Abstract
Pectins are complex polysaccharides that are essential components of the plant cell wall. In this study, a novel putative Arabidopsis S-adenosyl-L-methionine (SAM)-dependent methyltransferase, termed QUASIMODO 3 (QUA3, At4g00740), has been characterized and it was demonstrated that it is a Golgi-localized, type II integral membrane protein that functions in methylesterification of the pectin homogalacturonan (HG). Although transgenic Arabidopsis seedlings with overexpression, or knock-down, of QUA3 do not show altered phenotypes or changes in pectin methylation, this enzyme is highly expressed and abundant in Arabidopsis suspension-cultured cells. In contrast, in cells subjected to QUA3 RNA interference (RNAi) knock-down there is less pectin methylation as well as altered composition and assembly of cell wall polysaccharides. Taken together, these observations point to a Golgi-localized QUA3 playing an essential role in controlling pectin methylation and cell wall biosynthesis in Arabidopsis suspension cell cultures.
Keywords: Arabidopsis, cell wall, homogalacturonan, methyltransferase, pectin, plant suspension cultured cells, S-adenosyl-L-methionine
Introduction
The plant cell wall is vital in the support of the structure of the cell, and is also the first interface with the environment to sense numerous external signals including osmotic and mechanical stresses, pathogen attack, and hormones (Xu et al., 1995; Roberts, 2001; Dhugga, 2005). The mechanical and functional properties of the plant cell wall are sustained by the complex architecture of polysaccharides, which includes cellulose microfibrils, hemicellulose, and pectin (McNeil et al., 1984; Varner and Lin, 1989; Roberts, 2001; Dhugga, 2005; Harholt et al., 2010; Jolie et al., 2010; Keegstra, 2010).
Pectins are complex polysaccharides that play crucial roles in coordinating plant growth, development, and defence (Willats et al., 2001a; Bacic, 2006; Jensen et al., 2008; Mohnen, 2008). They are composed of a mixture of linear and branched polymers including homogalacturonan (HG), rhamnogalacturonan-I (RG-I), and rhamnogalacturonan-II (RG-II) that are enriched in galacturonic acid (GalA) (Varner and Lin, 1989; Carpita and Gibeaut, 1993; Willats et al., 2001a). HG, a major pectin component, is a linear homopolymer composed of 1,4-linked α-D-galacturonic acid residues, in a highly methylesterified form, secreted to the cell surface during cell wall assembly (Micheli, 2001; Bosch et al., 2005; Sterling et al., 2006). Methylesterification is the most widespread biosynthetic modification of HG in the regulation of cell wall composition and in effecting cell–cell connections (Carpita and Gibeaut, 1993; Micheli, 2001; Willats et al., 2001a; Krupkova et al., 2007; Mouille et al., 2007). The degree of methylesterification (DM) of pectin is thought to be largely controlled by the coordination of Golgi-localized pectin methyltransferase (MTase), which initially transfers the methyl groups from methyl donors to pectin (Vannier et al., 1992; Goubet and Mohnen, 1999a, b), with wall-associated pectin methylesterase (PME) that subsequently releases the methyl group from the polygalacturonic acid chain (Wen et al., 1999; YQ Li et al., 2002; Bosch et al., 2005). In addition, the carboxylate groups of HG could interact with either methyl groups or bivalent calcium ions. Unmethylesterified or demethylesterified sites within HG can be occupied by calcium to form cross-links, which result in the gelation of pectic polymers to stiffen the cell wall and thus affect cell growth and development (Carpita and Gibeaut, 1993; Willats et al., 2001a).
The Arabidopsis genome contains 29 putative genes for pectin MTase (Krupkova et al., 2007) and 66 putative genes for PME (Bosch and Hepler, 2005; Pina et al., 2005; Louvet et al., 2006). However, only a few have been characterized at the functional level, such as the pollen-specific PME genes, VANGUARD1 and QUARTET1, which regulate pollen development (Jiang et al., 2005; Francis et al., 2006), and a putative pectin MTase gene, QUA2/TSD2, that functions in controlling the development of the hypocotyl and cell adhesion in Arabidopsis (Krupkova et al., 2007; Mouille et al., 2007). Although pectin MTase activities have been detected in purified Golgi fractions from various plant species (Goubet et al., 1998; Goubet and Mohnen, 1999a, b; Ibar and Orellana, 2007), the biochemical methyltransferring activity of Arabidopsis pectin MTase remains to be demonstrated. Comprehensive investigation of mRNA and protein expression, and enzymatic activity of specific pectin MTase is crucial to understand the underling mechanisms involved in regulating cell wall modification. Here a novel S-adenosyl-L-methionine (SAM)-dependent MTase QUA3 has been characterized in Arabidopsis using a combination of cellular, molecular, and biochemical approaches.
Materials and methods
General methods for construction and characterization of recombinant plasmids, and maintenance of tobacco and Arabidopsis suspension-cultured cells have been described previously (Jiang and Rogers, 1998; Tse et al., 2004; Miao and Jiang, 2007; Miao et al., 2008; Lam et al., 2009; Suen et al., 2010; Wang et al., 2010). PSBL and PSBD are Arabidopsis suspension cultured cells growth under light and dark conditions, respectively. Arabidopsis seedlings were grown on MS agar plates in an environmental chamber prior to being transferred to soil and grown in the greenhouse, under 16 h light/8 h dark conditions at 22 °C.
Plant materials and transformation
Transgenic BY-2 cells were generated via Agrobacterium tumefaciens-mediated transformation using strain LBA4404 (Tse et al., 2004; Miao et al., 2006; Lam et al., 2007, 2008; Wang et al., 2010). Transgenic Arabidopsis (ecotype, Nossen) plants were generated via the Agrobacterium-mediated floral dip method (Clough and Bent, 1998). The qua3 (Pst13453) mutant was obtained from the RIKEN Genomic Science Center, Japan (Kuromori et al., 2004). Transient expression in Arabidopsis protoplasts was carried out as described previously (Miao and Jiang, 2007; Lam et al., 2009; Cai et al., 2011; Wang and Jiang, 2011).
Plasmid construction
Constructs used in this study were derived from either the PBI221 backbone plasmids for transient expression or the binary vector PBI121 for stable transformation. All constructs, containing the cauliflower mosaic virus (CaMV) 35S promoter and the NOS terminator, were checked by both restriction mapping and DNA sequencing (see Supplementary Fig. S3 available at JXB online). QUA3 cDNA (pda04138) was obtained from RIKEN (Seki et al., 2002) and subcloned into a CaMV expression cassette to generate QUA3-OE and QUA3–green fluorescent protein (GFP).
Chemicals
The chloride salt of SAM, Triton X-100, trypsin and trypsin inhibitor, trichloroacetic acid (TCA), bovine serum albumin (BSA), wortmannin, and polygalacturonic acid (HG) were purchased from Sigma (St Louis, MO, USA). [Methyl-14C]SAM (50–62 mCi mmol−1) was purchased from Amersham Pharmacia.
Antibodies
Synthetic peptides in between the transmembrane domain (TMD) and the DUF248 domain (CEDPRRNSQLSREMNFYR) were synthesized (GeneScript) and conjugated with keyhole limpet haemocyanin to immunize rabbits at the animal house of the Chinese University of Hong Kong (CUHK). The generated antibodies were further affinity purified with a CNBr-activated Sepharose (Sigma-Aldrich) column conjugated with the synthetic peptides as described previously (Rogers et al., 1997; Tse et al., 2004) and used at 4 μg ml−1 for western blot analysis. The production of antibodies specific for anti-Man1 and VSRat-1 was described previously (Tse et al., 2004). The Arabidopsis Sec23p and Sar1p antibodies (Yang et al., 2005) were kindly provided by Professor David Robinson (University of Heidelberg, Germany). The JIM5, JIM7, and LM7 antibodies were ordered from the Paul Knox cell wall lab (PlantProbes; University of Leeds, UK). Polyclonal Alexa fluor-568 anti-rabbit (Molecular Probes, USA) or monoclonal anti-rat secondary antibodies (Sigma, USA) were used for immunofluorescent detection.
Confocal immunofluorescence
Images of fluorescent signals were obtained from intact protoplasts in an incubation medium. Confocal fluorescent images were collected using a Bio-Rad Radiance 2100 system or an Olympus IX81 confocal microscope (Olympus). The settings for collecting the images within the linear range are described in Jiang and Rogers (1998). Images were processed using Adobe Photoshop software (San Jose, CA, USA) as previously described (Jiang and Rogers, 1998; Shen et al., 2011; Wang et al., 2011). All experiments were repeated at least twice, with similar results.
HG-MTase enzyme assay
The production of protoplasts from BY-2 tobacco cells was carried out as previously described (Miao and Jiang 2007). The medium containing the protoplasts (∼5 ml) was washed twice with 0.4 M mannitol and once with 13.7% (w/v) sucrose before being further lysed by passage four times through a 255/8 gauge needle, and then spun at 1000 g for 10 min. The supernatant was collected, loaded on top of 10 ml of 50% (w/v) sucrose in TM buffer (10 μM TRIS-HCl, pH 7.8, 2 mM MgCl2), and centrifuged at 110 000 g for 1 h. The middle layer in the tube (∼6 ml) was collected and loaded onto a step gradient of 8 ml of 8.5% and 40% (w/v) sucrose, and spun at 110 000 g for a further 1 h. The fraction collected from the interface was diluted with 1 vol. of TM buffer and centrifuged at 110 000 g for 15 min; enriched Golgi vesicles were subsequently resuspended in 150 μl of STM buffer (0.25 M sucrose, 10 mM TRIS-HCl, pH 7.8, 2 mM MgCl2).
The MTase assay was performed according to Ibar et al. (2007) with some modifications. Briefly, 25 μl of the enriched Golgi vesicles were incubated in a final volume of 50 μl of STM buffer (pH 7.8) containing 4 μM [methyl-14C]SAM (final concentration), 6 μM SAM (final concentration), 0.1% (v/v) Triton X-100, and 50 μg of HG at 30 °C for 2 h. The reaction was stopped by the addition of 1 vol. of 20% (w/v) TCA and 5 μl of 10% (w/v) BSA solution. The resulting suspension was centrifuged for 10 min at 4000 g. Unincorporated SAM was removed by washing the pellet twice with 200 μl of 2% (w/v) TCA. The washed pellets were dissolved in 0.1 N NaOH for 30 min before measurement of radioactivity incorporation with a liquid scintillation counter LS 6500 (Beckman, USA).
Immunofluorescence detection of HG
Fixation and preparation of tissues from hypocotyls of 2-week-old Arabidopsis seedlings for paraffin-embedded sections, antibody labelling, and subsequent analysis by confocal immunofluorescence have been described previously (Jiang and Rogers, 1998; Jiang et al., 2000, 2001; YB Li et al., 2002). Briefly, hypocotyls were cut and incubated in fixation solution containing 10% (v/v) formaldehyde, 50% (v/v) ethanol, and 5% (v/v) acetic acid for 24 h. Dehydration and infiltration of samples were performed with an Enclosed Tissue Processor (Leica TP-1050; Leica Microsystems), followed by embedding in paraffin blocks. Thin paraffin-embedded sections were deparaffinized and used for single antibody labelling using JIM5, JIM7, and LM7 at 1:10 dilution. At least three independent high-pressure freezing and subsequent substitution experiments were carried out for each set of electron microscopy (EM) studies. Immunolabellings using various antibodies were performed, respectively, on each set of ultrasectioned samples. The quantification of immunogold labelling was carried out essentially as described previously (Wang et al., 2009a, b; Barany et al., 2010). The density of gold particles from immuogold EM studies was determined by counting the numbers of gold particles from random transmission electron microscopy (TEM) images of identical size. The density of gold labelling was expressed as the average number of gold particles per square micrometre with standard errors.
EM of resin-embedded cells
The general procedures for TEM sample preparation and thin sectioning were performed as described (Ritzenthaler et al., 2002; Tse et al., 2004). For high-pressure freezing, cells were harvested by filtering and immediately frozen in a high-pressure freezing apparatus (EMP2, Leica, Bensheim, Germany). Substitution, resin infiltration, embedding, and polymerization were carried out as previously described (Lam et al., 2007; Wang et al., 2007) in an AFS freeze-substitution unit (Leica, Wetzlar, Germany). Immunolabelling on HM20 embedded sections was performed (Hohl et al., 1996; Tse et al., 2004) using QUA3 primary antibody at 100 μg ml−1, and gold-coupled secondary antibodies at 1:50 dilution. Aqueous uranyl acetate/lead citrate post-stained sections were examined with a JOEL JEM-1200 EXII (Joel, Tokyo, Japan) or a Hitachi H-7650 transmission electron microscope with a CCD camera (Hitachi High-Technologies Corporation, Japan) operating at 80 kV, as described previously (Tse et al., 2004; Wang et al., 2010).
Analysis of gene and protein expression
For total RNA isolation, different organs were collected for QUA3 expression analysis. RNA was extracted (RNeasy, Qiagen) and reverse transcribed (Superscript-II, Invitrogen). Quantitative real-time PCR (qRT-PCR) was performed using SYBR-Green in an iQ5 real-time PCR system (Bio-Rad Laboratories). Primers were designed by Primer Express 2.0 (Applied Biosystems) (Table 1). The tubulin β-3 gene was used as an amplification internal control. Protein extraction and western blot analysis were as previously described (Tse et al., 2004; Wang et al., 2007; Shen et al., 2011), and anti-tubulin (anti-TUB) antibodies (Roche Diagnostics, Switzerland) were used as an internal control.
Table 1.
Glycosyl residue composition of fractionated cell walls from Arabidopsis suspension-cultured PSBD cells and those of the RNAi line qua3i
| Residue | Sugarb | Sugarc | Ara (mol %) | Rha (mol %) | Fuc (mol %) | Xyl (mol %) | GalA (mol %) | Man (mol %) | Gal (mol %) | Glc (mol %) | |
| Cell wall fractiona | |||||||||||
| CDTA | PSBD | 34.5 | 8.6 | 37.1 | 2.9 | 1.5 | 1.2 | 41.8 | 0.5 | 11.9 | 3 |
| qua3i | 74 | 19.7 | 24.2 | 2.2 | 1.5 | 1.1 | 43.9 | 0.3 | 25.9 | 0.8 | |
| Na2CO3 | PSBD | 137.2 | 27.4 | 26.4 | 6.9 | 1.3 | 1.4 | 46.6 | 0.3 | 14.5 | 2.6 |
| qua3i | 145.1 | 38.7 | 32.3 | 8.6 | 1.6 | 1.4 | 44 | 0.2 | 9.3 | 2.6 | |
| KOH | PSBD | 248.9 | 62.2 | 9.6 | 2.6 | 1 | 6.7 | 7.6 | 0.1 | 3.8 | 68.5 |
| qua3i | 156 | 41.6 | 28.2 | 6.6 | 1.8 | 16 | 20.8 | 0.2 | 7.2 | 19.2 |
Cell walls were extracted from Arabidopsis suspension-cultured PSBD cells and its derived RNAi line qua3i, and sequentially fractionated using CDTA (sodium salt), Na2CO3, and finally 0.5–4 N KOH extraction. Soluble components were neutralized and dialysed against deionized water at 4 °C, and their trimethylsilane (TMS) derivatives were analysed by GC-MS for glycosyl residue composition.
The mass of total sugars recovered from each fraction (in micrograms).
The percentage of total sugars recovered from each fraction.
Analysis of QUA3 topology
Equal amounts of Golgi-enriched vesicles were incubated with 4 μg of trypsin in 25 μl of STM buffer for 1 h at 30 °C, in the presence or absence of 0.05% (v/v) Triton X-100. The reactions were stopped by the addition of 1.25 μl of 10 μg μl−1 soybean trypsin inhibitor, followed by incubation for 10 min at 30 °C. The samples were subsequently sonicated and boiled for western blot analysis using anti-QUA3.
Generation of QUA3 RNAi cell lines
For construction RNA interference (RNAi) lines, a 200 bp QUA3 fragment was amplified from the QUA3 coding sequence (257–456 bp relative to the 5′ ATG) using the primers qua3i-XhoI, qua3i-EcoRI, qua3i-XbaI, and qua3i-ClaI (Table S1 at JXB online). The PCR fragment generated with the Xba–Cla pair of primers (sense fragment) was cloned into the pHannibal vector (Wesley et al., 2001) to create pHannibal-QUA3-XC. The PCR fragment generated with the Xho–Eco pair of primers (antisense fragment) was cloned into this vector to create pHannibal-QUA3. An XbaI–XhoI RNAi fragment containing these two QUA3 sequences in opposing orientations separated by an 800 bp intron sequence was cloned into the binary vector pART27 (Gleave, 1992) containing the 35S promoter to create pART27-QUA3. Subsequently stable transformation was carried out in Arabidopsis suspension cell cultures using Agrobacterium C58C1Rif (pMP90M). Briefly, 3 ml of 2-day-old suspension cells were co-cultivated with 200 μl of pMP90M (OD600=1.0) in MS medium. After incubation at 130 rpm in an orbital shaker at 25 °C for 3 d, the cell cultures were subsequently washed three times with MS medium before 1 ml of the cells was transferred onto MS plates with kanamycin and cefotaxime for selection of resistant calli.
Extraction and fractionation of cell walls
Cell walls were extracted from freeze-dried culture cells (800 mg) by washing three times with 5 ml of homogenization buffer [40 mM HEPES-NaOH buffer, 10 mM imidazole, 1 mM benzamidine, 10 mM dithiothreitol, and 1 mM phenylmethylsulphonyl fluoride, methanol/chloroform (1:1), and acetone]. Air-dried pellets containing the cell walls were obtained by centrifugation at 10 000 g for 10 min and subjected to treatment with 30 U (1 mg ml−1) of protease (from Streptomyces griseus) in 50 mM TRIS-HCl buffer (pH 7.0) for 24 h at 37 °C. The deproteinated cell walls were centrifuged at 800 g for 10 min, dehydrated with acetone, and air dried. Subsequent fractionation of the cell walls for further analysis of glycosyl residue composition was performed as described by Persson et al. (2007) with modifications. Briefly, 100 mg of tissue were extracted with 50 mM cyclohexane-trans-1,2-diamine-NNN'N' tetra-acetate (CDTA; pH 6.5) at room temperature for 8 h and centrifuged at 2500 g for 5 min. The pellet was subsequently extracted with 50 mM Na2CO3 and 10 mM sodium borohydride for 16 h at 5 °C and then for 3 h at 20 °C. Finally, the pellet was sequentially extracted with 0.5, 1, and 4 M KOH, supplemented with 10 mM sodium borohydride, for 24 h at room temperature, and centrifuged at 2500 g for 5 min. The different fractions were acidified to pH 5.0 with glacial acetic acid and then dialysed for 3 d at 4 °C in water containing 0.05% chlorbutol.
Glycosyl residue composition analysis
Glycosyl composition analysis was performed by combined gas chromatography–mass spectrometry (GC-MS) of the per-O-trimethylsilyl (TMS) derivatives of the monosaccharide methyl glycosides produced by acidic methanolysis (York et al., 1985; Merkle and Poppe, 1994).
Results
Arabidopsis QUA3, a putative MTase, belongs to an evolutionarily conserved protein family unique to plants
The Golgi apparatus, trans-Golgi network (TGN), and pre-vacuolar compartment (PVC) have recently been isolated from Arabidopsis suspension-cultured cells for proteomic analysis (unpublished data). One of the identified proteins is At4g00740, a polypeptide of 600 amino acids with a predicted molecular mass of 67.5 kDa and pI of 9.06. At4g00740 shares 40% amino acid similarity with the Arabidopsis QUA2/TSD2 (Krupkova et al., 2007; Mouille et al., 2007) and 70% amino acid similarity in its MTase domain and C-terminal regions (Fig. 1A). Thus At4g00740 has been named after QUA2 as QUA3 in this study because of their sequence similarity. QUA3 is predicted (http://sparks.informatics.iupui.edu/Softwares-Services_files/thumbup.htm) to contain an N-terminal TMD, a DUF248 domain of unknown function, and an MTase domain belonging to a family of SAM-dependent MTases (Fig. 1B). Since SAM-dependent MTase is known to interact with both a methyl donor and acceptor (Martin and McMillan 2002), the high similarity between QUA3 and QUA2 in the MTase domains may indicate their similar function in plants. BLAST analysis (http://blast.ncbi.nl.m.nih.gov/Blast.cgi) showed that the plant contains at least 13 QUA3 homologues with a high amino acid similarity in various species; especially in their MTase domains (60% amino acid identity) (see Supplementary Fig. S1A at JXB online). The majority of MTase domains have a similar amino acid length, except for the rice homologue (Oryza sativa, CAH67036) (see Supplementary Fig. S1A). QUA3 and its plant homologues (see Supplementary Fig. S1B) also have no orthologue in animals or yeast, suggestive of a unique function for QUA3 in plants.
Fig. 1.
QUA3 structure and sequence alignment. (A) Sequence alignment of QUA2 and QUA3 proteins. Residues with identical amino acids or amino acids with similar chemical properties are shaded in black. The boxed region is the peptide sequence used for raising antibodies. (B) QUA3 protein structure. A transmembrane domain (TMD) is predicted at the N-terminal site. A C-terminal DUF248 domain is predicted to have a putative methyltransferase (MTase) function. The protein with an MTase domain belongs to the family of the S-adenosyl-L-methionine (SAM)-dependent MTases.
Subcellular localization of QUA3 in Arabidopsis
As a first step to determine the function of QUA3, its subcellular localization in plant cells was studied. QUA3 antibodies were generated using a synthetic peptide (CEDPRRNSQLSREMNFYR) for a region located at the N-terminal region of the DUF248 domain (Fig. 1B). Western blot analysis of Arabidopsis seedling protein extracts with affinity-purified QUA3 antibodies showed an expected QUA3 band at 67.5 kDa, in the membrane fraction (CM) but not the soluble protein fraction (CS) (Fig. 2). This is indicative that it is a membrane-associated protein. Similarly, a major single protein band of the same molecular mass was detected in Arabidopsis PSBL suspension-cultured cells. In addition, in transgenic tobacco BY-2 cells expressing a QUA3-GFP gene fusion, two major protein bands were detected representing the 67.5 kDa endogenous tobacco QUA3 homologue and the 95 kDa QUA3–GFP fusion protein, respectively (Fig. 2). Thus the QUA3 antibodies are highly specific for detecting both the endogenous QUA3 and the QUA3–GFP fusion proteins in Arabidopsis seedlings, Arabidopsis cultured cells, and transgenic tobacco BY-2 cells.
Fig. 2.
Peptide-derived QUA3 antibody specifically detects QUA3 in a membrane protein fraction obtained from Arabidopsis suspension-cultured cells. Membrane protein (CM) and soluble protein (CS) fractions of Arabidopsis 2-week-old seedlings, total protein of PSBL suspension cultures, and transgenic tobacco BY-2 cells expressing QUA3–GFP were extracted and examined by western blot using anti-QUA3. Asterisks indicate the expected size of QUA3 in Arabidopsis seedling and Arabidopsis PSBL suspension-cultureded cells, and QUA3 homologue and QUA3–GFP fusion proteins in tobacco BY-2 cells.
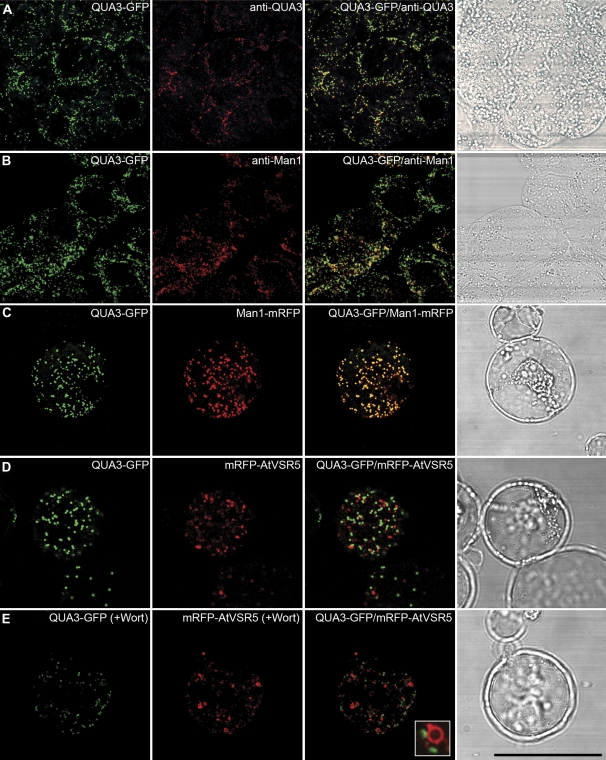
In transgenic tobacco BY-2 cells expressing the QUA3–GFP fusion, anti-QUA3 antibodies largely co-localized with the punctate QUA3–GFP in chemically fixed cells (Fig. 3A), again demonstrating the specificity of anti-QUA3 in detecting QUA3–GFP in transgenic BY-2 cells. Subsequent labelling experiments using various antibodies showed that QUA3–GFP co-localized with the Golgi marker anti-Man1 (Fig. 3B), but was separate from anti-VSR (a PVC marker), anti-AtSec23 (a COPII marker), and anti-AtSar1 (a COPI marker) (see Supplementary Fig. S2A–C at JXB online), indicative of the localization of QUA3–GFP in the Golgi of transgenic tobacco BY-2 cells. When QUA3–GFP was transiently co-expressed along with other known markers in Arabidopsis protoplasts, again QUA3–GFP co-localized with the Golgi marker Man1–mRFP (Fig. 3C), but not with the PVC marker monomeric red fluorescent protein (mRFP)–AtVSR5 in either wortmannin-untreated or -treated cells (Fig. 3D, E). Wortmannin, an effective inhibitor of phosphatidylinositol 3-kinase (PI3K), can induce specific dilation of plant PVCs, but has no obvious morphological effect on the Golgi apparatus (Tse et al., 2004; Miao et al., 2006, 2008).
Fig. 3.
Subcellular localization of a QUA3–GFP fusion protein in Arabidopsis suspension-cultured cells. (A and B) Co-localization of QUA3–GFP (green) is shown with anti-QUA3 (red, A) or anti-Man1 (red, B) in fixed transgenic BY-2 cells expressing QUA3–GFP as yellow signals. (C) QUA3–GFP (green) and Man1–mRFP (red) fusions were transiently co-expressed in protoplasts of Arabidopsis cells, and co-localized as a yellow signal. (D) QUA3–GFP (green) is separate from the PVC reporter mRFP–tVSR5 (red) in Arabidopsis protoplasts. (E) Arabidopsis protoplasts co-expressing QUA3–GFP (green) and mRFP–AtVSR5 (red) were treated with wortmannin (Wort) at 16.5 μM for 1 h before confocal imaging. The vacuolated PVCs marked by mRFP–AtVSR5 are separate from the unchanged green signal of QUA3–GFP. The corresponding DIC (differential interference contrast) images of the studied cells are included. Bar=50 μm.
Immunogold EM using anti-QUA3 was then used to determine QUA3 localization in Arabidopsis cells. As shown in Fig. 4, in ultra-thin sections prepared from high-pressure freezing/freeze substitution of Arabidopsis suspension PSBL cells, QUA3 antibodies specifically labelled the Golgi apparatus, with a relatively higher frequency of gold particles being associated with the stacks along the Golgi cisternae (Fig. 4). This detection was specific, because little labelling was observed in other organelles (Fig. 4D). Taken together, these results demonstrate that QUA3 and QUA3–GFP locate to the Golgi apparatus.
Fig. 4.
Immunogold labelling of QUA3 with anti-QUA3 in Arabidopsis suspension-cultured cells. Thin sections are prepared from high-pressure frozen/freeze-substituted PSBL suspension-cultured cells (A) and transgenic tobacco BY-2 cells overexpressing QUA3–GFP (B and C). QUA3 localization was detected by QUA3 (A, C) or GFP (B) antibodies. The Golgi apparatus is labelled specifically with these antibodies. (D) The distribution of immunogold as a percentage of each experiment was calculated (n=7). Bar in (A)=500 nm; in (B) and (C)=200 nm. Arrowheads indicate examples of gold particle labelling.
QUA3 is a type II integral membrane protein
QUA3 protein is predicted to contain a large C-terminal DUF248 domain, a SAM-dependent MTase domain, and a single TMD (Fig. 1). To find out the orientation of the functional domain of QUA3, transgenic BY-2 cells overexpressing QUA3 (QUA3-OE) (see Supplementary Fig. S3 at JXB online) were used to investigate the topology of the protein.
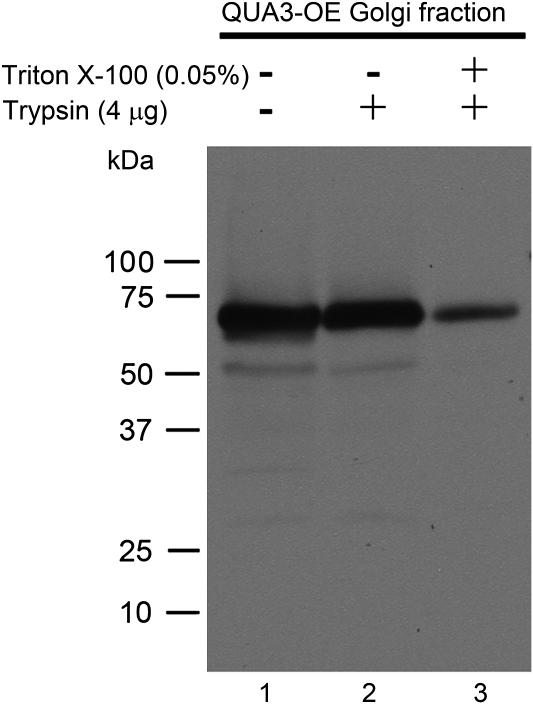
Protoplasts were first generated from QUA3-OE transgenic BY-2 cells by cellulose digestion, followed by isolation of Golgi-enriched vesicles using sucrose gradient fractionation. Isolated Golgi vesicles were then subjected to trypsin digestion in the presence or absence of Triton X-100, followed by SDS–PAGE and western blot detection using QUA3 antibodies. In the absence of both Triton X-100 and trypsin, a major 68 kDa band of QUA3 was detected (Fig. 5, lane 1). A similar, intense protein band was also detected in the Golgi fraction incubated with trypsin (Fig. 5, lane 2). In contrast, in the Golgi fraction pre-treated with both Triton X-100 and trypsin, the signal for the 68 kDa QUA3 protein band was greatly reduced (Fig. 5, lane 3). Since Triton X-100 treatment allows trypsin to access and digest the luminal proteins within the Golgi, these results demonstrate that QUA3 is a type II integral membrane protein with its C-terminal region inside the Golgi lumen.
Fig. 5.
Topological structure of QUA3 in the Golgi apparatus extracted from BY-2 cells. QUA3 orientation was determined by western blot analysis using anti-QUA3 on intact or Triton X-100-permeabilized Golgi vesicles of transgenic BY-2 cells overexpressing QUA3 proteins. Golgi fractions were treated (+) or not (–)with 0.05% (v/v) Triton X-100. Trypsin was used to digest the exposed protein fragments.
Substrate identification of QUA3
QUA3 belongs to a subfamily of SAM-dependent MTases in Arabidopsis, with 29 homologues (Krupkova et al., 2007; Mouille et al., 2007). Homologue QUA2/TSD2 has been suggested to be a putative pectin MTase that plays a critical role in cell adhesion (Krupkova et al., 2007; Mouille et al., 2007). However, the substrate of QUA2/TSD2 remains unknown.
Since QUA3 is a Golgi-localized, type II membrane protein similar to QUA2, it was hypothesized that it is a novel pectin MTase in Arabidopsis. QUA3 protein was thus expressed in tobacco cells and a methyl transfer assay was conducted using 14C-labelled methyl donor SAM to identify the substrate for this enzyme. To ensure plant-specific glycosylation of the recombinant QUA3 protein, Golgi-enriched fractions were prepared from wild-type (WT) and transgenic BY-2 cells expressing QUA3–GFP or QUA3-OE (see Supplementary Fig. S3 at JXB online) and used for enzymatic assays with HG as substrate. As shown in Fig. 6, the negative control of STM buffer alone showed ∼50–80 counts per minute (CPM) in the presence or absence of HG substrate (Fig. 6A, black columns). In contrast, WT tobacco BY-2 cells containing the endogenous tobacco pectin MTases showed activity of ∼200–240 CPM in the presence or absence of HG (Fig. 6A, white columns). Similar counts were obtained from BY-2 cells expressing the QUA3–GFP fusion (Fig. 6A, grey columns). In contrast, in tobacco BY-2 cells overexpressing QUA3-OE, relatively higher signals (∼350 CPM) compared with the WT cells were detected in the absence of HG, and activity increased substantially to 650 CPM in the presence of HG (Fig. 6A, dark grey columns), demonstrating the MTase activity of QUA3 on HG.
Fig. 6.
Identification of the pectin homogalacturonan as the QUA3 substrate in transgenic BY-2 cells in culture. (A) QUA3 enzymes are enriched in the Golgi fraction in extracts from transgenic BY-2 cells expressing QUA3 and QUA3–GFP, as shown using a discontinuous sucrose gradient. QUA3 activity was measured in the presence of 50 μg of polygalacturonic acid (HG), 4 μM [methyl-14C]SAM, 6 μM unlabelled SAM, and 0.1% (v/v) Triton X-100 in STM buffer. HG was precipitated in 20% (w/v) TCA and the radioactivity associated with the pellet was measured using a liquid scintillation counter. The measurements were performed in triplicate and the average activity plotted. (B) Loading of Golgi fractions of WT and transgenic BY-2 cells expressing QUA3 and QUA3–GFP. Equal loading of total Golgi proteins on the SDS–polyacrylamide gel was determined by Coomassie blue staining and on the western blot using Golgi markers anti-mannosidase 1 (anti-Man1) and anti-QUA3.
In the enzymatic assays equal amounts of proteins were used, as quantified by both Coomassie blue staining and western blot detection using the Golgi marker anti-Man1. Cells of overexpressed QUA3–GFP and QUA3-OE transgenic lines showed higher QUA3 activity than their QUA3 homologues in WT cells (Fig. 6B). However, transgenic tobacco BY-2 cells overexpressing QUA3–GFP proteins showed equal methyl transfer activity in the presence or absence of HG substrate when compared with the WT cells (Fig. 6A), indicating that the GFP fusion to the C-terminus of QUA3 abolished its enzymatic activity. In addition, the signal intensity of the introduced QUA3 protein in QUA3-OE BY-2 cells was higher than that of the endogenous protein in WT cells; this agrees with the methyl transfer activity results, accounting for the increased signals in QUA3-OE versus WT cells (Fig. 6A).
QUA3 regulates the degree of pectin methylesterification in Arabidopsis suspension cultures
The above enzyme activity assay demonstrated that QUA3 is a putative pectin MTase and transfers methyl groups to HG. Thus, the extent of methylation is expected to be correlated with that of expression of QUA3 in Arabidopsis cells. Western blot analysis demonstrated that Arabidopsis suspension cultures grown both in the dark (PSBD) and in the light (PSBL) have a similar QUA3 level (Fig. 7, lanes 1 and 2). To test the hypothesis that QUA3 functions in pectin methylesterification in these cells, a transgenic cell line expressing an RNAi silencing construct for QUA3 (termed qua3i) was next generated. This resulted in a greatly reduced amount of QUA3 mRNA transcripts (see Supplementary Fig. S4A at JXB online) and very little QUA3 protein compared with the WT PSBD cells (Fig. 7, lane 3 versus lane 4), indicating an effective knock-down of QUA3/QUA3 in the qua3i cell line.
Fig. 7.
Generation of an RNAi line of QUA3 in Arabidopsis suspension cell cultures. Anti-QUA3 reveals that QUA3 is expressed abundantly in Arabidopsis suspension cultures of PSBD or PSBL cells (1–2). The RNAi line of QUA3, qua3i, was generated from PSBD cells and is totally depleted of QUA3 expression (3–4). Anti-TUB was used as internal control to indicate equal protein loading.
To study further the differences in the degree of pectin methylesterification between WT PSBD and RNAi-containing qua3i cell lines, an immunogold labelling EM study was next performed on ultra-thin sections prepared from high-pressure freezing/frozen-substituted WT and qua3i cells using monoclonal antibodies JIM5, JIM7, and LM7, that recognize various degrees and patterns, respectively, of partially methylesterified HG in plant cell walls (Willats et al., 2000, 2001a; Clausen et al., 2003).
In sections labelled with JIM5 antibodies, the density of gold particles in qua3i cells was at least three times higher than in WT cells, especially in the middle lamella region that is enriched in pectin, which binds neighbouring cells together (Fig. 8A–E). However, no differences were observed between WT and qua3i cells labelled with either JIM7 (Fig. 8F–J) or LM7 antibodies (see Supplementary Fig. S5 at JXB online), which detect a different pattern and degree of pectin methylation than that detected by JIM5. No clear change in JIM7 and LM7 but only in JIM5 labelling also indicates that the methylation change is not due to an increase of pectic components. Cells expressing qua3i do not show any visible adhesion defects in the cell clumps (see Supplementary Fig. S6 at JXB online), which might be because the suspension-cultured cells already have a much looser connection than those of the intact plant. Taken together, these results demonstrate that depletion of QUA3 causes an obvious decrease of pectin methylation, leading to an increased number of unesterified pectic polymers.
Fig. 8.
qua3i shows decreased pectin methylesterification in suspension-cultured cells. (A) and (B) There is increased immunogold EM labelling with the monoclonal antibody, JIM5, in the cell walls of qua3i compared with those of WT PSBD cells. (C) and (D) Image magnification of (A) and (B), respectively. Monoclonal antibody JIM7 showed a similar density of immunogold labelling in WT PSBD cells (F) and qua3i (G). (H) and (I) Higher magnification of (F) and (G), respectively. The density of immunogold was analysed in both WT and qua3i cells (n=7), including the standard deviation on the graph (E, J). Bar in (A), (B), (F), and (G)=500 nm; in (C), (D), (H), and (I)=100 m.
Compositional analysis of fractionated cell wall materials of PSBD and qua3i
To determine if the polysaccharide composition was altered in qua3i cells, cell walls of 3-day-old qua3i and WT PSBD suspension cultures were isolated and sequentially separated into CDTA, Na2CO3, and KOH fractions for glycosyl composition analysis by GC-MS of trimethylsilyl derivatives, to quantify uronic acids and neutral sugars (York et al., 1985; Merkle and Poppe, 1994).
The CDTA and Na2CO3 fractions primarily contained galacturonic acid (GalA) and neutral sugars including arabinose (Ara), galactose (Gal), and rhamnose (Rha), but smaller amounts of xylose (Xyl), fucose (Fuc), mannose (Man), and glucose (Glc) (Table 1). Compared with the WT, no appreciable differences in GalA were observed in the cell walls of the qua3i cell lines, even though the CDTA and Na2CO3 fractions showed increased monosugars (Table 1). However, the qua3i-containing line showed increased Gal and decreased Ara in the CDTA fraction, and a reversed distribution of Gal and Ara in the Na2CO3 fraction, while other neutral sugars remained relatively constant in both fractions (Table 1).
The KOH fraction is composed of highly branched pectins (Stolle-Smits et al., 1999) and the majority of the hemicelluloses (Miedes and Lorences, 2004), and the present analysis showed that it mainly contains Glc, with moderate amounts of Xyl, Rha, Gal, and GalA, and a little Man and Fuc (Table 1). The Glc content obtained from the KOH fraction was much higher than that from the CDTA and Na2CO3 fractions. The total KOH-released glycan was decreased in qua3i cells, especially in Glc content, with an increase in other neutral sugars (Table 1); this is indicative of a possible change in cell wall structure in these cells. Since both CDTA and Na2CO3 fractions do not show increased GalA, Fuc, and Xyl in qua3i cell walls, only a subfraction of the pectin polymers is affected as a result of the decline in QUA3 activity.
Analysis of Arabidopsis knock-out mutant and overexpression plants
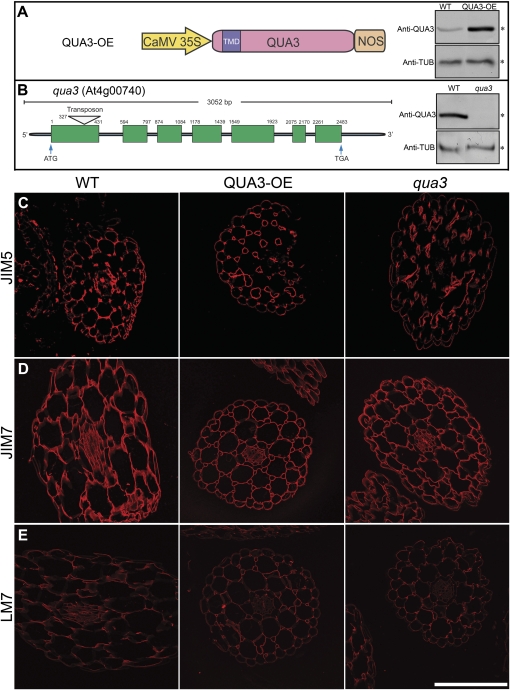
It was of interest to determine if this pattern of events in suspension cultures also occurs in intact tissues; hence the growing hypocotyl of Arabidopsis was used. Arabidopsis QUA3-OE plants (Fig. 9A) and the transposon insertional knock-out mutant (qua3) (Fig. 9B) were thus studied. The amount of QUA3 protein detected by anti-QUA3 antibody in QUA3-OE plants was much higher (∼10-fold) than in those of the WT (Fig. 9A); very little QUA3 protein was detected in the qua3 mutant (Fig. 9B) (as judged by equal loading of protein using anti-TUB).
Fig. 9.
Immunofluorescent labelling of transverse sections of qua3, QUA3-OE, and WT hypocotyls. (A) QUA3-OE was constructed under the control of the CaMV 35S promoter. Overexpressed QUA3 protein of transgenic Arabidopsis expressing QUA3-OE was compared with the WT, using tubulin antibody (anti-TUB) as the internal control. (B) The transposon, in the RIKEN line pst13453, was inserted into the first exon of qua3, which resulted in null QUA3 protein expression. (C–E) Immunofluorescent labelling of 6 μm thick transverse sections taken from hypocotyls of 2-week-old WT, QUA3-OE, and qua3 lines. Hypocotyls were labelled with monoclonal antibodies JIM5, JIM7, and LM7. Bar=200 μm.
To examine the possible effects of changes in QUA3 on pectin methylesterification, sections of the seedling hypocotyls of three Arabidopsis lines (WT, QUA3-OE, and qua3) were labelled with JIM5, JIM7, and LM7 (Willats et al., 2000, 2001a; Clausen et al., 2003). As shown in Fig. 9, no visible differences in labelling patterns or in signal intensity were observed in any line using anti-JIM5 (Fig. 9C), anti-JIM7 (Fig. 9D), or anti-LM7 (Fig. 9E). This result confirmed the immunogold EM studies using the same antibodies in ultra-thin sections of these lines (see Supplementary Fig. S7 at JXB online). In addition, no obvious phenotypic change was observed in either the QUA3-OE or qua3 seedlings as compared with the WT in terms of cell adhesion in the hypocotyls, the appearance of various organs using scanning EM, or in the length of the hypocotyls grown in the dark without sucrose supplementation (data not show).
Transcriptional profiles of the 29 QUA3 family members in Arabidopsis suspension cell cultures and seedlings
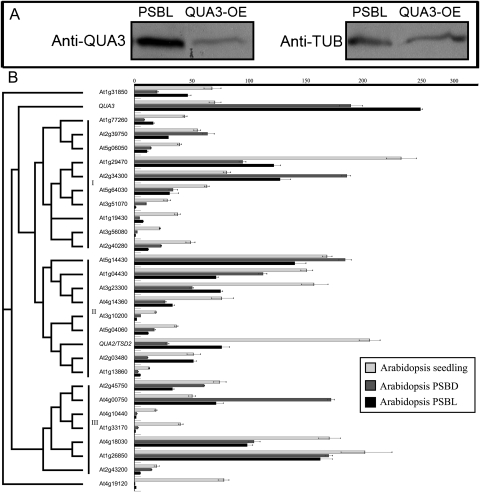
The above results indicate that QUA3 plays a role in Arabidopsis cell suspension cultures rather than in the intact seedling, so the reason for this was determined. Western blot analysis using anti-QUA3 showed that the amount of QUA3 protein was much higher in suspension-cultured cells than that in transgenic seedlings overexpressing this protein (QUA3-OE) (Fig. 10A). The Arabidopsis QUA3 family proteins can be classified into three clusters (I, II, and III), with QUA3 belonging to a unique branch outside of clusters I and II (Fig. 10B). qRT-PCR analysis of the 29 Arabidopsis QUA3 homologues showed that the expression of QUA3, At2g34300, and At4g00750 was much higher in suspension-cultured cells grown in the light or the dark (PSBD or PSBL) than in the seedlings (Fig. 10B). In addition, different tissues of the seedling all contained low amounts of QUA3 (see Supplementary Fig. S4B at JXB online). Also, transcripts of QUA3 were at least 2- to 3-fold higher in PSBD or PSBL suspension cells than in the seedling. Consistent with the critical role of QUA3 in controlling pectin methylation (Fig. 8) in suspension cell cultures, among all of the 29 homologues, the mRNA for QUA3 was almost the highest, especially in the PSBL cells. In contrast, most of the other QUA3 homologues, excluding At2g34300, At5g14430, At4g00750, and At1g26850, showed at least a 2-fold higher expression in the seedling than that in the PSBD or PSBL cells. Such a distribution of expression was especially obvious for QUA2/TSD2, one of the most highly expressed genes in the seedling, with only a moderate level of expression in the PSBD or PSBL cells (Fig. 10B), indicative of an important role for QUA2 in the developing seedling (Krupkova et al., 2007; Mouille et al., 2007). In addition, all of the highly expressed QUA3 homologues in suspension-cultured cells, At2g34300, At5g14430, At4g00750, and At1g26850, do not show perturbed mRNA expression in qua3i cells (data not shown), supporting the unique and specific regulatory role of QUA3 on pectin methylation in suspension cultures (Fig. 8).
Fig. 10.
qRT-PCR of 29 Arabidopsis QUA3 family members. (A) Arabidopsis PSBL suspension-cultured cells synthesize more QUA3 than transgenic Arabidopsis seedlings overexpressing QUA3 protein. (B) A phylogenetic consensus tree of 29 Arabidopsis QUA3 family members was constructed using the Neighbor–Joining algorithm with 1000 cycles of bootstrap resampling, and classified into three major clusters I, II, and III. Light grey, dark grey, and black bars represent mRNA expression levels determined using qRT-PCR, in 2-week-old Arabidopsis seedlings and 3-day-old Arabidopsis suspension-cultured PSBD and PSBL cells, respectively. The mRNA expression of Arabidopsis tubulin was used as an internal control. Bars indicate the average transcript level ±SE of three independent biological samples.
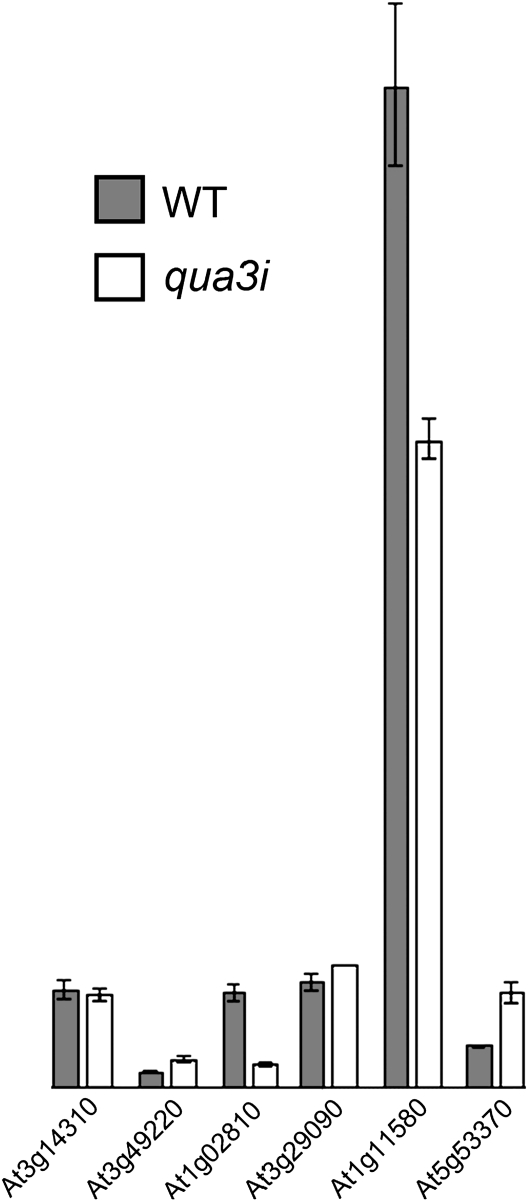
Since pectin MTase and PME play opposite roles in controlling pectin methylation, it is likely that the suspension-cultured cells contain corresponding PME genes working together with QUA3 in controlling the extent of pectin methylation. It is thus impossible to exclude the possibility that such increased unmethylesterified pectin is caused by an increase of PMEs in qua3i cells. To address this point and to find out if PMEs are increased to cause unesterification of qua3i pectin, among the 66 PME family members (Louvet et al., 2006), six seletive PME genes (At1g02810, At1g11580, AT3G29090, At3g14310, At3g49220, and At5g53370) that are the most highly expressed in suspension-cultured cells (https://www.genevestigator.com/gv/index.jsp) were analysed. As shown in Fig. 11, in comparison with the WT cells, both At1g02810 and the most highly expressed At1g11580 showed decreased mRNA expression in qua3i cells, while only At5g53370 showed a moderate increase in expression. Therefore, it is unlikely that the increased PME could cause the pectin phenotype of qua3i; rather, it is most probably the consequence of specific QUA3 depletion.
Fig. 11.
Expression of pectin methylesterase in Arabidopsis WT PSBD and qua3i suspension-cultured cells. The expression of the six selective PME genes (as indicated) was determined via qRT-PCR analysis using mRNA extracted from 3-day-old Arabidopsis suspension-cultured PSBD and qua3i cells, respectively.
Discussion
QUA3 is a Golgi-localized type II membrane pectin MTase
QUA3, containing an MTase domain, belongs to the superfamily of SAM-dependent MTases (Fig. 1), which can transfer methyl groups to various potential substrates including nucleotides, proteins, small molecules, and large polymers such as pectin polysaccharides (Martin and McMillan, 2002; Kozbial and Mushegian, 2005; Mouille et al., 2007). Arabidopsis QUA3 is unique in the plant kingdom in having a total of 15 homologues; there is no orthologue in humans or yeast (see Supplementary Fig. S1 at JXB online). QUA3 is thus predicted to have a function unique to plants. In addition, QUA3 has high amino acid similarity to the putative pectin MTase QUA2, with both MTase and C-terminus substrate interaction regions, and therefore is very likely top be involved in the biosynthesis and modification of cell wall pectins (Ibar and Orellana 2007). In this study, it has been shown that QUA3 is a Golgi-localized type II integral membrane protein in which the functional C-terminus contains an MTase domain facing the lumen (Figs 2–5), as expected of a protein with pectin modification properties located in this organelle (Dhugga, 2005; Ibar and Orellana, 2007; Krupkova et al., 2007; Mouille et al., 2007, Willats et al., 2001).
The transferase reactions that catalyse the synthesis and modification of cell wall pectins are all accomplished in the Golgi apparatus, in which HG becomes highly methylesterified by MTase prior to its secretion to the cell wall for assembly (Ridley et al., 2001; Willats et al., 2001a). HG was thus hypothesized to be the substrate of QUA3, which was shown in this study by enzymatic assay using Golgi fraction proteins isolated from transgenic tobacco BY-2 cells overexpressing QUA3 (QUA3-OE). Interestingly, transgenic BY-2 cells overexpressing a QUA3–GFP fusion did not show elevated methyl-transferring activity compared with the WT (Fig. 6), indicating that the presence of GFP affects the enzymatic activity. This could be due to a steric hindrance by the GFP fusion on the C-terminus of QUA3, thus affecting its ability to bind to either SAM or the HG substrate. However, such a negative effect on the QUA3 activity by GFP fusion did not affect the Golgi localization of QUA3–GFP as the GFP fusion fully co-localized with the endogenous QUA3 (Fig. 3A). These results thus indicate that GFP fusion only affects QUA3 activity probably via acting on either the substrate-binding domain or the methyl donor-binding domain, without altering its normal Golgi localization as the endogenous QUA3 proteins.
QUA3 plays a critical role in regulating pectin methylation in Arabidopsis suspension cell cultures
The extent of pectin methylesterification varies during plant growth and development (Schaumann et al., 1993; Goubet et al., 1998). This could be largely regulated by the methyl-transferring activities of pectin MTases in the Golgi apparatus (Vannier et al., 1992).
Ahigher amount of QUA3 protein was detected in Arabidopsis suspension cell cultures than in seedling hypocotyls (Fig. 10A), suggesting that QUA3 plays an important role in pectin methylation in the former but not in the latter. This is supported by the observation that there was less pectin methylation in the cell walls of RNAi knock-down suspension cells (qua3i), as detected by labelling with JIM5 antibody (Fig. 8). However, in the qua3i cells, in the absence of QUA3 there was neither an increase in cell aggregates nor looser cell adhesion, as was previously described for QUA2/TSD2 seedlings (Krupkova et al., 2007; Mouille et al., 2007). This could be due to several reasons: (i) the pectin reactive sites that are made available as a consequence of the lower amount of QUA3 might not be effectively occupied by calcium ions to stiffen the cell wall and to cause defects in cell adhesion; (ii) the increased cross-linking of the pectic polymers by calcium ions is not essential to cause cell aggregation; (iii) the walls of the suspension cells are already in a very loose state compared with those in the seedling; or (iv) the decline in pectin methylation in qua3i cells may be compensated for by one or several members of the other four highly expressed QUA3 homologue genes (At2g34300, At5g14430, At4g0750, and At1g26850) (Fig. 10). To find out the possible roles of QUA3 higher expression in suspension culture than in seedlings, western blot analysis with anti-QUA3 using proteins isolated from PSBL suspension-cultured cells that have been subjected to various treatments was also carried out. The results obtained indicate that cold treatment increased the QUA3 protein level by 50%, while both salt treatment using NaCl and dehydration treatment using polyethylene gloycol (PEG) reduced the QUA3 protein level by 50% (data not shown). Decreased expression of QUA3 under dehydration condition might be evidence accounting for the lower expression of QUA3 in seedlings than in culture cells.
The possible unique function of QUA3 in Arabidopsis suspension cells was further supported by analysis of the composition of the cell wall. Similarly to qua2/tsd2 cells (Krupkova et al., 2007; Mouille et al., 2007), the assembly and distribution of cell wall pectin in qua3i cells were also found to be altered (Table 1). For example, more sugar components were released by CDTA and Na2CO3 in qua3i cells compared with WT PSBD cells (Table 1). Since these fractions mainly contain pectic components, these results demonstrate that the pectic polymers in the qua3i cell walls is linked to a greater extent than in WT walls; this may be because the knock-down of QUA3 in qua3i cells causes more methylation sites to be exposed and these free sites could be partially occupied by calcium ions, linking together the neighbouring pectic polymers. Consistent with this scenario, structural EM studies show that qua3i cells have wider and more closely associated pectic polymer residues in the middle lamella region (e.g. Fig. 8). Furthermore, immunogold EM studies with JIM5 antibody, which detects a low degree of methylesterification of pectin, demonstrate that these types of polymers are abundant in pectin-enriched regions (Fig. 8). Thus, these increased residues in the primary walls of qua3i cells are probably due to an increase in pectic polymers or a decrease in the compactness of the pectin matrix. In fact, when compared with WT cells, the overall concentration of most sugars in the qua3i cell walls is increased, except for a decrease in Glc in the KOH fraction, which mainly contains branched pectin and hemicelluloses (Stolle-Smits et al., 1999; Miedes and Lorences, 2004). Total sugar yield of the WT is 420.6 μg while that of qua3i is 375.1 μg. The present analysis showed a 32% increase in total GalA content in qua3i compared with the WT, which could cause the corresponding 32% more JIM5 labelling in qua3i. However, such a percentage of GalA difference could be marginal by considering a 3-fold density difference of gold particles in immuno-EM data (Fig. 8). Taken together, these results demonstrate an increase in both non-branched high molecular mass pectins (HG) and branched neutral sugar-rich pectins (RG) in qua3i cell walls. Thus, the scaffolding between the HG polymers and other pectin polymers is eventually perturbed.
Interestingly, both JIM5 and LM7 recognize unesterified pectins (Willats et al., 2000, 2001a; Clausen et al., 2003), but only JIM5 shows a dramatic difference in labelling in WT versus qua3i cells (Fig. 8). LM7 recognizes better than JIM5 on more de-esterified pectin, whereas JIM5 bound weakly to totally de-esterified pectin but better on the pectin with methylation up to 40% on GalA (Willats et al., 2000, 2001a; Clausen et al., 2003). Even though the overall labelling efficiency of LM7 is low in the immunogold EM study (Fig. 8), the labelling difference between qua3i and WT by LM7 and JIM5 antibodies suggests that the pectin methylation of qua3i is only partially de-esterified. It is likely that other QUA3 homologues that are highly expressed in suspension cultures (Fig. 10) could play a functionally redundant or synergic role for QUA3 to control the pectin methylation in culture cells.
In conclusion, it is a challenge in future research to understand fully the functions of individual members of various family membrane proteins such as the SAM-dependent MTases, endomembrane proteins (EMPs), and the secretory carrier membrane proteins (SCAMPs) in plants (Law et al., 2011).
Supplementary data
Supplementary data is available at JXB online.
Figure S1. Phylogenetic analysis of QUA3 homologue proteins in different plant species.
Figure S2. Subcellular localization of QUA3–GFP in transgenic BY-2 cells.
Figure S3. Constructs used in this study.
Figure S4. mRNA expression of QUA3 in suspension-cultured cells and different Arabidopsis seedling tissues.
Figure S5. Immunogold labelling of qua3i suspension-cultured cells using LM7 antibodies.
Figure S6. Cell–cell connection of qua3i and WT PSBD suspension-cultured cells.
Figure S7. Immunogold labelling of hypocotyls of qua3, QUA3-OE, and WT Arabidopsis seedlings.
Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
This work was supported by grants from the Research Grants Council of Hong Kong (CUHK488707, CUHK465708, CUHK466309, and CUHK466610) and CUHK Schemes B/C, to LJ. This work was also supported in part by the Department of Energy-funded (DE-FG09-93ER-20097) Center for Plant and Microbial Complex Carbohydrates. We are grateful to the RIKEN Genomic Science Center (Yokohama, Japan) for the full-length cDNA and transposon insertion mutant pst13453. We thank Professor Paul Knox (Centre for Plant Sciences, University of Leeds) for the JIM5, JIM7, and LM7 monoclonal antibodies, and Professor Dirk Inze (Ghent University) for providing the PSBD, PSBL suspension cultures, and pMP90M Agrobacterium strain. We thank Professor Markus Pauly (University of California, Berkeley) for reading the manuscript and constructive comments. We also thank Professor J. Derek Bewley (University of Guelph, Canada) for reading and correcting the manuscript.
References
- Bacic A. Breaking an impasse in pectin biosynthesis. Proceedings of the National Academy of Sciences, USA. 2006;103:5639–5640. doi: 10.1073/pnas.0601297103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barany I, Fadon B, Risueno MC, Testillano PS. Cell wall components and pectin esterification levels as markers of proliferation and differentiation events during pollen development and pollen embryogenesis in Capsicum annuum L. Journal of Experimental Botany. 2010;61:1159–1175. doi: 10.1093/jxb/erp392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Cheung AY, Hepler PK. Pectin methylesterase, a regulator of pollen tube growth. Plant Physiology. 2005;138:1334–1346. doi: 10.1104/pp.105.059865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Hepler PK. Pectin methylesterases and pectin dynamics in pollen tubes. The Plant Cell. 2005;17:3219–3226. doi: 10.1105/tpc.105.037473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Jia T, Lam SK, Ding Y, Gao C, San Wan, Yan M, Pimpl P, Jiang L. Multiple cytosolic and transmembrane determinants are required for the trafficking of SCAMP1 via an ER–Golgi–TGN–PM pathway. The Plant Journal. 2011;65:882–896. doi: 10.1111/j.1365-313X.2010.04469.x. [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. The Plant Journal. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Clausen MH, Willats WG, Knox JP. Synthetic methyl hexagalacturonate hapten inhibitors of anti-homogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohydrate Research. 2003;338:1797–1800. doi: 10.1016/s0008-6215(03)00272-6. [DOI] [PubMed] [Google Scholar]
- Cleland R. Independence of effects of auxin on cell wall methylation and elongation. Plant Physiology. 1963;38:12–18. doi: 10.1104/pp.38.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AAF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dhugga KS. Plant golgi cell wall synthesis: from genes to enzyme activities. Proceedings of the National Academy of Sciences, USA. 2005;102:1815–1816. doi: 10.1073/pnas.0409649102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis KE, Lam SY, Copenhaver GP. Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiology. 2006;142:1004–1013. doi: 10.1104/pp.106.085274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AAP. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Molecular Biology. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Goubet F, Mohnen D. Subcellular localization and topology of homogalacturonan methyltransferase in suspension-cultured Nicotiana tabacum cells. Planta. 1999a;209:112–117. doi: 10.1007/s004250050612. [DOI] [PubMed] [Google Scholar]
- Goubet F, Mohnen D. Solubilization and partial characterization of homogalacturonan-methyltransferase from microsomal membranes of suspension-cultured tobacco cells. Plant Physiology. 1999b;121:281–290. doi: 10.1104/pp.121.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubet F, Council NL, Mohnen D. Identification and partial characterization of the pectin methyltransferase ‘homogalacturonan-methyltransferase’ from membranes of tobacco cell suspensions. Plant Physiology. 1998;117:337–347. doi: 10.1104/pp.121.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt J, Suttangkakul AA, Vibe Scheller H. Biosynthesis of pectin. Plant Physiology. 2010;153:384–395. doi: 10.1104/pp.110.156588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl I, Robinson DG, Chrispeels MJ, Hinz G. Transport of storage proteins to the vacuole is mediated by vesicles without a clathrin coat. Journal of Cell Science. 1996;109:2539–2550. doi: 10.1242/jcs.109.10.2539. [DOI] [PubMed] [Google Scholar]
- Ibar C, Orellana AA. The import of S-adenosylmethionine into the Golgi apparatus is required for the methylation of homogalacturonan. Plant Physiology. 2007;145:504–512. doi: 10.1104/pp.107.104679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen EF, Jang R. Pectic metabolism of growing cell walls. Plant Physiology. 1960;35:87–97. doi: 10.1104/pp.35.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JK, Sorensen SO, Harholt J, et al. Identification of a xylogalacturonan xylosyltransferase involved in pectin biosynthesis in Arabidopsis. The Plant Cell. 2008;20:1289–1302. doi: 10.1105/tpc.107.050906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Phillips TE, Hamm CA, Drozdowicz YM, Rea PA, Maeshima M, Rogers JC. The protein storage vacuole: a unique compound organelle. Journal of Cell Biology. 2001;155:991–1102. doi: 10.1083/jcb.200107012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Phillips TE, Rogers SW, Rogers JC. Biogenesis of the protein storage vacuole crystalloid. Journal of Cell Biology. 2000;150:755–770. doi: 10.1083/jcb.150.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Rogers JC. Integral membrane protein sorting to vacuoles in plant cells: evidence for two pathways. Journal of Cell Biology. 1998;143:1183–1199. doi: 10.1083/jcb.143.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Yang SL, Xie LF, Puah CS, Zhang XQ, Yang WC, Sundaresan V, Ye D. VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. The Plant Cell. 2005;17:584–596. doi: 10.1105/tpc.104.027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolie RP, Duvetter T, Van Loey AAM, Hendrickx ME. Pectin methylesterase and its proteinaceous inhibitor: a review. Carbohydrate Research. 2010;345:2583–2595. doi: 10.1016/j.carres.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Keegstra K. Plant cell walls. Plant Physiology. 2010;154:483–486. doi: 10.1104/pp.110.161240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozbial PZ, Mushegian AAR. Natural history of S-adenosylmethionine-binding proteins. BMC Structural Biology. 2005;5:19. doi: 10.1186/1472-6807-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupkova E, Immerzeel P, Pauly M, Schmulling T. The TUMOROUS SHOOT DEVELOPMENT2 gene of Arabidopsis encoding a putative methyltransferase is required for cell adhesion and co-ordinated plant development. The Plant Journal. 2007;50:735–750. doi: 10.1111/j.1365-313X.2007.03123.x. [DOI] [PubMed] [Google Scholar]
- Kuromori T, Hirayama T, Kiyosue Y, et al. A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. The Plant Journal. 2004;37:897–905. doi: 10.1111/j.1365.313x.2004.02009.x. [DOI] [PubMed] [Google Scholar]
- Lam SK, Cai Y, Hillmer S, Robinson DG, Jiang L. SCAMPs highlight the developing cell plate during cytokinesis in tobacco BY-2 cells. Plant Physiology. 2008;147:1637–1645. doi: 10.1104/pp.108.119925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SK, Cai Y, Tse YC, Jiang L. BFA-induced compartments from the Golgi apparatus and trans-Golgi network/early endosome are distinct in plant cells. The Plant Journal. 2009;60:865–881. doi: 10.1111/j.1365-313X.2009.04007.x. [DOI] [PubMed] [Google Scholar]
- Lam SK, Siu CL, Hillmer S, Jang S, An G, Robinson DG, Jiang L. Rice SCAMP1 defines clathrin-coated, trans-golgi-located tubular–vesicular structures as an early endosome in tobacco BY-2 cells. The Plant Cell. 2007;19:296–319. doi: 10.1105/tpc.106.045708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law AHY, Chow CM, Jiang L. 2011. Secretory carrier membrane proteins. Protoplasma. doi: 10.1007/s00709-011-0295-0. (in press) [DOI] [PubMed] [Google Scholar]
- Li YB, Rogers SW, Tse YC, Lo SW, Sun SS, Jauh GY, Jiang L. BP-80 and homologs are concentrated on post-Golgi, probable lytic prevacuolar compartments. Plant and Cell Physiology. 2002;43:726–742. doi: 10.1093/pcp/pcf085. [DOI] [PubMed] [Google Scholar]
- Li YQ, Mareck AA, Faleri C, Moscatelli AA, Liu Q, Cresti M. Detection and localization of pectin methylesterase isoforms in pollen tubes of Nicotiana tabacum L. Planta. 2002;214:734–740. doi: 10.1007/s004250100664. [DOI] [PubMed] [Google Scholar]
- Louvet R, Cavel E, Gutierrez L, Guenin S, Roger D, Gillet F, Guerineau F, Pelloux J. Comprehensive expression profiling of the pectin methylesterase gene family during silique development in Arabidopsis thaliana. Planta. 2006;224:782–791. doi: 10.1007/s00425-006-0261-9. [DOI] [PubMed] [Google Scholar]
- Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Current Opinion in Structural Biology. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- McNeil M, Darvill AAG, Fry SC, Albersheim P. Structure and function of the primary cell walls of plants. Annual Review of Biochemistry. 1984;53:625–663. doi: 10.1146/annurev.bi.53.070184.003205. [DOI] [PubMed] [Google Scholar]
- Merkle RK, Poppe I. Carbohydrate composition analysis of glycoconjugates by gas–liquid chromatography/mass spectrometry. Methods in Enzymology. 1994;230:1–15. doi: 10.1016/0076-6879(94)30003-8. [DOI] [PubMed] [Google Scholar]
- Miao Y, Jiang L. Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nature Protocols. 2007;2:2348–2353. doi: 10.1038/nprot.2007.360. [DOI] [PubMed] [Google Scholar]
- Miao Y, Li KY, Li HY, Yao X, Jiang L. The vacuolar transport of aleurain–GFP and 2S albumin–GFP fusions is mediated by the same pre-vacuolar compartments in tobacco BY-2 and Arabidopsis suspension cultured cells. The Plant Journal. 2008;56:824–839. doi: 10.1111/j.1365-313X.2008.03645.x. [DOI] [PubMed] [Google Scholar]
- Miao Y, Yan PK, Kim H, Hwang I, Jiang L. Localization of green fluorescent protein fusions with the seven Arabidopsis vacuolar sorting receptors to prevacuolar compartments in tobacco BY-2 cells. Plant Physiology. 2006;142:945–962. doi: 10.1104/pp.106.083618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli F. Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends in Plant Science. 2001;6:414–419. doi: 10.1016/s1360-1385(01)02045-3. [DOI] [PubMed] [Google Scholar]
- Miedes E, Lorences EP. Apple (Malus domestica) and tomato (Lycopersicum esculentum) fruits cell-wall hemicelluloses and xyloglucan degradation during Penicillium expansum infection. Journal of Agricultural and Food Chemistry. 2004;52:7957–7963. doi: 10.1021/jf048890f. [DOI] [PubMed] [Google Scholar]
- Mohnen D. Pectin structure and biosynthesis. Current Opinion in Plant Biology. 2008;11:266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Mouille G, Ralet MC, Cavelier C, et al. Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. The Plant Journal. 2007;50:605–614. doi: 10.1111/j.1365-313X.2007.03086.x. [DOI] [PubMed] [Google Scholar]
- Persson S, Caffall KH, Freshour G, Hilley MT, Bauer S, Poindexter P, Hahn MG, Mohnen D, Somerville C. The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. The Plant Cell. 2007;19:237–255. doi: 10.1105/tpc.106.047720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijo JAA, Becker JD. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiology. 2005;138:744–756. doi: 10.1104/pp.104.057935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley BL, O'Neill MAA, Mohnen D. Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry. 2001;57:929–967. doi: 10.1016/s0031-9422(01)00113-3. [DOI] [PubMed] [Google Scholar]
- Ritzenthaler C, Nebenfuhr AA, Movafeghi AA, Stussi-Garaud C, Behnia L, Pimpl P, Staehelin LAA, Robinson DG. Reevaluation of the effects of brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. The Plant Cell. 2002;14:237–261. doi: 10.1105/tpc.010237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts K. How the cell wall acquired a cellular context. Plant Physiology. 2001;125:127–130. doi: 10.1104/pp.125.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SW, Burks M, Rogers JC. Monoclonal antibodies to barley aleurain and homologs from other plants. The Plant Journal. 1997;11:1359–1368. doi: 10.1046/j.1365-313x.1997.11061359.x. [DOI] [PubMed] [Google Scholar]
- Schaumann AA, Bruyant-Vannier M-P, Goubet F, Morvan C. Pectic metabolism in suspension-cultured cells of flax, Linum usitatissimum. Plant and Cell Physiology. 1993;34:891–897. [Google Scholar]
- Seki M, Narusaka M, Kamiya AA, et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science. 2002;296:141–145. doi: 10.1126/science.1071006. [DOI] [PubMed] [Google Scholar]
- Shen Y, Wang J, Ding Y, Lo SW, Gouzerh G, Neuhaus JM, Jiang L. The rice RMR1 associates with a distinct organelle as a prevacuolar compartment for the protein storage vacuole pathway. Molecular Plant. 2011 doi: 10.1093/mp/ssr025. (in press) [DOI] [PubMed] [Google Scholar]
- Sterling JD, Atmodjo MAA, Inwood SE, Kumar Kolli VS, Quigley HF, Hahn MG, Mohnen D. Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proceedings of the National Academy of Sciences, USA. 2006;103:5236–5241. doi: 10.1073/pnas.0600120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolle-Smits T, Beekhuizen JG, Kok MT, Pijnenburg M, Recourt K, Derksen J, Voragen AAG. Changes in cell wall polysaccharides of green bean pods during development. Plant Physiology. 1999;121:363–372. doi: 10.1104/pp.121.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen PK, Shen J, Sun SS, Jiang L. Expression and characterization of two functional vacuolar sorting receptor (VSR) proteins, BP-80 and AtVSR4 from culture media of transgenic tobacco BY-2 cells. Plant Scence. 2010;179:68–76. [Google Scholar]
- Tse YC, Mo B, Hillmer S, Zhao M, Lo SW, Robinson DG, Jiang L. Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. The Plant Cell. 2004;16:672–693. doi: 10.1105/tpc.019703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier MP, Thoiron B, Morvan C, Demarty M. Localization of methyltransferase activities throughout the endomembrane system of flax (Linum usitatissimum L.) hypocotyls. Biochemical Journal. 1992;286:863–868. doi: 10.1042/bj2860863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varner JE, Lin LS. Plant cell wall architecture. Cell. 1989;56:231–239. doi: 10.1016/0092-8674(89)90896-9. [DOI] [PubMed] [Google Scholar]
- Wang H, Jiang L. Transient expression and analysis of fluorescent reporter proteins in plant pollens. Nature Protocols. 2011;6:419–426. doi: 10.1038/nprot.2011.309. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhuang XH, Hillmer S, Robinson DG, Jiang L. Vacuolar sorting receptor (VSR) proteins reach the plasma membrane in germinating pollen tubes. Molecular Plant. 2011 doi: 10.1093/mp/ssr011. (in press) [DOI] [PubMed] [Google Scholar]
- Wang J, Ding Y, Wang J, Hillmer S, Miao Y, Lo SW, Wang X, Robinson DG, Jiang L. EXPO: an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in plant cells. The Plant Cell. 2010;22:4009–4030. doi: 10.1105/tpc.110.080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li Y, Lo SW, Hillmer S, Sun SS, Robinson DG, Jiang L. Protein mobilization in germinating mung bean seeds involves vacuolar sorting receptors and multivesicular bodies. Plant Physiology. 2007;143:1628–1639. doi: 10.1104/pp.107.096263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JQ, Miao YS, Yi Cai Jiang L. Wortmannin induced homotypic fusion of prevacuolar compartment in plant cells. Journal of Experimental Botany. 2009a;60:3075–3083. doi: 10.1093/jxb/erp136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JQ, Suen PK, Xu ZF, Jiang L. A 64-kDa sucrose binding protein is membrane-associated and tonoplast-localized in developing mung bean seeds. Journal of Experimental Botany. 2009b;60:629–639. doi: 10.1093/jxb/ern308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F, Zhu Y, Hawes MC. Effect of pectin methylesterase gene expression on pea root development. The Plant Cell. 1999;11:1129–1140. doi: 10.1105/tpc.11.6.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CAA, Smith NAA, et al. Construct design for efficient, effective and high-throughput gene silencing in plants. The Plant Journal. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- Willats WG, Limberg G, Buchholt HC, van Alebeek GJ, Benen J, Christensen TM, Visser J, Voragen AA, Mikkelsen JD, Knox JP. Analysis of pectic epitopes recognised by hybridoma and phage display monoclonal antibodies using defined oligosaccharides, polysaccharides, and enzymatic degradation. Carbohydrate Research. 2000;327:309–320. doi: 10.1016/s0008-6215(00)00039-2. [DOI] [PubMed] [Google Scholar]
- Willats WG, McCartney L, Mackie W, Knox JP. Pectin: cell biology and prospects for functional analysis. Plant Molecular Biology. 2001a;47:9–27. [PubMed] [Google Scholar]
- Willats WG, Orfila C, Limberg G, et al. Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls. Implications for pectin methyl esterase action, matrix properties, and cell adhesion. Journal of Biological Chemistry. 2001b;276:19404–19413. doi: 10.1074/jbc.M011242200. [DOI] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J. Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. The Plant Cell. 1995;7:1555–1567. doi: 10.1105/tpc.7.10.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YD, Elamawi R, Bubeck J, Pepperkok R, Ritzenthaler C, Robinson DG. Dynamics of COPII vesicles and the Golgi apparatus in cultured Nicotiana tabacum BY-2 cells provides evidence for transient association of Golgi stacks with endoplasmic reticulum exit sites. The Plant Cell. 2005;17:1513–1531. doi: 10.1105/tpc.104.026757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York JY, Darvill AAG, McNeil M, Stevenson TT, Albersheim P. Isolation and characterization of plant cell walls and cell wall components. Methods in Enzymology. 1985;118:3–40. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.