major agricultural problem throughout the world (Munns and Tester, 2008). Consequently, to develop further insights into the means by which mannitol enhances resistance to stress, an extensive study of the effects of the M6PR transgene on growth and global gene expression in Arabidopsis in the presence and absence of salt stress was embarked upon. These studies indicate that there are certain commonalities in a number of complex responses related to the presence of the M6PR transgene, and presumably to mannitol. The analyses indicate that mannitol's effects are much more complicated than might be expected of an osmoticum or osmoprotectant. The presence of the M6PR transgene and thus mannitol appears to act as a signal, affecting genes responsive to both abiotic and biotic stresses and providing insights into global plant defence mechanisms.
Materials and methods
Plant materials and growth conditions
Two transgenic Arabidopsis thaliana L. (Heyn) lines (M2 and M5) with the M6PR gene under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Zhifang and Loescher, 2003), as well as Columbia-0 wild type (Col-WT), were used in this experiment. Seeds of both transgenic and WT plants were stratified at 4 °C for 4 d in the dark and then sown directly on soil in 26×26×6 cm pots filled with a standard planting medium (Baccto, Houston, TX, USA). Seeds of each line were pipetted onto the wet soil surface. Each pot was divided into 36 subsections with five seeds planted in each subsection. Plants were grown at 23/18 °C in the growth chamber with a 12 h light/12 h dark cycle at 350 μmol m−2 s−1 and 70% relative humidity. Plants were subirrigated. Fertilizer (half-strength Hoagland's solution) (Hoagland and Arnon, 1950) was applied once per week before the beginning of the salt treatment. At 14 days after sowing (DAS), the pots were thinned to one seedling per subsection.
Salt treatments
Salt treatments were initiated at 14 DAS. NaCl was dissolved in half-strength Hoagland's solution. Plants were watered from below to field capacity and then sprayed with 1.0 l of the same concentration of NaCl solution from the top, to ensure adequate leaching and prevent excess salinity. The NaCl concentrations were increased stepwise by 50 mM every 2 d for each line, to the indicated maximum (0, 100, or 200 mM). Plants were then watered every 2 d with or without NaCl at the indicated concentrations. The pots were rotated in the growth chamber daily to minimize the effect of environment. The experiment was repeated three times.
Measurement of growth parameters
Beginning 14 DAS, plants were examined every other day for the number of plants that were bolting or in bloom. Chlorosis/necrosis severity indices, numbers of leaves, and rosette diameters were measured every 6 d. Chlorosis/necrosis severity was rated as the following: 0, no yellow or purple leaves; 1, older leaves turning yellow or purple; 3, younger leaves turning yellow or purple; 5, some leaves dead; and 7, plants dead. Chlorosis/necrosis severity indices were then calculated according to the following equation: chlorosis/necrosis severity indices=Σ (no. of plants in each scale×scale value)/(total no. of plants×the highest scale value). At 32 DAS, plants were thinned to 18 per pot by removing seedlings from every other row. The plants were photographed at 50 DAS, and harvested at 62 DAS when most of them reached maturity. Seedling height and number of stalks were measured before harvest. Total dry mass and seed yields were measured after harvest. All data were analysed with SPSS 11 for Windows (SPSS Inc., Chicago, IL, USA). Mean separations were performed by Duncan's multiple range test. Differences at P ≤0.05 were considered to be significant.
Plant growth and salt treatment for the microarray experiment
Seeds of both transgenic and Col-WT plants were sown as described above with two replicate pots for each genotype and salt combination. Two replications were performed in different growth chambers on different dates with 36 plants/replicate pot for each genotype and salt combination. The plants used for these analyses were a subset of a larger experiment including additional transgenes and ecotypes; here the focus was on the effects of the M6PR transgene. Plants were grown at 23/18 °C in the growth chamber under a short-day cycle (10 h light/14 h dark) at 350 μmol m−2 s−1 and 70% relative humidity in order to increase seedling leaf area and growth while not promoting bolting. Salt treatments were initiated at 14 DAS and applied as described above. Sampling was performed at 20 DAS by collecting developed but not senescent leaves (∼0.5cm width×1.5 cm length) from at least 15 seedlings per treatment.
RNA isolation, preparation of biotin-labelled cRNA, and microarray hybridization
Total RNA was extracted and purified from leaves of at least 15 plants per sample for M2, M5, and Col-WT using a QIAGEN-RNeasy Mini Kit (QIAGEN, Valencia, CA, USA) according to guidelines specified by the manufacturer. Two biological replicates from different growth chambers were prepared for each genotype and salt combination. RNA was quantified using a Nanodrop spectrophotometer (ND-1000, NanoDrop Technologies, Wilmington, DE, USA) and RNA quality was assessed using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer's protocol.
For biotin labelling, equal amounts of total RNA from two transgenic lines (M2 and M5) were pooled. A 10 μg aliquot of total RNA from M6PR plants and Col-WT plants was converted to double-stranded cDNA using an Affymetrix One-Cycle cDNA Synthesis Kit (Affymetrix, Santa Clara, CA, USA) according to the manufacturer's instruction. Biotinylated complementary RNA (cRNA) was synthesized from 5 μg of cDNA by in vitro transcription using an Affymetrix GeneChip IVT Labeling Kit (Affymetrix). Labelled cRNA was purified using an Affymetrix GeneChip Sample Cleanup Module (Affymetrix) and was fragmented by heating at 94 °C for 35 min in a buffer containing 40 mM TRIS-acetate (pH 8.1), 100 mM KOAc, and 30 mM MgOAc to produce a distribution of RNA fragments ranging in size from ∼35 to 200 nucleotides.
Fragmented cRNAs (15 μg) were hybridized to a 24K GeneChip Arabidopsis ATH1 Genome Array (Affymetrix), in accordance with the standard protocol of the manufacturer. The arrays were scanned with a GeneChip® Scanner 3000 (Affymetrix) and raw image files were converted to probe set data (*.CEL files), using the Affymetrix GeneChip® Operating Software according to the manufacturer's instructions.
Microarray data analysis
All the analyses were performed using Bioconductor, a public source software for the analysis of genomic data rooted in the statistical computing environment R (Gentleman et al., 2004). The data were normalized by robust multiarray normalization of probe-level data with GCRMA. Subsequently, average expression values and their P-values (by t-test) are calculated using the affylmGUI package in R (Wettenhall et al., 2006). In order to minimize any potential statistical biases, modest threshold parameters were used to determine significant changes in gene expression. Transcript levels deemed significantly different were those with (i) ≥2-fold change; (ii) a P-value ≤0.05; and (iii) a detection call of ‘Present’ in duplicate with the Affymetrix GeneChip® Operating Software. For metabolic pathway analysis, no fold change cut-off was set (see below). The M6PR data have been submitted to the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE18217. The Col-WT type microarray data were previously deposited to the Gene Expression Omnibus (GEO) online database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE16765.
Biological category enrichment analysis
A total of 3239 (Col-100 mM versus Col-0 mM) and 2272 (M6PR-0 mM versus Col-0 mM) transcripts with P-values ≤0.05 and ≥2-fold change were loaded and annotated in the Classification SuperViewer Tool w/Bootstrap web database (http://bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer.cgi) (Provart and Zhu, 2003). The absolute values and normalized frequencies in the Arabidopsis genomic set of each functional category were then calculated automatically online. Normalized frequency was calculated as follows: (Number_in_Classinput_set/Number_Classifiedinput_set)/(Number_in_Classreference_set)/Number_Classifiedreference_set).
Metabolic pathway analysis
The effects of salt and the gene on metabolic pathways were analysed using MapMan software, which is a user-driven tool that paints gene expression data sets onto diagrams of metabolic pathways or other processes (Thimm et al., 2004). Transcripts with a P-value ≤0.05 and fold change ≥2 were loaded in MapMan and the numbers of transcripts changed in different pathways were counted.
Quantitative real-time PCR
Total RNA was extracted as described above and aliquots of 2 μg were reverse transcribed using a High Capacity RNA-to-cDNA Kit (Applied Biosystems, Carlsbad, CA, USA) under the conditions suggested by the manufacturer. Reverse transcription products were diluted 10 times in water prior to real-time quantitative PCR (qRT-PCR). Aliquots of cDNA were used as template for quantitative real-time PCRs. Reactions were set up with Power SYBR® Green PCR Master Mix (Applied Biosystems) according to the manufacturer's instructions in a total volume of 10 μl and 0.3 μM of each primer. The ABI PRISM® 7900HT Sequence Detection System (Applied Biosystems) was used to detect amplification levels and was programmed for an initial step at 95 °C for 10 min, followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. All reactions were run in duplicate or triplicate and average values were calculated. Quantification was performed with at least two independent experiments. The housekeeping F-actin gene (At3g05520) was used as an endogenous control. Relative expression levels of target genes and SD values were calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001).
Twenty-one genes with at least one significant sample difference from four comparisons based on microarray data were selected for qRT-PCR analysis, along with a single gene that did not (At3g63490). Log2 values for each replicate and their averages and standard errors were calculated. Primers were designed based on the probe sequences used by Affymetrix with a target size range between 280 bp and 320 bp, and sequences of forward and reverse primers and the sizes of the resulting fragments are listed in Supplementary Table S1 available at JXB online.
Results and Discussion
Growth of M6PR lines and Col-WT
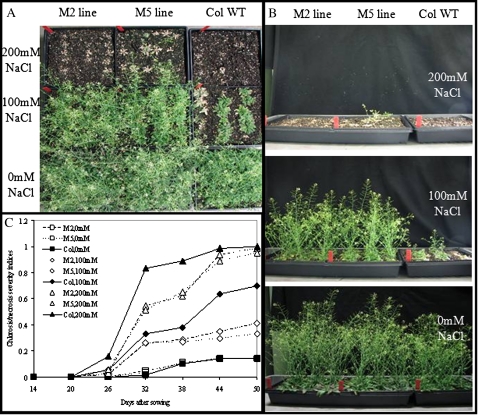
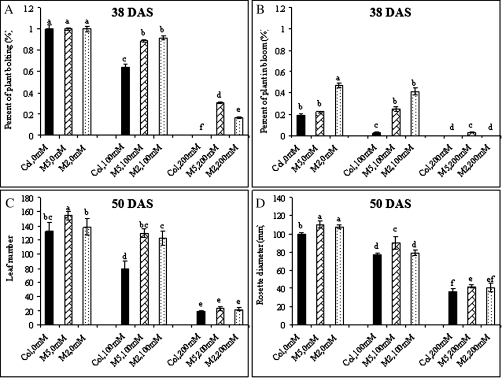
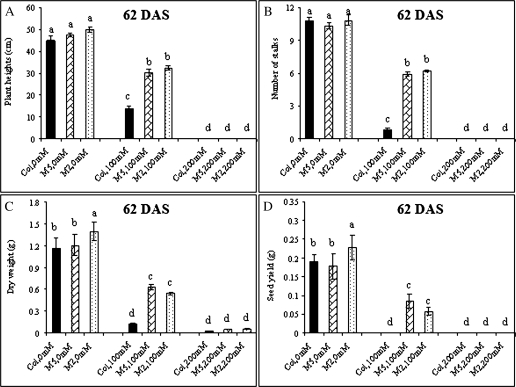
In the absence of salt, growth of the M6PR M2 and M5 lines was generally equivalent to that of Col-WT, with no indication of negative effects on growth rate, plant size, flowering time, dry matter, or seed production (Figs 1, 2). Under saline conditions, growth of both M6PR lines and Col-WT plants was inhibited (Fig. 1A, B, top and middle rows). All plants bolted and flowered later, and had lower leaf numbers, plant heights, stalk numbers, rosette diameters, seed yields, and dry weights when compared with the controls (Figs 2, 3). Treatment with 200 mM NaCl resulted in death of all genotypes 50 d after emergence (Fig. 1A, B, top row).
Fig. 1.
Salt effect on growth of transgenic M6PR lines and the WT. Salt treatments were initiated at 14 DAS and plants were photographed at 50 DAS. (A) Pictures were taken from the top. (B) Pictures were taken from the side. (C) Chlorosis/necrosis severity indices of transgenic M6PR lines and the WT under salinity conditions. Data were collected every 6 d. Scales of chlorosis/necrosis severity were described as below. 0, no yellow or purple leaves; 1, older leaves turn yellow or purple; 3, younger leaves turn yellow or purple; 5, some leaves die; and 7, plants die. Chlorosis/necrosis severity indices were then calculated according to the following equation: chlorosis/necrosis severity indices=Σ (no. of plants in each scale×scale value)/(total no. of plants×the highest scale value). (This figure is available in colour at JXB online.)
Fig. 2.
Percentage of plants bolting (A), percentage of plants in bloom (B), leaf numbers (C), and rosette diameters (D) of transgenic M6PR lines and the WT under various salinity conditions during growth. Growth parameters during the whole life cycle are indicated in Supplementary Fig. S1 at JXB online. The differences were analysed by a one-way ANOVA test. The same letters indicate significant differences with P <0.05. Plants were harvested at 62 DAS when most had reached maturity.
Fig. 3.
Plant height (A), stalk numbers (B), dry weights (C), and seed yields (D) of transgenic M6PR lines and the WT under salinity conditions at harvest (62 DAS). The differences were analysed by a one-way ANOVA test. Different letters indicate significant differences with P <0.05. Plants were harvested at 62 DAS when most had reached maturity. Plant heights were measured and stalk numbers were counted before harvest. Dry weights and seed yields were calculated after incubation at 60 °C for 2 d.
When compared with Col-WT, however, much less severe effects were observed for both M6PR lines (Fig. 1A, B). The WT was more sensitive to salt stress and showed an earlier and greater extent of leaf injury than M6PR lines, as illustrated by much higher chlorosis/necrosis severity indices with both 100 mM and 200 mM NaCl treatments, and greater decreases in leaf number (Figs 1C, 2C, D). Both M6PR lines produced more seed stalks than Col-WT when grown under 100 mM NaCl during the whole life cycle, and a few also bolted and produced inflorescences at 200 mM NaCl (Figs 2A, B, 3B). At harvest (62 DAS), both lines had significantly greater dry weight and seed yield than Col-WT after treatment with 100 mM NaCl (Fig. 3C, D). The M2 and M5 lines produced 57 mg and 85 mg of seeds per plant, respectively, but Col-WT produced only 2 mg (Fig. 3D).
Reproducibility and validation of microarray data
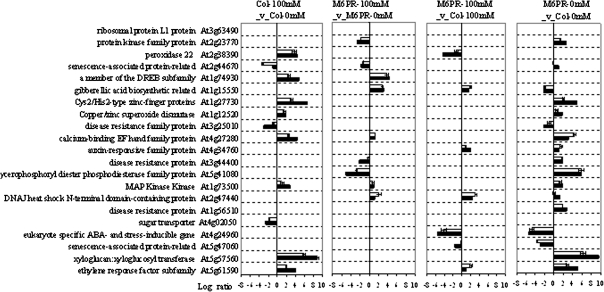
Transgenic M6PR and Col-WT plants were grown in the growth chamber in the presence and absence of salt stress as part of a larger experiment including several transgenes and ecotypes. Sampling was performed at 20 DAS by collecting developed but not senescent leaves from at least 15 seedlings per replicate tray. Microarray signal data from the two biological replicates were highly correlated (r ≥0.96 for all pairs of comparisons; Supplementary Fig. S2 at JXB online), indicating high reproducibility between the experiments. The microarray analyses were also partially verified using a qRT-PCR assay for a set of 21 genes selected for differential responses in different comparisons. The expression ratios measured by microarray and by qRT-PCR were highly correlated (r ≥0.93; Fig. 4).
Fig. 4.
Comparison of relative transcript abundance measured by qRT-PCR versus microarray analysis. Twenty-one genes with at least one significant sample difference (P-value ≤0.05 and fold change ≥2) from four comparisons based on microarray data were selected for qRT-PCR analysis, along with a single gene that did not (At3g05520). Gene names and accession numbers are indicated. Altogether, the expression ratios measured by microarray and by qRT-PCR were highly correlated (r >0.93). The trends of both increased and decreased expression for the comparison were similar. White bars, qRT-PCR; black bars, microarrays. Values are the mean ±SE (n >3).
General transcriptomic responses
Comparisons of global transcription levels after 6 d of salt treatment were performed to examine the salt effect on the Col-WT and M6PR lines, and the effect of the M6PR transgene in the absence and presence of salt. First, different threshold parameters were chosen to determine meaningful differences between samples in this study (Supplementary Table S2 at JXB online). The number of changed transcripts decreased with a more stringent cut-off P-value and fold change. In order to minimize any potential statistical biases, modest threshold parameters (fold difference ≥2 and P-value ≤0.05) were applied during data analysis as described in the Materials and methods. The full list of 4631 affected transcripts in all four comparisons is shown in Supplementary Table S3 at JXB online.
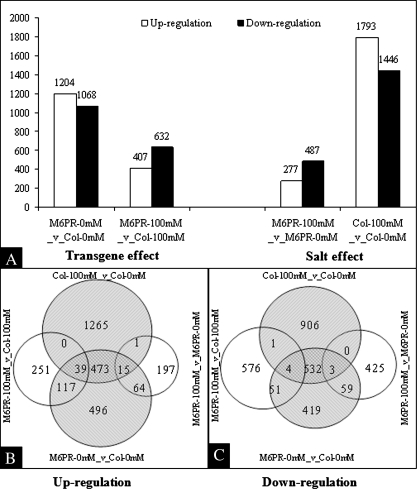
In the absence of salt stress, 1204 and 1068 transcripts were activated and repressed, respectively, by the M6PR transgene. In response to salt, however, many more transcripts were up- or down-regulated in Col-WT (1793 and 1446, respectively) than in the M6PR lines (277 and 487, respectively) (Fig. 5A). This indicates that the Col-WT plants were more affected by salt than the M6PR transgenic plants. These greater changes at the transcriptome level are consistent with the more severe growth and injury responses of Col-WT than M6PR lines under salt stress, and may represent a combination of adaptive responses and damage effects in the Col-WT plants. Relatively few transcript differences (407 up- and 632 down-regulated) were observed between WT and M6PR plants in the presence of salt (Fig. 5A). Comparisons of the transcripts affected by salt and M6PR showed that expression levels of 1166 transcripts (527 up- and 539 down-regulated) were affected by both salt stress and the M6PR transgene (Fig. 5B, C). This indicates that the stress tolerance of M6PR lines might be due, in part, to constitutive overexpression of several otherwise stress-inducible genes. However, only about half of the transcripts were in common between the salt effect and the M6PR transgene effect (Fig. 5B, C). Thus the M6PR transgene caused only some of the same responses as salt stress, as might be expected given the phenotypic differences observed between Col-WT and M6PR plants in response to salinity (Fig. 1).
Fig. 5.
Number of changed and overlapping transcripts (P-value ≤0.05 and fold change ≥2). (A) Total number of transcripts affected by the M6PR transgene and salt stress. (B) Overlapping transcripts displaying significantly increased levels of expression in response to the M6PR transgene or salt stress. (C) Overlapping transcripts displaying significantly decreased levels of expression in response to the M6PR transgene or salt stress. Comparisons are indicated around the circle.
Major transcriptomic responses common to both salt stress and the M6PR transgene
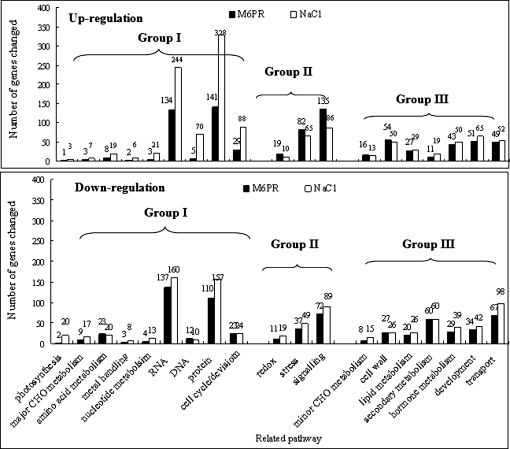
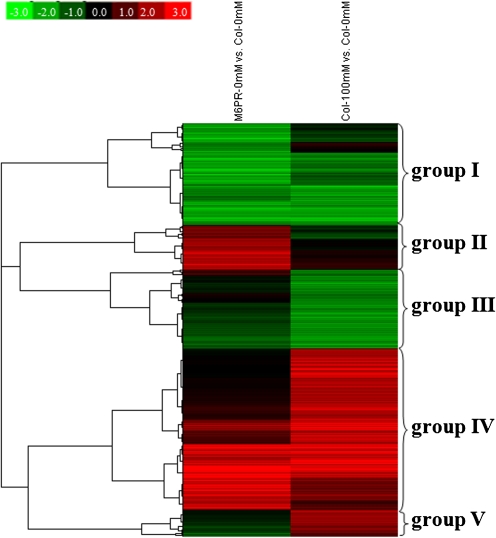
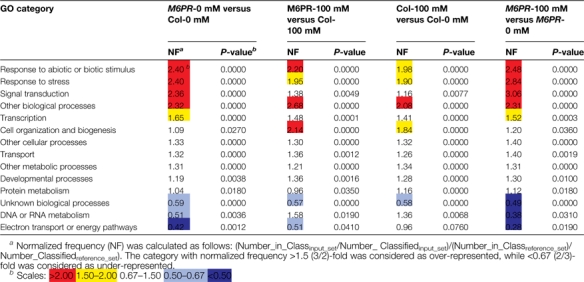
As a first approach to assigning possible gene function, salt- and M6PR transgene-responsive transcripts were characterized using MapMAN software. There were several similarities in the metabolic pathways affected by both salt stress and the M6PR transgene, i.e. many genes involved in minor carbohydrate metabolism, the cell wall, lipid metabolism, secondary metabolism, hormone metabolism, development, and transport (Fig. 6, group III). Cluster analysis revealed that several groups of genes were up- or down-regulated by both the M6PR transgene and salt stress (Fig. 7, groups I, III, and IV). Characterization of functional categories also indicated that three GO categories were over-represented by both salt stress and the M6PR transgene, including response to abiotic or biotic stimulus, response to stress, and other biological processes (Table 1).
Fig. 6.
Number of transcripts affected by salt stress and the M6PR transgene on different pathways using MapMan software. Transcripts with a P-value ≤0.05 and fold change ≥2 were loaded in MapMan and the numbers of transcripts changed in different pathways were counted. (A) Pathways mainly affected by salt stress; (B) pathways mainly activated by the M6PR transgene; (C) pathways affected by both salt stress and the M6PR transgene.
Fig. 7.
Cluster analyses of transcripts affected by the M6PR transgene and salt stress. Red, up-regulation; green, down-regulation; black, no change. Hierarchical cluster analysis was applied for differentially expressed transcripts (P ≤0.05 and fold change ≥2) with Cluster 3.0 software. The resulting tree figures were displayed using the software package, Java Treeview. The detailed fold changes are listed as Supplementary Table S3 at JXB online.
Table 1.
Biological enrichment analysis showed several categories were enriched by salt stress and the M6PR transgene
Transcripts affected by salt stress and the M6PR transgene (P ≤0.05 and fold change ≥2) were analysed with the Bio-Array Resource Classification SuperViewer. The detailed categories of gene and salt effects are listed in Supplementary Table S3 at JXB online.
 |
Major transcriptomic responses distinct to salt stress and the M6PR transgene
Although enriched categories analysis revealed commonalities between the effects of the M6PR transgene and salt stress, several categories were over- or under-represented only by salt stress or the M6PR transgene. Categories of transcription and signal transduction were only over-represented by the M6PR transgene in the absence of salt, but not by salt stress on Col. Two other categories (DNA or RNA metabolism and electron transport or energy pathways) were more broadly affected by salt stress, but less by the M6PR transgene in the absence of salt (i.e. under-represented) (Table 1). Further metabolic pathways analysis also indicated that salt stress extensively up-regulated many genes involved in pathways related to carbohydrate, DNA, protein, and cell cycle metabolism. However, genes involved in photosynthesis and major carbohydrate metabolism were mainly down-regulated by salt stress in Col-WT, not by the M6PR transgene (Fig. 6, group I). However, more genes involved in redox-, stress-, and signalling-related pathways were up-regulated by the M6PR transgene than by salt stress, indicating that these pathways were broadly affected by the M6PR transgene, possibly pre-conditioning the plants to stress (Fig. 6, group II).
Specific changes in gene expression resulting from salt stress or the M6PR transgene
Stress-related genes
Salt treatment altered the expression of many biotic stress-related genes (35 up-regulated and 27 down-regulated), including 24 genes encoding PR proteins (Supplementary Table S4 at JXB online), indicating possible interactions among abiotic and biotic stresses and their responses. A previous study indicated that constitutive expression of the celery MTD gene in transgenic tobacco enhances resistance to the mannitol-secreting fungus Alternaria alternata (Jennings et al., 2002). Consistent with these results, transgenic mannitol-producing Arabidopsis significantly activated more (58 by M6PR versus 35 by salt) and repressed fewer (15 by M6PR versus 27 by salt) transcripts involved in disease resistance than salt stress (Supplementary Table S4), indicating that the M6PR transgene not only increased abiotic stress tolerance, but may also induce pathogen defence responses in Arabidopsis.
Salt treatment altered expression of many heat stress-, low temperature-, and dehydration responsive-genes, including heat shock protein (HSP), drought-induced (DI), early-responsive to dehydration (ERD), and low-temperature induced (LTI) genes (Table 2A; Supplementary Table S4 at JXB online). Although the M6PR transgene also changed the expression of many genes involved in heat stress tolerance (Supplementary Table S4), fewer drought- and salt stress-related genes were changed by M6PR (eight) when compared with salt (15) (Table 2A). This may indicate that other mechanisms also contribute to increased salt tolerance of M6PR transgenic Arabidopsis besides the changes of abiotic stress-related genes.
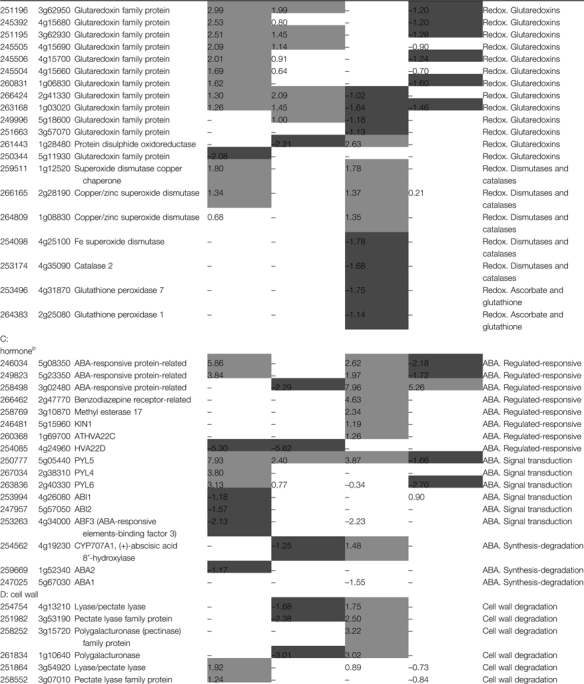
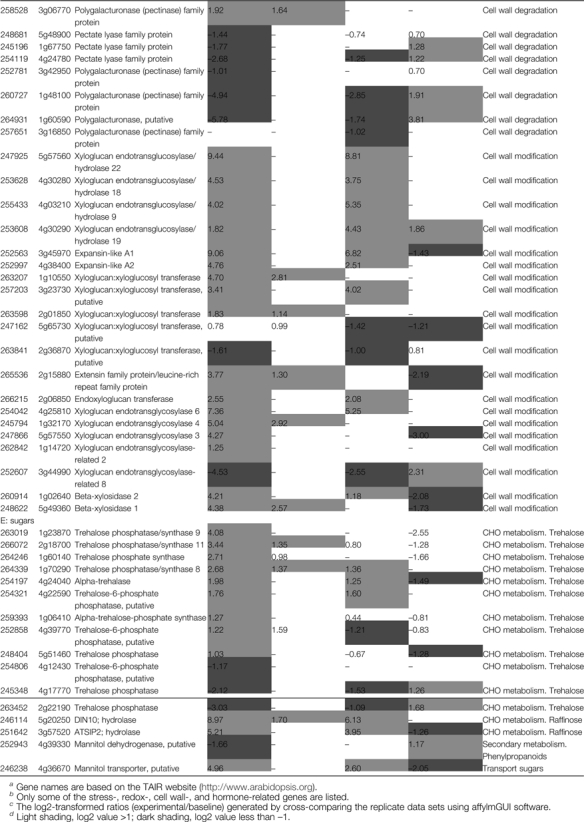
Table 2.
Specific transcripts affected by salt stress or M6PR transgene (P ≤0.05 and fold change ≥2)
|
Detailed information on transcripts affected by salt stress and M6PR is listed in Supplementary Table S4 at JXB online.
Gene names are based on the TAIR website (http://www.arabidopsis.org).
Only some of the stress-, redox-, cell wall-, and hormone-related genes are listed.
The log2-transformed ratios (experimental/baseline) generated by cross-comparing the replicate data sets using affylmGUI software.
Light shading, log2 value >1; dark shading, log2 value less than –1.
Redox-related genes
It has been previously reported that mannitol might act as a reactive oxygen quencher to suppress reactive oxygen-mediated plant defences (Smirnoff and Cumbes, 1989; Shen et al., 1997a, b). The presence of the M6PR transgene activated expression of many redox-related genes. The majority of these were glutaredoxin and thioredoxin family proteins. However, glutaredoxin-related genes were mainly inhibited by salt stress (Table 2B). Salt treatment also inhibited transcripts encoding catalase (CAT), Fe superoxide dismutase (SOD), and glutathione peroxidases (POD) which are directly involved in metabolism of reactive oxygen species (ROS). These genes were not changed by the M6PR transgene and were stable in M6PR lines after salt treatment (Table 2B). Inhibition of CAT, SOD, and POD transcript expression could result in loss of redox homeostasis in planta and further metabolic damage, which may explain why M6PR transgenic lines showed less severe salt effects than Col-WT.
ABA metabolism-related genes
After salt treatment, seven abscisic acid (ABA)-regulated or responsive genes were up-regulated in Col-WT, but only two were increased in M6PR transgenic lines and only one of them by the M6PR transgene in the presence of salt (Table 2C). That Col-WT exhibited a more severely stunted phenotype than M6PR lines under salinity stress may be due to activation of more ABA-regulated or responsive genes. Interestingly, there were major increases in genes for three recently identified ABA receptors (in the PYR/PYLs family of START proteins) that inhibit type 2C protein phosphatases (PP2Cs) involved in ABA signalling in the M6PR plants (Ma et al., 2009; Park et al., 2009) (14.0-fold for PYL4, 8.7-fold for PYL6, and, most dramatically, 244.3-fold for PYL5). Accordingly, expression of PP2C ABI1 (At4g26080) and ABI2 (At5g57050) was inhibited by the M6PR transgene (Table 2C). Moreover, ABA1, which functions in the first step of the biosynthesis of ABA (Nambara and Marion-Poll, 2005), was down-regulated 2.92-fold, and CYP707A1, which is involved in ABA catabolism (Kushiro et al., 2004), was up-regulated 2.78-fold by salt stress only in Col-WT
Cell wall-related genes
The plant cell wall is a complex network of cellulose, hemicelluloses, pectins (Zhong and Ye, 2003), and several structural proteins which are particularly rich in the amino acids hydroxyproline (hydroxyproline-rich glycoprotein; HPRG), proline (proline-rich protein; PRP), and glycine (glycine-rich protein; GRP) (Showalter, 1993; Jamet et al., 2006). Both the M6PR transgene and salt stress affected many cell wall-related genes involved in cell wall protein, cellulose synthesis, cell wall degradation, and cell wall modification pathways (Supplementary Table S4 at JXB online). Salt treatment resulted in up-regulation of several cell wall degradation-related genes which were not affected by the M6PR transgene, including two pectate lyase genes (At4g13210, 3.36-fold; At3g53190, 5.64-fold) and two polygalacturonase genes (At3g15720.9.33-fold; At1g10640, 8.08-fold). Although the expression levels of two pectate lyase genes (At3g54920 and At3g07010) and one polygalacturonase gene (At3g06770) were slightly increased (<3.8-fold) by the M6PR transgene, many other cell wall degradation-related genes were decreased in M6PR lines, especially two polygalacturonase genes (At1g60590 and At1g48100, down-regulated 55.0- and 30.8-fold, respectively) (Table 2D).
Further, transcripts encoding xyloglucan endotransglucosylase/hydrolase (XTH), responsible for cell wall construction in plants (Yokoyama and Nishitani, 2001), and expansin involved in cell growth (Cosgrove, 2000), were extensively increased by both salt stress and the M6PR transgene (Supplementary Table S4). One XTH gene was dramatically up-regulated 694-fold and one expansin gene 535-fold by the transgene (Table 2D). Expression of two xyloglucan:xyloglucosyl transferase (XXT) genes (At1g10550 and At2g01850), which are related to cell elongation (Hyodo et al., 2003), was enhanced 25.9- and 3.54-fold only by the M6PR transgene. In addition, two transcripts encoding xyloglucan endotransglycosylase-related proteins (XETs) were considerably increased only by M6PR by 4.0-fold (At1g32170) and 19-fold (At5g57550). XETs have been proposed to function in modification of a major component of the plant cell wall resulting in cell expansion (Pritchard, et al. 1993; Palmer and Davies, 1996; Campbell and Braam, 1999). Similarly, two β-xylosidases (BXLs) (At5g49360 and At1g02640) involved in secondary wall thickening (Arsovski et al., 2009) were up-regulated by 20- and 19-fold (Table 2D). These high level changes of XTH, XET, XXT, BXL, and expansin transcripts involved in cell wall strengthening and elongation suggested that expression of the M6PR transgene in Arabidopsis enhances the activity of cell growth or cell wall-strengthening functions.
Sugar metabolism-related genes
Nine trehalose biosynthesis-related genes (At1g23870, At2g18700, At1g60140, At1g70290, At4g24040, At4g22590, At1g06410, At4g39770, and At5g51460) were activated by M6PR, but only three were slightly up-regulated by salt stress (Table 2E). Trehalose is involved in abiotic stress, biotic stress, and plant growth (Schluepmann et al., 2004; van Dijken et al., 2004; Bae et al., 2005; Doehlemann et al., 2006). It is also notable that at least one hydrolase (At5g20250) involved in raffinose synthesis was up-regulated 70-fold by salt stress, but 503-fold by the M6PR transgene (Table 2E). These findings are consistent with other comprehensive studies showing that prolonged salt stress (3 d and 5 d) induces a strong accumulation of raffinose in Arabidopsis (Kempa et al., 2008). Recent research has indicated that raffinose constitutes a novel function to protect plants from oxidative damage caused by salinity or chilling (Nishizawa et al., 2008). In addition, the expression level of a putative mannitol transport gene (At4g36670) was increased 31.1-fold by the M6PR transgene, but only 6.1-fold by salt stress. One putative MTD (At4g39330) was slightly down-regulated by the M6PR transgene. All these data indicated the M6PR transgene activated several osmoprotectant-related genes to cope with the stress condition.
The M6PR transgene and mannitol biosynthesis might be expected also to affect the pools of hexose phosphates as they are utilized to produce mannose 6-phosphate and eventually mannitol; however, there were no significant changes in expression of any hexokinases. Hexokinases are known to be involved in sugar-sensing, hexose-dependent modulation of gene expression, and plant growth, but sugar signal transduction processes are relatively complex in plants and lack of an effect may reflect the multiple pathways or redundancy involved in these responses (Moore and Sheen, 1999). Alternatively, metabolic effects may result from changes in enzyme activities in the absence of transcriptional changes.
Conclusions
As evident in Figs 1–3, there was no apparent effect of the M6PR transgene on Arabidopsis growth and development in the absence of salt. Yet, the transgene clearly provided significant improvements in salt tolerance. Despite the lack of effects on phenotype in the absence of stress, genome-wide expression analyses indicated that expression levels of many genes were substantially altered (up and down) after introduction of the celery M6PR gene into Arabidopsis [i.e. 2272 changes (P-value ≤0.05 and fold change ≥2] (Fig. 5A). However, relatively few (764) additional genes were altered in salt-stressed M6PR transgenic plants, versus salt-stressed WT plants (3239), suggesting that M6PR may have caused pre-adaptive changes facilitating response to salt stress. The large number of changes in salt-stressed WT plants may represent both adaptive and damage responses. However, less than half of the transcripts affected by salt in WT plants overlapped with those affected by the transgene, and vice versa (Figs 5B, C, 7), indicating differences in the responses (i.e. the protective effect of the M6PR gene was different from the damage effects of salt stress). Given that salt tolerance is determined by several physiological components such as sodium transport and exclusion, tolerance to osmotic and oxidative stress, and tolerance to ion toxicity (Munns and Tester, 2008), the presence of the transgene could induce a cascade of protective effects. Similarly, salt stress could result in changes that include both protective (adaptive) responses and damage effects; however, as the data indicate, these responses can involve distinctly different genes.
Although early work with sugar alcohols such as mannitol suggested that their effects were primarily as compatible solutes or osmolytes (Yancey et al., 1982) or more recently as osmoprotectants (Shen et al., 1997a), the results here indicate that the presence of M6PR as a transgene has far-ranging effects on gene expression, and that many of these involve enhanced expression of stress resistance genes that may have much to do with the presumed osmoprotective effects of mannitol. These effects are quite evident in Fig. 6 which shows the numbers of genes affected by the M6PR transgene and salt stress. Many redox-, stress-, and cell wall-related genes are up-regulated by the presence of the transgene in the absence of stress, suggesting that the transgenic plant is predisposed to resist the effects of salt stress. Mannitol itself may have an effect on quenching ROS (Shen et al., 1997b), but the presence of the transgene also resulted in constitutive overexpression of a multitude of additional genes that deal with stress, both biotic and abiotic, and including a number that deal specifically with redox and the cell wall.
Changing expression levels of a single gene have previously been observed to result in alteration of expression of a large range of plant genes. For example, Sottosanto et al. (2004) found that a large spectrum of gene expression changes was the result of the absence of a vacuolar Na+/H+ antiporter gene (AtNHX1). Thus, in addition to the known role(s) of AtNHX1 on ion homeostasis, the vacuolar cation/proton antiporter appeared to play a significant role in intracellular vesicular trafficking, protein targeting, and other cellular processes. Likewise, microarray-based transcriptome analysis of transgenic rice engineered for drought stress resistance with a chloroplast-targeted choline oxidase gene for glycine betaine synthesis indicated altered expression of many genes involved in stress responses, signal transduction, gene regulation, hormone signalling, and cellular metabolism (Kathuria et al., 2009). Similarly selection for elevated glycine betaine levels in barley resulted in a broad array of effects on drought stress and solute potential (Grumet and Hanson, 1986).
Many plants have mechanisms to deal with mannitol derived from pathogen attack. Jennings et al. (2002) and others (Zamski et al., 2001) have proposed that certain plant pathogenic fungi may produce mannitol as a means of suppressing ROS-mediated plant defences. These defences are clearly important in the early hypersensitive response, and fungal mannitol secretion would thus appear to play a role in avoiding this well-defined resistance mechanism (Alvarez et al., 1998). Other reports suggest that biotic and abiotic signalling can share common components and have functional overlaps (Chini et al., 2004). The present data suggest that the presence of the M6PR transgene that results in biosynthesis of mannitol in turn elicits many of the same responses to pathogen attack, for example stress- and cell wall-related transcripts and the multitude of disease resistance genes (Fig. 6, Table 2B, D; Supplementary Table S3 at JXB online).
However, these pathogen-related responses may not completely explain the many increases in redox-related genes that are related to ROS-mediated plant defences. ROS are clearly involved in signalling in response to both infection and stress. They (e.g. the glutathione redox couple) appear to be essential for homeostatic adjustment of the cellular redox potential which in turn can affect the function of many proteins (Foyer et al., 2005), as well as developmental processes, for example root hair formation (Foreman et al., 2003) and stomatal behaviour (Kwak et al., 2003). Given a role for fungal-derived mannitol in quenching ROS, the plant genes normally involved in mediating low ROS levels may be down-regulated if part of the plant oxidative burst defence is to overcome the effect of mannitol as a quencher of ROS. Substantial differences in many genes associated with regulating ROS levels were found (Table 2B, Fig. 6); many glutaredoxin and thioredoxin family protein genes were up-regulated by the presence of the M6PR transgene and very few were down-regulated. These changes may reflect M6PR- (and mannitol) related perturbations in the redox potential of the cell, and the role of homeostatic mechanisms adjusting expression in response.
Interestingly, the M6PR transgene highly activated expression of ABA receptor genes (PYL4, PYL5, and PYL6) and inhibited that of PP2C genes (ABI1 and ABI2) (Table 2C). These changes resulted in activation of SNF1-related kinases (SnRK2s), and SnRK2s in turn activate downstream effectors to switch on stress response programmes (Cutler et al., 2010; Klingler et al., 2010).
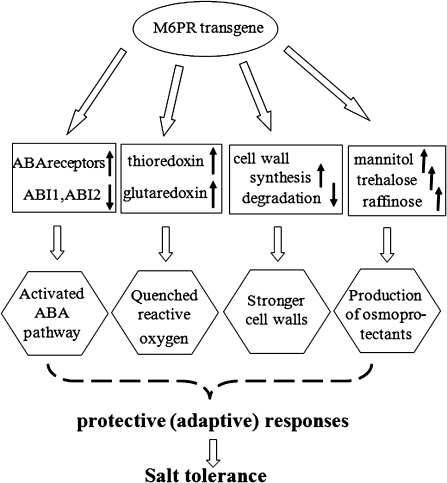
In conclusion, the present growth analyses indicate that mannitol clearly enhances salt tolerance, but the reasons for these effects are apparently much more complicated than what might be expected of an osmoticum or osmoprotectant. In a previous study, it was found that fructose, glucose, sucrose, and myo-inositol in M6PR lines all were lower than those in Col-WT under control conditions. However, in those plants treated with high salt concentrations, these carbohydrates increased to higher levels in M6PR lines than in Col-WT (Zhifang and Loescher, 2003). However, it was expected that mannitol synthesis in M6PR lines would result in changes in carbohydrate metabolism due to the impact of mannitol biosynthesis on hexose phosphate precursors. The expression data in this study indicate that the presence of the M6PR transgene, and thus mannitol, appears to act as a signal, affecting genes responsive to both biotic and abiotic stresses, which in turn suggests insights into global plant defence mechanisms. As depicted in Fig. 8, besides the production of osmoprotectants, the M6PR transgene also directly or indirectly activated expression of ABA receptor genes (PYL4, PYL5, and PYL6) and inhibited that of PP2C genes (ABI1 and ABI2) which resulted in activation of steps downstream of the ABA pathway. M6PR transgenic lines further exhibited increased ability to quench reactive oxygen and strengthen the cell wall, apparently through signals produced by mannitol and/or other metabolites. Thus, the gene expression data here indicate that stress tolerance of mannitol-producing Arabidopsis may be due at least in part to enhanced expression of a number of stress-inducible genes related to both biotic and abiotic stress tolerance. However, further studies need to be carried out to determine how the presence of mannitol and the M6PR gene induces specific responses. Separating mannitol's primary effects from secondary effects remains problematic.
Fig. 8.
Model depicting the mechanisms involved in salt tolerance of M6PR transgenic Arabidopsis. Besides the production of osmoprotectants, the M6PR transgene also directly or indirectly activated expression of ABA receptor genes (PYL4, PYL5, and PYL6) and inhibited that of PP2C genes (ABI1 and ABI2), which resulted in activation downstream of the ABA pathway. M6PR transgenic lines further showed increased ability to quench reactive oxygen and strengthen the cell wall.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Growth parameters of transgenic M6PR lines and the WT under salinity conditions during the whole life cycle.
Figure S2. Reproducibility of the microarray signal data from two independent biological replicates.
Table S1. Primers used for quantitative RT-PCR.
Table S2. Numbers of genes changed by salt stress or the M6PR transgene with different cut-off threshold parameters.
Table S3. Transcripts changed by salt stress and the M6PR transgene (fold change ≥2 and P-value ≤0.05)
Table S4. Specific genes affected by salt stress or the M6PR transgene.
Acknowledgments
This work was supported in part by USDA-BRAG #2005-39454-16516.
References
- Alvarez ME, Pennell RI, Meijer PJ, Ishikawa A, Dixon RA, Lamb C. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell. 1998;92:773–784. doi: 10.1016/s0092-8674(00)81405-1. [DOI] [PubMed] [Google Scholar]
- Arsovski AA, Popma TM, Haughn GW, Carpita NC, McCann MC, Western TL. AtBXL1 encodes a bifunctional β-d-xylosidase/α-l-arabinofuranosidase required for pectic arabinan modification in Arabidopsis thaliana mucilage secretory cells. Plant Physiology. 2009;150:1219–1234. doi: 10.1104/pp.109.138388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae H, Herman E, Bailey B, Bae HJ, Sicher R. Exogenous trehalose alters Arabidopsis transcripts involved in cell wall modification, abiotic stress, nitrogen metabolism, and plant defense. Physiologia Plantarum. 2005;125:114–126. [Google Scholar]
- Bieleski RL. Sugar alcohols. Plant carbohydrates I. Intercellular carbohydrates. In: Loewus FA, Tanner W, editors. Encyclopedia of plant physiology. New series. Vol. 13A. New York: Springer-Verlag; 1982. pp. 158–192. [Google Scholar]
- Bohnert HJ, Jensen RG. Strategies for engineering water-stress tolerance in plants. Trends in Biotechnology. 1996;14:89–97. [Google Scholar]
- Campbell P, Braam J. Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends in Plant Science. 1999;4:361–366. doi: 10.1016/s1360-1385(99)01468-5. [DOI] [PubMed] [Google Scholar]
- Chaturvedi V, Bartiss A, Wong B. Expression of bacterial mtlD in Saccharomyces cerevisiae results in mannitol synthesis and protects a glycerol-defective mutant from high-salt and oxidative stress. Journal of Bacteriology. 1997;179:157–162. doi: 10.1128/jb.179.1.157-162.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Silva H, Klessig DF. Active oxygen species in the induction of plant systemic acquired resistance by salicyclic acid. Science. 1993;262:1883–1885. doi: 10.1126/science.8266079. [DOI] [PubMed] [Google Scholar]
- Chini A, Grant JJ, Seki M, Shinozaki K, Loake GJ. Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. The Plant Journal. 2004;38:810–822. doi: 10.1111/j.1365-313X.2004.02086.x. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology. 2010;61:651–79. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- Doehlemann G, Molitor F, Hahn M. Trehalose metabolism is important for heat stress tolerance and spore germination of Botrytis cinerea. Microbiology. 2006;152:2625–2634. doi: 10.1099/mic.0.29044-0. [DOI] [PubMed] [Google Scholar]
- Everard JD, Gucci R, Kann SC, Flore JA, Loescher WH. Gas exchange and carbon partitioning in the leaves of celery (Apium graveolens L.) at various levels of root zone salinity. Plant Physiology. 1994;106:281–292. doi: 10.1104/pp.106.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JHF. Reactive oxygen species produced by NADPH oxidase regulated plant cell growth. Nature. 2003;422:442–446. doi: 10.1038/nature01485. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Gomez LD, van Heerden PDR. Glutathione. In: Smirnoff N, editor. Antioxidants and reactive oxygen species in plants. Oxford: Blackwell; 2005. pp. 1–24. [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumet R, Hanson AD. Genetic evidence for an osmoregulatory function of glycinebetaine accumulation in barley. Australian Journal of Plant Physiology. 1986;13:353–364. [Google Scholar]
- Hoagland DF, Arnon DI. University of California Agricultural Experiment Station, Circular 347. Berkeley, CA: 1950. The water-culture method for growing plants without soil. [Google Scholar]
- Hu L, Lu H, Liu QL, Chen XM, Jiang XN. Overexpression of mtlD gene in transgenic Populus tomentosa improves salt tolerance through accumulation of mannitol. Tree Physiology. 2005;25:1273–1281. doi: 10.1093/treephys/25.10.1273. [DOI] [PubMed] [Google Scholar]
- Hyodo H, Yamakawa S, Takeda Y, Tsuduki M, Yokota A, Nishitani K, Kohchi T. Active gene expression of a xyloglucan endotransglucosylase/hydrolase gene, XTH9, in inflorescence apices is related to cell elongation in Arabidopsis thaliana. Plant Molecular Biology. 2003;53:473–482. doi: 10.1023/a:1023904217641. [DOI] [PubMed] [Google Scholar]
- Jamet E, Canut H, Boudart G, Pont-Lezica RF. Cell wall proteins: a new insight through proteomics. Trends in Plant Science. 2006;11:33–39. doi: 10.1016/j.tplants.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Jennings DB, Daub ME, Pharr DM, Williamson JD. Constitutive expression of a celery mannitol dehydrogenase in tobacco enhances resistance to the mannitol-secreting fungal pathogen Alternaria alternata. The Plant Journal. 2002;32:41–49. doi: 10.1046/j.1365-313x.2001.01399.x. [DOI] [PubMed] [Google Scholar]
- Karakas B, Ozias-Akins P, Stushnoff C, Suefferheld M, Rieger R. Salinity and drought tolerance of mannitol-accumulating transgenic tobacco. Plant, Cell and Environment. 1997;20:609–616. [Google Scholar]
- Kathuria K, Giri J, Karaba N, Nataraja KN, Murata N, Udayakumar M, Tyagi AK. Glycinebetaine-induced water-stress tolerance in codA-expressing transgenic indica rice is associated with up-regulation of several stress responsive genes. Plant Biotechnology Journal. 2009;7:512–526. doi: 10.1111/j.1467-7652.2009.00420.x. [DOI] [PubMed] [Google Scholar]
- Kempa S, Krasensky J, Dal Santo S, Kopka J, Jonak C. A central role of abscisic acid in stress-regulated carbohydrate metabolism. PLoS ONE. 2008;3:e3935. doi: 10.1371/journal.pone.0003935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiedrowski S, Kawalleck P, Hahlbrock K, Somssich IE, Dangl JL. Rapid activation of a novel plant defense gene is strictly dependent on the Arabidopsis-Rpm1 disease resistance locus. EMBO Journal. 1992;11:4677–4684. doi: 10.1002/j.1460-2075.1992.tb05572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingler JP, Batelli G, Zhu J-K. ABA receptors: the START of a new paradigm in phytohormone signaling. Journal of Experimental Botany. 2010;61(12):3199–3210. doi: 10.1093/jxb/erq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8'-hydroxylases: key enzymes in ABA catabolism. EMBO Journal. 2004;23:1647–1656. doi: 10.1038/sj.emboj.7600121. doi:10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO Journal. 2003;22:2623–2633. doi: 10.1093/emboj/cdg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Moore BD, Sheen J. Plant sugar sensing and signaling—a complex reality. Trends in Plant Science. 1999;4:250. doi: 10.1016/s1360-1385(99)01433-8. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annual Review of Plant Biology. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Shigeoka S. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiology. 2008;147:1251–1263. doi: 10.1104/pp.108.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer SJ, Davies WJ. An analysis of relative elemental growth rate, epidermal cell size and xyloglucan endotransglycosylase activity through the growing zone of ageing maize leaves. Journal of Experimental Botany. 1996;47:339–347. [Google Scholar]
- Park SY, Fung P, Nishimura N, et al. Abscisic acid inhibits Type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharr DM, Stoop JMH, Williamson JD, Studer-Feusi ME, Massel MO, Conkling MA. The dual role of mannitol as osmoprotectant and photoassimilate in celery. Journal of the American Society for Horticultural Science. 1995;30:1182–1188. [Google Scholar]
- Pritchard J, Hetherington PR, Fry SC, Tomos AD. Xyloglucan endotransglycosylase activity, microfibril orientation and the profiles of cell wall properties along growing regions of maize roots. Journal of Experimental Botany. 1993;44:1281–1289. [Google Scholar]
- Provart NJ, Zhu T. A browser-based functional classification SuperViewer for Arabidopsis genomics. Currents in Computational Molecular Biology. 2003;2003:271–272. [Google Scholar]
- Schluepmann H, van Dijken A, Aghdasi M, Wobbes B, Paul M, Smeekens S. Trehalose mediated growth inhibition of Arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiology. 2004;135:879–890. doi: 10.1104/pp.104.039503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Jensen RG, Bohnert HJ. Increased resistance to oxidative stress in transgenic plants by targeting mannitol biosynthesis to the chloroplast. Plant Physiology. 1997a;113:1177–1183. doi: 10.1104/pp.113.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Jensen RG, Bohnert HJ. Mannitol protects against oxidation by hydroxyl radicals. Plant Physiology. 1997b;115:527–532. doi: 10.1104/pp.115.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheveleva E, Marquez S, Chmara W, Zegeer A, Jensen RG, Bohnert HJ. Sorbitol-6-phosphate dehydrogenase expression in transgenic tobacco. Plant Physiology. 1998;117:831–839. doi: 10.1104/pp.117.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM. Structure and function of plant cell wall proteins. The Plant Cell. 1993;5:9–23. doi: 10.1105/tpc.5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickler CM, Edwards GE, Kiirats O, Gao Z, Loescher W. Response of mannitol-producing Arabidopsis thaliana to abiotic stress. Functional Plant Biology. 2007;34:382–391. doi: 10.1071/FP06274. [DOI] [PubMed] [Google Scholar]
- Smirnoff N, Cumbes QJ. Hydroxyl radical scavenging activity of compatible solutes. Phytochemistry. 1989;28:612–619. [Google Scholar]
- Sottosanto JB, Gelli A, Blumwald E. DNA array analyses of Arabidopsis thaliana lacking a vacuolar Na+/H+ antiporter: impact of AtNHX1 on gene expression. The Plant Journal. 2004;40:752–771. doi: 10.1111/j.1365-313X.2004.02253.x. [DOI] [PubMed] [Google Scholar]
- Tarczynski MC, Jensen RG, Bohnert HJ. Expression of a bacterial mtlD gene in transgenic tobacco leads to production and accumlation of mannitol. Proceedings of the National Academy of Sciences, USA. 1992;89:2600–2604. doi: 10.1073/pnas.89.7.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Thomas JC, Sepahi M, Arendall B, Bohnert HJ. Enhancement of seed germination in high salinity by engineering mannitol expression in Arabidopsis thaliana. Plant, Cell and Environment. 1995;18:801–806. [Google Scholar]
- van Dijken AJH, Schluepmann H, Smeekens SCM. Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiology. 2004;135:969–977. doi: 10.1104/pp.104.039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall JM, Simpson KM, Satterley K, Smyth GK. affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics. 2006;22:897–899. doi: 10.1093/bioinformatics/btl025. [DOI] [PubMed] [Google Scholar]
- Williamson JD, Stoop JMH, Massel MO, Conkling MA, Pharr DM. Sequence analysis of a mannitol dehydrogenase cDNA from plants reveals a function for the pathogenesis related protein ELI3. Proceedings of the National Academy of Sciences, USA. 1995;92:7148–7152. doi: 10.1073/pnas.92.16.7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalpani N, Leon J, Lawton MA, Raskin I. Pathway of salicylic-acid biosynthesis in healthy and virus-inoculated tobacco. Plant Physiology. 1993;103:315–321. doi: 10.1104/pp.103.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: evolution of osmolyte systems. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K. A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant and Cell Physiology. 2001;42:1025–1033. doi: 10.1093/pcp/pce154. [DOI] [PubMed] [Google Scholar]
- Zamski E, Guo W-W, Yamamoto YT, Pharr DM, Williamson JD. Analysis of celery (Apium graveolens) mannitol dehydrogenase (Mtd) promoter regulation in Arabidopsis suggests roles for MTD in key environmental and metabolic responses. Plant Molecular Biology. 2001;47:621–631. doi: 10.1023/a:1012395121920. [DOI] [PubMed] [Google Scholar]
- Zhifang G, Loescher WH. Expression of a celery mannose 6-phosphate reductase in Arabidopsis thaliana enhances salt tolerance and induces biosynthesis of both mannitol and a glucosyl-mannitol dimer. Plant, Cell and Environment. 2003;26:275–283. [Google Scholar]
- Zhong R, Ye ZH. Unraveling the functions of glycosyltransferase family 47 in plants. Trends in Plant Science. 2003;8:565–568. doi: 10.1016/j.tplants.2003.10.003. [DOI] [PubMed] [Google Scholar]