Abstract
Sunflower (Helianthus annuus L.) seed viability is affected by moisture content (MC) during ageing and is related to accumulation of hydrogen peroxide and changes in energy metabolism. The aim of the present work was to investigate the effect of ageing on DNA alteration events by RAPD (random amplification of polymorphic DNA) analysis and to determine whether loss of seed viability might correspond to a controlled programmed cell death (PCD). Ageing of sunflower seeds was carried out at 35 °C for 7 d at different MCs. The higher the MC, the lower was the seed viability. RAPD analysis showed that DNA alterations occurred during ageing especially in seeds containing a high MC. In addition, PCD, as revealed by DNA fragmentation and TUNEL (terminal deoxynucleotide transferase-mediated dUTP nick-end labelling) assay, was detected in aged seeds at MCs which resulted in ∼50% seed viability. At the cellular level, TUNEL assay and propidium iodide staining showed that cell death concerns all the cells of the embryonic axis. The quantification of the adenylate pool highlights mitochondrial dysfunction in aged seeds containing a high MC. The involvement of oxidative burst, mitochondria dysfunction, and PCD in seed loss of viability is proposed.
Keywords: Ageing, DNA alteration, MC, PCD, seed, sunflower
Introduction
Ageing induces progressive seed deterioration ultimately leading to lethal damage and inability to germinate. In orthodox seeds (i.e. seeds which tolerate dehydration), accelerated ageing can be artificially induced at high temperature and high relative humidity (RH) (Priestley, 1986). In sunflower, for example, storage of seeds at 45 °C and 100% RH results in a progressive reduction of germination and induces abnormal seedling growth and then finally leads to death (Corbineau et al., 1988; Gay et al., 1991). However, the sensitivity of seeds to accelerated ageing is tightly dependent on temperature and on their water content. At a constant temperature, loss of seed viability is faster with increasing moisture content (MC), a key factor in seed longevity (Priestley, 1986; McDonald, 1999).
At the cellular level, seed ageing is associated with various alterations including loss of membrane integrity, reduced energy metabolism, impairment of RNA and protein synthesis, and DNA degradation (Vazquez-Ramos et al., 1988; Walters, 1998; McDonald, 1999; Corbineau et al., 2002; Kibinza et al., 2006). It has been known for a long time that a progressive fragmentation of embryonic nuclear DNA occurs during seed ageing (Cheah and Osborne, 1978; Osborne et al., 1981). DNA damage can be due to an uncontrolled degradation following extensive DNA oxidation (Slupphaug et al., 2003) or to DNA laddering, as is commonly observed in active and genetically controlled programmed cell death (PCD) (Stein and Hansen, 1999). In seeds, PCD-like DNA fragmentation was only detected in response to ageing in Pisum sativum (Kranner et al., 2006). These authors proposed that the increase of half-cell reduction potential of glutathione, presumably caused by reactive oxygen species (ROS), could be a part of a signalling cascade leading to PCD. ROS generation is indeed known to be implicated in plant and animal PCD that occurs during many stresses (Grant and Loake, 2000; Neill et al., 2002; Laloi et al., 2004).
ROS have been widely cited as being the main factor causing seed ageing during their prolonged storage (Priestley, 1986). In sunflower seeds, loss of viability during ageing has been shown to be associated with an accumulation of hydrogen peroxide (H2O2) and lipid peroxidation and with a decrease in the activity of antioxidant enzymes (Bailly et al., 1996; Kibinza et al., 2006). Due to the absence of free water in dry seeds, non-enzymatic mechanisms such as lipid peroxidation are likely to be involved in ROS accumulation during dry storage, but as soon as seeds imbibe, enzymatic mechanisms participate in ROS production. One of the major sources of ROS in metabolically active seeds is the mitochondrial respiratory chain (Bailly, 2004); however, any mitochondrial alteration leads to increased ROS production (Wei and Lee, 2002; Cash et al., 2007). ROS can moreover induce the opening of membrane permeability transition pores in mitochondria and release of cytochrome c and other apoptogenic factors into the cytoplasm, which leads to apoptosis (Tatton and Olanow, 1999). In animals, mitochondrial alterations have been proposed to be involved in, and possibly responsible for, cell death (Bras et al., 2005), and are often considered as being a central mechanism driving mammalian ageing (Kujoth et al., 2005).
Whether ROS-triggered PCD might participate in the loss of seed viability during seed ageing is largely unknown. It is proposed that the possible occurrence of PCD in seeds relies on the intracellular water availability which governs active metabolism, thus possibly allowing definition of a critical MC that drives this phenomenon. Therefore, the occurrence of such a mechanism of viability loss would be mainly involved in seed survival in the soil seed bank in which seed MC varies greatly with changing environmental conditions. In order to study the involvement of seed MC in seed longevity, an ageing protocol was designed using sunflower seeds which allowed the ageing mechanisms to be studied over a wide range of MCs. This protocol resulted in a progressive ageing rate that permitted determination of the sequences of cellular events involved in seed death and the MCs at which they occur. Using this model, the relationship between DNA damage intensity, a good marker of age-related damage, and seed MC during ageing was established. DNA alterations were revealed by random amplification of polymorphic DNA (RAPD) analysis which has already been described as a suitable method for the detection of DNA damage and mutational events (e.g. rearrangements, point mutation, small insert or deletions of DNA, and other possible changes such as structural distortion) in bacteria or animals (Atienzar et al., 2002; Atienzar and Jha, 2006), and also in seedlings (Liu et al., 2005) and seeds (Shatters et al., 1995; Vijay et al., 2009). DNA alterations were studied with regards to the possible occurrence of PCD during ageing. As PCD is related to a sequence of cell events that include impairment of mitochondria functioning or DNA fragmentation, the effect of seed MC on these two phenomena was studied. As DNA laddering is often hardly detectable, the terminal deoxynucleotide transferase-mediated dUTP nick-end labelling (TUNEL) reaction in situ was also used. This reaction labels the free 3′-OH DNA strand breaks and is widely accepted as a specific criterion of PCD (Danon et al., 2000).The results obtained allow a novel model to be proposed that presents a putative pattern of the cellular events occurring with increasing MC and which can play a role in seed death.
Materials and methods
Plant material and ageing treatments
Experiments were carried out with sunflower (Helianthus annuus L.) seeds harvested in 2004 and received from Monsanto-France (Peyrehorade, France). Seeds were stored for 3 months at 20 °C and 75% RH in order to break their dormancy (Corbineau et al., 1990) before ageing treatment. Ageing was performed according to Kibinza et al. (2006). Seeds were equilibrated for 24 h at 20 °C, in closed flasks with different amounts of water to obtain seed MCs ranging from 0.04 g H2O g−1 dry matter (DM) (dry seeds) to 0.48 g H2O g−1 DM, and then placed at 35 °C for 7 d. Flasks were regularly flushed with air to ensure seed oxygenation. After ageing treatment, germination assays were performed on whole seeds (whole embryo, i.e. embryonic axis plus two cotyledons, plus the seed coat) and embryonic axes (radicle plus gemmula) were isolated and frozen for DNA extraction or used fresh for adenosine phosphate assay and cytological experiments.
Germination assays
Germination assays were performed at 15 °C in darkness, in three replicates of 50 whole seeds placed in 9 cm diameter Petri dishes on a layer of cotton wool moistened with deionized water. Germination counts were made carried out for 7 d. The results presented correspond to the means of the germination percentages obtained after 7 d ±SD.
Water content determination
Water content determination was determined on 10 whole seeds according to Kibinza et al. (2006).
Adenosine phosphate measurement
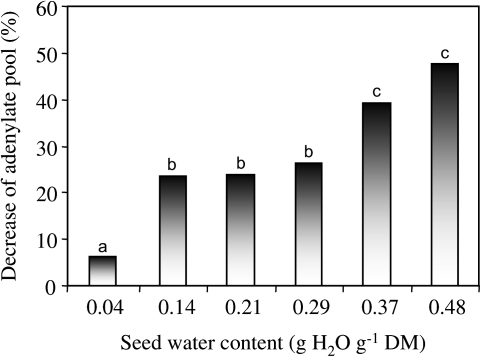
The adenylate pool (AP: ATP, ADP, and AMP) was extracted according to Olempska-Beer and Bautz-Freeze (1984) from three axes excised from seeds after 1 d of water equilibrium at different MCs at 20 °C and after a subsequent 7 d controlled deterioration at 35 °C. The components of the AP were measured using the bioluminescence method with a pico-ATP biophotometer (Jobin et Yvon, France), as described by Corbineau et al. (2002) and Kibinza et al. (2006). Values were calculated as nmol AP per mg DM and are the means of values obtained with 5–7 extracts. Figure 2 presents the percentage AP decrease which corresponds to the difference between the AP content before and after ageing. Data were subjected to an analysis of variance using Duncan's multiple range test at P ≤0.05.
Fig. 2.
Effect of 7 d of controlled deterioration at 35 °C of seeds equilibrated at a water content of 0.04, 0.14, 0.21, 0.29, 0.37, and 0.48 g H2O g−1 DM on the decrease of the adenylate pool (ATP, ADP, and AMP). The decrease corresponds to the difference between the values obtained after 1 d of water equilibrium of seeds at 20 °C and the values determined after controlled deterioration. Values are the means of three measurements. Columns having the same letter are not significantly different at the 0.05 probability level as determined by Duncan's test.
DNA extraction and analysis
Embryonic axes isolated from whole seeds using a sharp scalpel blade were immediately frozen in liquid nitrogen, and then stored at –80 °C. DNA was isolated from embryonic axes according to the method described by Goldenberger et al. (1995). A 0.2 g aliquot of sample was ground with liquid nitrogen by using a mortar and pestle for ∼5 min. The resultant fine powder was suspended in 2 ml of SDS extraction buffer (1% SDS, 200 mM TRIS-HCl, pH 7.5, 288 mM NaCl, and 25 mM EDTA) by vigorous vortexing for 2 min. Debris was removed by centrifugation at 10 000 g for 2 min. The supernatant was mixed with an equal volume of cold isopropanol and then the precipitate was separated by centrifugation (12 000 g, 7 min). The nucleic acids in the pellet were dried and then dissolved in 100 ml of TE buffer (100 mM TRIS-HCl, pH 7.5, and 1 mM EDTA).
DNA electrophoresis was performed to assess DNA fragmentation. DNA samples (10 μg lane−1) were loaded on a 1.5% agarose gel stained with 0.2 μg ml−1 ethidium bromide.
RAPD analysis
Random 10-base primers used correspond to Operon kit A primers, according to Sossey-Alaoui et al. (1998) (Table 1). Amplifications were carried out in 25 μl of reaction mixture containing 100 ng of genomic DNA, 120 ng of primer, 200 μM dNTPs (50 μM of each), 10× reaction buffer, and 0.5 U of Taq DNA polymerase. The RAPD protocol consisted of an initial denaturing step of 5 min at 94 °C, followed by 40 cycles at 94 °C for 1 min, 34 °C for 1 min, and 72 °C for 1 min, with an additional extension period of 10 min at 72 °C. RAPD products were analysed by electrophoresis in 1.5% agarose gels stained with 0.2 μg ml−1 ethidium bromide. The polymorphism presented is repeatable and corresponds to representative observations obtained with three different DNA preparations from independent biological replicates
Table 1.
Sequences of the 20 primers used in this study
| Primer no. | Primer sequences (5′→3′) |
| RAPD 1 | CAGCACCCAC |
| RAPD 2 | TTCCGAACCC |
| RAPD 3 | AGGTGACCGT |
| RAPD 4 | GTTGCGATCC |
| RAPD 5 | CAGGCCCTTC |
| RAPD 6 | AGTCAGCCAC |
| RAPD 7 | GGTCCCTGAC |
| RAPD 8 | GTGACGTAGG |
| RAPD 9 | TGCCGAGCTG |
| RAPD 10 | AATCGGGCTG |
| RAPD 11 | AGGGGTCTTG |
| RAPD 12 | GAAACGGGTG |
| RAPD 13 | GGGTAACGCC |
| RAPD 14 | GTGATCGCAG |
| RAPD 15 | CAATCGCCGT |
| RAPD 16 | TCGGCGATAG |
| RAPD 17 | AGCCAGCGAA |
| RAPD 18 | GACCGCTTGT |
| RAPD 19 | CAAACGTCGG |
| RAPD 20 | TCTGTGCTGG |
Paraffin embedding tissue TUNEL assay
Plant material was fixed in 4% (w/v) formaldehyde in 50 mM phosphate buffer (pH 7.5) (PBS) containing 150 mM NaCl for 2 h at room temperature. Tissue was then dehydrated and embedded in paraffin essentially as described by O'Brien and McCully (1981). Sections (6 μm) were cut with a Heidelberg (Jung Ag) microtome and mounted on polylysine-subbed slides. Paraffin was removed with two changes of Histoclear (Euromedex) during 10 min, and sections were hydrated in an ethanol/water series and then in 0.85% NaCl solution prior to TUNEL assay.
TUNEL was performed using the Promega in situ apoptosis detection kit according to the manufacturer's instructions.
DAPI and PI staining
Transverse hand-cut sections of embryonic axes were incubated in phosphate buffer (PBS) for 5 min at room temperature and an additional 10 min in propidium iodide (PI) at 5 μg ml−1 or 4',6-diamidino-2-phenylindole (DAPI) at 300 nM before a final step of washing with PBS. Sections were then placed on slides and observed with a Zeiss Axiophot fluorescence microscope.
Results
Effect of moisture content during seed deterioration at 35 °C on seed viability
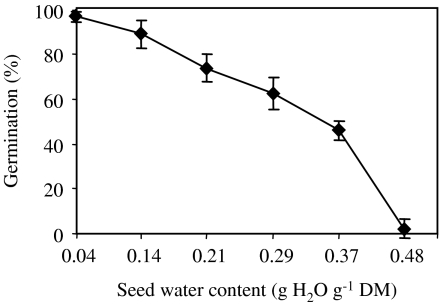
After 24 h of equilibrium at various MCs, seeds were aged at 35 °C for 7 d. Non-aged seeds (dry seeds, 0.04 g H2O g−1 DM) fully germinated within 7 d at 15 °C (data not shown). Controlled deterioration at 35 °C for 7 d resulted in a significant decrease of seed viability as evaluated by the germination percentages obtained after 7 d at 15 °C (Fig.1). The higher the water content was, the lower was the seed viability. An increase in seed water content up to 0.29 g H2O g−1 DM resulted in a significant reduction of seed viability. At an MC of 0.37 g H2O g−1 DM, 50% of the seed population remained viable, but all seeds were dead at 0.48 g H2O g−1 DM (Fig.1).
Fig. 1.
Effects of 7 d treatment at 35 °C of seeds previously equilibrated at a water content of 0.04, 0.14, 0.21, 0.29, 0.37, and 0.48 g H2O g−1 DM on subsequent germination percentages obtained after 7 days at 15 °C. Means of five measurements ±SD. Means are significantly different at the 0.05 probability level determined by the independent samples t- test.
Effect of ageing on energy metabolism as related to seed moisture content
The AP (ATP, ADP, and AMP) was measured in the axis before (upon seed equilibrium with various amount of water) and after 7 d of ageing at 35 °C. The decrease in the AP, which represents the difference between the AP content before and after ageing, is expressed as a percentage of the AP measured in unaged seeds equilibrated at various MCs (Fig. 2). This mode of calculation allows integration of the effect of increasing MCs on the AP (see also Kibinza et al., 2006). Seven days of controlled deterioration at 35 °C resulted in a decrease in the AP that was related to seed MC but not proportionally, thus allowing description of two levels of mitochondrial impairment. From 0.14 to 0.29 g H2O g−1 DM, the AP decreased by ∼25% after ageing (Fig. 2). When the seed MC was >0.37 g H2O g−1 DM, the pool decreased markedly and represented ∼40% of that found in unaged seeds (Fig. 2).
Relationship between seed moisture content and DNA alteration
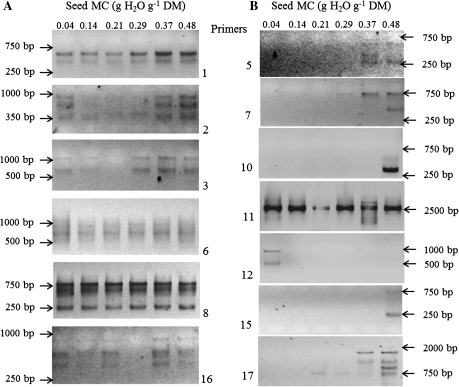
RAPD profiles obtained with DNA templates from seeds aged at various MCs are shown in Fig. 3. Among the 20 decamer oligonucleotide primers tested, only 13 gave specific and reproducible results. Several primers (1, 2, 3, 6, 8, and 16) gave a constant pattern of PCR products in all DNA samples, whatever the seed MC during ageing (Fig. 3A). In contrast, other primers showed polymorphism and gave different PCR products as a function of seed MC (Fig. 3B). Primer 12 amplified two bands only in dry seeds (0.04 g H2O g−1 DM) and did not gave PCR products at higher MCs (Fig. 3B). For primers 5, 7, 10, 11, 15, and 17, the greatest variability in the pattern of PCR products occurred when the seed MC was 0.37 or 0.48 g H2O g−1 DM (Fig. 3B). Primer 17 (Fig. 3B) allowed a slight amplification when the seed MC was ∼0.21 g H2O g−1 DM, but the number of bands increased to about three and four when the MC increased to 0.37 and 0.48 g H2O g−1 DM. Primer 11 amplified five bands only with the DNA sample of seeds at an MC of 0.37 g H2O g−1 DM. At the higher MCs, progressive and extensive DNA degradation probably induced a lack of sites for primer 11 (Fig. 3B) but a gain of sites for primers 10 and 15 (Fig. 3B). The general trend (i.e. six primers among the seven primers which give a variable pattern) was an increasing number and amount of products with increasing seed water content, and major changes in DNA occurred when the seed MC reached 0.37 g H2O g−1 DM.
Fig. 3.
Representative electrophoregrams of amplification products obtained from the DNAs of the axis of aged seeds containing a water content of 0.04, 0.14, 0.21, 0.29, 0.37, and 0.48 g H2O g−1 DM. (A) Consistent pattern obtained by amplification with RAPD primers 1, 2, 3, 6, 8, and 16. (B) Variable pattern obtained by amplification with RAPD primers 5, 7, 10, 11, 12, 15, and 17. RAPD-PCR experiments were performed three times for the 20 primers.
Detection of cell death: DNA laddering and TUNEL assay
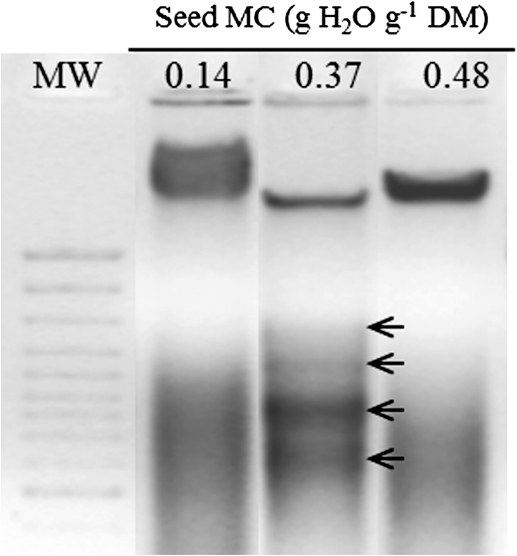
Electrophoresis of DNA on an agarose gel showed DNA fragmentation characterized by the appearance of bands in the case of the aged seeds equilibrated at 0.37 g H2O g−1 DM only (arrows Fig. 4). Laddering was not detected in seeds aged at 0.48 g H2O g−1 DM (Fig. 4) which could be explained by extensive DNA degradation including laddering fragment products.
Fig. 4.
Representative agarose gel electrophoresis of total DNA extracted from the axis isolated from seeds aged for 7 d at 35 °C at a water content of 0.14, 0.37, and 0.48 g H2O g−1 DM. A 10 μg aliquot of total DNA extracted from the axis was loaded on a 2% agarose gel, and stained with ethidium bromide. Four DNA bands of 600, 800, 1000, and 1200 bp are present in DNA of seeds at a water content of 0.37 g H2O g−1 DM (arrows). Laddering was detected in four independent experiments. MW: 100 PB DNA ladder (100–3000 bp) molecular weight (Euromedex).
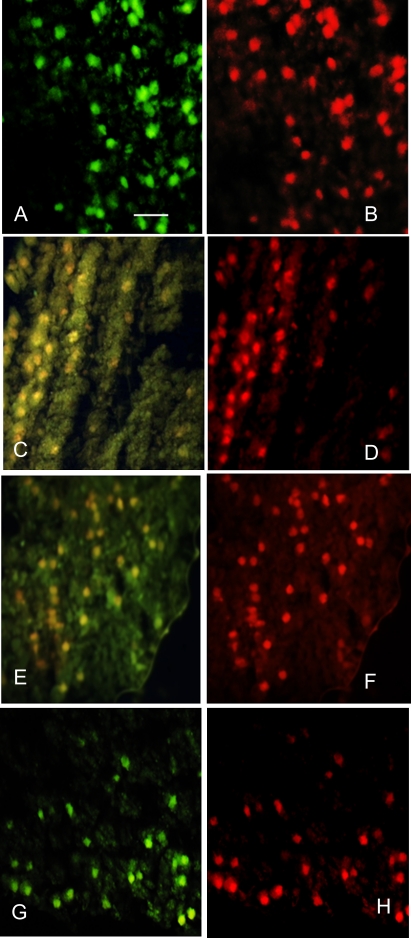
To confirm the finding of PCD in aged seeds, the TUNEL assay was used to detect apoptotic cells in sunflower axes. TUNEL assay was coupled with PI staining in order to co-localize nuclei of dead cells. Paraffin-embedded sections of sunflower embryonic axes were positive in the TUNEL assay as revealed by bright green fluorescent spots in Fig. 5G when seeds were equilibrated at 0.37 g H2O g−1 DM and aged for 7 d at 35 °C. PI staining confirmed the localization of the TUNEL reaction within the nuclei (Fig. 5H), and the use of a DNase showed that the TUNEL assay detected DNA fragmentation in embryonic axes (Fig. 5A, B). In unaged and aged seeds containing 0.14 g H2O g−1 DM, no TUNEL reaction was evidenced as shown by the absence of green fluorescence and the yellow tint of the nuclei in Fig. 5C and E.
Fig. 5.
Paraffin embryo axis sections labelled with TUNEL assay (A, C, E, and G) and corresponding PI staining (B, D, F, and H). (A and B) Positive control corresponding to a DNase-treated section. (C and D) Sections of unaged seeds. (E and F) Sections of seeds equilibrated at a water content of 0.14 g H2O g−1 DM and aged for 7 d at 35 °C. (G and H) Sections of seeds equilibrated at a water content of 0.37 g H2O g−1 DM and aged for 7 d at 35 °C.
DAPI and PI staining
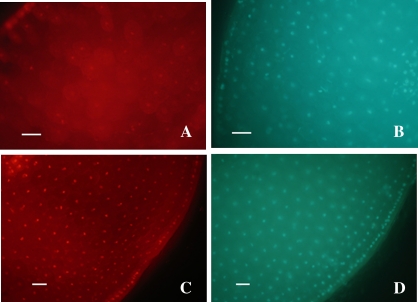
The viability of seeds treated at 35 °C for 7 d was examined at the cellular level using the fluorescent probes PI and DAPI. Successive hand-cut sections of each axis were stained with DAPI in order to visualize all nuclei, and with PI, which stains nuclei of dead cells only, PI being a marker of cell death (Nicoletti et al., 1991). PI staining of sections of seeds with a low level of hydration (from 0.04 to 0.29 g H2O g−1 DM) showed a very weak fluorescence, indicating the presence of living cells only (Fig. 6A), while the results shown in Fig. 6B confirmed the nuclear localization. Full loss of cell viability was observed in axes of seeds aged at 0.37 g H2O g−1 DM since all nuclei displayed PI staining (Fig. 6C, D). At 0.37 g H2O g−1 DM, 50% of the hand-cut sections observed showed an intense red fluorescence of nuclei due to PI, similar to that in Fig. 6C, while in 50% of the sections observed PI fluorescence was as weak as shown in Fig. 6A.
Fig. 6.
Fluorescence micrographs of sunflower seed sections stained with PI (A and C) and DAPI (B and D). Transverse hand-cut sections of an axis corresponding to viable dry seeds (0.04 g H2O g−1 DM) (A and B) and an axis from seeds previously equilibrated at a water content of 0.37 g H2O g−1 DM, and then aged at 35 °C for 7 d (C and D), were stained with PI (5 μg ml−1) and DAPI (300 nM) in PBS medium prior to image capture. Red nuclei correspond to dead cells. Bar=50 μm.
Discussion
Seed ageing is influenced by two environmental factors, RH and temperature. Accelerated ageing has been developed under laboratory conditions by exposure of seed to high temperatures (30–45 °C) and high humidity. Accelerated ageing has been recognized as a good predictor of the storability of seed lots (Priestley, 1986). It is associated with loss of vigour (i.e. the ability to germinate rapidly in a wide range of environmental conditions) and then with a progressive loss of viability. The loss of viability can be modulated by exposure of seeds at constant MC to different temperatures or by exposure of seeds equilibrated at different MCs to a constant temperature. In fact, 7 d ageing of sunflower seeds at a constant MC (0.29 g H2O g−1 DM) resulted in 90% germination when exposed at 25 °C, 80% at 35 °C, and 0% at 45 °C compared with non-aged seeds which displayed 98% germination (unpublished data). The present work shows that loss of seed viability during ageing at a constant temperature (35 °C) strongly depends on the MC (Fig. 1), as was previously described with seeds of the same species (Bailly et al., 1996; Kibinza et al., 2006). This behaviour allows a critical MC at which 50% of the seed population was dead (P50) to be defined. Under the present experimental conditions this value was 0.37 g H2O g−1 DM (Fig. 1).
It is shown that sunflower seed ageing is associated with increased DNA damage, as evidenced by RAPD analysis. These data are in agreement with previous publications which demonstrated that DNA deterioration is a key feature of seed ageing using either RAPD (Shatters et al., 1995; Vijay et al., 2009) or AFLP (amplified fragment length polymorphism; Chwedorzewska et al., 2002) techniques. Interestingly it is shown that the extent of DNA damage depends on seed MC, since polymorphism varies with increases in seed MC. This suggests that seed MC regulates the nature and/or the rate of DNA alteration, as shown by RAPD analysis (Fig. 3). A single primer (RAPD 12) produced a pattern of disappearance of bands when seed MC increased above 0.04 g H2O g−1 DM (Fig. 3B). DNA modification related to repair has been shown by Boubriak et al. (1997) in the early steps of seed imbibition, and this mechanism has been proposed as being a prerequisite for allowing the completion of the germination process. The most common kind of variability in the patterns of RAPD-PCR products nevertheless consisted of the appearance of new bands (primers 5, 7, 10, 11, 15, and 17 in Fig. 3). This trend has already been reported during ageing of soybean seeds (Shatters et al., 1995). New fragments can be amplified because some sites become accessible to the primer after structural changes in the DNA take place (Pietrasanata et al., 2000; Enan, 2006). This could be due to point mutations and/or extensive rearrangements of the DNA. A single point mutation within the primer site can generate significant changes in RAPD patterns (Williams et al., 1990). DNA alterations evidenced by polymorphism were observed when seed MC reached at least 0.37 g H2O g−1 DM since lower MCs had no effect on the pattern of bands (Fig. 3B). Interestingly, this is also the MC at which DNA fragmentation showing the typical pattern of laddering was evidenced (Fig. 4). This pattern is considered as being a hallmark of PCD (Hengartner, 2000), thus suggesting that this active mechanism of apoptosis is likely to take place during sunflower seed ageing under hydrated conditions. The involvement of PCD in loss of viability was confirmed using TUNEL assay and by studying the changes in the AP. The strong depletion of the AP that occurs at 0.37 g H2O g−1 DM suggests mitochondrial dysfunctioning at this MC which can result from changes in the mitochondrial ion channels and release of cytochrome c into the cytoplasm (Hengartner, 2000). A decrease in the AP was recorded from an MC of 0.14 g H2O g−1 DM, which suggests an impairment in mitochondrial metabolism at this ageing point corresponding to ∼90% germination. It has been reported that the impairment of energy metabolism is a process which occurs when cell viability is almost unaffected, which suggests that mitochondrial damage occurs in the early phase of PCD in plant and mammalian cells (Atlante et al., 1996, 1998; Vacca et al., 2004).
Primary events of PCD are triggered by ROS, and a threshold level of ROS is required to activate the signal transduction pathway that results in PCD (Desikan et al., 1998; Kranner et al., 2010). It was shown using the same model and ageing conditions that ROS and lipid peroxidation increased according to ageing intensity, namely MC (Kibinza et al., 2006). Oxidative stress and lipid peroxidation have been widely indicated as the major cause of deterioration of oilseeds during ageing (Priestly, 1986; Bailly et al., 1996; McDonald, 1999). Furthermore, changes in the half-cell reduction potential of glutathione [E(GSSG/2GSH)], which represents the major cellular antioxidant and redox buffer, were related to PCD during ageing in pee seeds (Kranner et al., 2006).
In situ observations (TUNEL and PI staining; Figs 5, 6) provide supplementary useful data because they allowed PCD to be studied at the level of individuals, whereas DNA fragmentation and mitochondrial failure were evidenced at the population level. These observations show that the whole cellular territory of seeds undergoes PCD in a synchronous manner when they have reached a critical point resulting from a combination of temperature, duration, and MC applied during the ageing treatment (close to 35 °C, 7 d, and 0.37 g H2O g−1 DM under the present conditions). If all cells simultaneously undergo PCD, as shown here, the seed will die and this mechanism cannot therefore be considered as a selective elimination of unwanted cells, as was shown in other developmental processes or in response to pathogens (Hengartner, 2000; Samejima and Earnshaw, 2005).
This set of data allows the proposal to be made that PCD participates in loss of seed viability in hydrated conditions. A similar observation was made by Kranner et al. (2006) who showed that DNA laddering occurred during ageing of pea seeds when their germination percentage was close to 50% (i.e. when 50% of the seeds were dead), but this was in dry seeds. The present results show that DNA laddering occurred when seed MC was close to 0.37 g H2O g−1 DM, an MC which also corresponds to 50% seed viability (Figs 1, 4). Similarly, RAPD-PCR assays also suggested that 50% seed viability was a key point for DNA alteration (Fig. 3). The time taken for germination to drop to 50% (P50) was already taken into account in orthodox seeds as an important parameter for considering seed deterioration during storage (Priestley, 1986). Here, it is demonstrated that the P50 could also be relevant for PCD and consequent loss of seed viability. The presence of <50% dead seeds does not allow the detection of PCD in a seed population. At the cellular level, this assessment has to be considered using the concept of ‘the point of no return’. The point of no return is defined as the point at which a cell that is undergoing PCD becomes irreversibly committed to die (Kroemer et al., 1995). Before this point of no return the deleterious events occurring in the cells are therefore reversible. The point of no return in fungi and animals has often been associated with lack of mitochondrial function (van Doorn, 2005). Recent studies have provided evidence to substantiate the importance of mitochondrial production of ROS in PCD during ageing in animal cells (Lee and Wei, 2007). Indeed, oxidative stress seems to be responsible for the decline of mitochondrial respiratory function during human ageing (Wei, 1998) and respiratory function could enhance ROS production in mitochondria resulting in ATP depletion, the initiation of the mitochondrial permeability transition, and PCD in plants (Tiwari et al., 2002). As mentioned previously, in the present system, the AP decreased during ageing whatever the MC, but stronger reduction began at an MC of 0.29–0.37 g H2O g−1 DM, suggesting the occurrence of mitochondrial dysfunction before PCD at P50 (Fig. 2).
The activities of detoxifying enzymes, superoxide dismutase (SOD), catalase (CAT), and glutathione reductase (GR), decreased rapidly, thus leading to ROS accumulation (Kibinza et al., 2006). Bailly et al. (1998) have demonstrated that priming could reverse the age-related damage of sunflower seeds, mainly by enhancing the activities of antioxidant enzymes and therefore allowing the cells to escape from oxidative stress. It is proposed here that the sustained ability of cells to withstand this oxidative stress would prevent them from reaching the point of no return.
These data combined with previous data using sunflower seeds (Bailly et al., 1996, 1998, 2002) allow the sequential involvement of the cellular events discussed above to be proposed, ROS being key players in loss of viability. When seed MC increases, lipoxygenase, an enzyme that produces free radicals, is activated (Bailly et al., 2002) and respiration resumes at ∼0.08 g H2O g−1 DM (Kibinza et al., 2006). Both events generate ROS that may serve, with lipid peroxidation by-products, as inducing factors for PCD through DNA alteration and mitochondrial dysfunctioning. Then mitochondrial activity is dramatically impaired and the cells pass the point of no return, leading to cell and seed death.
The present findings allow a model of seed ageing in hydrated conditions to be proposed which probably does not differ from that occurring in the soil seed bank, where seeds undergo hydration–rehydration cycles (Mickelson and Grey, 2006). Furthermore, this model is very close to the one involved in ageing of plant and animal organisms. It has already been suggested that plants, like mammalian cells, possess constitutively expressed PCD machinery in an inactive state waiting to receive activation signals and carry out cell death if necessary (Weil et al., 1996; Elbaz et al., 2002). In addition it is shown that seed death results from a synchronous loss of cell viability that occurs when degenerative processes reach the point of no return due to oxidative stress.
Glossary
Abbreviations
- DAPI
4′,6-diamidino-2-phenylindole
- DM
dry matter
- MC
moisture content
- PCD
programmed cell death
- PI
propidium iodide
- RAPD
random amplification of polymorphic DNA
- ROS
reactive oxygen species
References
- Atienzar FA, Jha AN. The random amplified polymorphic DNA (RAPD) assay and related techniques applied to genotoxicity and carcinogenesis studies: a critical review. Mutation Research. 2006;613:76–102. doi: 10.1016/j.mrrev.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Atienzar FA, Venier P, Jha AN, Depledge MH. Evaluation of the random amplified polymorphic DNA (RAPD) assay for the detection of DNA damage and mutations. Mutation Research. 2002;521:151–163. doi: 10.1016/s1383-5718(02)00216-4. [DOI] [PubMed] [Google Scholar]
- Atlante A, Gagliardi S, Marra E, Calissano P. Neuronal apoptosis in rats is accompanied by rapid impairment of cellular respiration and is prevented by scavengers of reactive oxygen species. Neuroscience Letters. 1998;245:127–130. doi: 10.1016/s0304-3940(98)00195-5. [DOI] [PubMed] [Google Scholar]
- Atlante A, Gagliardi S, Minervini GM, Marra E, Passarella S, Calissano P. Rapid uncoupling of oxidative phosporylation accompanies glutamate toxicity in rat cerebellar granule cells. Neuroreport. 1996;7:2519–2523. doi: 10.1097/00001756-199611040-00023. [DOI] [PubMed] [Google Scholar]
- Bailly C. Reactive oxygen species and antioxidants in seed biology. Seed Science Research. 2004;14:93–107. [Google Scholar]
- Bailly C, Benamar A, Corbineau F, Côme D. Changes in malondialdehyde content and in superoxide dismutase, catalase and glutathione reductase activities in sunflower seeds as related to deterioration during accelerated aging. Physiologia Plantarum. 1996;97:104–110. [Google Scholar]
- Bailly C, Benamar A, Corbineau C, Côme D. Free radical scavenging as affected by accelerated ageing and subsequent priming in sunflower seeds. Physiologia Plantarum. 1998;104:646–652. [Google Scholar]
- Bailly C, Bogatek-Leszczynska R, Côme D, Corbineau F. Changes in activities of antioxidant enzymes and lipoxygenase during growth of sunflower seedlings from seeds of different vigour. Seed Science Research. 2002;12:47–55. [Google Scholar]
- Boubriak I, Kargiolaki H, Lyne L, Osborne DJ. The requirement for DNA repair in desiccation tolerance of germinating embryos. Seed Science Research. 1997;7:97–105. [Google Scholar]
- Bras M, Queenan B, Susin SA. Programmed cell death via mitochondria: different modes of dying. Biochemistry. 2005;70:231–239. doi: 10.1007/s10541-005-0105-4. [DOI] [PubMed] [Google Scholar]
- Cash TP, Pan Y, Simon MC. Reactive oxygen species and cellular oxygen sensing. Free Radical Biology and Medicine. 2007;43:1219–1225. doi: 10.1016/j.freeradbiomed.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah KSE, Osborne DJ. DNA lesions occur with loss of viability in embryos of ageing rye seeds. Nature. 1978;272:593–599. doi: 10.1038/272593a0. [DOI] [PubMed] [Google Scholar]
- Chwedorzewska KJ, Bednarek PT, Puchalski J. AFLP-profiling of long-term stored and regenerated rye genebank samples. Cellular and Molecular Biology Letters. 2002;7:457–463. [PubMed] [Google Scholar]
- Corbineau F, Bagniol S, Côme D. Sunflower (Helianthus annuus L.) seed dormancy and its regulation by ethylene. Israel Journal of Botany. 1990;39:313–325. [Google Scholar]
- Corbineau F, Gay-Mathieu C, Vinel D, Côme D. Decrease in sunflower (Helianthus annuus) seed viability caused by high temperature as related to energy metabolism, membrane damage and lipid composition. Physiologia Plantarum. 2002;116:489–496. [Google Scholar]
- Corbineau F, Rudnicki RM, Côme D. Induction of secondary dormancy in sunflower seeds by high temperature. Possible involvement of ethylene biosynthesis. Physiologia Plantarum. 1988;73:368–373. [Google Scholar]
- Danon A, Delorme V, Mailhac N, Gallois P. Plant programmed cell death: a common way to die. Plant Physiology and Biochemistry. 2000;38:647–655. [Google Scholar]
- Desikan R, Reynolds A, Hancock JT, Neill SJ. Harpin and hydrogen peroxide both initiate programmed cell death but have differential effects on defence gene expression in Arabidopsis suspension cultures. Biochemical Journal. 1998;330:115–120. doi: 10.1042/bj3300115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz M, Avril A, Weil M. Constitutive caspase-like machinery executes programmed cell death in plant cells. Cell Death and Differentiation. 2002;9:726–733. doi: 10.1038/sj.cdd.4401030. [DOI] [PubMed] [Google Scholar]
- Enan MR. Application of random amplified polymorphic DNA (RAPD) to detect the genotoxic effect of heavy metals. Biotechnology and Applied Biochemistry. 2006;43:147–154. doi: 10.1042/BA20050172. [DOI] [PubMed] [Google Scholar]
- Gay C, Corbineau F, Côme D. Effects of temperature and oxygen on seed germination and seedling growth in sunflower (Helianthus annuus L) Environmental and Experimental Botany. 1991;31:193–200. [Google Scholar]
- Goldenberger A, Perschil I, Ritzler M, Altwegg M. A simple ‘universal’ DNA extraction procedure using SDS and proteinase K is compatible with direct PCR amplification. Genome Research. 1995;4:368–370. doi: 10.1101/gr.4.6.368. [DOI] [PubMed] [Google Scholar]
- Grant JJ, Loake GJ. Role of reactive oxygen intermediates and cognate redox signalling in disease resistance. Plant Physiology. 2000;124:21–29. doi: 10.1104/pp.124.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- Kibinza S, Vinel D, Côme D, Bailly C, Corbineau F. Sunflower seed deterioration as related to moisture content during ageing, energy metabolism and active oxygen species scavenging. Physiologia Plantarum. 2006;128:496–506. [Google Scholar]
- Kranner I, Birtic S, Anderson KM, Prichard HW. Glutathione half-cell reduction potential: a universal stress marker and modulator of programmed cell death. Free Radical Biology and Medicine. 2006;40:2155–2165. doi: 10.1016/j.freeradbiomed.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Kranner I, Minibayeva FV, Beckett RP, Seal CE. What is stress? Concepts, definitions and applications in seed science. New Phytologist. 2010;188:655–673. doi: 10.1111/j.1469-8137.2010.03461.x. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Petit P, Zamzami N, Vayssiere JL, Mignotte B. The biochemistry of programmed cell death. FASEB Journal. 1995;9:1277–1287. doi: 10.1096/fasebj.9.13.7557017. [DOI] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Laloi C, Apel K, Danon A. Reactive oxygen signalling: the latest news. Current Opinion in Plant Biology. 2004;7:323–328. doi: 10.1016/j.pbi.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Lee HC, Wei YH. Oxidative stress, mitochondrial DNA mutation, and apoptosis in aging. Experimental Biology and Medicine. 2007;232:592–606. [PubMed] [Google Scholar]
- Liu W, Li PJ, Qi XM, Zhou QX, Zheng L, Sun TH, Yang YS. DNA changes in barley (Hordeum vulgare) seedlings induced by cadmium pollution using RAPD analysis. Chemosphere. 2005;61:158–167. doi: 10.1016/j.chemosphere.2005.02.078. [DOI] [PubMed] [Google Scholar]
- McDonald MB. Seed deterioration: physiology, repair and assessment. Seed Science Technology. 1999;27:177–237. [Google Scholar]
- Mickelson JA, Grey WE. Effect of soil water content on wild oat (Avena fatua) seed mortality and seedling emergence. Weed Science. 2006;54:255–262. [Google Scholar]
- Neill S, Desikan R, Hancock J. Hydrogen peroxide signalling. Current Opinion in Plant Biology. 2002;5:388–395. doi: 10.1016/s1369-5266(02)00282-0. [DOI] [PubMed] [Google Scholar]
- Nicoletti IG, Migliorati MC Grignani PF, Riccardi C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. Journal of Immunological Methods. 1991;139:271–279. doi: 10.1016/0022-1759(91)90198-o. [DOI] [PubMed] [Google Scholar]
- O'Brien TP, McCully MF. The study of plant structure: principles and selected methods. Melbourne, Australia: Termacarphi Pty; 1981. [Google Scholar]
- Olempska-Beer Z, Bautz-Freeze E. Optimal extraction conditions for high-performance liquid chromatography determination of nucleotides in yeast. Analytical Biochemistry. 1984;140:236–245. doi: 10.1016/0003-2697(84)90159-3. [DOI] [PubMed] [Google Scholar]
- Osborne DJ, Sharon R, Ben-Ishai R. Studies on DNA integrity and DNA repair in germinating embryos of rye (Secale cereale) Israel Journal of Botany. 1981;29:259–272. [Google Scholar]
- Pietrasanta LI, Smith BL, MacLeod MC. A novel approach for analyzing the structure of DNA modified by benzo[a]pyrene diol epoxide at single-molecule resolution. Chemical Research in Toxicology. 2000;13:351–355. doi: 10.1021/tx9902035. [DOI] [PubMed] [Google Scholar]
- Priestley DA. Seed aging. Implications of seed storage and persistence in the soil. Ithaca, NY: Cornell University Press; 1986. [Google Scholar]
- Samejima K, Earnshaw WC. Trashing the genome: the role of nucleases during apoptosis. Nature Reviews Molecular Cell Biology. 2005;6:677–688. doi: 10.1038/nrm1715. [DOI] [PubMed] [Google Scholar]
- Shatters RG, Schweder ME, West SH, Abdelghany A, Smith RL. Environmentally-induced polymorphisms detected by RAPD analysis of soybean seed DNA. Seed Science Research. 1995;5:109–116. [Google Scholar]
- Slupphaug G, Kavli B, Krokan HE. The interacting pathways for prevention and repair of oxidative DNA damage. Mutation Research. 2003;531:231–251. doi: 10.1016/j.mrfmmm.2003.06.002. [DOI] [PubMed] [Google Scholar]
- Sossey-Alaoui K, Serieys H, Tersac M, Lambert P, Schilling E, Griveau Y, Kaan F, Berville A. Evidence for several genomes in Helianthus. Theoretical and Applied Genetics. 1998;97:422–430. [Google Scholar]
- Stein JC, Hansen G. Mannose induces an endonuclease responsible for DNA laddering in plant cells. Plant Physiology. 1999;121:71–80. doi: 10.1104/pp.121.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton WG, Olanow CW. Apoptosis in neurodegenerative diseases: the role of mitochondria. Biochimica et Biophysica Acta. 1999;1410:195–213. doi: 10.1016/s0005-2728(98)00167-4. [DOI] [PubMed] [Google Scholar]
- Tiwari BS, Belenghi B, Levine A. Oxidative stress increased respiration and generation of reactive oxygen species, resulting in ATP depletion, opening of mitochondrial permeability transition, and programmed cell death. Plant Physiology. 2002;128:1271–1281. doi: 10.1104/pp.010999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca RA, de PintoMC Valenti D, Passerella S, Marra E, De Gara L. Production of reactive oxygen species, alteration of cytosolic ascorbate peroxidase, and impairment of mitochondrial metabolism are early events in heat shock-induced programmed cell death in tobacco Bright-Yellow 2 cells. Plant Physiology. 2004;134:1100–1112. doi: 10.1104/pp.103.035956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn WG. Plant programmed cell death and the point of no return. Trends in Plant Science. 2005;10:478–483. doi: 10.1016/j.tplants.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Vazquez-Ramos JM, Lopez S, Vazquez E, Murillo E. DNA integrity and DNA polymerase activity in deteriorated maize embryo axes. Journal of Plant Physiology. 1988;133:600–604. [Google Scholar]
- Vijay D, Dadlani M, Kumar PA, Panguluri SK. Molecular marker analysis of differentially aged seeds of soybean and safflower. Plant Molecular Biology Reporter. 2009;27:282–291. [Google Scholar]
- Walters C. Understanding the mechanisms and kinetics of seed aging. Seed Science Research. 1998;8:223–244. [Google Scholar]
- Wei YH. Oxidative stress and mitochondrial DNA mutations in human aging. Proceedings of the Society for Experimental Biology and Medicine. 1998;217:53–63. doi: 10.3181/00379727-217-44205. [DOI] [PubMed] [Google Scholar]
- Wei YH, Lee HC. Oxidative stress, mitochondrial DNA mutation, and impairment of antioxidant enzymes in aging. Experimental Biology and Medicine. 2002;227:671–682. doi: 10.1177/153537020222700901. [DOI] [PubMed] [Google Scholar]
- Weil M, Jacobson MD, Coles HS, Davies TJ, Gardner RL, Raff KD, Raff MC. Constitutive expression of the machinery of programmed cell death. Journal of Cell Biology. 1996;133:1053–1059. doi: 10.1083/jcb.133.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J, Kubelik AR, Livak KJ, Rafalski JA, Tinger SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acid Research. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]