Abstract
The inactivation of starch branching IIb (SBEIIb) in rice is traditionally associated with elevated apparent amylose content, increased peak gelatinization temperature, and a decreased proportion of short amylopectin branches. To elucidate further the structural and functional role of this enzyme, the phenotypic effects of down-regulating SBEIIb expression in rice endosperm were characterized by artificial microRNA (amiRNA) and hairpin RNA (hp-RNA) gene silencing. The results showed that RNA silencing of SBEIIb expression in rice grains did not affect the expression of other major isoforms of starch branching enzymes or starch synthases. Structural analyses of debranched starch showed that the doubling of apparent amylose content was not due to an increase in the relative proportion of amylose chains but instead was due to significantly elevated levels of long amylopectin and intermediate chains. Rices altered by the amiRNA technique produced a more extreme starch phenotype than those modified using the hp-RNA technique, with a greater increase in the proportion of long amylopectin and intermediate chains. The more pronounced starch structural modifications produced in the amiRNA lines led to more severe alterations in starch granule morphology and crystallinity as well as digestibility of freshly cooked grains. The potential role of attenuating SBEIIb expression in generating starch with elevated levels of resistant starch and lower glycaemic index is discussed.
Keywords: CP/MAS NMR, crystalline polymorph, RNA interference, starch granules, XRD
Introduction
High amylose cereals are attracting considerable attention because of their potential health benefits, along with their industrial uses (Jobling, 2004; Morell and Myers, 2005; Rahman et al., 2007). High amylose maize (Shannon et al., 2009), wheat (Regina et al., 2006; Sestili et al., 2010), and barley (Morell et al., 2003; Regina et al., 2010) have so far been developed. The apparent amylose content (AAC) of these high amylose cereals ranges from 50% to 90% but, in rice, the highest reported AAC is only ∼30% for wild types (Juliano, 2003) and between 25% and 40% for chemical- and irradiation-induced mutants (Yano et al., 1985; Nishi et al., 2001; Kang et al., 2003; Yang et al., 2006). Recently, Wei et al. (2010a) have reported higher amylose contents, but the detailed characterization of the rice plants has yet to be described.
Amylose is the predominantly linear component of native cereal starches with a degree of polymerization (DP) <5000, whereas amylopectin is a very large (DP 5000–50 000) and highly branched biopolymer (Ball et al., 1998). When rice starch is debranched by isoamylase, amylopectin chains are in the range of DP 6–120, while amylose is in the range of DP 230–10 000 (Takeda et al., 2003; Ward et al., 2006; Fitzgerald et al., 2009). Granule-bound starch synthase I (GBSSI), encoded by the Waxy (Wx) gene, is essential for amylose biosynthesis, while amylopectin is synthesized by the combined action of several isoforms of starch synthases, starch branching enzymes, and starch debranching enzymes (Smith, 2001; Nakamura, 2002; Ball and Morell, 2003; Tetlow et al., 2004; Jeon et al., 2010).
In most wild-type cereal starches, amylose is usually 15–25% by weight and amylopectin is in the order of 75–85% (Ball et al., 1998; Fitzgerald, 2004). Screening a subset of the International Rice Research Institute's germplasm collection revealed that the range of amylose in wild and cultivated rice ranges from 0% to 30% (Butardo et al., 2008). To increase the levels of amylose further, one can overexpress a suitable Wx allele (Itoh et al., 2003; Hanashiro et al., 2008), but the more common method is to down-regulate the expression of enzymes involved in amylopectin biosynthesis to direct starch synthesis towards amylose production (Morell and Myers, 2005; Regina et al., 2006, 2010; Rahman et al., 2007).
The prime target for down-regulation to achieve high amylose is starch branching enzyme II (SBEII) based on elevated amylose mutants in maize, wheat, barley, and rice (Boyer and Preiss, 1978; Nishi et al., 2003; Regina et al., 2006, 2010; Wei et al., 2010a). The two isoforms of SBEII, SBEIIa and SBEIIb (Vandeputte and Delcour, 2004), share ∼80% sequence identity, but their expression patterns differ. In rice, SBEIIa (RBE4) is primarily expressed in the leaves while SBEIIb (RBE3) is primarily expressed in the grains (Yamanouchi and Nakamura, 1992; Ohdan et al., 2005; Yamakawa et al., 2007), although massively parallel sequencing data indicate that it is also expressed weakly in roots and seedlings (http://mpss.udel.edu/rice/mpss_index.php). In vitro studies suggest that rice SBEIIb acts preferentially on DP 6 and 7, while SBEIIa acts on a wider range of chain lengths of DP 6–15 from the outer chains of amylopectin and possibly amylose (Nakamura et al., 2010). High amylose rice and maize, which exhibit the amylose extender (ae) phenotype, result from the inactivation of SBEIIb (Boyer et al., 1980; Hedman and Boyer, 1982; Yano et al., 1985; Kim et al., 1998; Nishi et al., 2001). The ae mutants in rice have higher AAC than their wild-type parents, but only 35% AAC is found, in contrast to 50–75% in ae maize (Shannon et al., 2009).
Most high amylose mutants in rice have been obtained by chemical mutagenesis or by exposure to sublethal doses of radiation (Yano et al., 1985; Kim et al., 2005; Shu et al., 2006). Mutation of SBEIIb in rice (Yano et al., 1985; Asaoka et al., 1986) and in maize (Moore and Creech, 1972; Boyer et al., 1976; Garwood et al., 1976) was accomplished by directly mutating the genome, hence the expression of active SBEIIb in the entire plant is affected. Furthermore, genetically tightly linked but uncharacterized mutations may also be present in the genome. Possible effects on the whole plant from such genomic alterations can be avoided by RNA silencing using seed-specific promoters (Wang et al., 1998; Kawakatsu et al., 2008; Qu et al., 2008). RNA interference using hairpin RNA (hp-RNA) has been successfully demonstrated in wheat (Regina et al., 2006; Sestili et al., 2010) and in barley (Regina et al., 2010) endosperm. Another method that has been recently developed is RNA silencing by artificial microRNA (amiRNA) (Ossowski et al., 2008). Although this has not yet been successfully demonstrated in cereal endosperm, one study was able to silence the expression of three rice genes systemically in japonica (Nipponbare) and indica (IR64) backgrounds using an amiRNA driven by a ubiquitin promoter (Warthmann et al., 2008).
In the present study, the expression of SBEIIb in the endosperm has been reduced using both hp-RNA and amiRNA approaches. The amiRNA approach reduces yet further the possibility of non-specific targets, and this paper reports the first, highly effective, use of this technique in the grain endosperm. It is shown here that the ae phenotype in rice can be obtained by down-regulating the expression of SBEIIb alone, thereby further corroborating previous findings that this mutation is due to a defective SBEIIb. It is further demonstrated that the phenotype in a japonica background is due solely to the increased proportion of long amylopectin chains, not to an increase in ‘true’ amylose. Rice grains with different crystalline polymorphs and digestibility were obtained using the two different techniques although they only differed slightly in starch branch length distribution, and these starches are comprehensively characterized herein.
Materials and methods
Construction of RNA silencing expression vectors
The construction of SBEIIb hairpin RNA (hp-BEIIb) was based on previous methods (Regina et al., 2006, 2010). Briefly, a 397 bp SBEIIb fragment located at the 5' end of the SBEIIb gene (254–650 bp of LOC_Os02g32660 based on Oryza sativa MSU Osa1 Release 6.1 Annotation) was PCR amplified (Supplementary Table S1 available at JXB online) from Nipponbare cDNA and cloned into pGEM-T Easy (Promega) using Escherichia coli DH5α. The cloned SBEIIb fragment was inserted in forward and reverse orientations in an intermediate cloning vector containing a wheat high molecular weight glutenin (wHMWG) promoter and a nopaline synthase (NOS) 3' terminator (pBx17). The hairpin construct was then transferred into an Agrobacterium Ti binary expression vector (pVec8) containing a hygromycin resistance gene driven by a cauliflower mosaic virus (CaMV) 35S promoter (Wang et al., 1998). The binary vector containing the hp-BEIIb sequence was electroporated and maintained in Agrobacterium tumefaciens AGL1 using LB broth supplemented with 50 μg ml−1 rifampicin and spectinomycin.
The construction of SBEIIb artificial microRNA (ami-BEIIb) was based on a previous protocol (Warthmann et al., 2008), with modifications to express the amiRNA in the endosperm using pVec8. A 21 nucleotide microRNA (miRNA) target located at the middle of the SBEIIb gene (1258–1278 bp) was identified using Web MicroRNA Designer 2 (WMD2) (Ossowski et al., 2008). An amiRNA was selected from the list of potential amiRNAs based on its binding energy and specificity with the target gene. The secondary structure of ami-BEIIb in the osa-mir528 backbone was predicted using RNAfold (Hofacker et al., 1994).
The selected amiRNA (TTAATGCGTATCTGTACCATG) was synthesized by fusion PCR (Supplementary Table S1) using Expand Taq (Roche) and osa-mir528 (Liu et al., 2005) endogenous miRNA precursor as stem–loop backbone (Warthmann et al., 2008). After PCR purification using Wizard SV PCR Clean-up System (Promega), the amiRNA precursor (254 bp) was cloned into pGEM-T Easy (Promega) using E. coli DH5α. The resulting amiRNA (ami-BEIIb) was cloned in the forward orientation as described above.
Nipponbare transformation
Rice transformation was undertaken by standard procedures as previously described (Upadhyaya et al., 2000) but using 50 mg l−1 hygromycin to select for transformed calli. Regenerated hygromycin-resistant plants were acclimatized for 1 week inside a moist growth chamber before they were individually planted in pots (8 cm diameter) with soil supplemented with 1 g kg−1 Osmocote (Scotts Australia). The pots were maintained in submerged tanks inside a biosafety glasshouse with temperature maintained at 26.5±3.5 °C. The succeeding generations of transgenic and control plants were grown under similar conditions.
Genomic DNA analyses
Genomic DNA was extracted from 1-month-old leaves using a FastDNA Kit (Q-BIOgene). Initial screening for putative transformants was done using the hygromycin resistance gene (Supplementary Table S1 at JXB online). The putative transformants were verified using gene-specific primers that amplify a fragment containing a portion of the wHMWG promoter and a portion of the forward hp-SBEIIb or ami-SBEIIb fragment (Supplementary Table S1). PCR amplification was carried out using HotStar Taq (Qiagen) and products were resolved in 1% agarose in 1× TBE buffer using Hyper Ladder IV (Bio Line) as molecular weight standards.
Southern blot analysis was carried out as described (Lagudah et al., 1991) but using 6 M ammonium acetate to precipitate protein contaminants (QH Zhu personal communication) prior to the final precipitation with isopropanol. A total of 5 μg of DNA per sample was digested with the restriction enzymes BamHI/EcoRI/XbaI and resolved in 13% agarose, using the wHMWG promoter digested with BamHI/SphI as the molecular weight and positive control. Hybridization and wash conditions were as previously described (Rahman et al., 1997), using 25% formamide for hybridization.
Gene expression analyses
Total RNA from 10 dpa (days post-anthesis) rice grains was extracted using Trizol Reagent (Invitrogen). Long RNAs were purified using a Nucleospin miRNA extraction kit (Macherey-Nagel) and quantified using Nanodrop 1000 (Thermo Scientific). A total of 5 μg of long RNA template was used to synthesize cDNA using SuperScript III reverse transcriptase (Invitrogen). Quantitative real-time PCR (qRT-PCR) was done in a Rotor-Gene 6000 (Corbett) using 100 ng of cDNA template amplified using previously published branching enzyme and starch synthase primers (Hirose and Terao, 2004; Ohdan et al., 2005; Yamakawa et al., 2007). Real-time PCR amplification was conducted using Platinum Taq DNA polymerase (Invitrogen) and Sybr Green I (Invitrogen) reporter dye. Comparative quantitation was conducted using tubulin as a reference gene (Toyota et al., 2006), with data validation and melt curve analysis done using Rotor-Gene 6000 Series Real Time Rotary Analyzer Software (Corbett).
Protein expression analyses
Anti-rice SBEIIa was developed by conjugating the sequence IPAVAEASIKVVAED or AGAPGKVLVPG (GC was added to the C-terminal end of both peptides) to either keyhole limpet haemocyanin or ovalbumin. The antiserum raised in rabbits against AGAPGKVLVPG conjugated to ovalbumin was validated to be the most satisfactory and was used for the experiments described herein. On the other hand, anti-wheat SBEIIb rabbit polyclonal antibodies (Regina et al., 2005) which were shown by mass spectrometry to recognize SBEIIb in rice (unpublished data) were used for the western blot detection of SBEIIb.
Native soluble proteins were extracted as previously described (Regina et al., 2006). A total of 100 μg of protein quantified using Coomassie Protein Assay Reagent (Bio-Rad) was loaded into each lane. Two gels blotted separately were prepared to detect SBEIIa and SBEIIb individually using anti-SBEIIa and anti-SBEIIb antisera (1:2000 dilution). The immunoreactive proteins were probed by goat anti-rabbit immunoglobulins conjugated to horseradish peroxidase (Bio-Rad). Detection was carried out using ECL Western Blotting Detection Reagents (GE Healthcare) and Hyperfilm ECL chemiluminescence film (Amersham Biosciences). The film was developed using a CP 1000 automatic film processor (Agfa).
Branching enzyme zymograms were carried out as described (Nishi et al., 2001), with slight modifications. For each lane, 100 μg of total protein was loaded and resolved using native PAGE with 3.3% stacking and 5% separating layers. The gel was incubated overnight with gentle shaking at ambient temperature using a branching enzyme buffer (Nishi et al., 2003) containing 1 mM dithiothreitol (DTT) and 1.8 mM maltotriose. The addition of maltotriose was found to enhance the detection of branching enzyme activity (JP Ral, personal communication). The gel was stained with iodine solution (0.1% I2 and 1% KI) the following day. The reduction in branching frequency of debranched starch was determined based on a reducing end assay (Bernfeld, 1955) as modified by Regina et al. (2010).
Grain and starch granule analyses
Mature panicles were harvested and dried at 37 °C for at least 3 d. The seeds were then manually threshed and machine dehulled (Satake). Ten brown grains from selected lines were chosen and weighed in triplicate. Grain appearance and dimensions were determined using a SeedCount (SeedCount Australasia Pty Ltd), with the digital image analysis software module for medium grain rice. Opacity was measured using the chalkiness index for the Australian rice industry standard. Photomicrographs of whole rice grain samples were obtained using a Leitz M8 stereomicroscope.
Cross-sections of rice grains were observed uncoated with an environmental scanning electron microscope (Zeiss EVO LS15) under variable-pressure mode. Images of starch granules were taken with a back-scattered electron detector. Starch granules were isolated and viewed under a polarized light microscope to check for birefringence. The isolated starch granules were also stained with APTS (8-amino-1,3,6-pyrenetrisulphonic acid) and viewed under a fluorescence microscope as previously described (Wei et al., 2010a).
Characterization of starch crystallinity by X-ray diffraction (XRD) was carried out on a Panalytical X'Pert Pro diffractometer. The instrument was equipped with a Cu long fine focus tube, programmable incident beam divergence slit and diffracted beam scatter slit (both fixed at 0.125 °), and an X'celerator high speed detector. The samples were examined over the angular range of 4–40 ° with a step size of 0.0332 ° and a count time of 220 s per point. Crystallinity was determined using the crystal defect method as previously described (Lopez-Rubio et al., 2008).
Solid-state 13C cross-polarization/magic angle spinning (CP/MAS) nuclear magnetic resonance (NMR) experiments were performed at a 13C frequency of 75.46 MHz on a Bruker MSL-300 spectrometer. A standard of amorphous, regular maize starch was prepared by heating a starch suspension (1% w/v) for 30 min at 95 °C. The suspension was then lyophilized. Approximately 200 mg of rice flour was packed in a 4 mm diameter, cylindrical, partially stabilized zirconium oxide (PSZ) rotor with a KelF end cap. The rotor was spun at 5 kHz at the magic angle (54.7 °). The 90 ° pulse width was 5 μs and a contact time of 1 ms was used for all starches with a recycle delay of 3 s. The spectral width was 38 kHz, acquisition time 50 ms, time domain points 2 k, transform size 4 k, and line broadening 50 Hz. At least 1000 scans were accumulated for each spectrum. Spectra were referenced to external adamantane. Data fitting was carried out as previously described (Tan et al., 2007).
Carbohydrate analyses
Brown rice grains from selected lines were polished for 1 min using a fabricated machine with a circulating abrasive. Opaque or chalky seeds from each line were ground with a metal ball bearing for 30 s using an ESPE Capmix (3M). The amylose content of flour samples was determined by iodine colorimetry against a standard curve from five rice varieties whose actual amylose contents were determined by debranched size-exclusion chromatography (SEC; described below). Amylose estimations were done at two wavelengths: the traditional method using 620 nm (Juliano et al., 1981) and using 720 nm which was found to reduce the effect of long chain amylopectin on absorbance values (Fitzgerald et al., 2009). Peak gelatinization temperature (GT) was measured by differential scanning calorimetry (Cuevas et al., 2010).
The total starch content of rice flour (dry weight basis) was determined using a Megazyme total starch assay procedure (AACC Method 76.13) but adapted to a 96-microwell plate format. For samples which showed a reduction in total starch content compared with Nipponbare, resistant starch content was determined (AACC Method 32-40) using the 96-well plate format (Megazyme, Wicklow, Ireland). The resistant starch content was added to the amount of solubilized (non-resistant) starch to determine the actual total starch content.
Analysis of mixed-linkage β-glucan was conducted using a scaled-down standard method (Megazyme) with modifications, including using 50 mg of flour that was pre-washed in 1 ml of 70% ethanol (80 °C, 20 min) to remove free sugars before washing in 50% ethanol (80 °C, 5 min) and resuspending in 1 ml of sodium phosphate buffer (90 °C, 30 min). Subsequent incubations were performed at 42 °C, including with lichenase (2 U, 90 min) before a 20 μl aliquot of supernatant was taken (after 5 min centrifugation, 14 000 rpm) and addition of β-glucosidase (0.02 U μl−1, 30 min). After incubation with 200 μl of glucose oxidase/peroxidase reagent, the absorbance (510 nm) was used to calculate released glucose by blank subtraction. All flour incubations were shaken (1200 rpm) on a Thermomixer (Eppendorf).
Total pentosan content was determined colorimetrically as previously described by Bell (1985) with modifications. Following hydrolysis of samples with sulphuric acid (0.5 M, 30 min, 100 C), sugars were measured by adding 500 μl of freshly prepared reagent (55 ml of acetic acid, 1.1 ml of HCl, 0.6 ml of 0.7% glucose, and 2.4 ml of 25% phloroglucinol in ethanol) to 100 μl of sample containing 0–20 μg of pentose sugars (25 min, 100 °C); the mixture was then cooled for 5 min. The difference in absorbance (A552–A510) was used to calculate the pentosan content using a xylose standard and a conversion factor of 0.88 to express the pentose sugars in a polysaccharide form.
Determination of chain length distribution of amylopectin by fluorescence-activated capillary electrophoresis (FACE) was performed as previously described (O'Shea and Morell, 1996) with debranching as described by Ward et al. (2006). Molecular size distribution of debranched starch was determined by SEC as previously described (Ward et al., 2006; Cuevas et al., 2010). Two SEC columns were used: Ultrahydrogel 250 (Waters, Milford, MA, USA) which starts separating debranched starch at a DP of ∼2750, and Proteema (Polymer Standards Service GmbH, Mainz, Germany) which starts separating at DP ∼650. The molecular weight was estimated from the elution time using pullulan standards (Shodex P-82) calibrated with the Mark–Houwink–Sakaruda equation and universal calibration (Castro et al., 2005; Ward et al., 2006). A waxy rice was used to delineate the debranched amylose and amylopectin regions with a cut-off at DP 120 (Fitzgerald et al., 2009).
Resistant starch and glycaemic index predictions
The total resistant starch (RS) content and glycaemic index (GI) of freshly cooked polished rice grains were predicted using an in vitro incubation system which models the buccal, gastric, and pancreatic phases of food digestion as occurs in the human upper gastrointestinal tract. The methods have been extensively validated to have a high correlation with in vivo RS levels and GI values (AR Bird, S Usher, B Klingner, DL Topping, and MK Morell, unpublished data). Rice samples were prepared using the absorption method using a ratio of water to rice depending on amylose content (Supplementary Table S2 at JXB online). A total of 50 mg and 500 mg of available carbohydrates were used to predict GI and RS, respectively. For GI prediction, aliquots of supernatant were sampled at the designated time points for up to 5 h and the glucose concentration determined using an automated electrochemical procedure.
The total starch content of freshly cooked rice was determined based on a previously published protocol (McCleary et al., 1994) after freeze drying and milling into a fine powder. The predicted GI of the sample was calculated as the percentage of available carbohydrate converted to glucose and released during the time course of the incubation. For RS, the incubation period was extended to 16 h and the amount of starch remaining in the sample at that time was determined using conventional enzymatic and spectrophotometric techniques. The predicted RS content of the sample was calculated as the amount of starch remaining in the digest as a percentage of sample weight.
Neutral non-starch polysaccharides (NNSPs) were measured by a gas chromatographic technique using a slightly modified version of Theander et al. (1995) (AOAC 994.13). The insoluble and soluble NNSPs were separated by selective precipitation. The insoluble NNSPs were hydrolysed with 1 M sulphuric acid, while the soluble NNSPs were hydrolysed with 2 M trifluoroacetic acid.
Sampling and statistical analyses
Four generations of transformed lines (T0–T3) were selected for analysis. Negative segregants and empty vector Nipponbare callus regenerated by tissue culture were used as a negative controls for every generation. One-month-old leaves (T0 and T1) and mid-milk stage (10 dpa) grains (T1 and T2) were used for DNA and protein analyses, respectively. Homozygous T3 grains (10 dpa) were used for gene expression analyses. Developing T4 grains (5, 10, 15, and 20 dpa) were analysed by western blot and zymogram to compare the extent of protein down-regulation between ami-BEIIb and hp-BEIIb. Mature chalky to opaque grains were used for whole grain and starch analyses for three generations (T1, T2, and T3) planted at different seasons to assess stability of traits. At least three biological replicates from three independent transformed lines were used for every analysis, each with at least two technical replicates. IR36 and IR36ae were obtained from Yanco Agricultural Institute (NSW, Australia) and grown at the CSIRO Plant Industry (ACT, Australia).
Statistical analyses [one-way analysis of variance (ANOVA) with Tukey post-test, two-way ANOVA with Bonferroni post-test, and unpaired t-test] were done using GraphPad Prism Version 5.03. Standard error of the mean (SEM) was used to represent error values and error bars. Statistical significance was defined as P <0.05.
Results
Constructs and genomic DNA screening
Two RNA silencing constructs were cloned into pVec8 (Wang et al., 1998): an artificial microRNA (ami-BEIIb, Fig. 1A, B) and a hairpin RNA (hp-BEIIb, Fig. 1C), which were expressed in the rice endosperm using a wheat high molecular weight glutenin promoter. The selected amiRNA fragment has a predicted hybridization energy of –38.39 kcal mol−1, with a putative target cleavage site at positions 10 and 11, and mismatches at positions 1 and 21 (Fig. 1B).
Fig. 1.
Diagrammatic representation of the RNA silencing constructs (not drawn to scale). (A) A 21 nucleotide artificial microRNA (ami-BEIIb)-based osa-miR528 was synthesized by fusion PCR and cloned into Vec8, a Ti binary vector with a wheat high molecular weight glutenin promoter (wHMGPro) and nopaline synthase terminator (NOS). (B) The secondary structure of the osa-miR528 backbone as predicted by RNAfold, including information on ami-BEIIb (reverse complement). The predicted target cleavage site (arrow with sequence bold and highlighted) is located between positions 10 and 11 of the amiRNA, while the two mismatches (grey highlight) are located at positions 1 and 21. (C) The hairpin RNA (hp-BEIIb) was cloned in Vec8 by inserting a 397 bp BEIIb fragment in the sense (BEIIb→) and antisense (BEIIb←) orientations. The two fragments are flanked by two rice introns, Rint4 and Rint9, which form a hairpin loop. The amiRNA and hp-RNA fragments were directionally cloned using several restriction sites (H, HindIII; B, BamHI; K, KpnI; N, NotI; E EcoRI; S, SpeI).
PCR screening of T0 plants revealed that ∼75% of the putative transformants contained the constructs (Supplementary Fig. S1 at JXB online). The transgenic plants stably retained the hp-BEIIb and ami-BEIIb transgene in subsequent generations (T1–T4). Southern blot analyses of selected T1 plants revealed an insertion of one copy of each silencing construct for most of the hp-BEIIb and amiRNA plants tested (data not shown). Segregating PCR-positive T1 seeds (∼3:1 ratio based on grain appearance) were planted to obtain homozygous seeds. Four lines of hp-BEIIb and five lines of ami-BEIIb were homozygous in the T2 generation and this was verified up to T4. These lines were used in the following experiments.
Gene expression analyses
Gene expression analyses of homozygous T3 lines by real-time PCR revealed a >5-fold decrease in the expression of SBEIIb in ami-BEIIb lines, while only a 2-fold decrease was observed in transgenic lines harbouring hp-BEIIb (Fig. 2). The expression of transcripts for other starch branching isoforms (SBEI and SBEIIa) and major starch synthase isoforms (SSI, SSIIa, and SSIIIa) was unaffected (Fig. 2).
Fig. 2.
Gene expression analyses by quantitative RT-PCR. The expression of SBEIIb was reduced 5-fold in ami-BEIIb lines and only 2-fold in hp-BEIIb lines. The expression of other branching enzymes and major starch synthases was unaffected. The rice tubulin gene was used as a reference (Toyota et al., 2006) for comparative quantitation. Mean values with different letters are significantly different. Error bars indicate the SEM.
Enzyme expression and activity detection
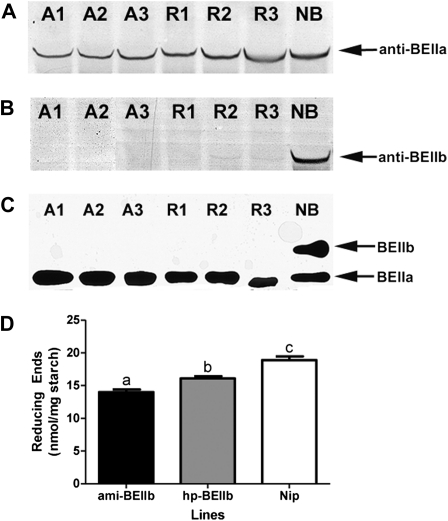
Western blot analyses showed trace levels of SBEIIb protein in the hp-BEIIb lines at 10 dpa, which was undetectable in the amiRNA lines (Fig. 3A), while the wild type had considerably more SBEIIb. The levels of SBEIIa remained similar to those of the wild-type Nipponbare for both the ami-BEIIb and the hp-BEIIb lines (Fig. 3B). This was verified by in-gel activity staining of branching enzymes, which showed that SBEIIb activity was only detectable in Nipponbare but not in the transgenic lines, while the SBEIIa activity remained unaffected (Fig. 3C). The activity of SBEI was unaffected (data not shown).
Fig. 3.
Enzyme expression and activity detection using western blots (A and B) and a zymogram (C) using 10 dpa grains. The levels of BEIIb (A) in rice endosperm are down-regulated to undetectable levels in ami-BEIIb (A1–A3) and to trace amounts in hp-BEIIb (R1–R3) lines. The levels of BEIIa (B) are similar for the transgenic and parental Nipponbare (NB) lines. Enzyme activity detection (C) shows that BEIIb activity is only detectable in NB but not in the transgenic lines, while BEIIa activity remains intact. This result is corroborated by reducing end assay (D) which shows a decreased concentration of reducing ends in the transgenic lines which is more pronounced in ami-BEIIb. Mean values with different letters are significantly different. Error bars indicate the SEM.
Western blot detection of developing T4 endosperm revealed that one homozygous line of ami-BEIIb had stably down-regulated and almost undetectable amounts of SBEIIb at 5, 10, 15, and 20 dpa, while one homozygous hp-BEIIb line had a down-regulated but higher concentration of SBEIIb (Supplementary Fig. S2 at JXB online). In contrast, the levels of SBEIIa in both lines were similar to those of the Nipponbare control at each stage (Supplementary Fig. S2). The decrease in branching enzyme activity due to SBEIIb down-regulation led to reduced branching frequency, with a decrease in the number of reducing ends per milligram of starch from ∼19 in Nipponbare wild type to 14 in the ami-BEIIb lines (Fig. 3D).
Grain and starch granule analyses
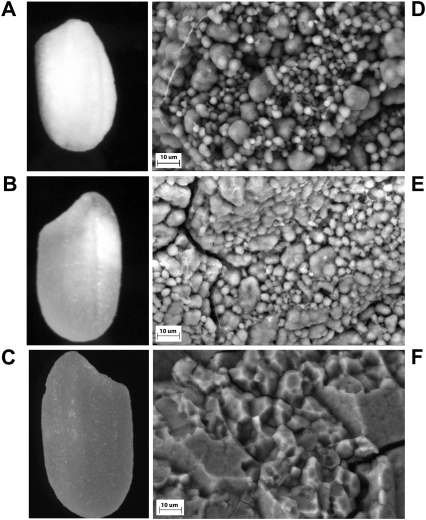
Polished grains of ami-BEIIb lines were opaque throughout (Fig. 4A), while polished grains of the hp-BEIIb lines were chalky, showing streaks of white along the translucent endosperm (Fig. 4B). In comparison, Nipponbare grain appeared uniformly translucent (Fig. 4C). Grain weight, width, and thickness of the transgenic lines were lower than those of Nipponbare (Table 1). Grain yield of the transgenic lines was also lower than for Nipponbare (data not shown).
Fig. 4.
Grain and starch granule morphology of transgenic lines. The polished grain of ami-BEIIb (A) appeared chalky, and that of hp-BEIIb (B) had some chalky character, compared with the translucent grains of Nipponbare tissue culture control (C). At ×1000 magnification, the starch granules of the transformed lines are loose and rounded (D and E), compared with the tight and angular granules in the control (F). Big and small rounded starch granules were observed in the transformed lines. The difference in starch granule morphology appears to be more pronounced in the ami-BEIIb lines (D). Actual grain dimensions are reported in Table 1.
Table 1.
Comparison of generated transgenic lines with their parent Nipponbare Values reported are means ± SEM. Mean values with different letters are significantly different
| Properties | ami-BEIIb lines | hp-BEIIb lines | Nipponbare |
| Ten brown grain weight (mg) | 140.3±8.5 a | 144.2±5.7 a | 205.2±2.3 b |
| Length (mm) | 5.2±0.1 | 5.1±0.0 | 5.2±0.0 |
| Width (mm) | 2.7±0.1a | 2.7±0.1 a | 2.9±0.0 b |
| Thickness (mm) | 2.0±0.0 a | 2.0±0.0 a | 2.2±0.0 b |
| Chalkiness (% per grain) | 75–100 | 25–50 | 0–10 |
| Total starch, flour samples (g 100 g−1) | 91.4±1.7* | 91.0±0.4 | 90.4±0.9 |
| Total starch, freshly cooked grains (g 100 g−1) | 86.9±0.6 | 86.0±0.4 | 85.5±0.0 |
| Apparent amylose content (%) at 620 nm | 41.2±0.5 a | 34.0±1.6 b | 19.6±0.7 c |
| Apparent amylose content (%) at 720 nm | 16.9±0.4 a | 13.7±0.8 b | 15.2±0.2 a,b |
| Pentosans (%) | 0.31±0.02 a | 0.25±0.01 a | 0.05±0.01 b |
| β-Glucan (%) | 0.07±0.01 | 0.05±0.01 | 0.05±0.01 |
| Neutral non-starch polysaccharides (g 100 g−1) | 1.5±0.2 a | 0.8±0.1 b | 0.8±0.0 b |
| Peak gelatinization temperature (°C) | 80.7±0.0 a | 75.0±1.2 b | 71.2±0.5 b |
*Of which, 9.5±0.8 g 100 g−1 are resistant starch.
Scanning electron micrographs of ami-BEIIb (Fig. 4D) and hp-BEIIb (Fig. 4E) lines revealed big and small rounded starch granules with large spaces in between, exhibiting a loss of compound granular organization. In contrast, the starch granules of Nipponbare appeared compact, compound, and angular, with fewer spaces in between (Fig. 4F). Changes in starch granule morphology were more pronounced in ami-BEIIb (Fig. 4D) than in hp-BEIIb (Fig. 4E). Isolated starch granules of hp-BEIIb and ami-BEIIb stained with APTS did not show any distinct difference from the wild type, and all showed normal birefringence under polarized light (data not shown).
XRD analyses showed that some hp-BEIIb lines retained the A-type starch crystalline polymorph (similar to the Nipponbare as well as another wild type, IR36) while others showed a mixture of A- and B-type crystallinity, also known as C-type starch (Fig. 5). In contrast, all ami-BEIIb lines fully shifted to B-type crystallinity, as also found in the SBEIIb mutant IR36ae (Fig. 5). These differences were also consistent with solid-state 13C CP/MAS NMR analyses (Supplementary Fig. S3 at JXB online).
Fig. 5.
XRD patterns of starches with down-regulated SBEIIb and its comparison with the wild type and the amylose extender mutant. Two hp-BEIIb lines (hp-BEIIb A-type) share the same A-type crystalline polymorph (peak at 18 °, but not 5 °, 2θ) with the wild-type rices (Nipponbare and IR36) while all four ami-BEIIb shifted to a B-type polymorph (peak at 5 °, but not 18 °, 2θ) similar to IR36ae. Two hp-BEIIb showed a crystalline structure that is intermediate between A- and B-type (hp-BEIIb C-type). Data are off set for clarity.
Starch structural analyses
The total starch content of hp-BEIIb and ami-BEIIb lines was comparable with that of the control at ∼90%, although the ami-BEIIb lines contain a greater proportion of starch resistant to enzymatic breakdown (Tables 1, 2). When measured at 620 nm, a >2-fold increase in AAC was observed in ami-BEIIb compared with Nipponbare (Table 1). The AAC levels of the hp-BEIIb lines were also significantly elevated compared with the control, but not as high as those of the ami-BEIIb lines (Table 1). However, when measured at 720 nm, the amylose content of ami-BEIIb lines was comparable with that of Nipponbare (Table 1). The hp-BEIIb lines showed a slight reduction, though this was not significant (Table 1).
Table 2.
Nutritional properties of freshly cooked grains Values reported are means ±SEM. Mean values with different letters are significantly different.
| Lines | Resistant starch cooked grains (g 100 g−1) | Predicted glycaemic index |
| Down-regulated BEIIb lines | ||
| ami-BEIIb | 4.8±0.2 a | 44±1 a |
| hp-BEIIb | 0.4±0.2 b | 79±8 b |
| Nipponbare | 0.2±0.0 b | 85±2 b |
| amylose extender mutant | ||
| IR36ae | 3.1±0.0 c | 54±2 a |
| IR36 | 0.7±0.4 d | 68±1 b |
Chain length distributions (CLDs) of debranched starches revealed a decrease in the proportion of amylopectin short chains (DP 6–12) with a concomitant increase in the longer chains (DP ≥14) in both the amiRNA (Fig. 6A) and hp-RNA (Fig. 6B) lines. The decrease in the ratio of short chains was more pronounced in the amiRNA lines than in the hp-BEIIb lines (>4% versus <3% for DP 9 and 10) (Fig. 6C). The same trend was observed for the increase of longer amylopectin chains (Fig. 6C). The increase reached up to 0.9% at DP 19 for the amiRNA lines, while the corresponding increase for the hp-BEIIb lines was 0.7%. The CLD of ami-BEIIb (but not hp-BEIIb) was very similar to the amylose extender mutant IR36ae (Supplementary Fig. S4 at JXB online).
Fig. 6.
Chain length distribution (CLD) profile of debranched starch (A and B) and mol% difference of ami-BEIIb and hp-BEIIb compared with Nipponbare (C). Compared with Nipponbare, ami-BEIIb (A) and hp-BEIIb (B) have reductions in DP 6–12 and an increase in DP 13 onwards. A difference plot (C) revealed that the reduction in short DP is more pronounced in ami-BEIIb (up to 4%) than in hp-BEIIb (up to 3%). Likewise, the increase in longer DPs is more pronounced in ami-BEIIb (up to 0.9%) than in hp-BEIIb (up to 0.7%). Error bars indicate the SEM.
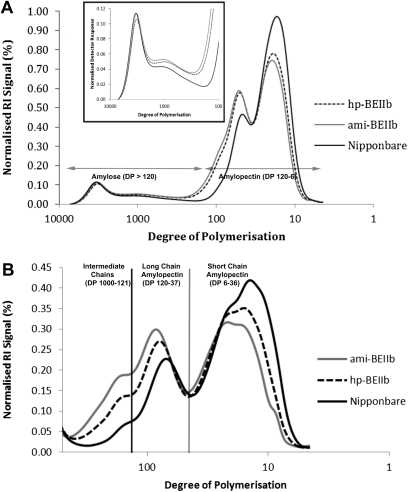
The molecular size distribution of debranched starch showed that the proportion of very long chains (DP >1000) characteristic of linear or lightly branched amylose was not altered in either hp-BEIIb or amiRNA lines (Fig. 7). The proportion of debranched amylose chains (Supplementary Table S3 at JXB online) roughly corresponds to the proportion of apparent amylose as measured at 720 nm (Table 1). Furthermore, an increase in the proportion of chains greater than DP ∼36 but <1000 in the transgenic lines compared with the wild type was observed and this was more pronounced for the ami-BEIIb lines (Fig. 7, Supplementary Table S3). There was also a reduction in debranched short amylopectin chains (DP 6–36) in the transgenic lines, which was again more pronounced in ami-BEIIb lines (Fig. 7, Supplementary Table S3). These results are consistent with the increase in the average chain length in the ami-BEIIb samples to DP ∼400 compared with DP ∼300 in the Nipponbare samples as deduced from the number of reducing ends per milligram of starch (Fig. 3D).
Fig. 7.
Debranched HP-SEC of transformed lines compared with Nipponbare using (A) Ultrahydrogel 250 and (B) Proteema columns. Intermediate length chains (apparent DP ≥37) were increased while the short amylopectin chains (DP ≤36) were decreased in both hp-SBEIIb and ami-BEIIb, with the changes more pronounced in the latter. No change in the proportion of long chain amylose peak (apparent DP ≥1000) was observed, while the mutants have elevated amounts of intermediate material—longer than traditionally found in amylopectin (DP 6–120) and shorter than classic long chain amylose (inset).
Peak GT was significantly increased in ami-BEIIb compared with the wild type (Table 1). The mean GT of hp-BEIIb was slightly elevated but the difference was not significant (Table 1).
Nutritional properties
The in vitro RS content of freshly cooked ami-BEIIb grains was 10-fold higher than that of Nipponbare (Table 2). In comparison, a 4-fold increase was observed for IR36ae compared with its parent IR36 (Table 2). Moreover, the RS content of ami-BEIIb grains was also considerably higher than that of IR36ae. On the other hand, the RS values obtained for hp-BEIIb were not significantly different from the control (Table 2).
The predicted GI for ami-BEIIb was reduced compared with Nipponbare and the SBEIIb mutant, IR36ae (Table 2). The predicted GI of IR36ae was also decreased compared with its parent IR36. The GI estimate for hp-BEIIb was also reduced, but this was not significantly different from the control (Table 2).
The NNSP content of ami-BEIIb lines was approximately twice that of hp-BEIIb and Nipponbare (Table 1), while the level of pentosans was significantly greater in ami-BEIIb and in hp-BEIIb lines compared with the control. On the other hand, the levels of β-glucans remained unchanged (Table 1).
Discussion
In the present study, the down-regulation of SBEIIb gene expression (Fig. 2) in the rice endosperm was achieved using an amiRNA (Fig. 1A, B) and a hp-RNA (Fig. 1C). This led to the reduction of SBEIIb protein, SBEIIb enzymatic activity, and the abundance of reducing ends present in debranched starch (Fig. 3). SBEIIa expression was unaffected (Fig. 3). The down-regulation of SBEIIb was further confirmed using endosperm at different stages of development, which also showed that SBEIIa expression was unaffected (Supplementary Fig. S2 at JXB online). The SBEIIa and SBEIIb isoforms are encoded by separate genes in maize, wheat, and rice (Fisher et al., 1996). In wheat, the down-regulation of SBEIIa leads to the down-regulation of SBEIIb (Regina et al., 2006). In the current work, the mRNA and protein levels as well as the enzymatic activity of SBEIIa were not affected (Figs 3, 4), similar to what is observed in an ae maize (Fisher et al., 1996; Gao et al., 1997) and transgenic barley with down-regulated BEIIb (Regina et al., 2010). Furthermore, the expression of BEI and other major starch synthase isoforms were also unaffected (Fig. 3). Even the expression of SSI, which is slightly down-regulated in an ae mutant of rice (Nishi et al., 2003). and corn (Boyer and Preiss, 1978), was not altered (Fig. 2), in either the hairpin or amiRNA lines. These results indicate that RNA silencing (Fig. 1) can be gene- and isoform-specific, given a unique target sequence and an accurate target-finding algorithm such as Web microRNA Designer (Ossowski et al., 2008).
Comparison of artificial microRNA and hairpin RNA techniques
RNA interference using hp-RNA was found to be effective in down-regulating branching enzymes in wheat (Regina et al., 2006; Sestili et al., 2010) and in barley (Regina et al., 2010), as also demonstrated in this study (Figs 1–4). This study also established that amiRNA (Fig 1A, B) is very efficient in attenuating SBEIIb expression in rice grains (Fig. 2), demonstrating the isoform specificity and efficacy of this technique in regulating gene expression in the rice endosperm.
The amiRNA technique (Fig. 1A, B) was more effective in reducing SBEIIb gene expression (Fig. 2) and in producing more extreme starch properties (Figs 5–7, Tables 1, 2) than the hp-RNA technique for the traits assessed here. The hp-BEIIb lines, especially those that retained the A-type polymorph (Fig. 5), might not have exceeded an RNA inhibition threshold necessary to achieve the phenotypes produced by the ami-BEIIb lines. This was supported by the observation that the amount of SBEIIb was more stably reduced in developing endosperm of an ami-BEIIb line compared with an hp-BEIIb line (Supplementary Fig. S2 at JXB online). Very high amylose was not achieved in potato because the necessary RNA inhibition threshold was also not exceeded (Safford et al., 1998).
One advantage of the amiRNA technique is that mismatches can be deliberately introduced in its 21 nucleotide sequence (Fig. 2) to facilitate not only mRNA cleavage (Kasschau et al., 2003; Bagga et al., 2005; Schwab et al., 2005) but also translational repression (Aukerman and Sakai, 2003; Chen, 2004). Such mRNA cleavage and translational repression is consistent with the observation of the 4-fold reduction in the gene expression of SBEIIb (Fig. 2) and the undetectable levels of SBEIIb protein and enzymatic activity (Fig. 3, and Supplementary Fig. S2 at JXB online).
Effects of down-regulating SBEIIb expression on starch structure
The ‘amylose’ content of both ami-BEIIb and hp-BEIIb lines was double that of the control when measured at 620 nm (Table 1), the wavelength routinely used for apparent amylose content estimation (Juliano et al., 1981). However, the molecular size distribution of debranched starch revealed that down-regulating BEIIb, at least in a japonica background like Nipponbare where GBSSI is less active, does not increase the amount of the very long amylose chains (Fig. 7, and Supplementary Table S3 at JXB online). Instead, the reduction in the amount of BEIIb alters starch properties by increasing the proportion of long amylopectin chains (DP 37–120) and decreasing the proportion of short amylopectin chains (DP 6–36) (Fig. 7, and Supplementary Table S3). ‘Long amylopectin’ chains here refer to the maximum DP spectrum observed in debranched starch in the absence of a functional GBSSI. Based on the result of Fitzgerald et al. (2009), debranched chains observed in the absence of GBSSI, as in waxy rice lines, have a maximum DP of ∼120. In the same study, much higher DPs are found in debranched starch of rice lines where GBSSI is catalytically active, which can reach beyond DP 7000. The general shift to longer branch lengths for ami-BEIIb compared with hp-BEIIb seen in the SEC data (Fig. 7) can also be observed in the FACE data (Fig. 6). Moreover, the proportion of intermediate chains from DP 120–1000 was also elevated (Fig. 7, and Supplementary Table S3), consistent with either significantly elevated levels of amylopectin B3 and B4 chains (Hizukuri, 1986) or the presence of short amylose branches. Longer average chain lengths and increased proportions of intermediate fractions were also observed in ae mutants of maize (Ikawa et al., 1978; Yeh et al., 1981).
The total proportion of long amylopectin and intermediate chains was 13% higher than Nipponbare for hp-BEIIb and 18% for ami-BEIIb (Supplementary Table S3 at JXB online). Whether these debranched intermediate chains—longer than traditionally found in amylopectin and shorter than classic long chain amylose—are attached to amylopectin or amylose, it is likely that they are the origin of the doubling of apparent amylose content when measured using 620 nm (Table 1). No difference in amylose levels was observed in the transgenic lines compared with Nipponbare when the estimation was done at 720 nm (Table 1). It is possible that these intermediate chains complex with iodine in a similar fashion to amylose, as they represent the main structural difference between the transgenic starches compared with the wild type (Figs 6, 7). If these intermediate chains connect clusters of branching density similar to those of the wild type, then little change in iodine binding would be expected due to the disruptive influence of branch points on the formation of the iodine complex. However, if these intermediate chains are lightly branched, they could complex with iodine, explaining the large increase in amylose content in the transgenic lines (Table 1). It is therefore likely that the increased proportion of intermediate chains is associated with lightly branched structures so that that iodine binding occurs similar to linear amylose.
Effects of amylopectin structure on starch granule morphology and crystallinity
The ae mutants in maize are often associated with slight reductions in grain dimensions and weight (Shannon et al., 2009), as also observed in this study (Table 1). Additionally, the opacity or chalkiness in grains (Fig. 4A, B, Table 1) is consistent with previous observations (Nishi et al., 2001; Sawada et al., 2009). The starch granules of the recombinant lines (Fig. 4D, E) appeared loose and spherical and seemed to have lost the higher level of organization into compound granules that is seen in the control (Fig. 5F). The increase in apparent amylose content also associates with decreases in the proportion of angular starch granules and increases in the proportion of elongated forms in maize (Banks et al., 1974). Furthermore, chains intermediate between amylose and amylopectin can distort starch granule morphology (Blennow et al., 2003). The differences in granular packing could therefore be due to the altered molecular structure of the transgenic starches (Figs 6, 7).
A-type crystallinity is commonly found in cereal starches, but mutations in SBEIIb often lead to B-type crystallinity (Gerard et al., 1999; Tanaka et al., 2004; Kubo et al., 2008). Interestingly, in the present study, the ami-BEIIb starches completely shifted to a B-type, while hp-BEIIb starches retained the A-type, or exhibited the intermediate C-type (Fig. 5, and Supplementary Fig. S3 at JXB online). The results obtained by Tanaka et al. (2004) are illuminating in this context. They reported that the absence of BEIIb in the mutant EM10 resulted in B-type crystallinity but when the mutation was complemented by the introduction of a BEIIb transgene, the starch crystallinity changed from B to C to A, depending on the strength of transgene expression. In the results of this present study, it is clear that the ami-BEIIb lines have more reduced SBEIIb expression than the hp-BEIIb lines (Fig. 3, and Supplementary Fig. S2). The reduction in the ami-BEIIb is sufficient to phenocopy the EM10 and 7.8 lines reported by Tanaka et al. (2004), whereas the reduction in the hp-BEIIb lines phenocopies lines 9.8 (C type) and 106-1 (A type). Furthermore, the difference in crystalline polymorphic form between ami-BEIIb and hp-BEIIb is consistent with the slight increase in longer amylopectin branches (Hizukuri et al., 1983) in the ami-BEIIb lines compared with hp-BEIIb lines, as this could cause a shift from the thermodynamically more stable A form to the kinetically trapped B form (Gidley, 1987). A difference in chains of DP 6–12 has also been associated with a shift from the A-type to B-type polymorph in several cereals (Hanashiro et al., 1996). Although it is not possible in this study to separate the effects of the changes in long amylopectin chains from the changes in the short amylopectin chains, it appears that rice starch exists at the boundary of the structural requirements for A- versus B-type crystallinity, and that only a relatively small change in amylopectin branch lengths of rice starch is sufficient for this transition.
Wei et al. (2010a) reported that simultaneous antisense inhibition of SBEI and SBEIIb (TRS line) only led to the intermediate C-type crystalline polymorph, which may indicate insufficient down-regulation of branching activity as also observed in hp-BEIIb lines (Figs 2, 3). However, alterations in starch granule morphology produced in the TRS line are more extreme that what was observed in this study (Wei et al., 2010a, b, c, d), maybe because the antisense inhibition was done in a high amylose background (Teqing). Elevated proportions of amylose chains might introduce additional perturbations in granule packing in this variety.
Effects of amylopectin structure on functional and predicted nutritional properties
It is known that ae starch granules are more resistant to thermal gelatinization than the wild type, at least in part due to alterations in amylopectin fine structure (Tanaka et al., 2004; Shannon et al., 2009). The decrease in short chains and the increase in longer chains (Figs 6, 7, and Supplementary Table S3 at JXB online) observed in the ami- and the hp-BEIIb lines is consistent with previous observations (Nishi et al., 2001) and with the proposed branching role of SBEIIb in the crystalline lamellae of the amylopectin cluster in the rice endosperm (Nakamura, 2002; Nakamura et al., 2010). Although B-type crystallites melt at lower temperature than A-type crystallites of the same chain length (Whittam et al., 1990), it is probable that the higher peak GT in ami-BEIIb is due to the elevated proportions of long amylopectin and intermediate chains as compared with hp-BEIIb (Fig. 7, and Supplementary Table S3). It was found that an ∼6% decrease in the proportion of chains of DP 6–12 relative to chains of DP 12–24 can lead to an increase in GT of at least 10 °C in waxy rices (Cuevas et al., 2010).
High amylose rice mutants, including the lines developed in this study, are interesting from a nutritional perspective because they are typically shown to contain higher levels of dietary fibre and/or resistant starch (Lee et al., 2006; Shu et al., 2006; Yang et al., 2006). The alterations in starch digestibility are consistent with the observed alterations in starch structure as it is known that linear amylose chains of DP ∼100 have the fastest rate of double helix formation and retrogradation in water of any starch polymer (Gidley and Bulpin, 1989). Increases in the proportion of DP 100 chains were observed (Fig. 7, and Supplementary Table S3 at JXB online).
In the lines described here, there is a 20-fold increase in the proportion of resistant starch in the amiRNA lines compared with hp-BEIIb and wild-type lines (Table 2). In fact the ami-BEIIb lines have higher RS content than the traditional amylose extender mutant IR36ae (Table 2). This is despite the fact that ami-BEIIb is in a japonica background with intermediate to low amylose content (Nipponbare), while IR36ae is in an indica background with a high amylose content (IR36). Likewise, the ami-BEIIb lines have lower GI estimates compared with IR36ae (Table 2), despite the former having a lower proportion of true amylose chains. This strongly suggests that aside from amylose, intermediate chains and long chain amylopectin molecules (Fig. 7) may play an important structural role in rendering the starch molecule less digestible. In particular, it appears that elevations in the proportion of intermediate chains may be associated with increase in the levels of resistant starch in rice.
Implications for starch biosynthetic pathways in cereals
While the rice mutants obtained by traditional mutagenesis techniques in previous studies as well as the recombinant lines obtained in this study have high apparent amylose contents compared with common cultivated varieties, these are significantly lower than apparent amylose contents that have been reported for analogous mutations in maize, wheat, and barley. In ae maize, lack of BEIIb is thought to be partially complemented by BEIIa and BEI (Liu et al., 2009). The same mechanism might also exist in rice. However, maize can produce up to 50–80% amylose when SBEIIb is altered, with levels increasing due to the presence of some modifier genes (Shannon et al., 2009). Another reason for this may be that in maize, the proportion of SBEIIb to SBEIIa in the endosperm is 50:1 (Gao et al., 1997), whereas in rice the ratio is closer to 5:1 (Regina et al., 2006).
In contrast to maize, down-regulation of SBEIIb alone did not significantly increase the levels of amylose in either wheat or barley (Regina et al., 2006, 2010). This is because SBEIIa is expressed at much higher levels in the grains of both wheat and barley (Regina et al., 2005, 2010), than in maize and even rice (Gao et al., 1997; Mizuno et al., 2001; Ohdan et al., 2005). The differences in expression levels of the SBEIIa and SBEIIb enzymes in cereals therefore appear to define which of these two enzymes should be down-regulated to generate high amylose starches. Furthermore, since down-regulating BEIIa in wheat produces high amylose (Regina et al., 2006) and down-regulating both BEIIa and BEIIb (Regina et al., 2010) or SSIIa alone (Morell et al., 2003) is necessary to produce high amylose in barley, it appears that producing very high amylose in rice might require modification of the expression of a different combination of target isoforms. Even the simultaneous down-regulation of SBEI and SBEIIb only resulted in a doubling of the apparent amylose content despite significant alterations in starch granule morphology (Wei et al., 2010a, b, c, d). The use of amiRNAs, which has been successfully demonstrated in this study to be effective in producing a more extreme starch phenotype than the conventional hp-RNA constructs, may be helpful in silencing other genes to produce very high amylose rice grains. Comparison of the efficacy of these techniques in down-regulating other genes in the cereal endosperm will be important to better understand the relative efficacy of the amiRNA and hp-RNA approaches.
Conclusion
The ae phenotype can be obtained by down-regulating the expression of SBEIIb in rice using RNA silencing. For the first time, the amiRNA technique was demonstrated to facilitate RNA silencing in rice endosperm and was better at producing a more extreme starch and nutritional phenotype than the hp-RNA technique in these experiments. For rice SBEIIb, the greater effectiveness of the amiRNA technique resulted in distinct differences in rice starch chemistry, crystallinity, and digestibility compared with the hp-RNA technique. However, in order to obtain very high amylose rice grains, the expression of multiple starch-synthesizing enzymes may need to be down-regulated simultaneously.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. PCR screening of transgenic lines.
Figure S2. Western blot detection of SBEIIa and SBEIIb in developing endosperms of ami-BEIIb and hp-BEIIb compared with Nipponbare at four developmental stages.
Figure S3. 13C NMR CP/MAS spectrum and deconvolution of an ordered subspectrum of rice flour samples.
Figure S4. Chain length distribution difference (mol%) of generated transgenic lines as compared with IR36ae.
Table S1. PCR primers used.
Table S2. Amylose classification and water to raw rice ratio used during the absorption method of resistant starch and glycaemic index estimations.
Table S3. Molecular size distribution of debranched amylose and amylopectin chains of transgenic lines compared with Nipponbare.
Acknowledgments
The authors thank Norman Warthmann and Detlef Weigl for providing the osa-miR528 amirRNA backbone. Ivan Moore produced the rice anti-BEIIa polyclonal antibodies. Robin Chapple is acknowledged for carrying out the β-glucan assay, Bradley Klingner for performing the in vitro RS and GI tests, and Rosa Paula Cuevas for running the DSC measurements. Mark Talbot is also acknowledged for his assistance during the scanning electron microscopy experiments. VMB is a recipient of an Australian Leadership Award Scholarship.
References
- Asaoka M, Okuno K, Sugimoto Y, Yano M, Omura T, Fuwa H. Characterization of endosperm starch from high-amylose mutants of rice (Oryza sativa L) Starch. 1986;38:114–117. [Google Scholar]
- Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. The Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Ball SG, Morell MK. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annual Review of Plant Biology. 2003;54:207–233. doi: 10.1146/annurev.arplant.54.031902.134927. [DOI] [PubMed] [Google Scholar]
- Ball SG, van de Wal M, Visser RGF. Progress in understanding the biosynthesis of amylose. Trends in Plant Science. 1998;3:462–467. [Google Scholar]
- Banks W, Greenwood CT, Muir DD. Studies on starches of high amylose content. Part 17. A review of current concepts. Starch. 1974;26:289–300. [Google Scholar]
- Bell BM. A rapid method of dietary fiber estimation in wheat products. Journal of the Science of Food and Agriculture. 1985;36:815–821. [Google Scholar]
- Bernfeld P. Amylases, alpha and beta. Methods in Enzymology. 1955;1:149–158. [Google Scholar]
- Blennow A, Hansen M, Schulz A, Jorgensen K, Donald AM, Sanderson J. The molecular deposition of transgenically modified starch in the starch granule as imaged by functional microscopy. Journal of Structural Biology. 2003;143:229–241. doi: 10.1016/j.jsb.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Boyer CD, Damewood PA, Matters GL. Effect of gene dosage at high amylose loci on the properties of the amylopectin fraction of the starches. Starch. 1980;32:217–222. [Google Scholar]
- Boyer CD, Daniels RR, Shannon JC. Abnormal starch granule formation in Zea mays L. endosperms possessing amylose extender mutant. Crop Science. 1976;16:298–301. [Google Scholar]
- Boyer CD, Preiss J. Evidence for independent genetic control of starch branching enzymes from developing maize kernels. Plant Physiology. 1978;61:39. doi: 10.1104/pp.67.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butardo V, Fitzgerald M, Rahman S, Gidley M. Efforts to capture high amylose in rice. 2008 AACC International Annual Meeting: Diversity of Grains. 2008;Vol. 53 Hawaii Convention Center, Honolulu, Hawaii, USA: Cereal Food World, A15. [Google Scholar]
- Castro JV, Ward RM, Gilbert RG, Fitzgerald MA. Measurement of the molecular weight distribution of debranched starch. Biomacromolecules. 2005;6:2260–2270. doi: 10.1021/bm050041t. [DOI] [PubMed] [Google Scholar]
- Chen XM. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas RP, Daygon VD, Corpuz HM, Nora L, Reinke RF, Waters DLE, Fitzgerald MA. Melting the secrets of gelatinisation temperature in rice. Functional Plant Biology. 2010;37:439–447. [Google Scholar]
- Fisher DK, Gao M, Kim KN, Boyer CD, Guiltinan MJ. Allelic analysis of the maize amylose-extender locus suggests that independent genes encode starch-branching enzymes IIa and IIb. Plant Physiology. 1996;110:611–619. doi: 10.1104/pp.110.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald MA. Starch. In: Champagne ET, editor. Rice chemistry and technology. St Paul, MN: AACC; 2004. [Google Scholar]
- Fitzgerald MA, Bergman CJ, Resurreccion AP, et al. Addressing the dilemmas of measuring amylose in rice. Cereal Chemistry. 2009;86:492–498. [Google Scholar]
- Gao M, Fisher DK, Kim KN, Shannon JC, Guiltinan MJ. Independent genetic control of maize starch-branching enzymes IIa and IIb. Isolation and characterization of a Sbe2a cDNA. Plant Physiology. 1997;114:69–78. doi: 10.1104/pp.114.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwood DL, Shannon JC, Creech RG. Starches of endosperms possessing different alleles at amylose-extender locus in Zea mays L. Cereal Chemistry. 1976;53:355–364. [Google Scholar]
- Gerard C, Planchot V, Buleon A, Colonna P. Crystalline structure and gelatinisation behaviour of genetically modified maize starches. Biopolymer Science: Food And Non Food Applications. 1999;91:59–63. [Google Scholar]
- Gidley MJ. Factors affecting the crystalline type (A–C) of native starches and model compounds: a rationalisation of observed effects in terms of polymorphic structures. Carbohydrate Research. 1987;161:301–304. [Google Scholar]
- Gidley MJ, Bulpin PV. Aggregation of amylose in aqueous systems: the effect of chain-length on phase-behavior and aggregation kinetics. Macromolecules. 1989;22:341–346. [Google Scholar]
- Hanashiro I, Abe J, Hizukuri S. A periodic distribution of the chain length of amylopectin as revealed by high-performance anion-exchange chromatography. Carbohydrate Research. 1996;283:151–159. [Google Scholar]
- Hanashiro I, Itoh K, Kuratomi Y, Yamazaki M, Igarashi T, Matsugasako JI, Takeda Y. Granule-bound starch synthase I is responsible for biosynthesis of extra-long unit chains of amylopectin in rice. Plant and Cell Physiology. 2008;49:925–933. doi: 10.1093/pcp/pcn066. [DOI] [PubMed] [Google Scholar]
- Hedman KD, Boyer CD. Gene dosage at the amylose-extender locus of maize—effects on the levels of starch branching enzymes. Biochemical Genetics. 1982;20:483–492. doi: 10.1007/BF00484699. [DOI] [PubMed] [Google Scholar]
- Hirose T, Terao T. A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.) Planta. 2004;220:9–16. doi: 10.1007/s00425-004-1314-6. [DOI] [PubMed] [Google Scholar]
- Hizukuri S, Kaneko T, Takeda Y. Measurement of the chain-length of amylopectin and its relevance to the origin of crystalline polymorphism of starch granules. Biochimica et Biophysica Acta. 1983;760:188–191. [Google Scholar]
- Hizukuri S. Polymodal distribution of the chain lengths of amylopectins, and its significance. Carbohydrate Research. 1986;147:342–347. [Google Scholar]
- Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatshefte fur Chemie. 1994;125:167–188. [Google Scholar]
- Ikawa Y, Glover DV, Sugimoto Y, Fuwa H. Amylose percentage and distribution of unit chain-length of maize starches having a specific genetic background. Carbohydrate Research. 1978;61:211–216. [Google Scholar]
- Itoh K, Ozaki H, Okada K, Hori H, Takeda Y, Mitsui T. Introduction of Wx transgene into rice wx mutants leads to both high- and low-amylose rice. Plant and Cell Physiology. 2003;44:473–480. doi: 10.1093/pcp/pcg068. [DOI] [PubMed] [Google Scholar]
- Jeon JS, Ryoo N, Hahn TR, Walia H, Nakamura Y. Starch biosynthesis in cereal endosperm. Plant Physiology and Biochemistry. 2010;48:383–392. doi: 10.1016/j.plaphy.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Jobling S. Improving starch for food and industrial applications. Current Opinion in Plant Biology. 2004;7:210–218. doi: 10.1016/j.pbi.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Juliano BO. Rice chemistry and quality. Munoz: PhilRice; [Google Scholar]
- Juliano BO, Perez CM, Blakeney AB, Castillo T, Kongseree N, Laignelet B, Lapis ET, Murty VVS, Paule CM, Webb BD. International cooperative testing on the amylose content of milled rice. Starch. 1981;33:157–162. [Google Scholar]
- Kang HJ, Hwang IK, Kim KS, Choi HC. Comparative structure and physicochemical properties of Ilpumbyeo, a high-quality japonica rice, and its mutant, Suweon 464. Journal of Agricultural and Food Chemistry. 2003;51:6598–6603. doi: 10.1021/jf0344946. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Xie ZX, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Developmental Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Yamamoto MP, Hirose S, Yano M, Takaiwa F. Characterization of a new rice glutelin gene GluD-1 expressed in the starchy endosperm. Journal of Experimental Botany. 2008;59:4233–4245. doi: 10.1093/jxb/ern265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KN, Fisher DK, Gao M, Guiltinan MJ. Molecular cloning and characterization of the amylose-extender gene encoding starch branching enzyme IIB in maize. Plant Molecular Biology. 1998;38:945–956. doi: 10.1023/a:1006057609995. [DOI] [PubMed] [Google Scholar]
- Kim KS, Hwang HG, Kang HT, Hwang IK, Lee YT, Choi HC. Ultrastructure of individual and compound starch granules in isolation preparation from a high-quality, low-amylose rice, ilpumbyeo, and its mutant, G2, a high-dietary fiber, high-amylose rice. Journal of Agricultural and Food Chemistry. 2005;53:8745–8751. doi: 10.1021/jf051194a. [DOI] [PubMed] [Google Scholar]
- Kubo A, Yuguchi Y, Takeniasa M, Suzuki S, Satoh H, Kitamura S. The use of micro-beam X-ray diffraction for the characterization of starch crystal structure in rice mutant kernels of waxy, amylose extender, and sugary1. Journal of Cereal Science. 2008;48:92–97. [Google Scholar]
- Lagudah ES, Appels R, McNeil D. The nor-d3 locus of Triticum tauschii: natural variation and genetic-linkage to markers in chromosome-5. Genome. 1991;34:387–395. [Google Scholar]
- Lee KW, Song KE, Lee HS, Kim YK, Lee SW, Kim DJ, Hwang WS, Choe SJ, Kim YS, Kim TY. The effects of Goami No. 2 rice, a natural fiber-rich rice, on body weight and lipid metabolism. Obesity. 2006;14:423–430. doi: 10.1038/oby.2006.56. [DOI] [PubMed] [Google Scholar]
- Liu B, Li PC, Li X, Liu CY, Cao SY, Chu CC, Cao XF. Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiology. 2005;139:296–305. doi: 10.1104/pp.105.063420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Makhmoudova A, Lee EA, Wait R, Emes MJ, Tetlow IJ. The amylose extender mutant of maize conditions novel protein–protein interactions between starch biosynthetic enzymes in amyloplasts. Journal of Experimental Botany. 2009;60:4423–4440. doi: 10.1093/jxb/erp297. [DOI] [PubMed] [Google Scholar]
- Lopez-Rubio A, Flanagan BM, Gilbert EP, Gidley MJ. A novel approach for calculating starch crystallinity and its correlation with double helix content: a combined XRD and NMR study. Biopolymers. 2008;89:761–768. doi: 10.1002/bip.21005. [DOI] [PubMed] [Google Scholar]
- McCleary BV, Gibson TS, Solah V, Mugford DC. Total starch measurement in cereal products. Interlaboratory evaluation of a rapid enzymatic test procedure. Cereal Chemistry. 1994;71:501–505. [Google Scholar]
- Mizuno K, Kobayashi E, Tachibana M, Kawasaki T, Fujimura T, Funane K, Kobayashi M, Baba T. Characterization of an isoform of rice starch branching enzyme, RBE4, in developing seeds. Plant and Cell Physiology. 2001;42:349–357. doi: 10.1093/pcp/pce042. [DOI] [PubMed] [Google Scholar]
- Moore CW, Creech RG. Genetic fine-structure analysis of amylose extender locus in Zea mays L. Genetics. 1972;70:611–619. doi: 10.1093/genetics/70.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell MK, Kosar-Hashemi B, Cmiel M, Samuel MS, Chandler P, Rahman S, Buleon A, Batey IL, Li ZY. Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. The Plant Journal. 2003;34:172–184. doi: 10.1046/j.1365-313x.2003.01712.x. [DOI] [PubMed] [Google Scholar]
- Morell MK, Myers AM. Towards the rational design of cereal starches. Current Opinion in Plant Biology. 2005;8:204–210. doi: 10.1016/j.pbi.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Nakamura Y. Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: rice endosperm as a model tissue. Plant and Cell Physiology. 2002;43:718–725. doi: 10.1093/pcp/pcf091. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Utsumi Y, Sawada T, Aihara S, Utsumi C, Yoshida M, Kitamura S. Characterization of the reactions of starch branching enzymes from rice endosperm. Plant and Cell Physiology. 2010;51:776–794. doi: 10.1093/pcp/pcq035. [DOI] [PubMed] [Google Scholar]
- Nishi A, Nakamura Y, Satoh H. Effect of starch-branching enzyme IIb on amylopectin structure and gelatinization property. Advances in rice genetics, Los Banos, Laguna, Philippines, 22–27 October 2000, 2003:459–461. [Google Scholar]
- Nishi A, Nakamura Y, Tanaka N, Satoh H. Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiology. 2001;127:459–472. [PMC free article] [PubMed] [Google Scholar]
- Ohdan T, Francisco PB, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. Journal of Experimental Botany. 2005;56:3229–3244. doi: 10.1093/jxb/eri292. [DOI] [PubMed] [Google Scholar]
- O'Shea MG, Morell MK. High resolution slab gel electrophoresis of 8-amino-1,3, 6–pyrenetrisulfonic acid (APTS) tagged oligosaccharides using a DNA sequencer. Electrophoresis. 1996;17:681–686. doi: 10.1002/elps.1150170410. [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D. Gene silencing in plants using artificial microRNAs and other small RNAs. The Plant Journal. 2008;53:674–690. doi: 10.1111/j.1365-313X.2007.03328.x. [DOI] [PubMed] [Google Scholar]
- Qu LQ, Xing YP, Liu WX, Xu XP, Song YR. Expression pattern and activity of six glutelin gene promoters in transgenic rice. Journal of Experimental Botany. 2008;59:2417–2424. doi: 10.1093/jxb/ern110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Abrahams S, Abbott D, Mukai Y, Samuel M, Morell M, Appels R. A complex arrangement of genes at a starch branching enzyme I locus in the D-genome donor of wheat. Genome. 1997;40:465–474. doi: 10.1139/g97-062. [DOI] [PubMed] [Google Scholar]
- Rahman S, Bird A, Regina A, Li Z, Philippe Ral J, McMaugh S, Topping D, Morell M. Resistant starch in cereals: exploiting genetic engineering and genetic variation. Journal of Cereal Science. 2007;46:251–260. [Google Scholar]
- Regina A, Bird A, Topping D, Bowden S, Freeman J, Barsby T, Kosar-Hashemi B, Li ZY, Rahman S, Morell M. High-amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proceedings of the National Academy of Sciences, USA. 2006;103:3546–3551. doi: 10.1073/pnas.0510737103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regina A, Kosar-Hashemi B, Li ZY, Pedler A, Mukai Y, Yamamoto M, Gale K, Sharp P, Morell MK, Rahman S. Starch branching enzyme IIb in wheat is expressed at low levels in the endosperm compared to other cereals and encoded at a non-syntenic locus. Planta. 2005;222:899–909. doi: 10.1007/s00425-005-0032-z. [DOI] [PubMed] [Google Scholar]
- Regina A, Kosar-Hashemi B, Ling S, Li Z, Rahman S, Morell M. Control of starch branching in barley defined through differential RNAi suppression of starch branching enzyme IIa and IIb. Journal of Experimental Botany. 2010;61:1469–1482. doi: 10.1093/jxb/erq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safford R, Jobling SA, Sidebottom CM, Westcott RJ, Cooke D, Tober KJ, Strongitharm BH, Russell AL, Gidley MJ. Consequences of antisense RNA inhibition of starch branching enzyme activity on properties of potato starch. Carbohydrate Polymers. 1998;35:155–168. [Google Scholar]
- Sawada T, Francisco PB, Aihara S, Utsumi Y, Yoshida M, Oyama Y, Tsuzuki M, Satoh H, Nakamura Y. Chlorella starch branching enzyme II (BEII) can complement the function of BEIIb in rice endosperm. Plant and Cell Physiology. 2009;50:1062–1074. doi: 10.1093/pcp/pcp058. [DOI] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of MicroRNAs on the plant transcriptome. Developmental Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Sestili F, Janni M, Doherty A, Botticella E, D'Ovidio R, Masci S, Jones HD, Lafiandra D. Increasing the amylose content of durum wheat through silencing of the SBEIIa genes. BMC Plant Biology. 2010;10:144. doi: 10.1186/1471-2229-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon JC, Garwood DL, Boyer CD. Genetics and physiology of starch development. In: BeMiller J, Whistler RL, editors. Starch: chemistry and technology. New York: Academic Press; 2009. pp. 23–82. [Google Scholar]
- Shu XL, Jiao G, Fitzgerald MA, Yang CZ, Shu QY, Wu DX. Starch structure and digestibility of rice high in resistant starch. Starch. 2006;58:411–417. [Google Scholar]
- Smith AM. The biosynthesis of starch granules. Biomacromolecules. 2001;2:335–341. doi: 10.1021/bm000133c. [DOI] [PubMed] [Google Scholar]
- Takeda Y, Shibahara S, Hanashiro I. Examination of the structure of amylopectin molecules by fluorescent labeling. Carbohydrate Research. 2003;338:471–475. doi: 10.1016/s0008-6215(02)00488-3. [DOI] [PubMed] [Google Scholar]
- Tan I, Flanagan BM, Halley PJ, Whittaker AK, Gidley MJ. A method for estimating the nature and relative proportions of amorphous, single, and double-helical components in starch granules by C-13 CP/MAS NMR. Biomacromolecules. 2007;8:885–891. doi: 10.1021/bm060988a. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Fujita N, Nishi A, Satoh H, Hosaka Y, Ugaki M, Kawasaki S, Nakamura Y. The structure of starch can be manipulated by changing the expression levels of starch branching enzyme llb in rice endosperm. Plant Biotechnology Journal. 2004;2:507–516. doi: 10.1111/j.1467-7652.2004.00097.x. [DOI] [PubMed] [Google Scholar]
- Tetlow IJ, Morell MK, Emes MJ. Recent developments in understanding the regulation of starch metabolism in higher plants. Journal of Experimental Botany. 2004;55:2131–2145. doi: 10.1093/jxb/erh248. [DOI] [PubMed] [Google Scholar]
- Theander O, Aman P, Westerlund E, Andersson R, Petersson D. Total dietary fiber determined as neutral sugar residues, uronic acid residues, and Klason Lignin (The Uppsala method): collaborative study. Journal of AOAC International. 1995;78:1030–1044. [PubMed] [Google Scholar]
- Toyota K, Tamura M, Ohdan T, Nakamura Y. Expression profiling of starch metabolism-related plastidic translocator genes in rice. Planta. 2006;223:248–257. doi: 10.1007/s00425-005-0128-5. [DOI] [PubMed] [Google Scholar]
- Upadhyaya NM, Surin B, Ramm K, Gaudron J, Schunmann PHD, Taylor W, Waterhouse PM, Wang MB. Agrobacterium-mediated transformation of Australian rice cultivars Jarrah and Amaroo using modified promoters and selectable markers. Australian Journal of Plant Physiology. 2000;27:201–210. [Google Scholar]
- Vandeputte GE, Delcour JA. From sucrose to starch granule to starch physical behaviour: a focus on rice starch. Carbohydrate Polymers. 2004;58:245–266. [Google Scholar]
- Wang MB, Li ZY, Matthews PR, Upadhyaya NM. Improved vectors for Agrobacterium tumefaciens-mediated transformation of monocot plants. In: Drew RA, editor. International Symposium on Biotechnology of Tropical and Subtropical Species – Part Ii. 1998. Leuven 1: International Society Horticultural Science, 401–407. [Google Scholar]
- Ward RM, Gao QY, de Bruyn H, Gilbert RG, Fitzgerald MA. Improved methods for the structural analysis of the amylose-rich fraction from rice flour. Biomacromolecules. 2006;7:866–876. doi: 10.1021/bm050617e. [DOI] [PubMed] [Google Scholar]
- Warthmann N, Chen H, Ossowski S, Weigel D, Herve P. Highly specific sene silencing by artificial miRNAs in rice. PLoS ONE. 2008;3:e1829. doi: 10.1371/journal.pone.0001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CX, Qin FL, Zhou WD, Chen YF, Xu B, Wang YP, Gu MH, Liu QQ. Formation of semi-compound C-type starch granule in high-amylose rice developed by antisense RNA inhibition of starch-branching enzyme. Journal of Agricultural and Food Chemistry. 2010c;58:11097–11104. doi: 10.1021/jf1024533. [DOI] [PubMed] [Google Scholar]
- Wei CX, Qin FL, Zhou WD, Yu HG, Xu B, Chen C, Zhu LJ, Wang YP, Gu MH, Liu QQ. Granule structure and distribution of allomorphs in C-type high-amylose rice starch granule modified by antisense RNA inhibition of starch branching enzyme. Journal of Agricultural and Food Chemistry. 2010d;58:11946–11954. doi: 10.1021/jf103412d. [DOI] [PubMed] [Google Scholar]
- Wei CX, Qin FL, Zhu LJ, Zhou WD, Chen YF, Wang YP, Gu MH, Liu QQ. Microstructure and ultrastructure of high-amylose rice resistant starch granules modified by antisense RNA inhibition of starch branching enzyme. Journal of Agricultural and Food Chemistry. 2010a;58:1224–1232. doi: 10.1021/jf9031316. [DOI] [PubMed] [Google Scholar]
- Wei CX, Xu B, Qin FL, Yu HG, Chen C, Meng XL, Zhu LJ, Wang YP, Gu MH, Liu QQ. C-type starch from high-amylose rice resistant starch granules modified by antisense RNA inhibition of starch branching enzyme. Journal of Agricultural and Food Chemistry. 2010b;58:7383–7388. doi: 10.1021/jf100385m. [DOI] [PubMed] [Google Scholar]
- Whittam MA, Noel TR, Ring SG. Melting behaviour of A- and B-type crystalline starch. International Journal of Biological Macromolecules. 1990;12:359–362. doi: 10.1016/0141-8130(90)90043-a. [DOI] [PubMed] [Google Scholar]
- Yamakawa H, Hirose T, Kuroda M, Yamaguchi T. Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiology. 2007;144:258–277. doi: 10.1104/pp.107.098665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanouchi H, Nakamura Y. Organ specificity of isoforms of starch branching enzyme (Q-enzyme) in rice. Plant and Cell Physiology. 1992;33:985–991. [Google Scholar]
- Yang CZ, Shu XL, Zhang LL, Wang XY, Zhao HJ, Ma CX, Wu DX. Starch properties of mutant rice high in resistant starch. Journal of Agricultural and Food Chemistry. 2006;54:523–528. doi: 10.1021/jf0524123. [DOI] [PubMed] [Google Scholar]
- Yano M, Okuno K, Kawakami J, Satoh H, Omura T. High amylose mutants of rice, Oryza sativa L. Theoretical and Applied Genetics. 1985;69:253–257. doi: 10.1007/BF00662436. [DOI] [PubMed] [Google Scholar]
- Yeh JY, Garwood DL, Shannon JC. Characterization of starch from maize endosperm mutants. Starch. 1981;33:222–230. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.